Abstract
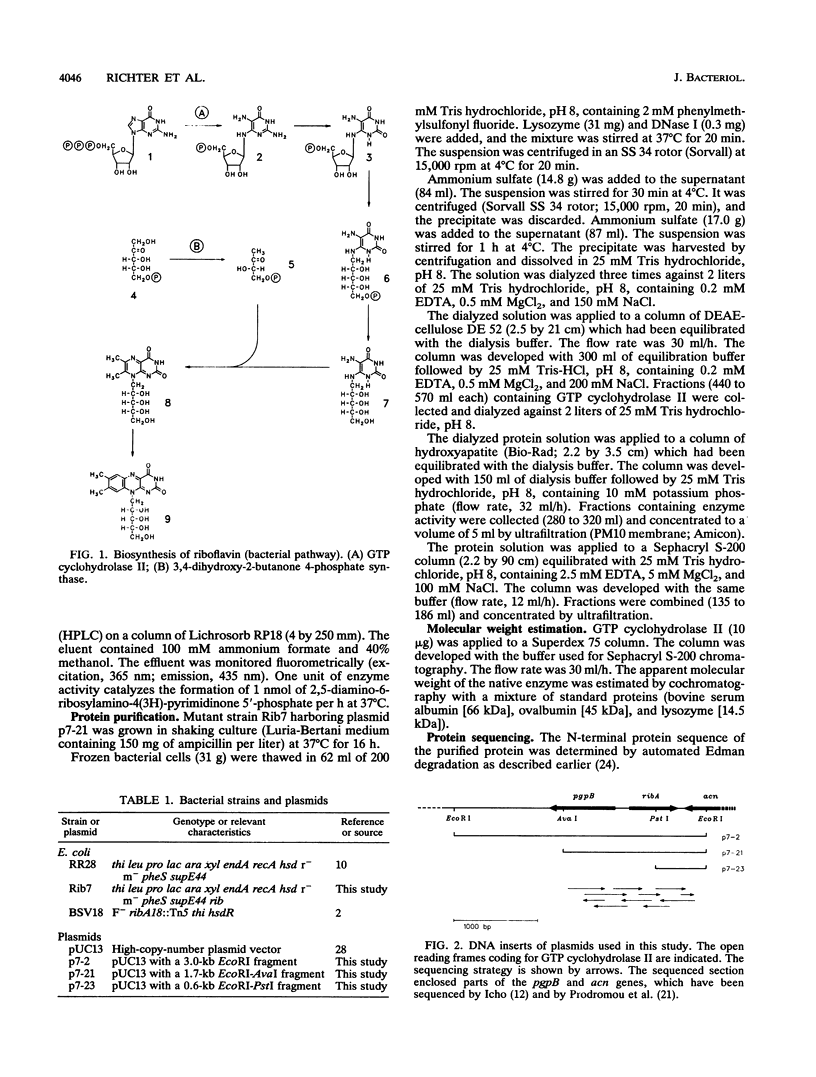
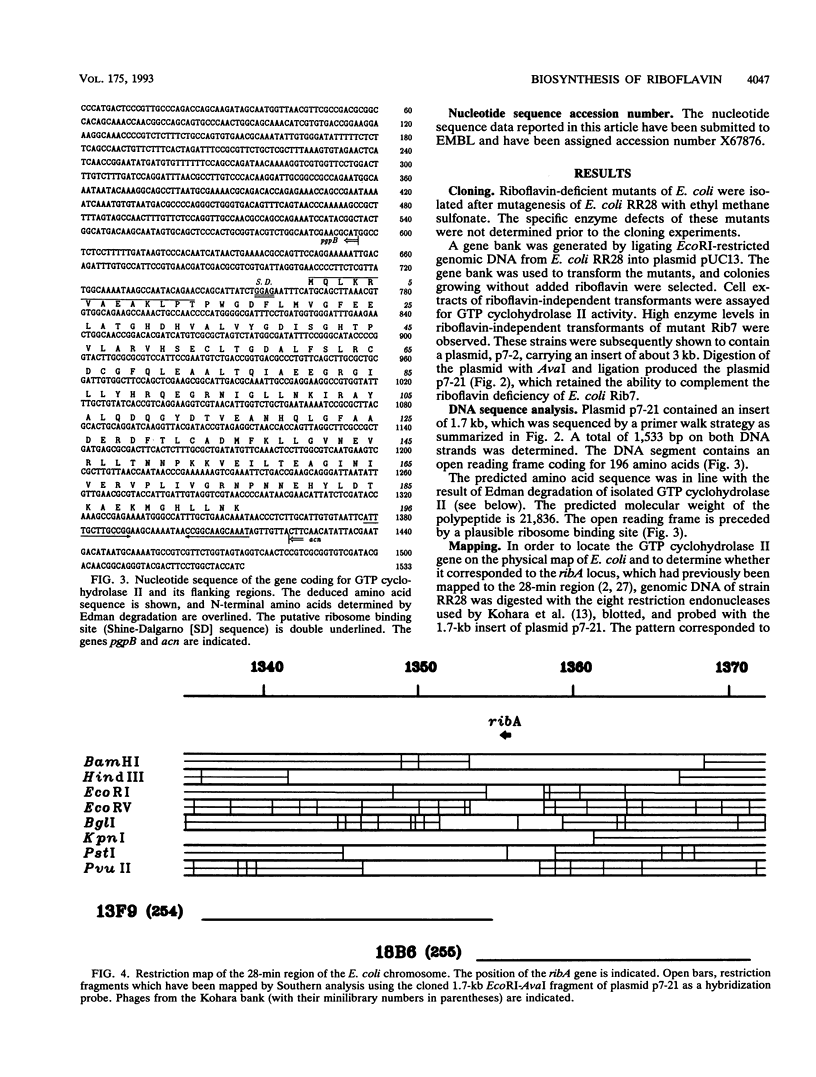
GTP cyclohydrolase II catalyzes the first committed step in the biosynthesis of riboflavin. The gene coding for this enzyme in Escherichia coli has been cloned by marker rescue. Sequencing indicated an open reading frame of 588 bp coding for a 21.8-kDa peptide of 196 amino acids. The gene was mapped to a position at 28.2 min on the E. coli chromosome and is identical with ribA. GTP cyclohydrolase II was overexpressed in a recombinant strain carrying a plasmid with the cloned gene. The enzyme was purified to homogeneity from the recombinant strain. The N-terminal sequence determined by Edman degradation was identical to the predicted sequence. The sequence is homologous to the 3' part of the central open reading frame in the riboflavin operon of Bacillus subtilis.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bandrin S. V., Rabinovich P. M., Stepanov A. I. Tri gruppy stsepleniia genov biosinteza riboflavina Escherichia coli. Genetika. 1983 Sep;19(9):1419–1425. [PubMed] [Google Scholar]
- Chikindas M. L., Luk'ianov E. V., Rabinovich P. M., Stepanov A. I. Izuchenie oblasti 210 gradusov khromosomy Bacillus subtilis s pomoshch'iu rekombinantnykh plazmid. Mol Gen Mikrobiol Virusol. 1987 Feb;(2):20–24. [PubMed] [Google Scholar]
- Foor F., Brown G. M. Purification and properties of guanosine triphosphate cyclohydrolase II from Escherichia coli. J Biol Chem. 1975 May 10;250(9):3545–3551. [PubMed] [Google Scholar]
- Godson G. N., Vapnek D. A simple method of preparing large amounts of phiX174 RF 1 supercoiled DNA. Biochim Biophys Acta. 1973 Apr 11;299(4):516–520. doi: 10.1016/0005-2787(73)90223-2. [DOI] [PubMed] [Google Scholar]
- Harzer G., Rokos H., Otto M. K., Bacher A., Ghisla S. Biosynthesis of riboflavin. 6,7-Dimethyl-8-ribityllumazine 5'-phosphate is not a substrate for riboflavin synthase. Biochim Biophys Acta. 1978 Apr 19;540(1):48–54. doi: 10.1016/0304-4165(78)90433-6. [DOI] [PubMed] [Google Scholar]
- Hennecke H., Günther I., Binder F. A novel cloning vector for the direct selection of recombinant DNA in E. coli. Gene. 1982 Sep;19(2):231–234. doi: 10.1016/0378-1119(82)90011-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Icho T. Membrane-bound phosphatases in Escherichia coli: sequence of the pgpB gene and dual subcellular localization of the pgpB product. J Bacteriol. 1988 Nov;170(11):5117–5124. doi: 10.1128/jb.170.11.5117-5124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Meighen E. A. The lux genes in Photobacterium leiognathi are closely linked with genes corresponding in sequence to riboflavin synthesis genes. Biochem Biophys Res Commun. 1992 Jul 31;186(2):690–697. doi: 10.1016/0006-291x(92)90802-r. [DOI] [PubMed] [Google Scholar]
- Prodromou C., Artymiuk P. J., Guest J. R. The aconitase of Escherichia coli. Nucleotide sequence of the aconitase gene and amino acid sequence similarity with mitochondrial aconitases, the iron-responsive-element-binding protein and isopropylmalate isomerases. Eur J Biochem. 1992 Mar 1;204(2):599–609. doi: 10.1111/j.1432-1033.1992.tb16673.x. [DOI] [PubMed] [Google Scholar]
- Read S. M., Northcote D. H. Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal Biochem. 1981 Sep 1;116(1):53–64. doi: 10.1016/0003-2697(81)90321-3. [DOI] [PubMed] [Google Scholar]
- Richter G., Volk R., Krieger C., Lahm H. W., Röthlisberger U., Bacher A. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J Bacteriol. 1992 Jun;174(12):4050–4056. doi: 10.1128/jb.174.12.4050-4056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott K., Kellermann J., Lottspeich F., Bacher A. Riboflavin synthases of Bacillus subtilis. Purification and amino acid sequence of the alpha subunit. J Biol Chem. 1990 Mar 15;265(8):4204–4209. [PubMed] [Google Scholar]
- Swartzman E., Miyamoto C., Graham A., Meighen E. Delineation of the transcriptional boundaries of the lux operon of Vibrio harveyi demonstrates the presence of two new lux genes. J Biol Chem. 1990 Feb 25;265(6):3513–3517. [PubMed] [Google Scholar]
- Tesliar G. E., Shavlovskii G. M. Lokalizatsiia genov, kodiruiushchikh GTF-tsiklogidrolazu II i riboflavinsintazu na khromosome Escherichia coli K-12. Tsitol Genet. 1983 Sep-Oct;17(5):54–56. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Volk R., Bacher A. Studies on the 4-carbon precursor in the biosynthesis of riboflavin. Purification and properties of L-3,4-dihydroxy-2-butanone-4-phosphate synthase. J Biol Chem. 1990 Nov 15;265(32):19479–19485. [PubMed] [Google Scholar]



