Abstract
Aspartate aminotransferase (AAT) is an important enzyme in aspartate catabolism and biosynthesis and, by converting tricarboxylic acid cycle intermediates to amino acids, AAT is also significant in linking carbon metabolism with nitrogen metabolism. To examine the role of AAT in symbiotic nitrogen fixation further, plasmids encoding three different aminotransferases from Rhizobium meliloti 104A14 were isolated by complementation of an Escherichia coli auxotroph that lacks three aminotransferases. pJA10 contained a gene, aatB, that coded for a previously undescribed AAT, AatB. pJA30 encoded an aromatic aminotransferase, TatA, that had significant AAT activity, and pJA20 encoded a branched-chain aminotransferase designated BatA. Genes for the latter two enzymes, tatA and batA, were previously isolated from R. meliloti. aatB is distinct from but hybridizes to aatA, which codes for AatA, a protein required for symbiotic nitrogen fixation. The DNA sequence of aatB contained an open reading frame that could encode a protein 410 amino acids long and with a monomer molecular mass of 45,100 Da. The amino acid sequence of aatB is unusual, and AatB appears to be a member of a newly described class of AATs. AatB expressed in E. coli has a Km for aspartate of 5.3 mM and a Km for 2-oxoglutarate of 0.87 mM. Its pH optimum is between 8.0 and 8.5. Mutations were constructed in aatB and tatA and transferred to the genome of R. meliloti 104A14. Both mutants were prototrophs and were able to carry out symbiotic nitrogen fixation.
Full text
PDF



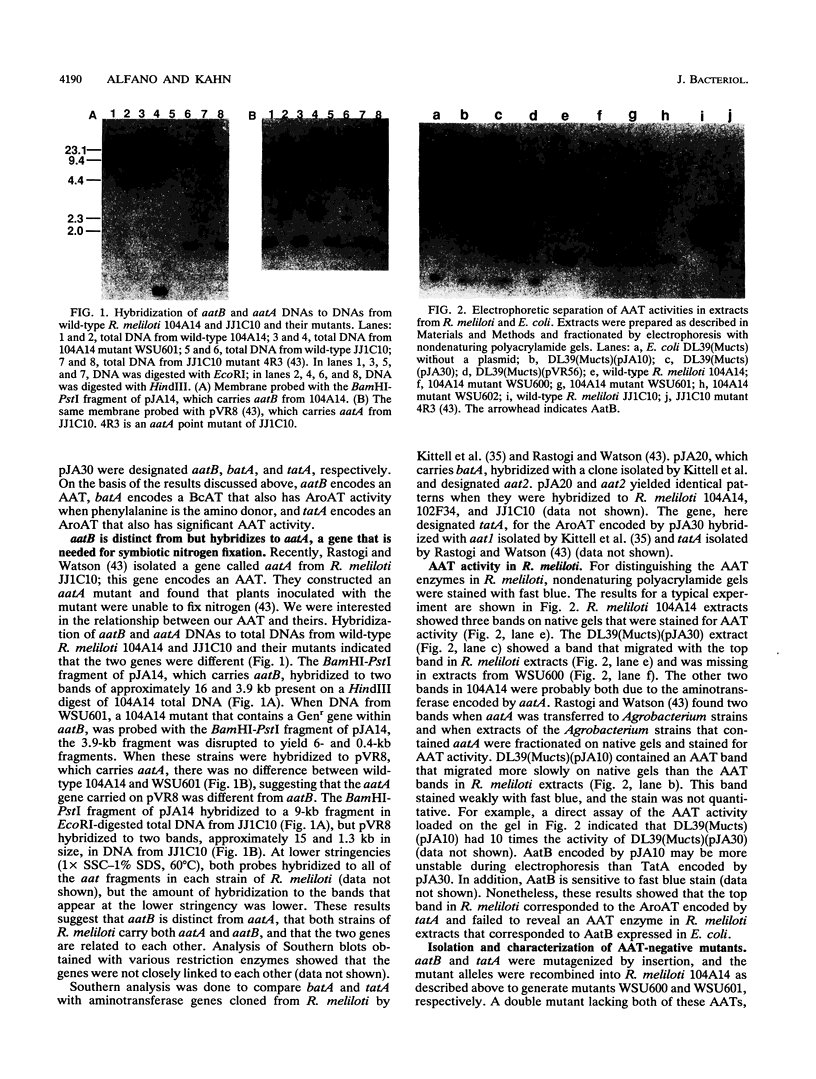





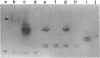
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appels M. A., Haaker H. Glutamate Oxaloacetate Transaminase in Pea Root Nodules : Participation in a Malate/Aspartate Shuttle between Plant and Bacteroid. Plant Physiol. 1991 Mar;95(3):740–747. doi: 10.1104/pp.95.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg C. M., Wang M. D., Vartak N. B., Liu L. Acquisition of new metabolic capabilities: multicopy suppression by cloned transaminase genes in Escherichia coli K-12. Gene. 1988 May 30;65(2):195–202. doi: 10.1016/0378-1119(88)90456-8. [DOI] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C., Fickett J. W., Goad W. B., Lewitter F. I., Rindone W. P., Swindell C. D., Tung C. S. The GenBank genetic sequence databank. Nucleic Acids Res. 1986 Jan 10;14(1):1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birolo L., Arnone M. I., Cubellis M. V., Andreotti G., Nitti G., Marino G., Sannia G. The active site of Sulfolobus solfataricus aspartate aminotransferase. Biochim Biophys Acta. 1991 Nov 15;1080(3):198–204. doi: 10.1016/0167-4838(91)90002-h. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cubellis M. V., Rozzo C., Nitti G., Arnone M. I., Marino G., Sannia G. Cloning and sequencing of the gene coding for aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. Eur J Biochem. 1989 Dec 8;186(1-2):375–381. doi: 10.1111/j.1432-1033.1989.tb15219.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Wood J. M., Jordan D. C. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol. 1983 Jun;154(3):1403–1413. doi: 10.1128/jb.154.3.1403-1413.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford G. C., Eichele G., Jansonius J. N. Three-dimensional structure of a pyridoxal-phosphate-dependent enzyme, mitochondrial aspartate aminotransferase. Proc Natl Acad Sci U S A. 1980 May;77(5):2559–2563. doi: 10.1073/pnas.77.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotheringham I. G., Dacey S. A., Taylor P. P., Smith T. J., Hunter M. G., Finlay M. E., Primrose S. B., Parker D. M., Edwards R. M. The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K12. Comparison of the primary structures of the aspartate aminotransferase and aromatic aminotransferase of E. coli with those of the pig aspartate aminotransferase isoenzymes. Biochem J. 1986 Mar 15;234(3):593–604. doi: 10.1042/bj2340593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougère F., Le Rudulier D., Streeter J. G. Effects of Salt Stress on Amino Acid, Organic Acid, and Carbohydrate Composition of Roots, Bacteroids, and Cytosol of Alfalfa (Medicago sativa L.). Plant Physiol. 1991 Aug;96(4):1228–1236. doi: 10.1104/pp.96.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J. S., Larson R. J., Farnham M. W., Pathirana S. M., Miller S. S., Vance C. P. Aspartate aminotransferase in effective and ineffective alfalfa nodules : cloning of a cDNA and determination of enzyme activity, protein, and mRNA levels. Plant Physiol. 1992 Mar;98(3):868–878. doi: 10.1104/pp.98.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange T., Guénet C., Dietrich J. B., Chasserot S., Fromont M., Befort N., Jami J., Beck G., Pictet R. Complete complementary DNA of rat tyrosine aminotransferase messenger RNA. Deduction of the primary structure of the enzyme. J Mol Biol. 1985 Jul 20;184(2):347–350. doi: 10.1016/0022-2836(85)90386-9. [DOI] [PubMed] [Google Scholar]
- Griffith S. M., Vance C. P. Aspartate aminotransferase in alfalfa root nodules : I. Purification and partial characterization. Plant Physiol. 1989 Aug;90(4):1622–1629. doi: 10.1104/pp.90.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm G. H., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1986 Jan 10;14(1):5–9. doi: 10.1093/nar/14.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M. D., Heldt H. W. Synthesis, storage, and stability of [4-14C]oxaloacetic acid. Anal Biochem. 1985 Mar;145(2):393–397. doi: 10.1016/0003-2697(85)90379-3. [DOI] [PubMed] [Google Scholar]
- Huynh Q. K., Sakakibara R., Watanabe T., Wada H. The complete amino acid sequence of mitochondrial glutamic oxaloacetic transaminase from rat liver. J Biochem. 1981 Sep;90(3):863–875. doi: 10.1093/oxfordjournals.jbchem.a133543. [DOI] [PubMed] [Google Scholar]
- Kahn M. L., Ziermann R., Dehò G., Ow D. W., Sunshine M. G., Calendar R. Bacteriophage P2 and P4. Methods Enzymol. 1991;204:264–280. doi: 10.1016/0076-6879(91)04013-e. [DOI] [PubMed] [Google Scholar]
- Kahn M., Hanawalt P. Size distribution of DNA replicative intermediates in bacteriophage P4 and in Escherichia coli. J Mol Biol. 1979 Mar 15;128(4):501–525. doi: 10.1016/0022-2836(79)90290-0. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kahn M. L. Symbiotic phenotypes of auxotrophic mutants of Rhizobium meliloti 104A14. J Gen Microbiol. 1988 Apr;134(4):913–919. doi: 10.1099/00221287-134-4-913. [DOI] [PubMed] [Google Scholar]
- Kittell B. L., Helinski D. R., Ditta G. S. Aromatic aminotransferase activity and indoleacetic acid production in Rhizobium meliloti. J Bacteriol. 1989 Oct;171(10):5458–5466. doi: 10.1128/jb.171.10.5458-5466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu S., Ogawa T., Ogawa H., Kagamiyama H. Branched-chain amino acid aminotransferase of Escherichia coli: nucleotide sequence of the ilvE gene and the deduced amino acid sequence. J Biochem. 1985 Apr;97(4):993–999. doi: 10.1093/oxfordjournals.jbchem.a135176. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Okuno S., Ogawa T., Ogawa H., Kagamiyama H. Aspartate aminotransferase of Escherichia coli: nucleotide sequence of the aspC gene. J Biochem. 1985 Apr;97(4):1259–1262. doi: 10.1093/oxfordjournals.jbchem.a135173. [DOI] [PubMed] [Google Scholar]
- Marino G., Nitti G., Arnone M. I., Sannia G., Gambacorta A., De Rosa M. Purification and characterization of aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. J Biol Chem. 1988 Sep 5;263(25):12305–12309. [PubMed] [Google Scholar]
- McDermott T. R., Kahn M. L. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J Bacteriol. 1992 Jul;174(14):4790–4797. doi: 10.1128/jb.174.14.4790-4797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P. K., Hale T. I., Christen P. Evolutionary relationships among aminotransferases. Tyrosine aminotransferase, histidinol-phosphate aminotransferase, and aspartate aminotransferase are homologous proteins. Eur J Biochem. 1989 Dec 8;186(1-2):249–253. doi: 10.1111/j.1432-1033.1989.tb15202.x. [DOI] [PubMed] [Google Scholar]
- Obaru K., Nomiyama H., Shimada K., Nagashima F., Morino Y. Cloning and sequence analysis of mRNA for mouse aspartate aminotransferase isoenzymes. J Biol Chem. 1986 Dec 25;261(36):16976–16983. [PubMed] [Google Scholar]
- Rastogi V. K., Watson R. J. Aspartate aminotransferase activity is required for aspartate catabolism and symbiotic nitrogen fixation in Rhizobium meliloti. J Bacteriol. 1991 May;173(9):2879–2887. doi: 10.1128/jb.173.9.2879-2887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibach P. H., Streeter J. G. Evaluation of active versus passive uptake of metabolites by Rhizobium japonicum bacteroids. J Bacteriol. 1984 Jul;159(1):47–52. doi: 10.1128/jb.159.1.47-52.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibach P. H., Streeter J. G. Metabolism of C-labeled photosynthate and distribution of enzymes of glucose metabolism in soybean nodules. Plant Physiol. 1983 Jul;72(3):634–640. doi: 10.1104/pp.72.3.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rej R. A convenient continuous-rate spectrophotometric method for determination of amino acid substrate specificity of aminotransferases: application to isoenzymes of aspartate aminotransferase. Anal Biochem. 1982 Jan 1;119(1):205–210. doi: 10.1016/0003-2697(82)90687-x. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Lyttleton P., Robertson J. G. C(4)-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4284–4288. doi: 10.1073/pnas.78.7.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl L., Vance C. P., Pedersen W. B. Products of Dark CO(2) Fixation in Pea Root Nodules Support Bacteroid Metabolism. Plant Physiol. 1990 May;93(1):12–19. doi: 10.1104/pp.93.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Quandt J., Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene. 1989 Aug 1;80(1):161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- Somerville J. E., Kahn M. L. Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J Bacteriol. 1983 Oct;156(1):168–176. doi: 10.1128/jb.156.1.168-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers M. D. Carbon metabolism in Rhizobium species. Annu Rev Microbiol. 1985;39:89–108. doi: 10.1146/annurev.mi.39.100185.000513. [DOI] [PubMed] [Google Scholar]
- Streeter J. G. Carbohydrate, organic Acid, and amino Acid composition of bacteroids and cytosol from soybean nodules. Plant Physiol. 1987 Nov;85(3):768–773. doi: 10.1104/pp.85.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M. H., Tanizawa K., Tanaka H., Kuramitsu S., Kagamiyama H., Hirotsu K., Okamoto A., Higuchi T., Soda K. Thermostable aspartate aminotransferase from a thermophilic Bacillus species. Gene cloning, sequence determination, and preliminary x-ray characterization. J Biol Chem. 1991 Feb 5;266(4):2567–2572. [PubMed] [Google Scholar]
- Udvardi M. K., Kahn M. L. Isolation and analysis of a cDNA clone that encodes an alfalfa (Medicago sativa) aspartate aminotransferase. Mol Gen Genet. 1991 Dec;231(1):97–105. doi: 10.1007/BF00293827. [DOI] [PubMed] [Google Scholar]
- Watson R. J. Analysis of the C4-dicarboxylate transport genes of Rhizobium meliloti: nucleotide sequence and deduced products of dctA, dctB, and dctD. Mol Plant Microbe Interact. 1990 May-Jun;3(3):174–181. doi: 10.1094/mpmi-3-174. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Rastogi V. K. Cloning and nucleotide sequencing of Rhizobium meliloti aminotransferase genes: an aspartate aminotransferase required for symbiotic nitrogen fixation is atypical. J Bacteriol. 1993 Apr;175(7):1919–1928. doi: 10.1128/jb.175.7.1919-1928.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Olson M. E., Kahn M. L., Hurlbert R. E. Characterization of Tn5-Induced Mutants of Xenorhabdus nematophilus ATCC 19061. Appl Environ Microbiol. 1991 Apr;57(4):1173–1180. doi: 10.1128/aem.57.4.1173-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T., Kagamiyama H., Nozaki M., Soda K. Glutamate-aspartate transaminase from microorganisms. Methods Enzymol. 1985;113:83–89. doi: 10.1016/s0076-6879(85)13020-x. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]