Abstract
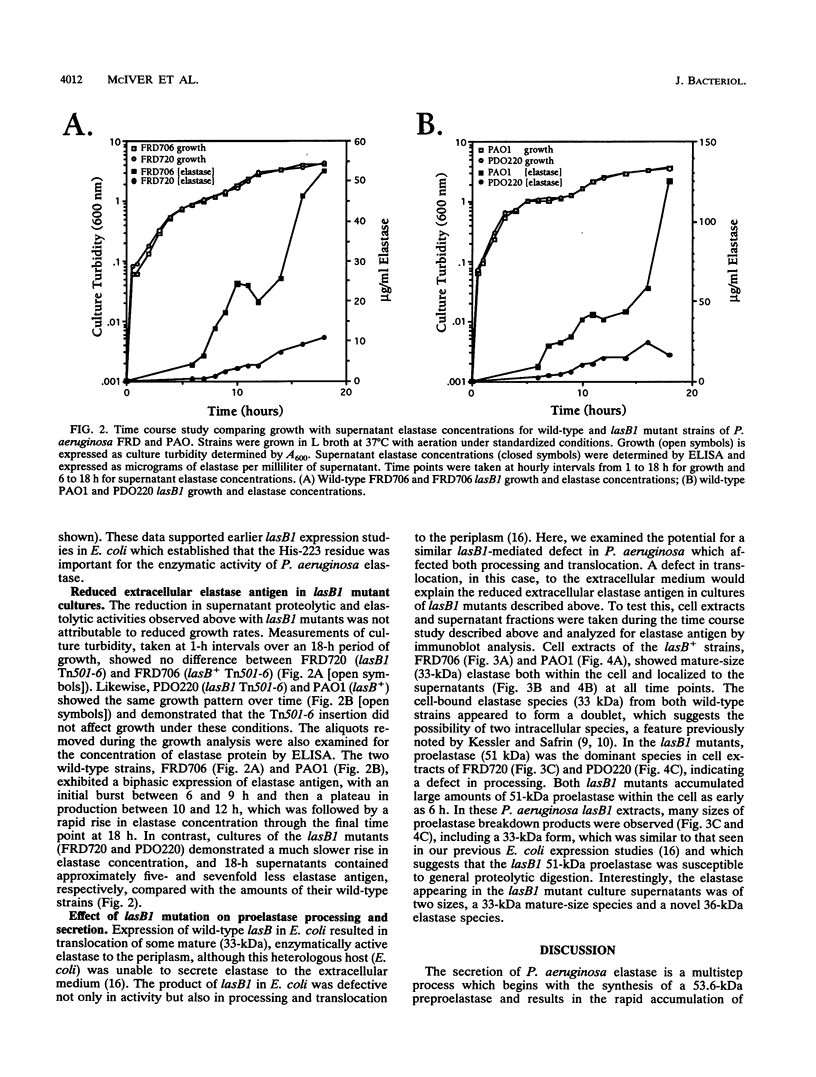
Pseudomonas aeruginosa secretes elastase in a multistep process which begins with the synthesis of a preproelastase (53.6 kDa) encoded by lasB, is followed by processing to proelastase (51 kDa), and concludes with the rapid accumulation of mature elastase (33 kDa) in the extracellular environment. In this study, mutants of P. aeruginosa were constructed by gene replacement which expressed lasB1, an allele altered in vitro at an active-site His-223-encoding codon. The lasB1 allele was exchanged for chromosomal lasB sequences in two strain backgrounds, FRD2 and PAO1, through a selectable-cassette strategy which placed a downstream Tn501 marker next to lasB1 and provided the selection for homologous recombination with the chromosome. Two lasB1 mutants, FRD720 and PDO220, were characterized, and their culture supernatants contained greatly reduced proteolytic (9-fold) and elastolytic (14- to 20-fold) activities compared with their respective parental lasB+ strains. This was primarily due to the effect of His-223 substitution on substrate binding by elastase and thus its proteolytic activity. However, the concentration of supernatant elastase antigen was also reduced (five- to sevenfold) in the mutant strains compared with the parental strains. An immunoblot analysis of cell extracts showed a large accumulation of 51-kDa proelastase within lasB1 mutant cells which was not seen in wild-type cell extracts. A time course study showed that production of extracellular elastase was inefficient in the lasB1 mutants compared with that of parental strains. This showed that expression of an enzymatically defective elastase inhibits proper processing of proelastase and provides further evidence for autoproteolytic processing of proelastase in P. aeruginosa. Unlike the parental strains, culture supernatants of the lasB1 mutants contained two prominent elastase species that were 33 and 36 kDa in size. Extracellular 51-kDa proelastase was barely detectable, even though it accumulated to high concentrations within the lasB1 mutant cells. These data suggest that production of an enzymatically defective elastase affects proper secretion because autoproteolytic processing of proelastase is necessary for efficient localization to the extracellular milieu. The appearance of reduced amounts of extracellular elastase and their sizes of 33 and 36 kDa suggest that lasB1-encoded elastase was processed by alternate, less-efficient processing mechanisms. Thus, proelastase must be processed by removal of nearly all of the 18-kDa propeptide before elastase is a protein competent for extracellular secretion.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bever R. A., Iglewski B. H. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J Bacteriol. 1988 Sep;170(9):4309–4314. doi: 10.1128/jb.170.9.4309-4314.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring G., Obernesser H. J., Botzenhart K. Extrazelluläre Toxine von Pseudomonas aeruginosa. II. Einwirkung zweier gereinigter Proteasen auf die menschlichen Immunoglobuline IgG, IgA und sekretorisches IgA. Zentralbl Bakteriol A. 1981 Mar;249(1):89–98. [PubMed] [Google Scholar]
- Döring G., Pfestorf M., Botzenhart K., Abdallah M. A. Impact of proteases on iron uptake of Pseudomonas aeruginosa pyoverdin from transferrin and lactoferrin. Infect Immun. 1988 Jan;56(1):291–293. doi: 10.1128/iai.56.1.291-293.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. L., Ohman D. E. Use of a gene replacement cosmid vector for cloning alginate conversion genes from mucoid and nonmucoid Pseudomonas aeruginosa strains: algS controls expression of algT. J Bacteriol. 1988 Jul;170(7):3228–3236. doi: 10.1128/jb.170.7.3228-3236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima J., Yamamoto S., Morihara K., Atsumi Y., Takeuchi H., Kawamoto S., Okuda K. Structural gene and complete amino acid sequence of Pseudomonas aeruginosa IFO 3455 elastase. J Bacteriol. 1989 Mar;171(3):1698–1704. doi: 10.1128/jb.171.3.1698-1704.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck L. W., Morihara K., Abrahamson D. R. Degradation of soluble laminin and depletion of tissue-associated basement membrane laminin by Pseudomonas aeruginosa elastase and alkaline protease. Infect Immun. 1986 Oct;54(1):149–153. doi: 10.1128/iai.54.1.149-153.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck L. W., Morihara K., McRae W. B., Miller E. J. Specific cleavage of human type III and IV collagens by Pseudomonas aeruginosa elastase. Infect Immun. 1986 Jan;51(1):115–118. doi: 10.1128/iai.51.1.115-118.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E., Israel M., Landshman N., Chechick A., Blumberg S. In vitro inhibition of Pseudomonas aeruginosa elastase by metal-chelating peptide derivatives. Infect Immun. 1982 Nov;38(2):716–723. doi: 10.1128/iai.38.2.716-723.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E., Safrin M. Partial purification and characterization of an inactive precursor of Pseudomonas aeruginosa elastase. J Bacteriol. 1988 Mar;170(3):1215–1219. doi: 10.1128/jb.170.3.1215-1219.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E., Safrin M., Peretz M., Burstein Y. Identification of cleavage sites involved in proteolytic processing of Pseudomonas aeruginosa preproelastase. FEBS Lett. 1992 Mar 16;299(3):291–293. doi: 10.1016/0014-5793(92)80134-3. [DOI] [PubMed] [Google Scholar]
- Kessler E., Safrin M. Synthesis, processing, and transport of Pseudomonas aeruginosa elastase. J Bacteriol. 1988 Nov;170(11):5241–5247. doi: 10.1128/jb.170.11.5241-5247.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McIver K., Kessler E., Ohman D. E. Substitution of active-site His-223 in Pseudomonas aeruginosa elastase and expression of the mutated lasB alleles in Escherichia coli show evidence for autoproteolytic processing of proelastase. J Bacteriol. 1991 Dec;173(24):7781–7789. doi: 10.1128/jb.173.24.7781-7789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morihara K., Tsuzuki H., Oda K. Protease and elastase of Pseudomonas aeruginosa: inactivation of human plasma alpha 1-proteinase inhibitor. Infect Immun. 1979 Apr;24(1):188–193. doi: 10.1128/iai.24.1.188-193.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Cryz S. J., Iglewski B. H. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980 Jun;142(3):836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., West M. A., Flynn J. L., Goldberg J. B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985 Jun;162(3):1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J. C., Ohman D. E. Efficient production and processing of elastase and LasA by Pseudomonas aeruginosa require zinc and calcium ions. J Bacteriol. 1992 Jun;174(12):4140–4147. doi: 10.1128/jb.174.12.4140-4147.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. R., Miller K. D. Elastase of Pseudomonas aeruginosa: inactivation of complement components and complement-derived chemotactic and phagocytic factors. Infect Immun. 1974 Jul;10(1):128–135. doi: 10.1128/iai.10.1.128-135.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E., Kawamoto S., Fukushima J., Hamajima K., Onishi H., Miyagi Y., Inami S., Morihara K., Okuda K. Detection of elastase production in Escherichia coli with the elastase structural gene from several non-elastase-producing strains of Pseudomonas aeruginosa. J Bacteriol. 1991 Oct;173(19):6153–6158. doi: 10.1128/jb.173.19.6153-6158.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M. M., Flaherty K. M., McKay D. B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J Biol Chem. 1991 Feb 15;266(5):2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- Toder D. S., Gambello M. J., Iglewski B. H. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol Microbiol. 1991 Aug;5(8):2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wolz C., Hellstern E., Haug M., Galloway D. R., Vasil M. L., Döring G. Pseudomonas aeruginosa LasB mutant constructed by insertional mutagenesis reveals elastolytic activity due to alkaline proteinase and the LasA fragment. Mol Microbiol. 1991 Sep;5(9):2125–2131. doi: 10.1111/j.1365-2958.1991.tb02142.x. [DOI] [PubMed] [Google Scholar]