Abstract
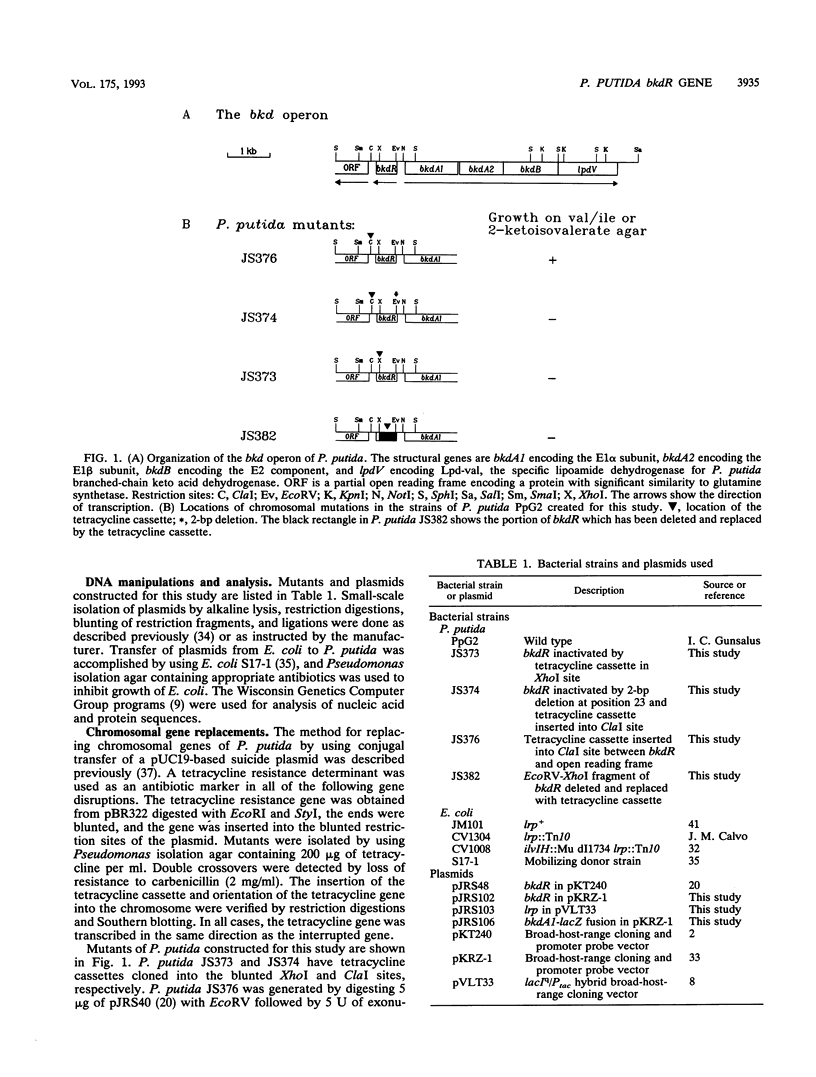
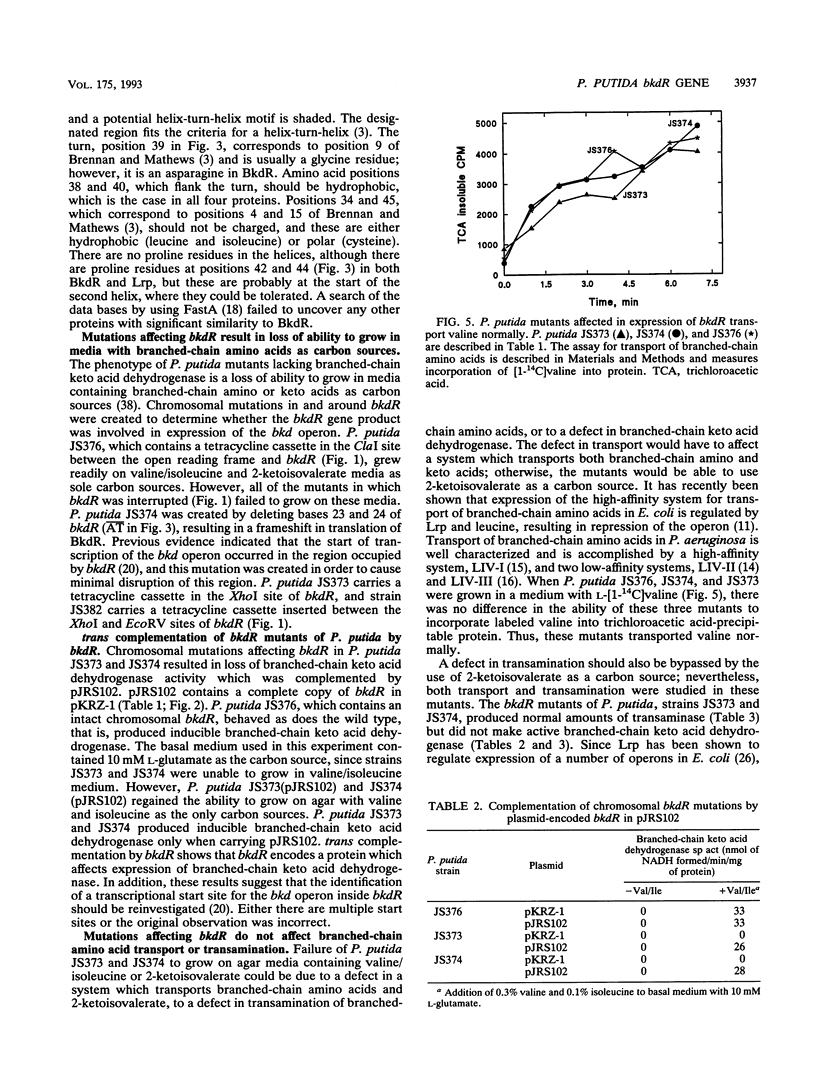
Branched-chain keto acid dehydrogenase is a multienzyme complex which is required for the metabolism of the branched-chain amino acids in Pseudomonas putida. The structural genes encoding all four proteins of the bkd operon have been cloned, and their nucleotide sequences have been determined (G. Burns, K. T. Madhusudhan, K. Hatter, and J. R. Sokatch, p. 177-184 in S. Silver, A. M. Chakrabarty, B. Iglewski, and S. Kaplan [ed.], Pseudomonas: Biotransformations, Pathogenesis, and Evolving Biotechnology, American Society for Microbiology, Washington D.C., 1990). An open reading frame which encoded a protein with 36.5% amino acid identity to the leucine-responsive regulatory protein (Lrp) of Escherichia coli was found immediately upstream of the bkd operon. Chromosomal mutations affecting this gene, named bkdR, resulted in a loss of ability to use branched-chain amino acids as carbon and energy sources and failure to produce branched-chain keto acid dehydrogenase. These mutations were complemented in trans by plasmids which contained intact bkdR. Mutations affecting bkdR did not have any effect on transport of branched-chain amino acids or transamination. Therefore, the bkdR gene product must affect expression of the bkd operon and regulation must be positive. Mutations affecting bkdR could also be complemented by plasmids containing lrp of E. coli. This is the first instance of a Lrp-like protein demonstrated to regulate expression of an operon outside of E. coli.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Airas R. K. Branched-chain-amino-acid aminotransferase assay using radioisotopes. Biochim Biophys Acta. 1989 Sep 15;992(3):397–399. doi: 10.1016/0304-4165(89)90103-7. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M. M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Burns G., Brown T., Hatter K., Idriss J. M., Sokatch J. R. Similarity of the E1 subunits of branched-chain-oxoacid dehydrogenase from Pseudomonas putida to the corresponding subunits of mammalian branched-chain-oxoacid and pyruvate dehydrogenases. Eur J Biochem. 1988 Sep 15;176(2):311–317. doi: 10.1111/j.1432-1033.1988.tb14283.x. [DOI] [PubMed] [Google Scholar]
- Burns G., Brown T., Hatter K., Sokatch J. R. Comparison of the amino acid sequences of the transacylase components of branched chain oxoacid dehydrogenase of Pseudomonas putida, and the pyruvate and 2-oxoglutarate dehydrogenases of Escherichia coli. Eur J Biochem. 1988 Sep 1;176(1):165–169. doi: 10.1111/j.1432-1033.1988.tb14264.x. [DOI] [PubMed] [Google Scholar]
- Burns G., Brown T., Hatter K., Sokatch J. R. Sequence analysis of the lpdV gene for lipoamide dehydrogenase of branched-chain-oxoacid dehydrogenase of Pseudomonas putida. Eur J Biochem. 1989 Jan 15;179(1):61–69. doi: 10.1111/j.1432-1033.1989.tb14521.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein R. S., Miller R. H., Hoganson G., Harper A. E. Phylogenetic comparisons of the branched-chain alpha-ketoacid dehydrogenase complex. Comp Biochem Physiol B. 1990;97(4):719–726. doi: 10.1016/0305-0491(90)90113-8. [DOI] [PubMed] [Google Scholar]
- Haney S. A., Platko J. V., Oxender D. L., Calvo J. M. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli. J Bacteriol. 1992 Jan;174(1):108–115. doi: 10.1128/jb.174.1.108-115.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hoshino T., Kose-Terai K., Uratani Y. Isolation of the braZ gene encoding the carrier for a novel branched-chain amino acid transport system in Pseudomonas aeruginosa PAO. J Bacteriol. 1991 Mar;173(6):1855–1861. doi: 10.1128/jb.173.6.1855-1861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Kose K. Cloning and nucleotide sequence of braC, the structural gene for the leucine-, isoleucine-, and valine-binding protein of Pseudomonas aeruginosa PAO. J Bacteriol. 1989 Nov;171(11):6300–6306. doi: 10.1128/jb.171.11.6300-6306.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Kose K. Cloning, nucleotide sequences, and identification of products of the Pseudomonas aeruginosa PAO bra genes, which encode the high-affinity branched-chain amino acid transport system. J Bacteriol. 1990 Oct;172(10):5531–5539. doi: 10.1128/jb.172.10.5531-5539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T. Transport systems for branched-chain amino acids in Pseudomonas aeruginosa. J Bacteriol. 1979 Sep;139(3):705–712. doi: 10.1128/jb.139.3.705-712.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling R., Lother H. AsnC: an autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli. J Bacteriol. 1985 Oct;164(1):310–315. doi: 10.1128/jb.164.1.310-315.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lowe P. N., Hodgson J. A., Perham R. N. Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxo acids in Bacillus subtilis. Biochem J. 1983 Oct 1;215(1):133–140. doi: 10.1042/bj2150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudhan K. T., Huang G., Burns G., Sokatch J. R. Transcriptional analysis of the promoter region of the Pseudomonas putida branched-chain keto acid dehydrogenase operon. J Bacteriol. 1990 Oct;172(10):5655–5663. doi: 10.1128/jb.172.10.5655-5663.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall V. D., Sokatch J. R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972 Jun;110(3):1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. R., Marshall V. D., Sokatch J. R., Unger L. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J Bacteriol. 1973 Jul;115(1):198–204. doi: 10.1128/jb.115.1.198-204.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully V., Burns G., Sokatch J. R. Resolution of branched-chain oxo acid dehydrogenase complex of Pseudomonas aeruginosa PAO. Biochem J. 1986 Feb 1;233(3):737–742. doi: 10.1042/bj2330737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman E. B., D'Ari R., Lin R. T. The leucine-Lrp regulon in E. coli: a global response in search of a raison d'être. Cell. 1992 Feb 21;68(4):617–619. doi: 10.1016/0092-8674(92)90135-y. [DOI] [PubMed] [Google Scholar]
- Odessey R. Purification of rat kidney branched-chain oxo acid dehydrogenase complex with endogenous kinase activity. Biochem J. 1982 Apr 15;204(1):353–356. doi: 10.1042/bj2040353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton R., Harris R. A. Isolation of rabbit liver branched chain alpha-ketoacid dehydrogenase and regulation by phosphorylation. J Biol Chem. 1982 Dec 10;257(23):14433–14439. [PubMed] [Google Scholar]
- Perham R. N., Packman L. C., Radford S. E. 2-Oxo acid dehydrogenase multi-enzyme complexes: in the beginning and halfway there. Biochem Soc Symp. 1987;54:67–81. [PubMed] [Google Scholar]
- Pettit F. H., Yeaman S. J., Reed L. J. Purification and characterization of branched chain alpha-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platko J. V., Willins D. A., Calvo J. M. The ilvIH operon of Escherichia coli is positively regulated. J Bacteriol. 1990 Aug;172(8):4563–4570. doi: 10.1128/jb.172.8.4563-4570.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmel R. K., Shinabarger D. L., Parsek M. R., Aldrich T. L., Chakrabarty A. M. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA-binding site by hydroxyl-radical footprinting. J Bacteriol. 1991 Aug;173(15):4717–4724. doi: 10.1128/jb.173.15.4717-4724.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Roberts C. M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele M. I., Lorenz D., Hatter K., Park A., Sokatch J. R. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding methylmalonate-semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J Biol Chem. 1992 Jul 5;267(19):13585–13592. [PubMed] [Google Scholar]
- Sykes P. J., Menard J., McCully V., Sokatch J. R. Conjugative mapping of pyruvate, 2-ketoglutarate, and branched-chain keto acid dehydrogenase genes in Pseudomonas putida mutants. J Bacteriol. 1985 Apr;162(1):203–208. doi: 10.1128/jb.162.1.203-208.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins D. A., Calvo J. M. In vitro transcription from the Escherichia coli ilvIH promoter. J Bacteriol. 1992 Dec;174(23):7648–7655. doi: 10.1128/jb.174.23.7648-7655.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins D. A., Ryan C. W., Platko J. V., Calvo J. M. Characterization of Lrp, and Escherichia coli regulatory protein that mediates a global response to leucine. J Biol Chem. 1991 Jun 15;266(17):10768–10774. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Eltis L., Kessler B., Timmis K. N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993 Jan 15;123(1):17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]




