Abstract
What is already known about this subject
Gliclazide is a widely used oral hypoglycaemic agent.
The major metabolites of gliclazide formed in vivo have been identified.
However, the cytochrome P450 enzymes catalysing the rate-limiting pathways of gliclazide elimination are unknown.
What this study adds
CYP2C9 is the major enzyme involved in the various hydroxylation pathways of gliclazide, although a contribution of CYP2C19 to tolymethylhydroxylation, the major metabolic route, cannot be discounted.
Factors known to influence CYP2C9 activity will provide the main source of variability in gliclazide pharmacokinetics.
Aims
To identify the human cytochrome P450 (CYP) enzymes responsible for the formation of the 6β-hydroxy (6β-OHGz), 7β-hydroxy (7β-OHGz) and hydroxymethyl (MeOH-Gz) metabolites of gliclizide (Gz).
Methods
6β-OHGz, 7β-OHGz and MeOH-Gz formation by human liver microsomes and a panel of recombinant human P450s was measured using a high-performance liquid chromatography procedure, and the kinetics of metabolite formation was determined for each pathway. Effects of prototypic CYP enzyme selective inhibitors were characterized for each of the microsomal metabolic pathways.
Results
Microsomes from six human livers converted Gz to its 6β-OHGz, 7β-OHGz, and MeOH-Gz metabolites, with respective mean (± SD) Km values of 461 ± 139, 404 ± 143 and 334 ± 75 µm and mean Vmax values of 130 ± 55, 82 ± 31 and 268 ± 115 pmol min−1 mg−1, respectively. Vmax/Km ratios for the microsomal reactions parallelled relative metabolite formation in vivo. Sulfaphenazole inhibited microsomal 6β-OHGz, 7β-OHGz and MeOH-Gz formation by 87, 83 and 64%, respectively, whereas S-mephenytoin caused significant inhibition (48%) of only MeOH-Gz formation. Recombinant CYP2C9, CYP2C18 and CYP2C19 catalysed all hydroxylation pathways, whereas CYP2C8 formed only 6β-OHGz and 7β-OHGz.
Conclusion
Taken together, the results indicate that CYP2C9 is the major contributor to Gz metabolic clearance, although CYP2C19 may also be involved in MeOH-Gz formation (the major metabolic pathway). Factors known to influence CYP2C9 activity will provide the main source of variability in Gz pharmacokinetics.
Keywords: CYP2C18, CYP2C19, CYP2C9, cytochrome P450, gliclazide, reaction phenotyping
Oral hypoglycaemic agents remain the cornerstone of the treatment of Type 2 diabetes in patients not responsive to diet, exercise and weight reduction [1]. Of the various oral hypoglycaemic agents available, sulphonylureas and biguanides are considered the first-line treatment. Amongst the sulphonylureas, gliclazide (Gz) is widely used. Almost 4 and 1.2 million prescriptions for Gz were dispensed in the UK (http://www.ic.nhs.uk/pubs/precostanalysis2005/final/file) and Australia (http://www.medicareaustralia.gov.au/providers/health_statistics/statistical_reporting/pbs.htm), respectively, in 2005. Considerable interindividual variability in metabolic clearance is a feature of the sulphonylureas [2, 3] and differences in elimination are believed to contribute to therapeutic outcome and the occurrence of adverse effects [3]. Despite the widespread use of Gz, factors that contribute to pharmacokinetic variability have received less attention than for other sulphonylureas [3].
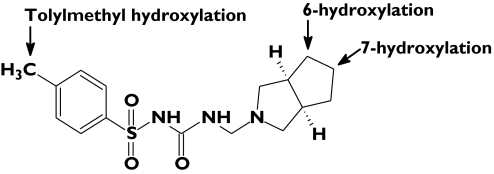
Gz is extensively metabolized in humans and multiple urinary metabolites have been identified [4, 5]. Like the structurally related 4-methylsulphonylureas tolbutamide [6, 7] and torsemide (used clinically as a diuretic) [8], tolylmethyl hydroxylation (to form MeOH-Gz) followed by oxidation to the corresponding carboxylic acid is the major metabolic pathway. However, Gz is also hydroxylated in the azabicyclooctyl ring with the potential to form seven monohydroxylated metabolites (Figure 1). Of these, four monohydroxy-azabicyclooctyl metabolites (at the 6α-, 6β-, 7α- and 7β- positions) have been identified in human urine [4, 5]. Based on metabolite excretion in urine to 96 h postdose, MeOH-Gz (as this metabolite plus the derived carboxylic acid), 6α-OHGz, 6β-OHGz, 7α-OHGz and 7β-OHGz accounted for 59, 1, 20, 6 and 14% of the recovered dose, respectively [4]. Thus, 6β-, 7β- and tolylmethyl- hydroxylation represent the rate-liming pathways of Gz elimination.
Figure 1.
Positions of gliclazide hydroxylation
Although the in vivo fate of Gz has been elucidated, no systematic investigation has been undertaken to identify the human cytochrome P450 (CYP) enzymes responsible for the various pathways of Gz hydroxylation. The aim of this study was to characterize Gz hydroxylation by human liver microsomes (HLM) and, using CYP enzyme selective inhibitors and recombinant P450s, identify the human enzymes responsible for the formation of the individual hydroxylated metabolites in order to gain insights into factors likely to influence Gz elimination and response.
Materials and methods
Chemicals and reagents
Gliclazide (Gz, 1-(3-azabicyclo [3.3.0]oct-3-yl)-3-(4-methylphenylsulphonyl)urea), 6α-hydroxygliclazide (6α-OHGz; 1-(3-aza-6-hydroxybicyclo [3.3.0]oct-3-yl)-3-(4-methylphenylsulphonyl)urea) as the enantiomeric mixture (1R,5S,6S and 1S,5R,6R), 6β-hydroxygliclazide (6β-OHGz; 1-(3-aza-6-hydroxybicyclo [3.3.0]oct-3-yl)-3-(4-methylphenylsulphonyl)urea) as the enantiomeric mixture (1R,5S,6R and 1S,5R,6S), 7β-hydroxygliclazide (7β-OHGz; (1R,5S,7β)-1-(3-aza-7-hydroxybicyclo [3.3.0]oct-3-yl)-3-(4-methylphenylsulphonyl)urea) and hydroxymethylgliclazide (MeOH-Gz; 1-(3-azabicyclo [3.3.0]oct-3-yl)-3-(4-hydroxymethylphenylsulphonyl)urea) were gifts from Technologie Servier (Melbourne, Australia). Furafylline was obtained from Hoffman La Roche (Basel, Switzerland), whereas coumarin, quinidine hydrochloride monohydrate (S)-(+)-mephenytoin, troleandomycin triacetate, trimethoprim, sulfaphenazole, glucose-6-phosphate, glucose-6-phosphate dehydrogenase and β-nicotinamide adenine dinucleotide phosphate (NADP) were purchased from Sigma-Aldrich (Sydney, Australia). All other chemicals and reagents were of analytical reagent grade.
Human liver microsomes and recombinant CYP enzymes
Human livers (H6, H7, H10, H12, H13 and H40) were obtained from the human liver ‘bank’ of the Department of Clinical Pharmacology, Flinders Medical Centre. Approval was obtained from the Flinders Medical Centre Clinical Investigation Committee and from the donor next-of-kin for the procurement and use of human liver tissue in xenobiotic metabolism studies. Microsomes were prepared from human livers by differential centrifugation according to Bowalgaha et al. [9].
Recombinant human CYP enzymes and rat NADPH-cyctochrome P450 reductase were coexpressed in Escherichia coli following published methods; CYP1A2 according to Polasek et al. [10], CYP2A6, CYP2D6 and CYP2E1 as described by Parikh et al. [11], CYP2C8 and CYP2C9 by the method of Boye et al. [12] and CYP2C18 and CYP2C19 as reported by Kinobe et al. [13] and Cuttle et al. [14], respectively.
Kinetic and inhibitor studies
Incubation mixtures, in a total volume of 0.5 ml, contained HLM (0.5 mg of microsomal protein) or recombinant CYP [10 pmol, except for CYP2C8 (20 pmol)], Gz (50–750 µm), acetonitrile (1% v/v) and NADPH generating system (1 mm NADP, 100 mm glucose-6-phosphate, 1 IU glucose-6 phosphate dehydrogenase and 5 mm MgCl2) in phosphate buffer (0.1 m, pH 7.4). Incubations were performed in air at 37°C in a shaking water bath. After a 5-min preincubation, reactions were initiated by the addition of NADPH generating system. Reactions were terminated after 90 min (HLM) or 60 min (recombinant enzymes) by the addition of perchloric acid (5 µl, 11.6 m). Mixtures were vortex mixed, cooled on ice and then centrifuged at 3000 g for 10 min to precipitate microsomal protein. Sodium hydroxide (2 m, 5 µl) was added to a 200-µl aliquot of the supernatant fraction to raise the pH to 6 before injection onto the high-performance liquid chromatography (HPLC) column for analysis.
6α-OHGz, 6β-OHGz, 7β-OHGz, 7β-OHGz and MeOH-Gz formation was quantified by reversed phase HPLC using an Agilent 1100 series HPLC system (Agilent Technologies, Sydney, Australia), comprising a quaternary solvent delivery module with in-line degasser, autoinjector and variable-wavelength ultraviolet/visible detector. The HPLC was fitted with a Beckman ODS column (250 × 4 mm i.d., 5 µm particle size) (Beckman, Fullerton, CA, USA). The mobile phase consisted of an acetate buffer (5 mm, pH 4.3) containing 20% v/v acetonitrile (A) and acetonitrile (B) gradient delivered at a flow rate of 1.5 ml min−1: initial conditions were 96% A:4% B held for 3 min, changing to 76% A:24% B over 7 min then held for 2 min, changing to 40% A:60% B over 1 min, which was held for 0.5 min before returning to starting conditions. Gz and its hydroxy metabolites were detected at 235 nm. Retention times for 6α-OHGz, 6β-OHGz, 7β-OHGz, 7β-OHGz, MeOH-Gz and Gz, determined by reference to authentic standards, were 8.1, 8.5, 9.2, 10.2 and 15.5 min, respectively. Concentrations of the individual metabolites present in incubations were determined by comparison of the peak areas with calibration curves constructed over the concentration range 0.2–10 µm for each metabolite. Calibration curves were linear with r2 values >0.99. The lower limit of quantification was 0.045 µm (equivalent to rates of product formation of 0.50 pmol min−1 mg−1 for incubations with HLM as the enzyme source and 0.025 pmol min−1 per pmol CYP for incubations with recombinant CYP enzymes).
Incubation conditions were optimized to ensure linearity with respect to protein concentration and time; substrate depletion was <10%. Overall assay within-day precision was assessed at a single Gz concentration (400 µm) by measuring 6β-OHGz, 7β-OHGz and MeOH-Gz formation (see Results) in nine separate incubations with the same batch of HLM. The within-day coefficients of variation were 3.8, 3.2 and 4.1% for 6β-OHGz, 7β-OHGz and MeOH-Gz, respectively. Inhibition studies with CYP enzyme selective inhibitors were performed at a Gz concentration of 500 µm using pooled HLM (equal protein amounts from each liver). Concentrations of inhibitors used were: furafylline (10 µm, CYP1A2), coumarin (2.5 µm, CYP2A6), trimethoprim (100 µm, CYP2C8), sulfaphenazole (2.5 µm, CYP2C9) (S)-(+)-mephenytoin (100 µm, CYP2C19), quinidine (2.5 µm, CYP2D6) and troleandomycin (50 µm, CYP3A). The mechanism-based inhibitors furafylline and troleandomycin were preincubated with HLM for 10 min in the presence of NADPH generating system prior to the addition of Gz.
Kinetic constants for the various pathways of Gz hydroxylation were determined by fitting untransformed experimental data to the Michaelis–Menten equation using EnzFitter (Biosoft, Cambridge, UK), a nonlinear least squares fitting program. It should be noted that there was no deviation from hyperbolic kinetics for any of the hydroxylation pathways characterized. Data points represent the mean of duplicate determinations (<10% variance).
Results
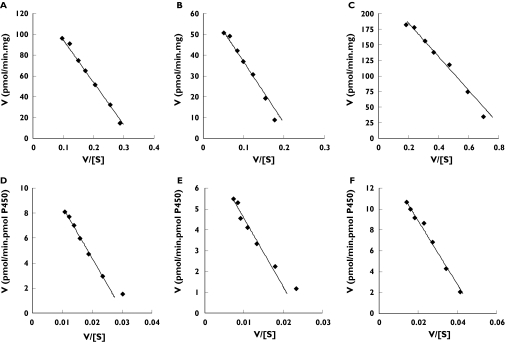
Microsomes from all six human livers studied hydroxylated Gz. The principal metabolites detected were 6β-OHGz, 7β-OHGz and MeOH-Gz. A small peak corresponding to 6α-OHGz was observed only at the highest Gz concentration (750 µm), while 7α-OHGz was not detected across the substrate concentration range employed. Formation of hydroxylated Gz metabolites was not observed in the absence of NADPH generating system. 6β-OHGz, 7β-OHGz and MeOH-Gz formation exhibited Michaelis–Menten kinetics with microsomes from the six livers (Figure 2). It should be noted that the poor solubility of Gz limited the concentration range studied (50–750 µm) and all incubations contained 1% by volume acetonitrile to assist solubilization of substrate. Mean apparent Km (± SD, with 95% CIs in parentheses) values for 6β-OHGz, 7β-OHGz and MeOH-Gz were 461 ± 139 µm (356–620 µm), 404 ± 143 µm (269–576 µm) and 334 ± 75 µm (270–422 µm), respectively. The mean Vmax (± SD, with 95% CIs in parentheses) values for 6β-OHGz, 7β-OHGz and MeOH-Gz were 130 ± 55 pmol min−1 mg−1 (70–195 pmol min−1 mg−1), 82 ± 31 pmol min−1 mg−1 (48–119 pmol min−1 mg−1) and 268 ± 115 pmol min−1 mg−1 (135–369 pmol min−1 mg−1), respectively. Based on intrinsic clearance (Vmax/Km) values, the predominant pathway is the formation of MeOH-Gz (through oxidation of the tolylmethyl group); the Vmax/Km ratio for MeOH-Gz (0.81 µl min−1 mg−1) formation was 2.8 and 3.9 times higher than the Vmax/Km ratios for 6β-OHGz (0.29 µl min−1 mg−1) and 7β-OHGz (0.21 µl min−1 mg−1), respectively.
Figure 2.
Eadie–Hofstee plots for gliclazide 6β-hydroxylation (A), 7β-hydroxylation (B) and tolylmethyl hydroxylation (C) by microsomes from liver H40, and gliclazide tolylmethyl hydroxylation by CYP2C9 (D), CYP2C18 (E) and CYP2C19 (F)
The effect of CYP selective inhibitors on the formation of Gz metabolites by HLM is shown in Table 1. Sulfaphenazole, a selective inhibitor of CYP2C9, decreased MeOH-Gz formation by approximately two-thirds and the 6β-OHGz and 7β-OHGz pathways by > 80%. (S)-(+)-mephenytoin inhibited the OHMe-Gz pathway by 48%, but exhibited only 11 and 17% inhibition of 6β-OHGz and 7β-OHGz formation, respectively. Interestingly, activation of MeOH-Gz formation (by 16–27%) occurred in the presence of coumarin, trimethoprim and quinidine, although these compounds caused a comparatively minor reduction in the formation of 6β-OHGz and 7β-OHGz. Similarly, furafylline and troleandomycin had a minor effect on Gz hydroxylation.
Table 1.
Effect of CYP enzyme selective inhibitors on the hydroxylation of gliclazide by pooled human liver microsomes
| Inhibitor and enzyme | Inhibitor concentration (µm) | Percent control activity | |||
|---|---|---|---|---|---|
| 6β-OHGz | 7β-OHGz | MeOH-Gz | |||
| Furafylline | (CYP1A2) | 10 | 92 | 94 | 93 |
| Coumarin | (CYP2A6) | 2.5 | 88 | 81 | 116 |
| Trimethoprim | (CYP2C8) | 100 | 89 | 90 | 118 |
| Sulfaphenazole | (CYP2C9) | 2.5 | 13 | 17 | 36 |
| (S)-(+)-mephenytoin | (CYP2C19) | 100 | 89 | 83 | 52 |
| Quinidine | (CYP2D6) | 2.5 | 80 | 73 | 127 |
| Troleandomycin | (CYP3A4) | 50 | 93 | 93 | 83 |
Data represent mean of duplicate estimations using pooled human liver microsomes (see ‘Kinetic and inhibitor studies’).
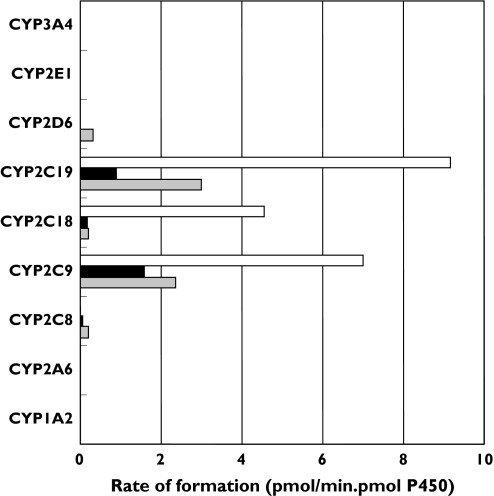
A panel of recombinant P450s was screened to determine the capacity of the major drug-metabolizing CYP enzymes to catalyse Gz hydroxylation at a substrate concentration of 500 µm. Results are shown in Figure 3. All members of the CYP2C subfamily hydroxylated Gz. Of the remaining P450s, only CYP2D6 hydroxylated Gz (at the 6β position). Kinetic parameters were determined with recombinant CYP2C8, CYP2C9, CYP2C18 and CYP2C19 as the enzyme sources (Table 2 and Figure 2). MeOH-Gz formation was the predominant metabolic pathway for CYP2C9, CYP2C18 and CYP2C19. However, CYP2C8 formed only 6β-OHGz and 7β-OHGz. The largest differences in Km occurred for 6β-hydroxylation, with a threefold difference between CYP2C8 and CYP2C19. Generally, however, differences in intrinsic clearances (i.e. Vmax/Km) between and within metabolic pathways arose predominately from variability in Vmax.
Figure 3.
Formation of 6β-, 7β- and tolylmethyl- hydroxy gliclazide by recombinant human CYP enzymes (at a substrate concentration of 500 µm). MeOHGz (□); 7β-OHGz (▪); 6β-OHGz ( )
)
Table 2.
Derived kinetic parameters for gliclazide hydroxylation by recombinant CYP2C enzymes
| 6β-OHGz | 7β-OHGz | MeOH-Gz | Pathway ratio* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | 6β-OHGz | 7β-OHGz | MeOH-Gz |
| CYP2C8 | 984 | 0.63 | 0.0006 | 346 | 0.06 | 0.0002 | NA† | NA | NA | 1.0 | 0.3 | 0 |
| CYP2C9 | 471 | 4.5 | 0.0096 | 327 | 2.6 | 0.0080 | 415 | 13 | 0.0306 | 1.0 | 0.8 | 3.2 |
| CYP2C18 | 794 | 0.53 | 0.0007 | 524 | 0.37 | 0.0007 | 328 | 7.8 | 0.0239 | 1.0 | 1.1 | 35.8 |
| CYP2C19 | 321 | 5.0 | 0.0155 | 434 | 1.7 | 0.0039 | 304 | 15 | 0.0493 | 1.0 | 0.2 | 3.2 |
Units: Km, µm; Vmax, pmol min−1 pmol CYP−1; Vmax/Km, µl min−1 pmol CYP−1.
Based on Vmax/Km ratios.
No measurable activity.
Discussion
Based on metabolite recovery data, tolylmethyl hydroxylation (to form MeOH-Gz) and subsequent oxidation to the corresponding aromatic carboxylic acid, is responsible for approximately 60% of Gz metabolic clearance in humans [4]. Hydroxylation in the azabicyclooctyl ring, at the 6α-, 6β-, 7α- and 7β- positions, accounts for the remainder of Gz metabolism in vivo, although 6α- and 7α- hydroxylation represent minor biotransformation pathways (≤6% of the recovered dose) [4]. Consistent with the in vivo observations, human liver microsomes formed MeOH-Gz, 6β-OHGz and 7β-OHGz. Vmax/Km ratios for the microsomal reactions paralleled relative urinary metabolite ratios in vivo.
Inhibition of the human liver microsomal metabolism of a compound by CYP enzyme selective inhibitors is considered to provide the most reliable indication of the relative contributions of P450s to a metabolic pathway [15]. The effect of the prototypic CYP2C9 inhibitor sulfaphenazole [6, 7] demonstrates that this enzyme is responsible for half to two-thirds of MeOH-Gz formation, while inhibition by (S)-(+)-mephenytoin indicates that CYP2C19 accounts for the remainder of tolylmethyl hydroxylation. Based on sulfaphenazole inhibition, CYP2C9 is responsible for > 80% of Gz 6β- and 7β- hydroxylation. Activity screening and kinetic experiments with recombinant P450s support a major contribution of CYP2C9 and CYP2C19 to hepatic Gz tolylmethyl hydroxylation. Of the hepatically expressed enzymes (i.e. CYP2C8, CYP2C9 and CYP2C19), highest Vmax/Km values were observed with CYP2C9 and CYP2C19. In contrast to the ‘gold standard’ microsomal inhibition data, however, derived kinetic constants obtained for recombinant P450s suggest significant involvement of CYP2C19 in Gz 6β- and 7β- hydroxylation. Similar observations have been reported for chlorpropamide 2-hydroxylation [16]; comparable activities were obtained with recombinant CYP2C9 and CYP2C19 despite near complete inhibition of the human liver microsomal reaction by sulfaphenazole and the absence of an effect of CYP2C19 genotype on chlorpropamide clearance and response in vivo. These examples highlight the difficulties associated with data interpretation when reaction phenotyping is conducted with recombinant P450s alone. However, Km values for the various pathways of Gz hydroxylation by recombinant CYP2C enzymes were comparable in value, consistent with the monophasic kinetics observed for the human liver microsomal reactions.
CYP2C9 exhibits genetic polymorphism and the six livers used in this study were previously genotyped [17] for the most common variant alleles, namely CYP2C9*2 and CYP2C9*3. None carried the Arg144Cys mutation associated with CYP2C9*2 and one was heterozygous for CYP2C9*3. CYP2C9*3 encodes a protein with leucine at residue 359, compared with isoleucine in the wild-type enzyme [17, 18]. The leucine-359 variant is associated with lower activity towards all known CYP2C9 substrates [2, 3, 17–20]. Consistent with a major involvement of CYP2C9 in the Gz hydroxylations, Vmax/Km values observed for 6β-OHGz, 7β-OHGz and MeOH-Gz formation by microsomes from the CYP2C9*3 heterozygous liver were four- to fivefold lower than the mean Vmax/Km values for the corresponding pathways obtained using microsomes from the five CYP2C9*1 homozygous livers. The predominant involvement of CYP2C9 in the Gz hydroxylations is also consistent with the known major contribution of this enzyme to the metabolism of other sulphonylureas, and a previous report linking MeOH-Gz formation to the tolbutamide hydroxylase activity of rat liver microsomes [21]. Reaction phenotyping studies in vitro and/or investigation of the effects of polymorphic variants on drug disposition in vivo indicate that CYP2C9 activity is the principal determinant of the metabolic clearance for tolbutamide [6, 7, 22, 23], chlorpropamide [16], glipizide [24], glimepiride [25] and glibenclamide [25–27]. Similarly, CYP2C9 is responsible for the tolylmethyl hydroxylation of the structurally related sulphonylurea torsemide [8, 28], used clinically as a diuretic. Although CYP2C19 has been implicated in tolbutamide and chlorpropamide hydroxylation in vitro [16, 29], phenotype- and genotype-based clinical studies exclude a significant contribution of this enzyme to drug elimination in vivo [26, 30]. However, the concordance between (S)-(+)-mephenytoin inhibition of human liver microsomal Gz tolylmethyl hydroxylation and MeOH-Gz formation by recombinant CYP2C19 observed here suggests a potentially significant role for CYP2C19 in Gz metabolic clearance, given that tolylmethyl hydroxylation represents the major elimination pathway in vivo. Although the contribution of CYP2B6 to Gz hydroxylation was not investigated here, this enzyme appears to have a minor role in drug metabolism and the sulfaphenazole and (S)-(+)-mephenytoin inhibition data preclude significant involvement of another P450 in hepatic Gz hydroxylation.
Regioselectivity was observed for the various pathways of Gz hydroxylation by the individual CYP2C enzymes. The relative formation of 6β- and 7β-OHGz (based on Vmax/Km ratios) differed between CYP2C9 and CYP2C19, while CYP2C18 preferentially formed MeOH-Gz. Unlike other CYP2C enzymes, CYP2C8 did not catalyse Gz tolylmethyl hydroxylation. Previous work has similarly shown an approximately 60-fold difference in torsemide tolylmethyl hydroxylation by CYP2C8 and CYP2C9 [28].
Of further interest was the observation that the Vmax/Km ratio for Gz tolylmethyl hydroxylation by CYP2C18 was similar to the ratios determined for CYP2C9 and CYP2C19. In contrast to CYP 2C8, 2C9 and 2C19, CYP2C18 appears to be expressed only to a minor extent in liver [31, 32]. However, it is the most abundant CYP2C enzyme in skin and lung and is also found in brain, duodenum, kidney, mammary gland and uterus [33–35]. CYP2C18 has previously been shown to metabolize torsemide, phenytoin, diclofenac, tienilic acid and tolbutamide, other predominantly CYP2C9 substrates [13, 36–39]. Thus, there appears to be overlapping substrate selectivity between the two enzymes. At this stage the functional significance of CYP2C18-catalysed biotransformation in extrahepatic tissues remains unknown, but a role in phenytoin hypersensitivity has been postulated [13].
In summary, CYP2C9 is the principal enzyme involved in the 6β-, 7β- and tolylmethyl- hydroxylation of Gz. Numerous factors are known to affect CYP2C9 activity, including genetic polymorphism and drug–drug interactions [2, 19, 20], and these may contribute to dose variability and the development of adverse effects during therapy with sulphonylureas [3]. Indeed, an association between the incidence of severe hypoglycaemia and CYP2C9 poor metabolizer genotypes has been reported in patients receiving sulphonylureas [40]. CYP2C19, another polymorphic enzyme, may also contribute to Gz tolylmethyl hydroxylation, introducing an additional potential source of variability in Gz pharmacokinetics.
Acknowledgments
Competing interests: None declared.
This work was supported in part by a grant from the National Health and Medical Research Council of Australia.
References
- 1.Inzucchi SE. Oral antihyperglycemic therapy for type 2 diabetes. JAMA. 2002;287:360–72. doi: 10.1001/jama.287.3.360. [DOI] [PubMed] [Google Scholar]
- 2.Kirchheiner J, Brockmoller J. Clinical consequences of cytochrome P4502C9 polymorphisms. Clin Pharmacol Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Kirchheiner J, Roots I, Goldammer M, Rosenkranz B, Brockmoller J. Effect of genetic polymorphisms in cytochrome P450 (CYP) 2C9 and CYP2C8 on the pharmacokinetics of oral antidiabetic drugs – clinical relevance. Clin Pharmacokinet. 2005;44:1209–25. doi: 10.2165/00003088-200544120-00002. [DOI] [PubMed] [Google Scholar]
- 4.Oida T, Yoshida K, Kagemoto A, Sekine Y, Higashijima T. The metabolism of gliclazide in man. Xenobiotica. 1985;15:87–96. doi: 10.3109/00498258509045338. [DOI] [PubMed] [Google Scholar]
- 5.Taylor AR, Brownsill RD, Grandon H, Lefoulon F, Petit A, Luijten W, Kopelman P, Walther B. Synthesis of putative metabolites and investigation of the metabolic fate of gliclazide [1-(3-azabicyclo(3,3,0)oct-3-yl)-3-(4-methylphenylsulfonyl) urea] in diabetic patients. Drug Metab Dispos. 1996;24:55–64. [PubMed] [Google Scholar]
- 6.Miners JO, Smith KJ, Robson RA, McManus ME, Veronese ME, Birkett DJ. Tolbutamide hydroxylation by human liver microsomes – kinetic characterization and relationship to other cytochrome-P-450 dependent xenobiotic oxidations. Biochem Pharmacol. 1988;37:1137–44. doi: 10.1016/0006-2952(88)90522-9. [DOI] [PubMed] [Google Scholar]
- 7.Miners JO, Birkett DJ. Use of tolbutamide as a substrate probefor human hepatic cytochrome P450 2C9. Methods Enzymol. 1996;272:139–45. doi: 10.1016/s0076-6879(96)72017-7. [DOI] [PubMed] [Google Scholar]
- 8.Miners JO, Rees DLP, Valente L, Veronese ME, Birkett DJ. Human hepatic cytochrome-P450 2C9 catalyzes the rate-limiting pathway of torsemide metabolism. J Pharmacol Exp Ther. 1995;272:1076–81. [PubMed] [Google Scholar]
- 9.Bowalgaha K, Elliot DJ, Mackenzie PI, Knights KM, Swedmark S, Miners JO. S-Naproxen and desmethylnaproxen glucuronidation by human liver microsomes and recombinant human UDP-glucuronosyltransferases (UGT): role of UGT2B7 in the elimination of naproxen. Br J Clin Pharmacol. 2005;60:423–33. doi: 10.1111/j.1365-2125.2005.02446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polasek TM, Elliot DJ, Somogyi AA, Gillam EMJ, Lewis BC, Miners JO. An evaluation of potential mechanism-based inactivation of human drug metabolizing cytochromes P450 by monoamine oxidase inhibitors, including isoniazid. Br J Clin Pharmacol. 2006;61:570–84. doi: 10.1111/j.1365-2125.2006.02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parikh A, Gillam EMJ, Guengerich FP. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nature Biotechnol. 1997;15:784–8. doi: 10.1038/nbt0897-784. [DOI] [PubMed] [Google Scholar]
- 12.Boye SL, Kerdpin O, Elliot DJ, Miners JO, Kelly L, McKinnon RA, Bhasker CR, Yoovathaworn K, Birkett DJ. Optimizing bacterial expression of catalytically active human cytochromes P450: comparison of CYP2C8 and CYP2C9. Xenobiotica. 2004;34:49–60. doi: 10.1080/00498250310001636868. [DOI] [PubMed] [Google Scholar]
- 13.Kinobe RT, Parkinson OT, Mitchell DJ, Gillam EMJ. P4502C18 catalyzes the metabolic bioactivation of phenytoin. Chem Res Toxicol. 2005;18:1868–75. doi: 10.1021/tx050181o. [DOI] [PubMed] [Google Scholar]
- 14.Cuttle L, Munns AJ, Hogg NA, Scott JR, Hooper WD, Dickinson RG, Gillam EMJ. Phenytoin metabolism by human cytochrome P450: involvement of P450 3A and 2C forms in secondary metabolism and drug–protein adduct formation. Drug Metab Dispos. 2000;28:945–50. [PubMed] [Google Scholar]
- 15.Miners JO, Veronese ME, Birkett DJ. In-vitro approaches for the prediction of human drug metabolism. Annu Reports Med Chem. 1994;272:307–16. [Google Scholar]
- 16.Shon JH, Yoon YR, Kim MJ, Kim KA, Lim YC, Liu KH, Shin DH, Lee CH, Cha IJ, Shin JG. Chlorpropamide 2-hydroxylation is catalysed by CYP2C9 and CYP2C19 in vitro: chlorpropamide disposition is influenced by CYP2C9, but not by CYP2C19 genetic polymorphism. Br J Clin Pharmacol. 2005;59:552–63. doi: 10.1111/j.1365-2125.2005.02364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhasker CR, Miners JO, Coulter S, Birkett DJ. Allelic and functional variability of cytochrome P4502C9. Pharmacogenetics. 1997;7:51–8. doi: 10.1097/00008571-199702000-00007. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang Z-Y, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–9. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 19.Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–38. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rettie AE, Jones JP. Clinical and toxicological relevance of CYP2C9: drug–drug interactions and pharmacogenetics. Annu Rev Pharmacol Toxicol. 2005;45:477–94. doi: 10.1146/annurev.pharmtox.45.120403.095821. [DOI] [PubMed] [Google Scholar]
- 21.Rieutord A, Stupans I, Shenfield GM, Gross AS. Gliclazide hydroxylation by rat liver microsomes. Xenobiotica. 1995;25:1345–54. doi: 10.3109/00498259509061922. [DOI] [PubMed] [Google Scholar]
- 22.Veronese ME, Mackenzie PI, Doecke CJ, McManus ME, Miners JO, Birkett DJ. Tolbutamide and phenytoin hydroxylations by cDNA-expressed human liver cytochrome-P4502C9. Biochem Biophys Res Commun. 1991;175:1112–8. doi: 10.1016/0006-291x(91)91680-b. [DOI] [PubMed] [Google Scholar]
- 23.Veronese ME, Doecke CJ, Mackenzie PI, McManus ME, Miners JO, Rees DLP, Gasser R, Meyer UA, Birkett DJ. Site-directed mutation studies of human liver cytochrome P-450 isoenzymes in the CYP2C subfamily. Biochem J. 1993;289:533–8. doi: 10.1042/bj2890533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidd RS, Straughn AB, Meyer MC, Blaisdell J, Goldstein JA, Dalton JT. Pharmacokinetics of chlorpheniramine, phenytoin, glipizide and nifedipine in an individual homozygous for the CYP2C9*3 allele. Pharmacogenetics. 1999;9:71–80. doi: 10.1097/00008571-199902000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Niemi M, Cascorbi I, Timm R, Kroemer HK, Neuvonen PJ, Kivisto KT. Glyburide and glimepiride pharmacokinetics in subjects with different CYP2C9 genotypes. Clin Pharmacol Ther. 2002;72:326–32. doi: 10.1067/mcp.2002.127495. [DOI] [PubMed] [Google Scholar]
- 26.Kirchheiner J, Brockmoller J, Meineke I, Bauer S, Rohde W, Meisel C, Roots I. Impact of CYP2C9 amino acid polymorphisms on glyburide kinetics and on the insulin and glucose response in healthy volunteers. Clin Pharmacol Ther. 2002;71:286–96. doi: 10.1067/mcp.2002.122476. [DOI] [PubMed] [Google Scholar]
- 27.Yin OQP, Tomlinson B, Chow MSS. CYP2C9, but not CYP2C19, polymorphisms affect the pharmacokinetics and pharmacodynamics of glyburide in Chinese subjects. Clin Pharmacol Ther. 2005;78:370–7. doi: 10.1016/j.clpt.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Kerdpin O, Elliot DJ, Boye SL, Birkett DJ, Yoovathaworn K, Miners JO. Differential contribution of active site residues in substrate recognition sites 1 and 5 to cytochrome p450 2C8 substrate selectivity and regioselectivity. Biochemistry. 2004;43:7834–42. doi: 10.1021/bi0496844. [DOI] [PubMed] [Google Scholar]
- 29.Wester MR, Lasker JM, Johnson EF, Raucy JL. CYP2C19 participates in tolbutamide hydroxylation by human liver microsomes. Drug Metab Dispos. 2000;28:354–9. [PubMed] [Google Scholar]
- 30.Shon JH, Yoon YR, Kim KA, Lim YC, Lee KJ, Park JY, Chi IJ, Flockhart DA, Shin JG. Effects of CYP2C19 and CYP2C9 genetic polymorphisms on the disposition of and blood glucose lowering response to tolbutamide in humans. Pharmacogenetics. 2002;12:111–9. doi: 10.1097/00008571-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 31.Furuya H, Meyer UA, Gelboin HV, Gonzalez FJ. Polymerase chain reaction directed identification, cloning, and quantification of human CYP2C18 messenger RNA. Mol Pharmacol. 1991;40:375–82. [PubMed] [Google Scholar]
- 32.Richardson TH, Griffin KJ, Jung F, Raucy JL, Johnson EF. Targeted antipeptide antibodies to cytochrome P450 2C18 based on epitope mapping of an inhibitory monoclonal antibody to P450 2C5. Arch Biochem Biophys. 1997;338:157–64. doi: 10.1006/abbi.1996.9817. [DOI] [PubMed] [Google Scholar]
- 33.Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. 1997;17:2985–93. doi: 10.1128/mcb.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mace K, Bowman ED, Vautravers P, Shields PG, Harris CC, Pfeifer AMA. Characterisation of xenobiotic-metabolising enzyme expression in human bronchial mucosa and peripheral lung tissues. Eur J Cancer. 1998;34:914–20. doi: 10.1016/s0959-8049(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 35.Klose TS, Blaisdell JA, Goldstein JA. Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Molec Toxicol. 1999;13:289–95. doi: 10.1002/(sici)1099-0461(1999)13:6<289::aid-jbt1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 36.Miners JO, Coulter S, Birkett DJ, Goldstein JA. Torsemide metabolism by CYP2C9 variants and other human CYP2C subfamily enzymes. Pharmacogenetics. 2000;10:267–70. doi: 10.1097/00008571-200004000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Mancy A, Antignac M, Minoletti C, Dijols S, Mouries V, Duong NTH, Battioni P, Dansette PM, Mansuy D. Diclofenac and its derivatives as tools for studying human cytochromes P450 active sites. Particular efficiency and regioselectivity of P4502Cs. Biochemistry. 1999;38:14264–70. doi: 10.1021/bi991195u. [DOI] [PubMed] [Google Scholar]
- 38.Jean P, LopezGarcia P, Dansette P, Mansuy D, Goldstein JL. Oxidation of tienilic acid by human yeast-expressed cytochromes P-450 2C8, 2C9, 2C18 and 2C19 – evidence that this drug is a mechanism-based inhibitor specific for cytochrome P-450 2C9. Eur J Biochem. 1996;241:797–804. doi: 10.1111/j.1432-1033.1996.00797.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhuge J, Yu YN, Qian YL, Li X. Establishment of a transgenic cell line stably expressing human cytochrome P4502C18 and identification of a CYP2C18 clone with exon 5 missing. World J Gastroenterol. 2002;8:888–92. doi: 10.3748/wjg.v8.i5.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holstein A, Plaschke A, Ptak M, Egberts EH, El-Din J, Brockmoller J, Kircheiner J. Association between CYP2C9 slow metabolizer genotypes and severe hypoglycaemia on medication with sulphonylurea hypoglycaemic agents. Br J Clin Pharmacol. 2005;60:103–6. doi: 10.1111/j.1365-2125.2005.02379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]