Abstract
Pre-mRNA splicing requires a large number of RNA-binding proteins that have one or more RNA-recognition motifs (RRMs). Among these is the SR protein family, whose members are essential for splicing and are able to commit pre-mRNAs to the splicing pathway with overlapping but distinct substrate specificity. Some SR proteins, such as SC35, contain an N-terminal RRM and a C-terminal arginine/serine-rich (RS) domain, whereas others, such as SF2/ASF, also contain a second, atypical RRM. Although both the RRMs and the RS domain of SR proteins are required for constitutive splicing, it is unclear which domain(s) defines their substrate specificity, and whether two RRMs in a given SR protein function independently or act coordinately. Using domain swaps between SC35 and SF2/ASF and a functional commitment assay, we demonstrate that individual domains are functional modules, RS domains are interchangeable, and substrate specificity is defined by the RRMs. The atypical RRM of SF2/ASF does not appear to function alone in splicing, but can either activate or suppress the splicing specificity of an N-terminal RRM. Therefore, multiple RRMs in SR proteins act coordinately to achieve a unique spectrum of pre-mRNA substrate specificity.
Pre-mRNA splicing is a critical step in the posttranscriptional regulation of gene expression and requires both small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP factors. These factors assemble on pre-mRNA into a large complex known as a spliceosome, in which splicing takes place (for review, see ref. 1). A family of arginine/serine-rich non-snRNP splicing factors called SR proteins mediate various steps of spliceosomal assembly (for reviews, see refs. 2 and 3). In constitutive splicing, binding of SR proteins is sufficient to commit pre-mRNA to the splicing pathway (4), probably by facilitating U1 snRNP binding to a functional 5′ splice site (5), stabilizing complex assembly at the 3′ splice site (6), and bridging complexes assembled at the 5′ and 3′ splice sites (7–9). Additionally, SR proteins can modulate splice site selection, which is consistent with their involvement in early steps of splice site recognition (for review, see ref. 10).
SR proteins are essential splicing factors, but they have overlapping functions, at least in vitro, because all SR proteins can complement the splicing-deficient S100 cytoplasmic extract (11, 12). However, individual SR proteins clearly perform distinct activities in splicing different pre-mRNA substrates, and bind to different RNA sequence elements (for reviews, see refs. 2 and 3). It has been shown that the SR protein B52/SRp55 is essential for Drosophila development, although splicing of a number of transcripts examined in mutant larvae was not affected, indicating that these transcripts are not dependent on B52/SRp55 (13, 14). More recently, another SR protein, SF2/ASF, was analyzed by targeted gene disruption in a chicken B-cell line and shown to be essential for cell viability, indicating that this SR protein also has at least one nonredundant function in vivo (15).
Although SR proteins have distinct functions, little is known about the structural basis of their observed splicing specificity. The family of SR proteins is characterized by the presence of an N-terminal RNA recognition motif (RRM) and a C-terminal arginine/serine-rich (RS) domain. A subset of SR protein family members have a unique central domain separating the N-terminal canonical RRM and the C-terminal RS domain that is common to all family members. This central domain is an atypical RRM that lacks conserved residues present in the RNP2- and RNP1-submotifs of canonical RRMs (for review, see ref. 16). RNA-binding studies of SF2/ASF revealed that each of its RRMs can bind to RNA, but two RRMs together seem to bind better (17, 18) and with distinct binding specificity (19). On the other hand, in other RNA-binding proteins containing multiple RRMs, such as the U1-associated A (U1A) protein, the N-terminal RRM appears to bind specifically to U1 snRNA (20, 21), whereas the second RRM may independently bind to pre-mRNA sequences (22).
In this paper, we addressed the specificity and cooperation of individual domains in SR proteins using a functional splicing commitment assay. Our results demonstrate that the RS domains are functionally interchangeable and that splicing specificity is determined by the RRMs in SR proteins. Most interestingly, we found that the second, atypical RRM in SF2/ASF appears to be inactive in splicing as the sole RRM, but is able to suppress or activate the splicing specificity of an adjacent canonical RRM.
MATERIALS AND METHODS
Construction of Chimeric SC35 and SF2/ASF Proteins.
The SC35 coding sequence (23) was subcloned from pSP73-SC35 as a NarI-PvuI fragment into PharMingen’s pAcG2T baculovirus vector at the SmaI site to generate pAcG2T-SC35. The SF2/ASF coding sequence (24) was isolated from pBSK-SF2/ASF by PCR amplification using the upstream primer F1 (5′-GCCGGATCCGGAGGTGGTGTG-3′), which contains a BamHI site (underlined) and anneals to the SF2/ASF coding region corresponding to amino acids 2–6, and a downstream primer complementary to a plasmid sequence. The amplified cDNA was subcloned into pAcG2T at the BamHI and EcoRI sites to produce pAcG2T-SF2/ASF. Construction of individual chimeric proteins was as follows.
C1F2Frs.
The SC35 RRM coding sequence was amplified from pSP73-SC35 using an upstream primer in the plasmid and the downstream primer C1 (CGGCTCCGAGCTCCGTAGCCA), which contains a SacI site (underlined) and anneals to the region corresponding to SC35 amino acids 113–116. The amplified product was digested with StuI and SacI and used to replace the StuI-SacI fragment in pBSK-SF2/ASF. The hybrid cDNA insert was isolated by digestion with NarI, followed by filling-in with Klenow fragment, and then with EcoRI. The fragment was subcloned into pAcG2T at the SmaI and EcoRI sites.
F1C1Frs.
The SC35 RRM cDNA was amplified from pSP73-SC35 using the upstream primer C2 (CTCAGAGCTCTGAGCTACGGCCGC), which contains a SacI site (underlined) and anneals to the region corresponding to SC35 amino acids 1–5, and the downstream primer C3 (GGGGCCCGTAGCGCGCCATTTGC), which contains an ApaI site (underlined) and anneals to SC35 amino acids 87–93. The amplified cDNA was digested with SacI and ApaI and used to replace the SacI-ApaI fragment in pAcG2T-SF2/ASF.
F1F2Crs.
A cDNA encoding both RRMs of SF2/ASF was amplified from pAcG2T-SF2/ASF with a pAcG2T upstream primer and the downstream primer F2 (CCATACCTAGGACTTCTGGGCCCATC), which contains an AvrII site (underlined) and anneals to the region corresponding to SF2/ASF amino acids 195–203. The cDNA was subcloned into pAcG2T-SC35 using BamHI and AvrII.
C1F2Crs.
The cDNA encoding RRM2 of SF2/ASF was amplified from pAcG2T-SF2/ASF using the upstream primer F3 (CCCCACCTAGGCGGTCTGAAAC), which anneals to the region corresponding to SF2/ASF amino acids 114–121, and the downstream primer F2, both of which contain an AvrII site (underlined). The cDNA was subcloned into pAcG2T-SC35 at the AvrII site.
C1F1Crs.
The SF2/ASF RRM1 coding region was amplified from pBSK-SF2/ASF using the upstream primer F4 (GTCACCCCTAGGTCGGGAGGT), which anneals to the region corresponding to SF2/ASF amino acids 1–4, and the downstream primer F5 (ACGGCCTGTCCTAGGGCCGCT), which anneals to the region corresponding to SF2/ASF amino acids 121–127. The PCR product was digested with AvrII at the underlined sites in both primer sequences and the cDNA was inserted into the AvrII site in pAcG2T-SC35.
F2Frs.
The upstream primer F6 (CGGTCTGGATCCAGAGTGGTTG), which contains a BamHI site (underlined) and anneals to the region corresponding SF2/ASF amino acids 148–154, and a downstream primer for pBSK sequence were used to amplify the coding region for RRM2 and the RS domain of SF2/ASF from pBSK-SF2/ASF. The PCR product was digested with BamHI and EcoRI and the DNA was inserted into pAcG2T at the corresponding sites.
F1Frs.
The cDNA encoding the SF2/ASF RS domain was amplified using the upstream primer F7 (CCCAGAGCTCCAAGTTATGG), which contains a SacI site (underlined) and anneals to the region corresponding to SF2/ASF amino acids 197–203, and a downstream primer complementary to a plasmid sequence in pAcG2T-SF2/ASF. The cDNA product was digested with SacI and EcoRI and used to replace the SacI-EcoRI fragment in pAcG2T-SF2/ASF.
F1Crs.
The cDNA encoding the SC35 RS domain was amplified using primer C4 (TGGCTACGGAGCTCGGAG), which contains a SacI site (underlined) and anneals to the region corresponding to SC35 amino acids 113–119, and a downstream primer complementary to a plasmid sequence in pAcG2T-SC35. The amplified product was digested with SacI and EcoRI and used to replace the SacI-EcoRI fragment in pAcG2T-SF2/ASF.
Baculovirus Expression of Recombinant Proteins.
All constructs were sequenced prior to transfection. The PharMingen Baculogold System was used to express wild-type and chimeric proteins in Sf9 cells. Following initial transfection, the viral titer was increased through three rounds of amplification over 16 days. Final protein production was performed using 1–2 ml of virus stock (about 1 × 108 pfu/ml) to infect 2 × 107 Sf9 cells in one 150-mm culture dish. In general, 10 dishes were infected with each construct. Infected cells were harvested, resuspended in 10 ml of PBS with 1% Triton X-100, and lysed by sonication. Cellular debris was removed by centrifugation for 30 min at 35,000 rpm in a Beckman SW41 rotor, and the supernatant was added to Pharmacia Sepharose 4B glutathione beads equilibrated with PBS. Proteins were allowed to bind at 4°C for 1 hr to overnight. Following three washes with PBS, proteins were eluted with 15 mM reduced glutathione in BC100 buffer (100 mM KCl/20 mM Hepes·HCl, pH 7.6/10% glycerol). The protein concentration was normalized to about 0.1 mg/ml by Coomassie blue staining after SDS/PAGE, using BSA as a standard. The samples were aliquoted and stored at −80°C until they were used.
Splicing Commitment Assay.
The assay was carried out as described (4). Briefly, individual splicing mixtures were set up containing 0.2 μg of each SR protein (the volume was adjusted to 12 μl by adding BC100), 2 μl labeled pre-mRNA substrate (105 cpm, 0.05 pmol), 1 μl ATP (12.5 mM), 1 μl phosphocreatine (0.5 M, Sigma), 1 μl MgCl2 (80 mM), 0.25 μl DTT (0.2 M), 0.25 μl RNAsin (40 units per μl, Promega), and 2.5 μl H2O. After incubation at 30°C for 10 min, a mixture containing 3 μl nuclear extract plus 2 μl competitor RNA (1 pmol/μl; the unlabeled competitor is the 5′ half of human β-globin pre-mRNA transcribed from plasmid T7Hβ linearized with XhoI in the middle of the intron, see ref. 25) was added to each tube and incubation was continued for 2 hr. Products of the splicing reactions were resolved on a 6% denaturing polyacrylamide gel. Quantitation was obtained using a scan analysis program, and splicing efficiency was expressed as the ratio of spliced mRNA/unspliced pre-mRNA.
RESULTS
Construction and Expression of SF2/ASF and SC35 Chimeric Proteins.
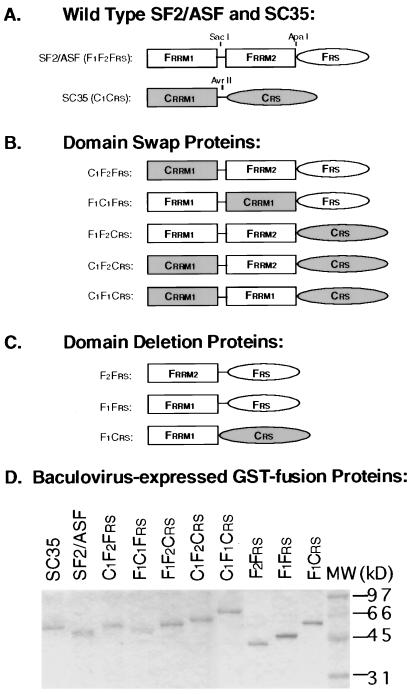
We chose two SR proteins, SF2/ASF (which has the additional RRM) and SC35 (which lacks it), as models to investigate the specificity contributed by individual domains of SR proteins in constitutive splicing. The splicing activity of wild-type and chimeric proteins was measured by a commitment assay, in which a labeled pre-mRNA is preincubated with a specific SR protein, followed by addition of a nuclear extract whose ability to splice by itself is blocked by excess competitor RNA (4). We have previously shown that SC35 commits human β-globin pre-mRNA to splicing more efficiently than SF2/ASF, whereas SF2/ASF, but not SC35, commits HIV tat pre-mRNA to splicing (ref. 4; see also Fig. 2). To investigate which domains of SF2/ASF contribute to tat pre-mRNA splicing, we constructed and expressed a series of chimeric glutathione S-transferase (GST)-fusion SR proteins (Fig. 1). GST-fusion proteins were purified in a single step from baculovirus-infected cells (Fig. 1D). To ensure that the GST-fusion proteins function in splicing with the same specificity as authentic proteins, we compared untagged SF2/ASF and SC35 with the GST-fusion proteins in splicing of both human β-globin and tat pre-mRNA. The results confirmed that the GST-fusion proteins have the same substrate specificity as native SR proteins (A.M. et al., unpublished data).
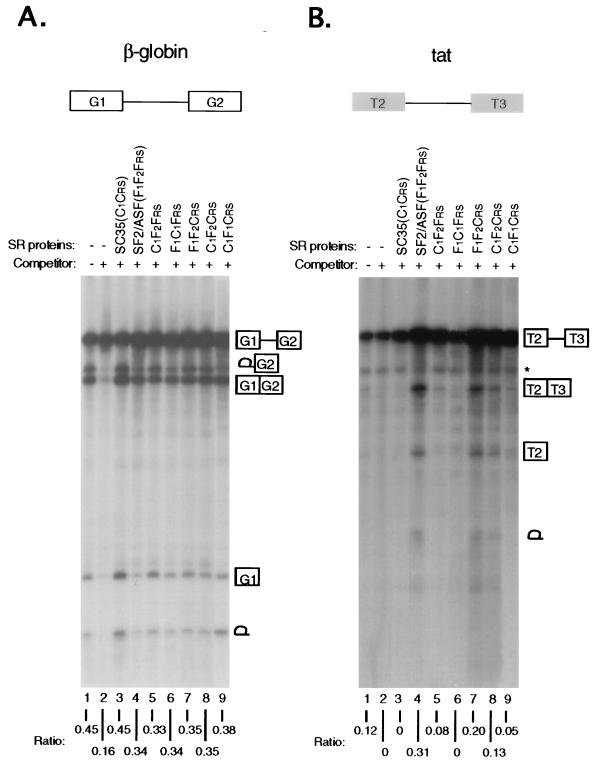
Figure 2.
Activities of domain-swap proteins in a splicing commitment assay. (A) Human β-globin pre-mRNA (transcribed from a minigene, pSP73–HβΔ6, which comprises exons G1 and G2 and intron 1) was spliced in the absence (lane 1) or the presence (lanes 2–9) of unlabeled competitor RNA corresponding to the 5′ half of the human β-globin pre-mRNA (4, 25). In each lane (from 2 to 9), 0.2 μg of the indicated protein was preincubated with the labeled pre-mRNA, followed by the addition of 3 μl of HeLa cell nuclear extract blocked with 2 pmol of unlabeled competitor RNA. (B) HIV tat pre-mRNA (transcribed from a minigene, pSP73-tat, which contains exons T2 and T3 and a truncated intron 1) was spliced under the same conditions. The bands corresponding to splicing intermediates and products are indicated. ∗, aberrant tat pre-mRNA cleavage product unrelated to splicing as previously described (11). Splicing efficiency is shown as the ratio of spliced to unspliced RNA underneath each lane.
Figure 1.
Construction and expression of SF2/ASF-SC35 chimeric proteins. (A) Wild-type SF2/ASF and SC35. Domains of SF2/ASF and SC35 are shown by open and shaded boxes, respectively (not to scale). Three restriction sites shown in the wild-type constructs were used to swap domains between the two proteins. The nomenclature is based on the origin of each domain: F, a domain from SF2/ASF; C, a domain from SC35. RRM, RNA-recognition motif. RS, arginine/serine-rich domain. (B) Domain-swap proteins. (C) Domain-deletion proteins. (D) Purified GST-fusion proteins resolved by 12.5% SDS/PAGE. Molecular weight markers are indicated on the right.
RRMs, but Not RS Domains, Determine Splicing Specificity.
To determine whether the splicing specificity of SF2/ASF with the tat pre-mRNA is defined by its RRMs or by its RS domain, we first substituted individual domains of SF2/ASF with the corresponding domains of SC35 to generate C1F2Frs, F1C1Frs, and F1F2Crs (Fig. 1B). All three chimeric GST-fusion proteins were active in committing human β-globin pre-mRNA to the splicing pathway, indicating that the chimeric proteins fold properly to function in constitutive splicing (Fig. 2A). In contrast, striking differences in the activities of the same proteins were observed in tat pre-mRNA splicing commitment (Fig. 2B). The F1F2Crs mutant, in which the RS domain of SF2/ASF was replaced with that of SC35, was as active as wild-type SF2/ASF, suggesting that the RS domains of SF2/ASF and SC35 are functionally interchangeable, and therefore, do not contribute to tat splicing specificity. This result is reminiscent of the previous observation that the RS domains from the Drosophila splicing regulators Tra and SWAP are interchangeable in vivo (26). In contrast, replacing the N-terminal RRM (RRM1) of SF2/ASF with that of SC35 (C1F2Frs) dramatically reduced, but did not completely abolish, its ability to splice the tat pre-mRNA. Replacing the second RRM (RRM2) of SF2/ASF with the RRM of SC35 (F1C1Frs), however, completely abolished tat splicing. These findings demonstrate that both RRMs of SF2/ASF contribute to tat splicing specificity.
The Atypical RRM2 of SF2/ASF Activates SC35 for tat Splicing.
To further evaluate the contributions of RRM1 and RRM2 of SF2/ASF in defining tat substrate specificity, we constructed two additional chimeric proteins, C1F1Crs and C1F2Crs, by inserting individual RRMs from SF2/ASF into SC35 (Fig. 1B). Both proteins were highly active in β-globin splicing commitment (Fig. 2A). When tested with the tat pre-mRNA, the insertion of RRM2 into SC35 conferred tat splicing specificity more efficiently than the insertion of RRM1 (compare lanes 8 and 9 in Fig. 2B). These data suggest that the atypical RRM2 of SF2/ASF plays a major role in defining tat pre-mRNA specificity by positively modulating an upstream canonical RRM from either SF2/ASF or SC35, although the natural combination of RRM1 and RRM2 in wild-type SF2/ASF results in maximal synergy for tat pre-mRNA splicing (compare lanes 4, 8, and 9 in Fig. 2B). The results of these functional studies are consistent with the previous observations that RRM1 and RRM2 of SF2/ASF cooperate for general binding to RNA (17, 18) and for high affinity binding to specific splicing enhancer elements (19), similar to those present in tat pre-mRNA (27, 28).
The Canonical RRM1 of SF2/ASF Can Function in Splicing in the Absence of the Atypical RRM2.
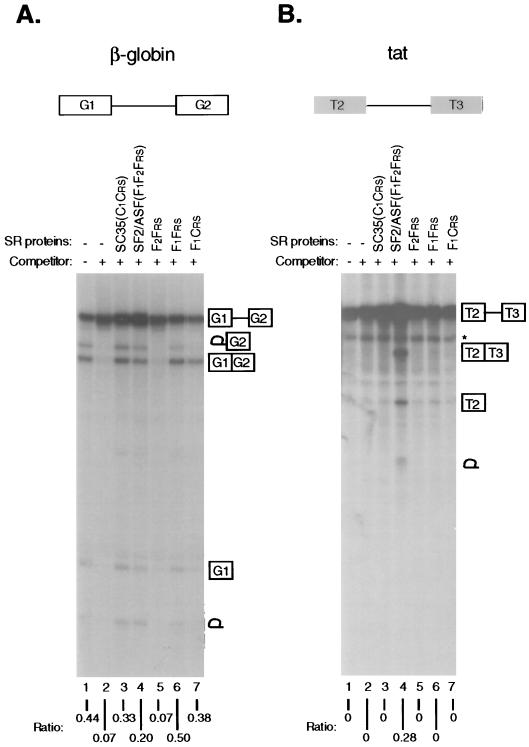
The observation that domains from two SR proteins can be interchanged to generate chimeric proteins that are active in both β-globin and tat splicing commitment suggests that individual domains in SR proteins are functional modules, which can act either independently or cooperatively. In a number of other RNA-binding proteins with multiple RRMs, it has been observed that individual RRMs function independently in binding to RNA (20–22, 29). The RRMs of SF2/ASF also appear to function independently in the formation of a ternary complex with pre-mRNA and U1 snRNP, since either RRM1 or RRM2, in combination with the RS domain, is sufficient to promote binding of purified U1 snRNP to a 5′ splice site (30). To extend this observation in the context of the complete splicing reaction, we deleted RRM1 or RRM2 from SF2/ASF, or fused RRM1 of SF2/ASF with the RS domain of SC35, to produce F2Frs, F1Frs, and F1Crs, respectively (Fig. 1C). As expected from the requirement for both RRMs of SF2/ASF in tat pre-mRNA splicing, all three domain-deletion mutants were inactive in tat splicing (Fig. 3B). We observed no activity of F2Frs in β-globin splicing (Fig. 3A), indicating either that RRM2 of SF2/ASF cannot function alone, or that the deletion mutant is misfolded and therefore inactive in the commitment assay. We also tested an RS domain deletion mutant of SF2/ASF (referred to as RRM/ψRRM by Cáceres and Krainer, see ref. 17) in the commitment assay. This mutant was previously shown to be inactive in constitutive splicing by S100 extract complementation, but capable of affecting alternative splice site selection in nuclear extracts (17). We detected no activity of the mutant protein in either β-globin or tat splicing, confirming that the RS domain is also essential for constitutive splicing in nuclear extracts (data not shown). In contrast, both F1Frs, and F1Crs were functional in committing β-globin pre-mRNA to splicing (Fig. 3A). Therefore, the N-terminal canonical RRM1 of SF2/ASF can function in splicing without the atypical RRM2, in a structural arrangement that resembles that of authentic SC35. In this single RRM context, the RS domains are again interchangeable between the two SR proteins.
Figure 3.
Activities of domain-deletion mutants in the splicing commitment assay. (A) Human β-globin pre-mRNA. (B) HIV tat pre-mRNA. Each pre-mRNA was spliced in the absence (lane 1) or presence (lanes 2–7) of unlabeled competitor RNA after preincubation under splicing conditions with buffer control (lanes 1 and 2) or with wild-type and domain-deletion mutants (lanes 3–7), as indicated above the autoradiogram.
The Atypical RRM2 of SF2/ASF Suppresses Its Activity in IgM Splicing.
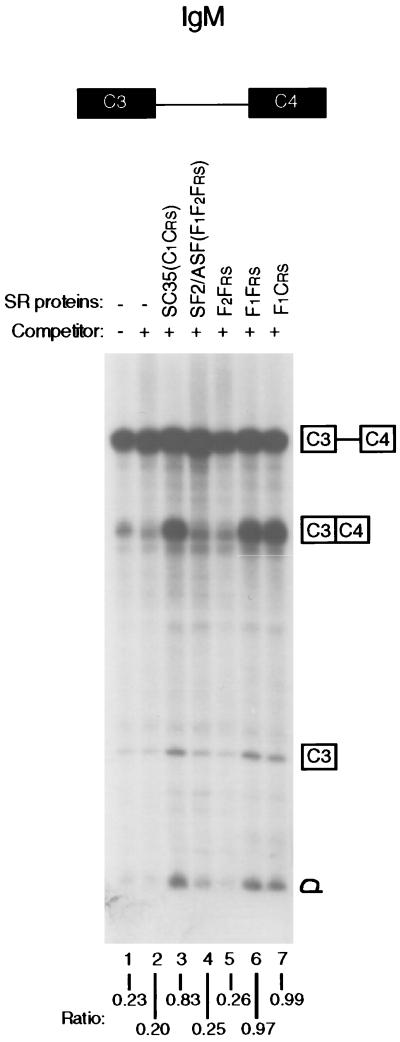
Recently, it was observed by S100 complementation that a constitutive splicing event in a human IgM pre-mRNA comprising exons C3 and C4 depends on SC35, but not on SF2/ASF (A.M. et al., unpublished results). Because F1Frs and SC35, both of which consist of a single RRM joined to an RS domain, behaved similarly in β-globin splicing, we asked whether the lack of activity of wild-type SF2/ASF in IgM splicing was due to the suppression of IgM-specific splicing through negative modulation by the atypical RRM2 of SF2/ASF. As shown in Fig. 4, wild-type SC35, but not SF2/ASF, committed IgM pre-mRNA to splicing. Remarkably, deletion of RRM2 from SF2/ASF allowed commitment of IgM pre-mRNA to splicing (F1Frs). As was seen for β-globin and tat pre-mRNAs, the RRM2 of SF2/ASF as the sole RRM could not function in IgM splicing (F2Frs), and the RS domains from SF2/ASF and SC35 were interchangeable (F1Crs). This experiment clearly shows that RRM2 negatively modulates the N-terminal RRM1 of native SF2/ASF to suppress its activity in committing IgM pre-mRNA to the splicing pathway.
Figure 4.
Activities of domain-deletion mutants in commitment of an IgM pre-mRNA. The splicing commitment assay was the same as in Fig. 3, except that an IgM pre-mRNA (transcribed from a mini-gene, pμC3-C4) was used as the splicing substrate (31), which comprises the constitutive exons C3 and C4 and intron 3.
DISCUSSION
The present studies illustrate the modular nature of SR proteins and show that, although the RS domains are essential for constitutive splicing, they are interchangeable between SC35 and SF2/ASF for the substrates tested. These observations suggest that the RS domain of SR proteins may function as an effector domain to activate splicing, but plays no role in defining substrate specificity. However, in light of the phylogenetic conservation of RS domains in individual SR proteins (16), and the proposed involvement of this domain in specific protein–protein interactions (5, 8) and in regulation of splicing or localization by phosphorylation (2, 3), individual RS domains are probably not functionally equivalent in vivo, as they may engage in specific interactions to determine the efficiency, regulation, or localization of individual members of the SR family of splicing factors.
Our findings on the function and cooperation of RRMs in SR proteins determined by a functional splicing assay extend previous structure–function studies on SF2/ASF. These studies showed that a deletion within RRM2 (18) or the complete removal of the domain (17) abolished both the in vitro splicing and 5′ splice site switching activities associated with SF2/ASF. In addition, the high-affinity consensus RNA elements selected with the wild-type SF2/ASF are distinct from that selected with the RRM1 of SF2/ASF alone (19). These observations have led to the conclusion that the individual RNA recognition motifs of SF2/ASF act synergistically in RNA binding and splicing. Our studies extend this view of domain cooperation by showing that the SF2/ASF-SC35 chimeric proteins are highly active in human β-globin splicing. Thus, the RRM1 of SF2/ASF need not be coupled with the adjacent RRM2 to function as a general splicing factor. Our observation emphasizes that the individual domains of SF2/ASF may act independently as functional modules, which confirms and extends previous binding studies with SF2/ASF. First, the individual RRMs of SF2/ASF can each bind to RNA (17–19). Second, either RRM1 or RRM2 of SF2/ASF in combination with its RS domain is capable of interacting with U1 snRNP and the 5′ splice site to form ternary complexes (30). Thus, it appears that a single RRM plus an RS domain is sufficient for 5′ splice site recognition.
This finding appears to contradict previous reports that the RRM2 of SF2/ASF is required for β-globin pre-mRNA splicing in a splicing-deficient S100 cytosolic extract, which contains only trace amounts of SR proteins (17, 18). This discrepancy may result from differences in the assay systems used (S100 extract complementation vs. commitment in nuclear extract) or in the way recombinant proteins were obtained (his-tagged proteins expressed in Escherichia coli vs. GST-fusions expressed in baculovirus) or both. We therefore tested our baculovirus-expressed GST-fusion proteins by S100 complementation. All chimeric proteins (Fig. 1B) gave results identical to those (as shown in Fig. 2) obtained by the commitment assay (A.M. et al., unpublished results), indicating that both assays provide equivalent measures of the substrate specificity of SR proteins. In contrast, however, we detected only a trace amount of activity with both F1Frs and F1Crs in β-globin splicing by S100 complementation (data not shown). Therefore, it appears that the discrepancy results largely from the nature of the two assays. The commitment assay (see Materials and Methods) may facilitate specific RNA–protein interactions during preincubation of the SR protein with the pre-mRNA. In addition, this assay employs competitor RNA-blocked nuclear extracts, which may contain a higher concentration of splicing factors for cooperation than the S100 extract.
Our results demonstrate that, in the case of SF2/ASF, the atypical RRM2 does not appear to function alone in splicing, but can either positively or negatively modulate splicing activity in conjunction with a canonical N-terminal RRM1. The mechanism for such modulation is not known and may operate in one of the following ways. First, individual RRMs in an SR protein may have their own RNA-binding specificity and interact independently with distinct RNA elements in pre-mRNA, which can lead to the stimulation or suppression of spliceosome assembly. Although there is no experimental evidence to support this model for SF2/ASF, other RNA-binding proteins, such as U1A, appear to function in such a manner (22). Second, the RNA-binding properties of adjacent RRMs may be mutually affected such that two RRMs together select a distinct set of optimal RNA sequences present in splicing substrates. This possibility is consistent with the observations that the RRM2 of SF2/ASF is a functional RNA-binding domain (17, 18), but the two-RRM configuration in native SF2/ASF has a different RNA-binding specificity (19). Finally, RRM2 of SF2/ASF may be a domain involved in both protein–protein and protein–RNA interactions, thereby modulating splicing specificity during spliceosome assembly. Such a mechanism has been shown to operate for other DNA- and RNA-binding proteins. For example, in the case of two snRNP-associated proteins (U1A and U2B”), whereas the U1A protein can recognize U1 snRNA alone, the U2B” protein binds specifically to U2 snRNA only in the presence of a second protein, U2A′ (20, 21). Further studies are required to distinguish among these possibilities.
Acknowledgments
We thank Drs. A. Watakabe and Y. Shimura for the gift of the IgM mini-gene plasmid. S.D.C. and J.M.Y. are supported by National Institutes of Health pre- and postdoctoral fellowships, respectively. This work was supported by grants from the National Institutes of Health to X.-D.F. and A.R.K.; X.-D.F. is a Searle Scholar.
ABBREVIATIONS
- RRM
RNA-recognition motifs
- RS
arginine/serine-rich
- snRNP
small nuclear ribonucleoprotein particles
References
- 1.Krämer A. Annu Rev Biochem. 1996;65:367–490. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 2.Fu X-D. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 3.Manley J L, Tacke R. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 4.Fu X-D. Nature (London) 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 5.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Nature (London) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 6.Tarn W Y, Steitz J A. Proc Natl Acad Sci USA. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu X-D, Maniatis T. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J Y, Maniatis T. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 9.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horowitz D S, Krainer A R. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 11.Krainer A R, Conway G C, Kozak D. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 12.Zahler A M, Lane W S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 13.Ring H Z, Lis J T. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng X, Mount S M. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Manley J L. Genes Dev. 1996;6:837–847. [Google Scholar]
- 16.Birney E, Kumar S, Krainer A R. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cáceres J F, Krainer A R. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo P, Manley J L. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tacke R, Manley J L. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherly D, Boelens W, Dathan N A, van Venrooij W J, Mattaj I W. Nature (London) 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 21.Bentley R C, Keene J D. Mol Cell Biol. 1991;11:1829–1839. doi: 10.1128/mcb.11.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutz C S, Alwine J C. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 23.Fu X-D, Maniatis T. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 24.Krainer A R, Mayeda A, Kozak D, Binns G. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 25.Reed R, Griffith J, Maniatis T. Cell. 1988;53:949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Bingham P M. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 27.Amendt B A, Hesslein D, Chang L J, Stoltzfus C M. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staffa A, Cochrane A. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sachs A B, Davis R W, Kornberg R D. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamison S F, Pasman Z, Wang J, Will C, Lührmann R, Manley J L, Garcia-Blanco M A. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watakabe A, Inoue K, Sakamoto H, Shimura Y. Nucleic Acids Res. 1989;17:8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]