SUMMARY
Metabolic fluxes estimated from stable-isotope studies provide a key to understanding cell physiology and regulation of metabolism. A limitation of the classical method for metabolic flux analysis (MFA) is the requirement for isotopic steady state. To extend the scope of flux determination from stationary to nonstationary systems, we present a novel modeling strategy that combines key ideas from isotopomer spectral analysis (ISA) and stationary MFA. Isotopic transients of the precursor pool and the sampled products are described by two parameters, D and G parameters, respectively, which are incorporated into the flux model. The G value is the fraction of labeled product in the sample, and the D value is the fractional contribution of the feed for the production of labeled products. We illustrate the novel modeling strategy with a nonstationary system that closely resembles industrial production conditions, i.e. fed-batch fermentation of E. coli that produces 1,3-propanediol (PDO). Metabolic fluxes and the D and G parameters were estimated by fitting labeling distributions of biomass amino acids measured by GC/MS to a model of E. coli metabolism. We obtained highly consistent fits from the data with 82 redundant measurements. Metabolic fluxes were estimated for 20 time points during course of the fermentation. As such we established, for the first time, detailed time profiles of in vivo fluxes. We found that intracellular fluxes changed significantly during the fed-batch. The intracellular flux associated with PDO pathway increased by 10%. Concurrently, we observed a decrease in the split ratio between glycolysis and pentose phosphate pathway from 70/30 to 50/50 as a function of time. The TCA cycle flux, on the other hand, remained constant throughout the fermentation. Furthermore, our flux results provided additional insight in support of the assumed genotype of the organism.
Keywords: Instationary fluxes, 13C flux analysis, elementary metabolite units (EMU), gas chromatography mass spectrometry (GC/MS), statistical analysis
1 Introduction
Isotopic tracer experiments are routinely used to quantify fluxes in biochemical networks (Stephanopoulos, 1999). In a typical carbon-13 labeling experiment a labeled substrate, e.g. [1-13C]glucose, is introduced to the metabolic system where it is taken up and metabolized by the cells. Atoms of the specifically labeled substrate are biochemically rearranged generating molecules with specific labeling patterns that can be detected by nuclear magnetic resonance (NMR) and mass spectrometry (MS) (Des Rosiers et al., 2004; Szyperski, 1995). The labeling patterns of cellular components provide rich information for the estimation of metabolic fluxes. The goal of metabolic flux analysis (MFA) is to extract as much flux information as possible from stable isotope measurements and from external flux measurements. Currently, MFA requires that the system is at metabolic and isotopic steady state, i.e. that labeling of the substrate and measured metabolite pools are equilibrated. For example, this condition is approximated in continuous culture experiments after five or more residence times. The isotopic steady state assumption simplifies the computational problem of MFA from a problem involving ordinary differential equations (ODE) to a problem involving only algebraic equations (Schmidt et al., 1997). However, analysis of nonstationary systems is important because many systems of industrial and medical significance never reach isotopic steady state (Drysch et al., 2003; Kelleher, 2001). In this contribution, we extend the scope of MFA to nonstationary systems, i.e. systems that do not approximate isotopic steady state, without increasing the complexity of computations (i.e. we only solve algebraic equations). To account for isotopic transients we have developed a novel modeling strategy that combines key ideas from isotopomer spectral analysis (ISA) and stationary MFA. ISA was initially introduced as a general method for modeling polymerization biosynthesis reactions in systems that do not approximate isotopic steady state (Kelleher and Masterson, 1992). In analogy with the ISA method we introduce two parameters to account for isotopic transients. These parameters are the fraction of labeled products in the sample (the G parameter), and the fractional contribution of the feed to the production of labeled products (the D parameter). We illustrate this modeling strategy with a nonstationary system that closely resembles industrial production conditions, i.e. microbial fed-batch fermentation of E. coli that overproduces 1,3-propanediol (PDO). In this experiment neither the labeling of the substrate, nor the labeling of the biomass were at isotopic steady state. Metabolic fluxes and their confidence intervals were successfully estimated in this system using the novel modeling framework. We obtained an overdetermined data set consisting of labeling profiles of biomass amino acids (measured by GC/MS) and external flux measurements that were fitted to a detailed model of E. coli metabolism including the additional D and G parameters. We illustrate that both D and G parameters are required to obtain statistically acceptable fits. The estimated fluxes provided detailed time profiles of intracellular fluxes during the course of the fed-batch fermentation and as such provided valuable insight into the physiology of industrial overproduction of PDO that could not be obtained from classical stationary MFA. Thus, the novel modeling strategy allows us to study changes in metabolism that develop over periods of hours in nonstationary systems, which are of major industrial and medical importance.
2 Materials and methods
2.1 Materials
[1-13C]Glucose (99 At% 13C) and [U-13C]glucose (99 At% 13C) were purchased from IsoTec (Miamisburg, OH) and Cambridge Isotope Laboratories (Andover, MA), respectively. The defined growth medium contained (per liter medium): 2.8g KH2PO4, 2.0 g citric acid monohydrate, 1.1 g MgSO4·7H2O, 0.2 g FeSO4·7H2O, 0.2 g CaCl2·2H2O, 0.5 g Yeast Extract (BBL), 1 mL Balch’s Modified Trace Metals (100x), 0.5 mL sulfuric acid (98%), 1.5 mL phosphoric acid (85%), 0.4 mL Mazu DF204 antifoam, appropriate vitamin B12, and 2 mL spectinomycin (50 mg/mL). The natural feed contained 46.2 wt% natural glucose (not enriched in 13C) as the main carbon source. The 13C-labeled feed was chemically identical, and contained 75 wt% [1-13C]glucose and 25 wt% [U-13C]glucose.
2.2 Strain and growth conditions
We used an E. coli K12 strain that was metabolically engineered to overproduce 1,3-propanediol (Nakamura and Whited, 2003). The cultivation was performed as a fed-batch fermentation. E. coli was cultured in an aerobic fermentor with a working volume of 1 L. The pH was controlled at 6.8 ± 0.04 by addition of NH4OH, the temperature was controlled at 34 °C, and the dissolved oxygen was controlled at 10% ± 0.7 of saturation by adjusting the stirrer speed. The aeration rate was constant at 0.5 standard liters per minute (SLPM). Batch phase was initiated with 45.1 g of natural glucose medium. Glucose feed was initiated after 16.3 hr, and controlled such that glucose concentration in the medium was maintained at 45 ± 5 mM. After 18.6 hr, natural glucose feed was replaced with 13C-labeled glucose feed. After 30.0 hr, the labeled glucose feed was discontinued and replaced with natural glucose feed; the fermentation was continued for an additional 14.6 hr.
2.3 Off-gas analysis
Molar percentages of oxygen, carbon dioxide and nitrogen in the inlet and outlet air streams were monitored online by Prima 600S mass spectrometer (VG Gas, Manchester, UK). The flow rate of the inlet air stream was measured in standard liters per minute (SLPM, defined at 1 Atm and 25°C) using Brooks 5850 series mass flow controller. Fractional labeling of CO2 in the off-gas was determined from the relative intensities of 12CO2 (m/z 44) and 13CO2 (m/z 45).
2.4 Sampling and sample processing
Medium samples were periodically taken during the fed-batch fermentation for HPLC and GC/MS analysis. In total 21 samples of 11-21 mL were collected between time points 15.4 hr and 44.6 hr. Biomass concentration was determined by measuring the optical density at 550 nm (OD550), assuming 3.0 g/L/OD550 cell dry weight and 25.3 g/C-mol for molecular weight of dry biomass. Culture samples were centrifuged and the supernatant separated from the biomass pellet. The biomass pellet was stored at -80 °C and the supernatant at -20 °C prior to analysis.
2.5 HPLC analysis
Concentrations of glucose, glycerol, 1,3-propanediol (PDO), acetate, citrate, and pyruvate in the medium samples were measured by high-performance liquid chromatography (Waters HPLC, Shodex SH1011 sugar column, RI detector).
2.6 Gas chromatography / mass spectrometry
Gas chromatography / mass spectrometry (GC/MS) analysis was performed on a HP 5890 Series II GC (Gas Chromatograph) equipped with a DB-1701 (30 m × 0.25 mm i.d., 0.25 μm-phase thickness; Agilent J&W Scientific) capillary column, connected to HP 5971 quadrupole MSD (Mass Selective Detector) operating under ionization by electron impact (EI) at 70 eV. The mass spectrometer was calibrated using the ‘Max Sensitivity Autotune’ setting.
2.7 GC/MS analysis of biomass amino acids
GC/MS analysis of TBDMS derivatized amino acids was performed as described by Antoniewicz et al. (Antoniewicz et al., 2006a). In short, about 20 mg of wet biomass pellet was transferred to 700 μL of 6 N HCl and heated at 110 °C for 24 hr in a closed vacuum hydrolysis tube. After cooling to room temperature the solvent was evaporated and the residue dissolved in 150 μL of distilled water, which was then filtered through a 0.2 μm pore size filter to remove cell debris. The filtrate was evaporated to dryness and dissolved in 50 μL of pyridine, followed by addition of 70 μL of N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide (MTBSTFA). The mixture was heated at 60 °C for 30 min and transferred to an injection vial for GC/MS analysis. The injection volume was 1 μL and samples were injected in purged splitless mode. The amount of sample analyzed was controlled by varying the purge activation time between 1 sec and 1.5 min. Helium flow was maintained at 0.74 mL/min via electronic pressure control. The injection port temperature was 270 °C. The temperature of the column was started at 100 °C for 1.5 min, increased to 130 °C at 20 °C/min and increased to 220 °C at 10 °C/min and held for 3 min. The temperature was then increased to 280 °C at 5 °C/min and held for 3 min. The interface temperature was maintained at 300 °C. Mass spectra were acquired in the mass range m/z 195-445 at 2.7 scans/s. Measured mass isotopomer abundances were corrected for the concentration effect as described previously (Antoniewicz et al., 2006a). Mass isotopomer values for each fragment were expressed as fractional abundances, i.e. for each fragment the sum of all mass isotopomers equals one.
2.8 GC/MS analysis of glucose
Labeling of glucose was determined by GC/MS analysis of the aldonitrile pentapropionate derivative of glucose. About 20 μL of medium sample was deproteinized by addition of 300 μL of acetone (~4 °C). The mixture was centrifuged and the supernatant evaporated to dryness. 50 μL of 2 wt% hydroxylamine hydrochloride in pyridine was added to the dry residue and the mixture was heated at 90 °C for 60 min. This was followed by addition of 100 μL of propionic anhydride and heating at 60 °C for another 30 min. After cooling, the sample was evaporated to dryness, dissolved in 100 μL of ethyl acetate and transferred to an injection vial for GC/MS analysis. The injection volume was 1 μL and samples were injected in purged splitless mode. Helium flow was maintained at 0.88 mL/min via electronic pressure control. The injection port temperature was 250 °C. The temperature of the column was started at 80 °C for 1 min, increased to 280 °C at 20 °C/min, and held for 4 min. The interface temperature was maintained at 300 °C. Mass spectra were acquired in the mass range m/z 150-450 at 2.3 scans/s. The labeling of glucose was determined from the ion fragment at m/z 370 (C17H24O8N), which contains carbon atoms C1-C5 of glucose. Measured mass isotopomer distributions were corrected for natural isotope enrichments as described by Fernandez et al. (Fernandez et al., 1996). The corrected intensity at m/z 370 corresponds to the fraction of natural glucose, the intensity at m/z 371 to the fraction of [1-13C]glucose, and the sum of intensities at m/z 374 and 375 corresponds to the fraction of [U-13C]glucose (i.e., the m/z 374 peak was ~3% of m/z 375 peak resulting from incomplete labeling of [U-13C]glucose tracer).
2.9 Calculation of external fluxes
Cumulative consumption of glucose and citrate, and cumulative production of biomass, glycerol, PDO and acetate were calculated from measured concentrations and fermentor weight, after accounting for losses due to sampling. Rates of consumption and production were determined by fitting a smooth curve through the data points. Rates of total oxygen uptake (TOUR) and total carbon dioxide evolution (TCER) were calculated from off-gas analysis as follows:
| (1) |
| (2) |
These equations assume that nitrogen is not consumed, produced or accumulated in the system. As such, these equations account for any differences in temperature and pressure between the inlet and outlet air streams.
2.10 Metabolic network model
A detailed network model for E. coli metabolism was constructed. The network model contains reactions for glycolysis, pentose phosphate pathway, Entner-Doudoroff pathway, TCA cycle, PDO biosynthesis pathway, amphibolic reactions, one-carbon metabolism and amino acid biosynthesis reactions (Appendix A). The model is comprised of 75 reactions (with corresponding carbon transitions) utilizing 74 metabolites, with 5 substrates (i.e. glucose, citrate, O2, NH3, SO4), 5 products (i.e. PDO, biomass, CO2, acetate, and ATP), and 63 balanced intracellular metabolites. Glycerol was allowed to be either a substrate or a product depending on the direction of the external flux. Reversible reactions were modeled as separate forward and backward fluxes. Net and exchange fluxes were determined as follows: vnet = vf − vb; vexch = min(vf, vb). All forward and backward fluxes were required to be non-negative at the final solution. The model also included D and G parameters to account for isotopic transients (see section 3.6).
2.11 Simulation of labeling distributions using elementary metabolite units (EMUs)
Mass isotopomer distributions of amino acids were simulated using the EMU modeling framework (Antoniewicz et al., 2007). The EMU method is based on a highly efficient decomposition algorithm that identifies the minimum amount of information that is needed for isotopic simulations using the knowledge of atomic transitions occurring in the network reactions. It was shown previously that EMU models are equivalent to corresponding isotopomer models, however, require significantly fewer variables and computation time to simulate isotopic labeling in a system. In this case, EMU modeling of the E. coli network resulted in 95% reduction in the number of variables, i.e. 223 EMUs vs. 4612 isotopomers.
2.12 Flux determination and statistical analysis
Metabolic fluxes were estimated by fitting mass isotopomer abundances of biomass amino acid and 7 external fluxes to the model of E. coli (including the D and G parameters). Flux determination and statistical analysis were performed as described previously (Antoniewicz et al., 2006b). In short, the variance-weighted sum of squared differences between simulated and measured data was minimized using an algorithm based on successive quadratic programming. At convergence, accurate confidence intervals of fluxes were calculated by evaluating the sensitivity of the objective function with respect to individual fluxes. Validation of the fit was accomplished by a statistical test for the goodness-of-fit (i.e. chi-square test for model adequacy), and a normality test for the weighted residuals. To ensure that a global optimum was found, flux estimation was repeated at least ten times starting with random initial values for all fluxes. Sensitivity analysis was used to determine the relative importance of specific measurements for the estimation of individual fluxes.
3 Results
3.1 External fluxes
From the HPLC concentration measurements and off-gas analysis data external fluxes were calculated, i.e. net consumption and production rates (see Supplementary Materials). Figure 1A shows the external fluxes (mmol/h) as a function of fermentation time. The overall carbon balance closed within 2%, and the degree of reduction balance closed within 4% (Appendix B), indicating that there were no gross errors in the measurements. Figure 1B shows the rates expressed as biomass specific fluxes (mmol/h/gDW), which significantly decreased during the fermentation indicating that the cells were either becoming metabolically less active in time, or that the fraction of metabolically active cells decreased (i.e., the fraction of dead cells increased). We did not measure cell viability in this experiment, and thus the exact cause for the decrease in specific fluxes remains uncertain. External fluxes normalized to glucose uptake rate are shown in Figure 1C. Glucose flux to biomass decreased significantly from 80.3 C-mol/h at 18.3 hr to 14.6 C-mol/h at 40.7 hr, while the efflux of PDO gradually increased from 78.6 mol/h at 18.3 hr to 140.1 mol/h at 40.7 hr. Note that glycerol was initially produced by the cells (i.e. positive flux), but after 25 hrs glycerol was taken up by the cells (i.e. negative flux). The rates of acetate production and citrate uptake were negligible compared to the other fluxes (see Supplementary Materials).
Figure 1.
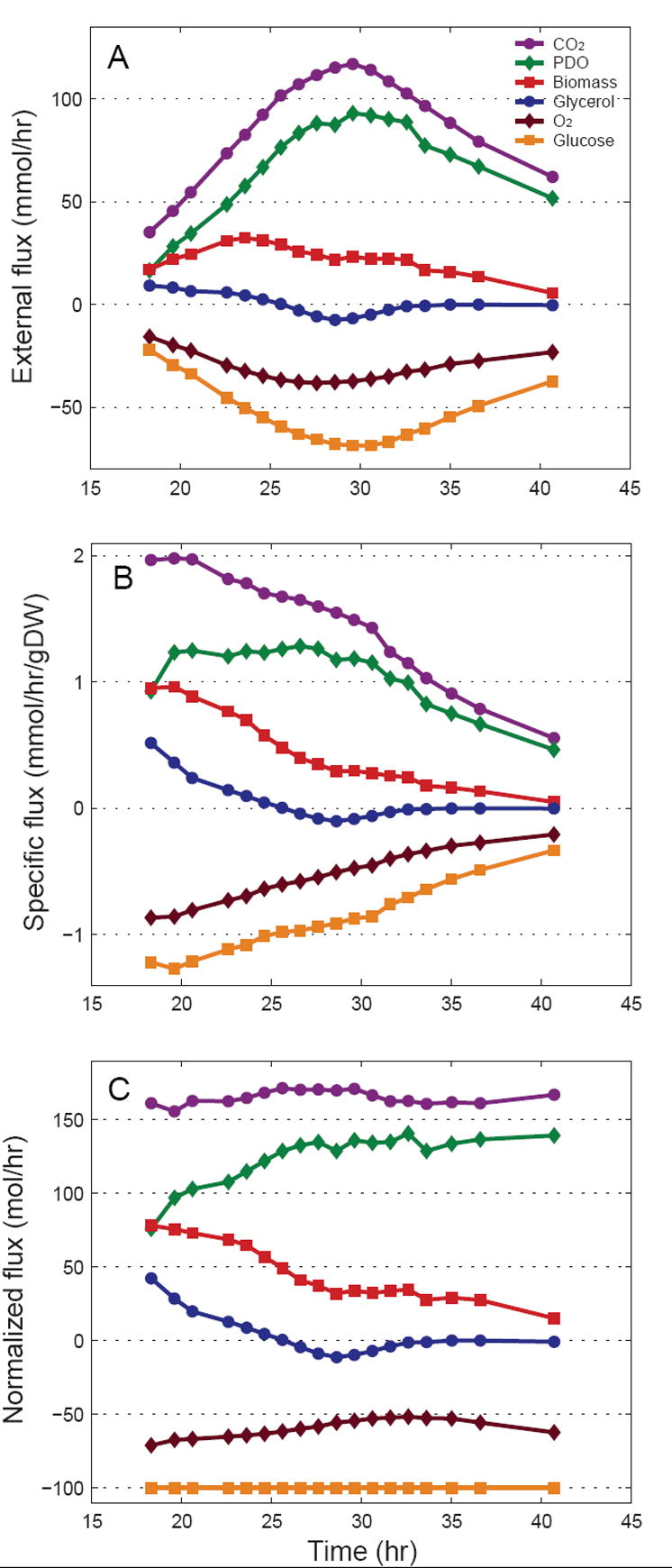
Characterization of external fluxes as a function of the fermentation time. (A) Calculated net uptake and consumption rates (mmol/h). Negative fluxes correspond to consumption rates. (B) Biomass specific external fluxes (mmol/h/gDW). (C) Fluxes normalized to glucose uptake rate.
3.2 Characterization of glucose feed
Based on preliminary analysis of the metabolic network model we determined that the optimal labeling of glucose in this study would be 75% [1-13C]glucose and 25% [U-13C]glucose. First, we validated the isotopic purity of the purchased tracers. We measured 99.7 ± 0.2 At% isotopic purity for [1-13C]glucose, and 99.4 ± 0.1 At% isotopic purity for [U-13C]glucose, i.e. both slightly higher than the 99 At% isotopic purity according to manufactures’ specifications. The two tracers were then mixed, 105 g [1-13C]glucose + 35 g [U-13C]glucose (i.e. 75/25 wt/wt, or 75.5/24.5 mol/mol), and the composition of the labeled glucose feed was validated by GC/MS analysis. The measured composition was 75.4 ± 0.2 mol% [1-13C]glucose and 24.6 ± 0.2 mol% [U-13C]glucose.
3.3 Dynamics of glucose labeling
Figure 2 shows the measured isotopic composition of medium glucose as a function of fermentation time. Labeled glucose feed was first introduced at 18.6 hr and continued until 30.0 hr, when it was replaced with natural glucose feed. Glucose labeling shows a transient phase of about 5 hr after the introduction of the tracer (18.6 to 23.6 hr), and a second transient phase of about 5 hr after the switch to natural glucose feed (30.0 to 35.0 hr). The isotopic composition for samples taken between 23.6 and 29.6 hr was constant, with 75.4% [1-13C]glucose and 24.6% [U-13C]glucose (measured by GC/MS). The length of isotopic transients depends on the glucose concentration in the fermentor and rates of glucose supply and uptake. In this experiment, glucose concentration was maintained at 45 ± 5 mM, i.e. much higher than the typical limiting levels of glucose achieved in continuous cultures. To describe the observed labeling transients quantitatively we constructed a material balance model assuming ideal mixing. The balance equations for total glucose and glucose labeling are:
Figure 2.

Isotopic composition of glucose in the fermentor as a function of fermentation time measured by GC/MS. 13C-Labeled glucose feed was introduced between 18.6 and 30.0 hr. The composition of glucose reached isotopic steady state for samples between 23.6 and 29.6 hr, with 75.4 mol% [1-13C]glucose and 24.6 mol% [U-13C]glucose. The dashed lines correspond to predicted composition of glucose based on Eq. 7.
| (3) |
| (4) |
where F (L/hr) is the flow rate of the feed, cgluc (mmol/L) is the glucose concentration in the fermentor, cgluc,in (mmol/L) is the glucose concentration in the feed, V (L) is the fermentor volume, rgluc (mmol/hr) is the glucose uptake rate, and x and xin are the molfraction of labeled glucose in the fermentor and feed, respectively. In this experiment, glucose concentration and fermentor volume were relatively constant during the observed isotopic transients; however, the feed rate and glucose uptake changed significantly. Assuming constant glucose concentration and fermentor volume Eq. 3 simplifies to:
| (5) |
Substitution of Eq. 5 in Eq. 4 yields:
| (6) |
During the first transient phase, glucose concentration and fermentor volume were 43 mmol/L and 0.98 L, respectively, and glucose uptake rate increased linearly from 21.7 mmol/h at 18.3 hr to 50.2 mmol/hr at 23.6 hr (rgluc = 5.40·t − 77; linear fit with R2=0.99). During the second transient phase, glucose concentration and fermentor volume were 48 mmol/L and 1.06 L, respectively, and glucose uptake rate decreased linearly from 68.7 mmol/h at 30.6 hr to 54.7 mmol/h at 35.0 hr (rgluc = −3.25·t + 169; linear fit with R2=0.99). Integration of Eq. 6 assuming linear function for glucose uptake as a function of time (rgluc=α+β·t) yields the following expression for the labeling of glucose as a function of time:
| (7) |
The dashed lines in Figure 2 correspond to the predicted isotopic composition of glucose based on Eq. 7. We found very good agreement between the predicted and observed isotopic transients.
3.4 GC/MS analysis of biomass amino acids
Proteinogenic amino acids from hydrolyzed biomass samples were derivatized to their respective tert-butyldimethylsilyl (TBDMS) derivatives and analyzed by electron impact GC/MS. We detected 15 of the 20 amino acids, i.e. cysteine and tryptophan were lost in hydrolysis due to oxidation, and glutamine and asparagine were deamidated to glutamate and aspartate, respectively. Histidine was not detected. Previously, we validated the accuracy and precision of GC/MS analysis of TBDMS derivatized amino acid (Antoniewicz et al., 2006a), and reported a protocol to measure mass isotopomer distributions of 29 amino acid ion fragments with an accuracy of 0.4 mol% or better and precision of 0.2 mol%, about one order-of-magnitude more accurate than previous reports (Dauner and Sauer, 2000; Klapa et al., 2003) and similar to Wittmann et al. (Wittmann et al., 2002). For example, for a natural biomass sample taken at 18.3 hr the measured mass isotopomer abundances deviated less than 0.4 mol% from theoretical abundances. Table 1 shows the 29 measured amino acid fragments. These fragments provide an overdetermined set of 112 independent constraints for metabolic flux analysis (i.e. after accounting for natural isotope enrichments).
Table 1.
Fragments of TBDMS derivatized amino acids used for flux analysis. The identity of amino acid ion fragments and the accuracy of measured mass isotopomer distributions were verified previously (Antoniewicz, 2006a). Assignment of precursor metabolites reflects the metabolic network model of E. coli in Appendix A.
| Amino
acid |
Monitored
ions |
Amino acid
carbon atoms |
Fragmentation | Precursor metabolite(s) |
|---|---|---|---|---|
| Ala | 232 – 239 | 2-3 | M – C5H9O | Pyr(2-3) |
| Ala | 260 – 267 | 1-2-3 | M – C4H9 | Pyr(1-2-3) |
|
| ||||
| Gly | 218 – 223 | 2 | M – C5H9O | multiple sources* |
| Gly | 246 – 252 | 1-2 | M – C4H9 | multiple sources* |
|
| ||||
| Val | 260 – 268 | 2-3-4-5 | M – C5H9O | Pyr(2-3) + Pyr(2-3) |
| Val | 288 – 297 | 1-2-3-4-5 | M – C4H9 | Pyr(1-2-3) + Pyr(2-3) |
|
| ||||
| Leu | 274 – 283 | 2-3-4-5-6 | M – C5H9O | AcCoA(2) + Pyr(2-3) + Pyr(2-3) |
|
| ||||
| Ile | 200 – 208 | 2-3-4-5-6 | M – C5H9O | OAC(2-3-4) + Pyr(2-3) |
| Ile | 274 – 283 | 2-3-4-5-6 | M – C5H9O | OAC(2-3-4) + Pyr(2-3) |
|
| ||||
| Ser | 288 – 294 | 2-3 | M – C7H15O2Si | multiple sources* |
| Ser | 362 – 369 | 2-3 | M – C5H9O | multiple sources* |
| Ser | 390 – 398 | 1-2-3 | M – C4H9 | multiple sources* |
|
| ||||
| Thr | 376 – 382 | 2-3-4 | M – C5H9O | OAC(2-3-4) |
| Thr | 404 – 413 | 1-2-3-4 | M – C4H9 | OAC(1-2-3-4) |
|
| ||||
| Met | 218 – 226 | 2-3-4-5 | M – C5H9O | OAC(2-3-4) + C-1 |
| Met | 292 – 298 | 2-3-4-5 | M – C5H9O | OAC(2-3-4) + C-1 |
| Met | 320 – 327 | 1-2-3-4-5 | M – C4H9 | OAC(1-2-3-4) + C-1 |
|
| ||||
| Phe | 234 – 242 | 2-3-4-5-6-7-8-9 | M – C5H9O | PEP(2-3) + PEP(2-3) + E4P(1-2-3-4) |
| Phe | 302 – 307 | 1-2 | M – C7H7 | PEP(1-2) |
| Phe | 308 – 316 | 2-3-4-5-6-7-8-9 | M – C5H9O | PEP(2-3) + PEP(2-3) + E4P(1-2-3-4) |
| Phe | 336 – 345 | 1-2-3-4-5-6-7-8-9 | M – C4H9 | PEP(1-2-3) + PEP(2-3) + E4P(1-2-3-4) |
|
| ||||
| Asp | 302 – 308 | 1-2 | M – C8H17O2Si | OAC(1-2) |
| Asp | 376 – 382 | 1-2 | M – C6H11O | OAC(1-2) |
| Asp | 390 – 397 | 2-3-4 | M – C5H9O | OAC(2-3-4) |
| Asp | 418 – 427 | 1-2-3-4 | M – C4H9 | OAC(1-2-3-4) |
|
| ||||
| Glu | 330 – 336 | 2-3-4-5 | M – C7H15O2Si | AKG(2-3-4-5) |
| Glu | 404 – 408 | 2-3-4-5 | M – C5H9O | AKG(2-3-4-5) |
| Glu | 432 – 442 | 1-2-3-4-5 | M – C4H9 | AKG(1-2-3-4-5) |
|
| ||||
| Tyr | 302 – 305 | 1-2 | M – C13H21OSi | PEP(1-2) |
Abbreviations: 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; E4P, erythrose-4-phosphate; AKG, α-ketoglutarate; OAC, oxaloacetate; AcCoA, acetyl coenzyme-A; R5P, ribose-5-phosphate; C-1, one-carbon unit.
Glycine and serine are produced by multiple pathways (see Appendix A). Hence, the labeling of these fragments depends on the relative contribution from multiple sources.
3.5 Labeling dynamics of fed-batch fermentation
Figure 3 shows time profiles of isotopic enrichment for four metabolite pools in the pathway from glucose feed to biomass. The first panel shows the labeling of glucose feed, which was changed instantaneously from natural to 13C-labeled glucose at 18.6 hr, and visa versa at 30.0 hr. The second panel shows the observed labeling profile of glucose in the fermentor, which was discussed in detail in section 3.3. The characteristic time that describes isotopic transients is given by the ratio of the pool size (mmol/L) relative to the turnover rate of the pool (mmol/L/hr). Metabolite pools with short characteristic times relative to the length of the experiment will reach isotopic steady state quickly, whereas pools with long characteristic times may never reach isotopic steady state. The characteristic time for glucose was about 1 hr. The third panel shows the observed enrichment of carbon dioxide in the off-gas, which is an indirect measure of the labeling state of intracellular metabolites, i.e. carbon dioxide is produced in the pentose pathway and TCA cycle by decarboxylation of intracellular metabolites. We found that CO2 enrichment in the off-gas follows closely the profile of glucose labeling in the medium. This suggests that intracellular metabolite pools equilibrated quickly, as could be expected based on the relatively small size of intracellular pools. Note also that after the second transient the CO2 labeling dropped to almost zero, which indicated that biomass turnover was not significant. The estimated characteristic time for intracellular metabolites was less than 5 minutes (i.e., the short lag time between glucose and CO2 labeling profiles). The last panel in Figure 3 shows the enrichment of biomass amino acids as a function of time. Here, we plotted time profiles of four representative mass isotopomers. Similar profiles were observed the other amino acid fragments. Biomass amino acids never reached isotopic steady state during the experiment, and we estimated a characteristic time of about 5 to 10 hr.
Figure 3.
Time profiles of isotopic enrichment of four metabolite pools in the pathway from glucose feed to biomass. 13C-Labeled glucose was introduced between 18.6 hr and 30.0 hr. The first isotopic transient is observed for glucose in the fermentor with a characteristic time of about 1 hr. The labeling of carbon dioxide in the off-gas followed closely the labeling of glucose, which suggested pseudo steady-state for intracellular metabolites. The labeling of biomass amino acids never reached isotopic steady state. The characteristic time for biomass amino acids was about 5 to 10 hr.
3.6 Nonstationary model for fed-batch fermentation
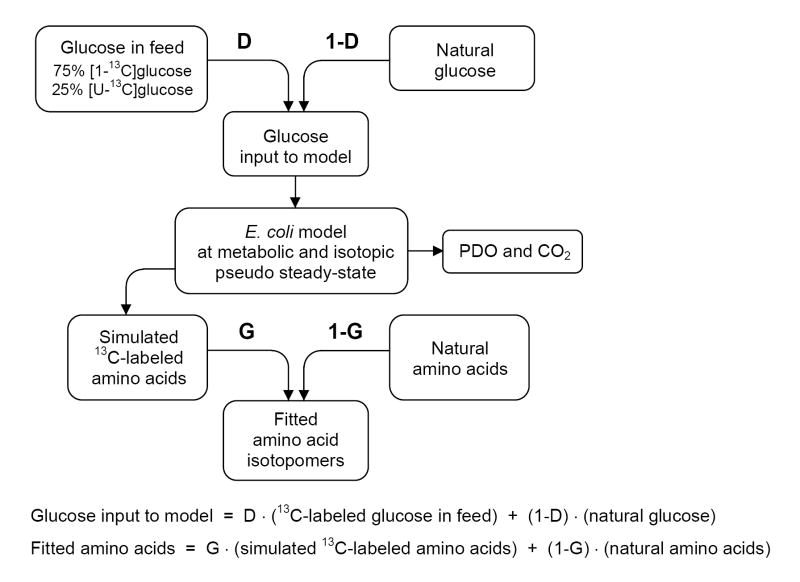
Based on these observations we propose the following model for the analysis of this nonstationary system. The model shown in Figure 4 builds on the classical stationary MFA with the addition of two dilution parameters, i.e. D and G parameters, which were initially proposed by Kelleher and Masterson (Kelleher and Masterson, 1992) in the isotopomer spectral analysis (ISA) framework. ISA is a general method for modeling polymerization biosynthesis reactions in systems that do not approximate isotopic steady state. Here, we identify two dilution effects, i.e. dilution of the tracer (i.e. glucose), and dilution of the product (i.e. biomass amino acids). In our framework, the D parameter describes the fractional contribution of the feed to the production of labeled products. Hence, the D value is zero if there are no tracers in the system, and equals unity if glucose reaches isotopic steady state quickly during the labeling phase. The G parameter describes the fraction of labeled amino acids in the biomass sample, and 1-G corresponds to the fraction of natural biomass. In theory, we could introduce separate G parameters for each measured amino acid, or alternatively use one G parameter for all biomass components. In our initial model we used separate G parameters for each amino acid. Note that the D and G parameters are easily incorporated into the classical stationary MFA framework as additional fluxes that will be estimated together with the other fluxes. Hence, we can apply the already developed flux estimation tools for stationary MFA to estimate all fluxes and D and G parameters without increasing the complexity of calculations. An additional application of this model is the analysis of chemostat data prior to isotopic steady state. In that case the D value would be one, because of the vanishing low residual glucose concentration during carbon limitation, and G value would represent the approach to isotopic steady state. As such we avoid errors due to extrapolation of isotopomer data to isotopic steady state by fitting measured data directly to the model.
Figure 4.
Model for the analysis of nonstationary tracer experiments. We identify two dilution effects, i.e. dilution of the tracer (glucose), and dilution of the sampled pools (biomass amino acids). The labeling of glucose during the labeling period is a mixture of 13C-labeled glucose feed and natural glucose. The D parameter reflects the contribution of feed glucose to intracellular glucose. The G parameter corresponds to the fraction of 13C-labeled amino acids in the biomass samples, and 1-G is the fraction of natural biomass.
3.7 Metabolic flux analysis
Metabolic fluxes were estimated for 20 time points during the fermentation, corresponding to biomass samples taken between 18.3 hr and 44.6 hr. Metabolic fluxes and their confidence intervals were determined by fitting 112 independent mass isotopomer abundances and 7 external fluxes to the model. Fluxes were estimated separately for each time point. The minimized objective function was the variance-weighted sum of squared residuals (SSRES). In total 37 independent flux parameters were estimated for each time point (including the D and G parameters), i.e. there were 112+7-37 = 82 redundant measurements for each time point. At convergence, the goodness-of-fit was assessed by statistically evaluating SSRES, which is a χ2-stochastic with 82 degrees of freedom. The minimum and maximum allowed values for SSRES were 59 and 109, respectively, at 95% confidence level. To illustrate that both D and G parameters were required to fit the data, we performed metabolic flux analysis using network models with and without the D and G parameters. The SSRES values for models without D or G parameters were statistically not acceptable (Figure 5). Only the model that included both parameters produced statistically acceptable fits for all sample points. Table 2 shows the optimally-fitted mass isotopomer distributions for sample #12 (taken at 29.6 hr). We found excellent agreement between the observed and predicted mass isotopomer abundances with SSRES = 85.8. The maximum deviation between the measured and fitted mass isotopomer abundances was 0.3 mol%. Figure 6 show the statistical evaluation of the standard-deviation weighted residuals, which are expected to be normally distributed. The normal probability plot (Figure 6B) indicated that the weighted residuals were indeed normally distributed. Accurate nonlinear confidence intervals for all estimated fluxes were determined as described previously (Antoniewicz et al., 2006b).
Figure 5.

Evaluation of goodness-of-fit for models with and without D and G parameters. Shown is the minimized variance-weighted sum of squared residuals (SSRES) for 20 sample points. The shaded area indicates the statistically acceptable 95% confidence region for SSRES. Models lacking the D or G parameters were statistically not acceptable. Only the model that included both parameters produced statistically acceptable fits for all sample points.
Table 2.
Measured and fitted mass isotopomer abundances of biomass amino acids. Shown are the uncorrected mass isotopomer abundances (molar percentages, mol%) for biomass sample #12 (taken at 29.6 hr). The measured values (‘Exp’; mean ± SD) and optimally fitted values (‘Calc’) corresponded well indicating a good quality of fit.
| Fragment | M+0 | M+1 | M+2 | M+3 | M+4 | M+5 | M+6 | M+7 | M+8 | M+9 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ala-232 | Exp | 49.0 ± 0.2 | 24.3 ± 0.2 | 20.7 ± 0.2 | 4.5 ± 0.2 | 1.4 ± 0.2 | 0.2 ± 0.1 | |||||
| Calc | 49.0 | 24.3 | 20.6 | 4.5 | 1.4 | 0.2 | ||||||
|
| ||||||||||||
| Ala-260 | Exp | 46.6 ± 0.2 | 24.4 ± 0.2 | 10.8 ± 0.2 | 14.1 ± 0.2 | 2.9 ± 0.2 | 1.1 ± 0.2 | 0.1 ± 0.1 | ||||
| Calc | 46.7 | 24.5 | 10.6 | 14.2 | 2.9 | 1.1 | 0.1 | |||||
|
| ||||||||||||
| Gly-218 | Exp | 62.0 ± 0.3 | 27.4 ± 0.2 | 8.3 ± 0.2 | 2.0 ± 0.2 | 0.3 ± 0.1 | ||||||
| Calc | 61.8 | 27.6 | 8.3 | 2.0 | 0.3 | |||||||
|
| ||||||||||||
| Gly-246 | Exp | 56.9 ± 0.3 | 20.0 ± 0.2 | 18.1 ± 0.2 | 3.7 ± 0.2 | 1.2 ± 0.2 | 0.2 ± 0.1 | |||||
| Calc | 57.2 | 19.9 | 18.0 | 3.6 | 1.2 | 0.2 | ||||||
|
| ||||||||||||
| Val-260 | Exp | 35.8 ± 0.2 | 22.5 ± 0.2 | 23.2 ± 0.2 | 11.1 ± 0.2 | 5.8 ± 0.2 | 1.3 ± 0.2 | 0.4 ± 0.1 | ||||
| Calc | 36.0 | 22.6 | 23.1 | 11.0 | 5.7 | 1.3 | 0.3 | |||||
|
| ||||||||||||
| Val-288 | Exp | 34.9 ± 0.2 | 22.2 ± 0.2 | 17.4 ± 0.2 | 13.9 ± 0.2 | 6.4 ± 0.2 | 4.1 ± 0.2 | 0.9 ± 0.1 | 0.3 ± 0.1 | |||
| Calc | 34.7 | 22.3 | 17.6 | 13.8 | 6.4 | 4.1 | 0.9 | 0.3 | ||||
|
| ||||||||||||
| Leu-274 | Exp | 28.3 ± 0.2 | 22.6 ± 0.2 | 21.8 ± 0.2 | 15.4 ± 0.2 | 7.8 ± 0.2 | 3.1 ± 0.2 | 0.7 ± 0.1 | 0.2 ± 0.1 | |||
| Calc | 28.4 | 22.4 | 21.9 | 15.5 | 7.8 | 3.1 | 0.7 | 0.2 | ||||
|
| ||||||||||||
| Ile-200 | Exp | 31.6 ± 0.2 | 22.7 ± 0.2 | 21.5 ± 0.2 | 14.6 ± 0.2 | 6.8 ± 0.2 | 2.4 ± 0.2 | 0.4 ± 0.1 | 0.1 ± 0.1 | |||
| Calc | 31.7 | 22.6 | 21.5 | 14.7 | 6.7 | 2.4 | 0.4 | 0.1 | ||||
|
| ||||||||||||
| Ile-274 | Exp | 28.5 ± 0.2 | 22.6 ± 0.2 | 21.8 ± 0.2 | 15.3 ± 0.2 | 7.8 ± 0.2 | 3.1 ± 0.2 | 0.7 ± 0.1 | 0.2 ± 0.1 | |||
| Calc | 28.5 | 22.5 | 21.9 | 15.5 | 7.8 | 3.1 | 0.7 | 0.2 | ||||
|
| ||||||||||||
| Ser-288 | Exp | 45.5 ± 0.2 | 29.8 ± 0.2 | 18.5 ± 0.2 | 4.8 ± 0.2 | 1.3 ± 0.2 | 0.2 ± 0.1 | |||||
| Calc | 45.3 | 29.7 | 18.7 | 4.8 | 1.3 | 0.2 | ||||||
|
| ||||||||||||
| Ser-362 | Exp | 40.6 ± 0.2 | 29.8 ± 0.2 | 20.3 ± 0.2 | 6.6 ± 0.2 | 2.2 ± 0.2 | 0.5 ± 0.1 | 0.1 ± 0.1 | ||||
| Calc | 40.8 | 29.7 | 20.3 | 6.6 | 2.2 | 0.5 | 0.1 | |||||
|
| ||||||||||||
| Ser-390 | Exp | 38.8 ± 0.2 | 27.2 ± 0.2 | 15.6 ± 0.2 | 12.7 ± 0.2 | 3.9 ± 0.2 | 1.5 ± 0.2 | 0.3 ± 0.1 | 0.1 ± 0.1 | |||
| Calc | 38.7 | 27.1 | 15.8 | 12.6 | 3.9 | 1.5 | 0.3 | 0.1 | ||||
|
| ||||||||||||
| Thr-376 | Exp | 31.7 ± 0.2 | 28.9 ± 0.2 | 22.1 ± 0.2 | 11.9 ± 0.2 | 4.0 ± 0.2 | 1.2 ± 0.2 | 0.3 ± 0.1 | ||||
| Calc | 31.7 | 28.8 | 22.0 | 12.0 | 4.0 | 1.2 | 0.3 | |||||
|
| ||||||||||||
| Thr-404 | Exp | 27.9 ± 0.2 | 26.0 ± 0.2 | 21.8 ± 0.2 | 14.0 ± 0.2 | 7.2 ± 0.2 | 2.3 ± 0.2 | 0.7 ± 0.1 | 0.2 ± 0.1 | |||
| Calc | 27.8 | 26.0 | 21.8 | 14.0 | 7.1 | 2.3 | 0.7 | 0.2 | ||||
|
| ||||||||||||
| Met-218 | Exp | 30.9 ± 0.2 | 26.6 ± 0.2 | 22.7 ± 0.2 | 13.3 ± 0.2 | 5.1 ± 0.2 | 1.2 ± 0.2 | 0.3 ± 0.1 | ||||
| Calc | 30.8 | 26.5 | 22.9 | 13.3 | 5.1 | 1.2 | 0.3 | |||||
|
| ||||||||||||
| Met-292 | Exp | 27.4 ± 0.2 | 25.9 ± 0.2 | 23.4 ± 0.2 | 14.5 ± 0.2 | 6.4 ± 0.2 | 1.9 ± 0.2 | 0.6 ± 0.1 | 0.1 ± 0.1 | |||
| Calc | 27.7 | 25.8 | 23.4 | 14.4 | 6.2 | 1.9 | 0.5 | 0.1 | ||||
|
| ||||||||||||
| Met-320 | Exp | 25.1 ± 0.2 | 22.9 ± 0.2 | 22.2 ± 0.2 | 15.6 ± 0.2 | 9.1 ± 0.2 | 3.7 ± 0.2 | 1.1 ± 0.2 | 0.3 ± 0.1 | 0.1± 0.1 | ||
| Calc | 24.9 | 22.7 | 22.4 | 15.8 | 9.1 | 3.7 | 1.1 | 0.3 | 0.1 | |||
|
| ||||||||||||
| Phe-234 | Exp | 30.5 ± 0.2 | 17.9 ± 0.2 | 17.3 ± 0.2 | 12.1 ± 0.2 | 9.6 ± 0.2 | 6.1 ± 0.2 | 4.1 0.2 | 1.7 ± 0.2 | 0.7± 0.1 | 0.1 ± 0.1 | |
| Calc | 30.8 | 17.7 | 17.4 | 12.1 | 9.6 | 6.0 | 4.0 | 1.7 | 0.7 | 0.1 | ||
|
| ||||||||||||
| Phe-302 | Exp | 58.1 ± 0.3 | 18.4 ± 0.2 | 18.0 ± 0.2 | 4.0 ± 0.2 | 1.3 ± 0.2 | 0.2 ± 0.1 | |||||
| Calc | 57.9 | 18.4 | 18.1 | 4.1 | 1.3 | 0.2 | ||||||
|
| ||||||||||||
| Phe-308 | Exp | 27.5 ± 0.2 | 18.1 ± 0.2 | 17.8 ± 0.2 | 12.6 ± 0.2 | 10.1 ± 0.2 | 6.5 ± 0.2 | 4.4 ± 0.2 | 2.0 ± 0.2 | 0.9± 0.1 | 0.2 ± 0.1 | |
| Calc | 27.7 | 18.0 | 17.8 | 12.6 | 10.1 | 6.5 | 4.4 | 2.0 | 0.9 | 0.2 | ||
|
| ||||||||||||
| Phe-336 | Exp | 27.2 ± 0.2 | 18.1 ± 0.2 | 14.2 ± 0.2 | 13.1 ± 0.2 | 10.4 ± 0.2 | 7.6 ± 0.2 | 4.7 ± 0.2 | 2.8 ± 0.2 | 1.3± 0.2 | 0.6 ± 0.1 | |
| Calc | 26.9 | 17.9 | 14.4 | 13.1 | 10.4 | 7.7 | 4.8 | 2.8 | 1.3 | 0.6 | ||
|
| ||||||||||||
| Asp-302 | Exp | 45.6 ± 0.2 | 28.3 ± 0.2 | 19.5 ± 0.2 | 5.0 ± 0.2 | 1.4 ± 0.2 | 0.2 ± 0.1 | |||||
| Calc | 45.6 | 28.3 | 19.5 | 5.0 | 1.4 | 0.2 | ||||||
|
| ||||||||||||
| Asp-376 | Exp | 41.0 ± 0.2 | 28.5 ± 0.2 | 20.9 ± 0.2 | 6.8 ± 0.2 | 2.3 ± 0.2 | 0.5 ± 0.1 | 0.1 ± 0.1 | ||||
| Calc | 41.0 | 28.4 | 20.9 | 6.8 | 2.3 | 0.5 | 0.1 | |||||
|
| ||||||||||||
| Asp-390 | Exp | 31.7 ± 0.2 | 28.9 ± 0.2 | 22.0 ± 0.2 | 11.9 ± 0.2 | 4.0 ± 0.2 | 1.2 ± 0.2 | 0.3 ± 0.1 | 0.1 ± 0.1 | |||
| Calc | 31.7 | 28.8 | 22.0 | 12.0 | 4.0 | 1.2 | 0.3 | 0.1 | ||||
|
| ||||||||||||
| Asp-418 | Exp | 27.8 ± 0.2 | 26.0 ± 0.2 | 21.9 ± 0.2 | 14.0 ± 0.2 | 7.2 ± 0.2 | 2.4 ± 0.2 | 0.7 ± 0.1 | 0.2 ± 0.1 | |||
| Calc | 27.8 | 26.0 | 21.9 | 14.0 | 7.2 | 2.4 | 0.7 | 0.2 | ||||
|
| ||||||||||||
| Glu-330 | Exp | 29.5 ± 0.2 | 26.1 ± 0.2 | 23.1 ± 0.2 | 13.5 ± 0.2 | 5.9 ± 0.2 | 1.6 ± 0.2 | 0.4 ± 0.1 | 0.1 ± 0.1 | |||
| Calc | 29.3 | 26.0 | 23.3 | 13.5 | 5.8 | 1.5 | 0.4 | 0.1 | ||||
|
| ||||||||||||
| Glu-404 | Exp | 26.4 ± 0.2 | 25.3 ± 0.2 | 23.6 ± 0.2 | 14.7 ± 0.2 | 7.0 ± 0.2 | 2.3 ± 0.2 | 0.7 ± 0.1 | 0.1 ± 0.1 | |||
| Calc | 26.4 | 25.4 | 23.7 | 14.6 | 7.0 | 2.3 | 0.6 | 0.1 | ||||
|
| ||||||||||||
| Glu-432 | Exp | 23.5 ± 0.2 | 22.1 ± 0.2 | 22.3 ± 0.2 | 16.7 ± 0.2 | 9.4 ± 0.2 | 4.3 ± 0.2 | 1.4 ± 0.2 | 0.4 ± 0.1 | 0.1± 0.1 | ||
| Calc | 23.6 | 22.1 | 22.3 | 16.7 | 9.3 | 4.2 | 1.3 | 0.4 | 0.1 | |||
|
| ||||||||||||
| Tyr-302 | Exp | 57.7 ± 0.3 | 18.4 ± 0.2 | 18.2 ± 0.2 | 4.1 ± 0.2 | 1.4 ± 0.2 | 0.2 ± 0.1 | |||||
| Calc | 57.7 | 18.4 | 18.3 | 4.1 | 1.4 | 0.2 | ||||||
Figure 6.
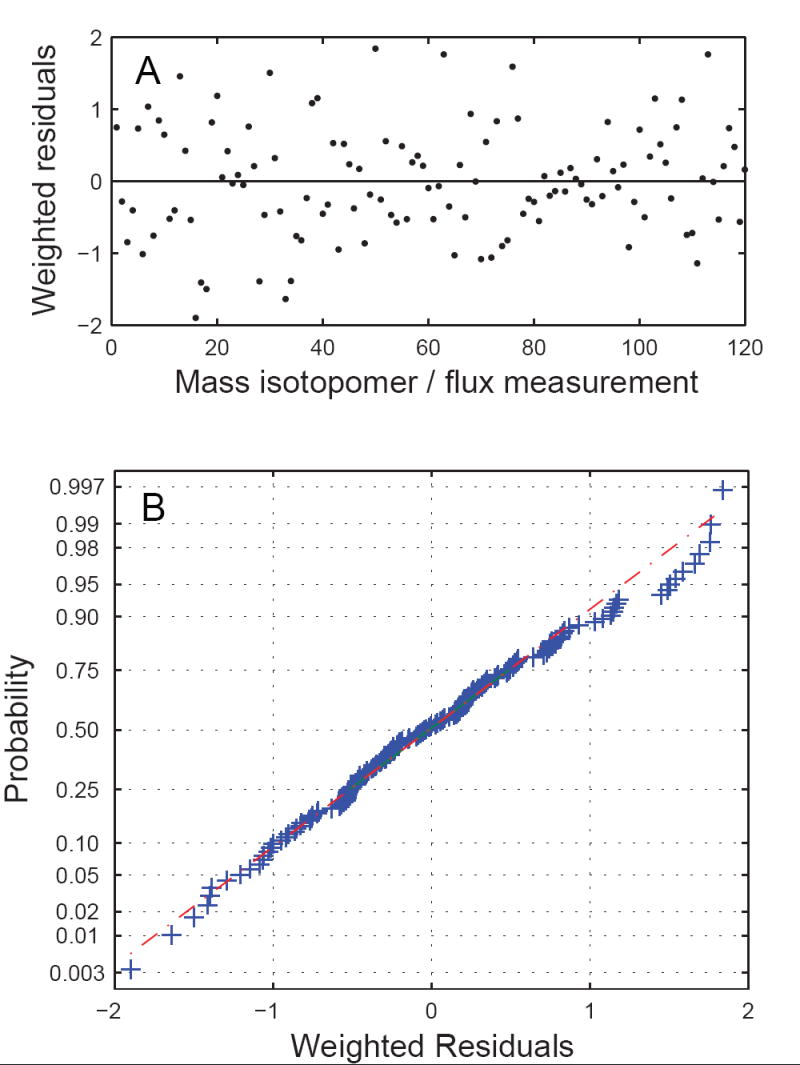
Statistical evaluation of the standard-deviation weighted residuals for sample #12 (taken at 29.6 hr). (A) Weighted residuals of mass isotopomer and external flux measurements. (B) Normal probability plot indicated that the weighted residuals were normally distributed.
3.8 Evaluation of estimated metabolic fluxes
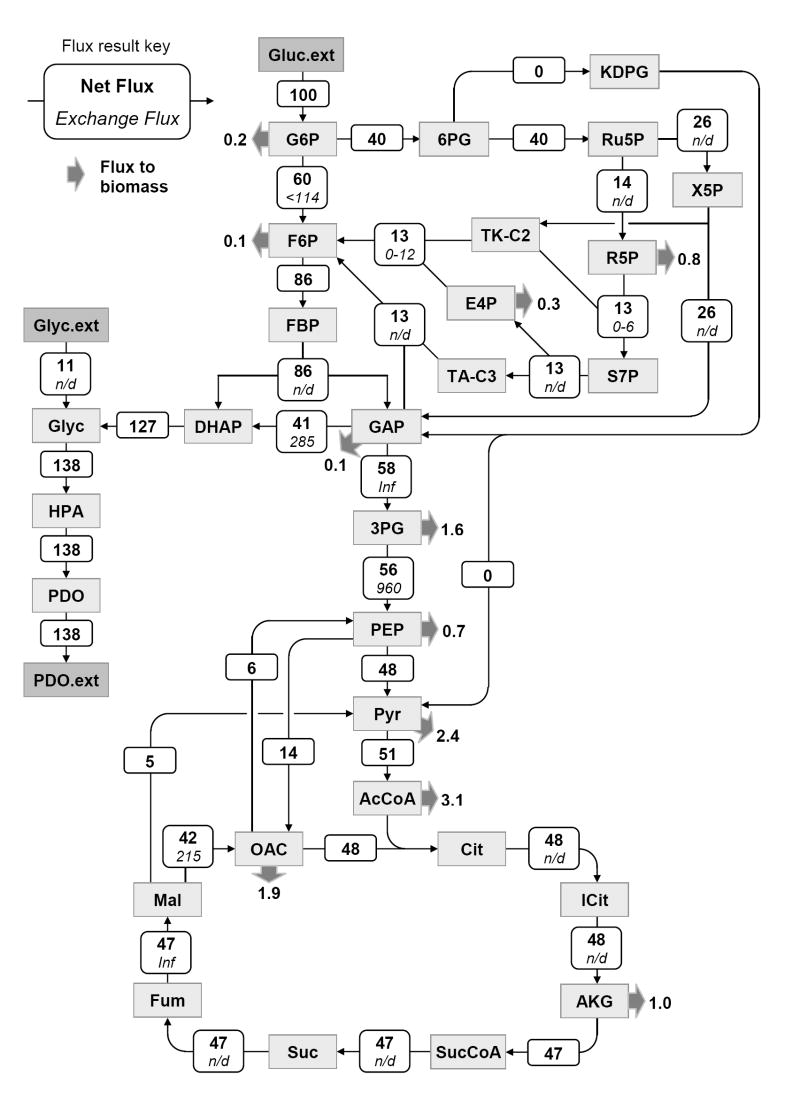
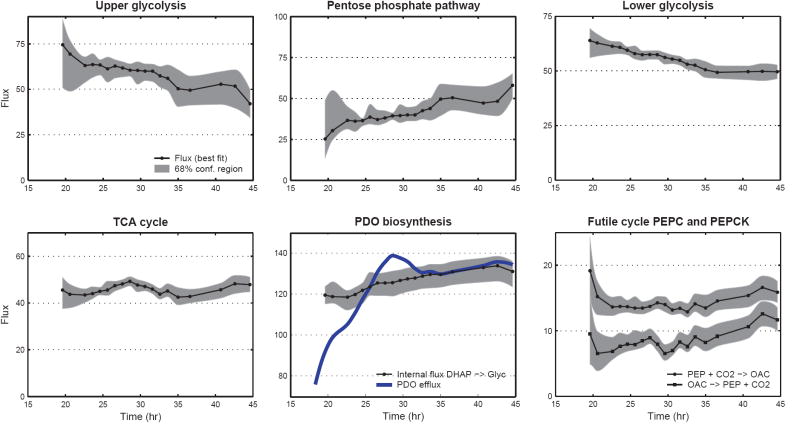
Figure 7 shows the estimated fluxes for sample point #12 (see Supplementary Materials for further details). Fluxes were normalized to glucose uptake rate, which was given the value 100. Similarly, metabolic fluxes were estimated for the other time points, and as such we established for the first time detailed time profiles of intracellular fluxes during a fed-batch culture. Figure 8 shows the time profiles for selected intracellular fluxes. The optimally-fitted flux value and the 68% confidence interval (i.e. flux ± SD) are plotted as a function of fermentation time. The time profiles in Figure 8 clearly illustrate that intracellular fluxes changed during the fed-batch fermentation. The split ratio between glycolysis and pentose phosphate pathway decreased from 70/30 at 20 hr to about 50/50 at 43 hr. Similarly, the flux of lower glycolysis (GAP→3PG) decreased from 64±3 to 50±2. An interesting finding was that the TCA flux, i.e. the main energy producing pathway, remained constant at 46±2 throughout the fermentation. The intracellular flux towards PDO and glycerol (DHAP→glycerol 3-phosphate) increased by about 10% from 120±6 to 132±6. This was in contrast to the large fluctuations observed for the efflux of PDO, i.e. PDO efflux increased from 78 (at 18.6 hr) to 138 (at 28.6 hr), and then decreased to 130 (at 40.7 hr). Our results further indicated that the Entner-Doudoroff pathway was inactive, i.e. the estimated flux of 0.0±0.5 was not statistically different from zero for all sample points. This result confirms the genotype of this strain of E. coli, where the phosphogluconate dehydratase gene was knocked-out, a key enzymes of the Entner-Doudoroff pathway. Our flux results further revealed the presence of a futile cycle between oxaloacetate and phosphoenolpyruvate. The estimated phosphoenolpyruvate carboxylase (PEPC) and phosphoenolpyruvate carboxykinase (PEPCK) fluxes were 14±1 and 8±1, respectively. Simultaneous activity of these two reactions created a futile cycle where 1 ATP was lost at each turn of the cycle. It is not yet clear what the physiological role of this cycle is. The total activity of malic enzyme was estimated at 5±1. We could not distinguish between the two isoforms of malic enzyme, i.e. NADH and NADPH dependent malic enzyme. Thus, only the combined malic enzyme flux was determined. A slightly net positive transhydrogenase flux (i.e. NADH → NADPH) was estimated at 20±15, which was not statistically different from zero, indicating that this strain of E. coli may not posses transhydrogenation activity. Finally, we estimated a significant net production of ATP of 176±32 not accounted for by the ATP consuming reactions in our model. Potential sinks for this ATP are cell maintenance, transport of metabolites across cell membrane, and futile cycles. We should note that our model assumed a theoretical P/O ratio of 3. Assuming a more realistic P/O ratio of 2 we estimated 123±32 excess ATP production.
Figure 7.
Metabolic fluxes of E. coli grown in carbon-limited fed-batch culture. Shown are the estimated fluxes for sample point #12 (at 29.6 hr), normalized to glucose uptake rate. Metabolic fluxes were determined by fitting 112 mass isotopomer abundances and 7 external fluxes to a network model of E. coli. The top number denotes the estimated net flux and the bottom is the estimated exchange flux (in italics); ‘n/d’ indicates that the exchange flux was not observable. Thick arrows represent drain of metabolites to biomass.
Figure 8.
Time profiles of selected intracellular fluxes. The best fit (solid line) and the 68% confidence interval (i.e., flux ±SD; shaded area) are shown as a function of fermentation time. Fluxes were normalized to glucose uptake rate. The upper glycolysis flux (G6P→F6P) and lower glycolysis flux (GAP→3PG) decreased in time, while the pentose phosphate pathway flux (G6P→Ru5P) increased. The TCA flux was constant at 46 ± 2. The intracellular flux towards glycerol and PDO (DHAP→glycerol 3-phosphate) increased by 10% from 120±6 to 132±6, whereas the efflux of PDO fluctuated significantly. The simultaneous activity of PEPC and PEPCK created a futile cycle where 1 ATP was lost at each turn of the cycle.
3.9 Evaluation of estimated D and G parameters
The key to successful flux determination in this nonstationary system was the introduction of two dilution parameters, the D parameter to account for the dilution of the tracer and the G parameter to account for the dilution of biomass components. Values for both parameters were estimated together with the other fluxes. Time profiles of the estimated D and G parameters are shown in Figure 9. Our model contained 12 individual G parameters, i.e. one G parameter for each of the 12 measured amino acids. The estimated G values for the 12 amino acids were not significantly different from one another, indicating that one G value could be used for all biomass components. Biomass produced before the introduction of tracers is natural, and thus the G value is zero. The observed increase in G value after 18.6 hr reflects the production of 13C-labeled biomass, i.e. the fraction of 13C-labeled biomass increases. The G value decreased after tracers were depleted reflecting the production of natural biomass. Note that there was a short delay between the switch to natural feed at 30.0 h and the first notable decrease in the G value at 31.6 hr. This delay of ~1.6 hr reflects the fact that 13C-labeled glucose was still present in the fermentor for several hours after the feed was switched to natural glucose feed (see section 3.3). The D value describes the fractional contribution of labeled feed as a precursor for 13C-labeled biomass. Figure 9 shows that the D value first increased at 18.6 hr, but never reached unity due to a small fraction of 13C-labeled biomass that was produced from partially labeled glucose. After the tracer was removed the D value first dropped slightly reflecting the second glucose transient (i.e. 13C-labeled biomass is produced from diluted glucose), and then remained constant at about 0.9. From then on, only natural biomass was produced, already accounted for by the G parameter, and thus the D value remained constant. We can set up the following theoretical model for the D and G parameters.
Figure 9.
Time profiles of the estimated and predicted D and G parameters. The dots represent estimated D and G parameters from MFA. The G values for individual amino acids (dashed lines in right panel) were not significantly different from one another, indicating that one G value could be used for all biomass amino acids. The solid line represents the predicted parameter values based on Eqs. 8 and 9. The good agreement between the predicted and estimated parameter values supports the validity of our modeling approach.
| (8) |
| (9) |
| (10) |
where L(t) is the fractional labeling of glucose in the medium, μ(t) is the growth rate, and ϕ(t) formally defines ‘the labeling phase’, i.e. the time when fractional labeling of glucose is at least 20% of the isotopic steady state value. This definition was needed, otherwise the D and G parameters became highly correlated at low glucose enrichments. In this experiment, ϕ(t) was unity between 19.0 h and 31.3 h, and zero elsewhere. We used Eqs. 8 and 9 to predict the time profiles of the theoretical D and G parameters from the measured labeling of glucose and growth rate as a function time. The predicted time profiles are also shown in Figure 9 (solid lines). We found good agreement between the predicted and MFA estimated D and G parameters.
4 Discussion
Metabolic flux analysis (MFA) is a powerful tool for the analysis of metabolic fluxes in vivo, however, it is currently limited by the isotopic steady state requirement. On the other hand, isotopomer spectral analysis (ISA) is a widely used method to study polymerization biosynthesis reactions in systems that do not approximate isotopic steady state (Kelleher and Masterson, 1992). ISA has been successfully applied to study cholesterol biosynthesis, lipogenesis, and gluconeogenesis in human subjects and other models of human metabolic disease (Brunengraber et al., 1997; Lindenthal et al., 2002; Yoo et al., 2004). In this paper, we present a novel modeling strategy for metabolic flux analysis in nonstationary systems that combines key ideas from MFA and ISA. Isotopic transients of the precursor pool and sampled products were captured by two dilution parameters, i.e. D and G parameters, respectively, that were included in the classical MFA model as additional fluxes. We applied this modeling strategy to estimate metabolic fluxes of E. coli in a fed-batch fermentation. Fluxes were determined by fitting an over-determined data set of mass isotopomer distributions of biomass amino acids and external flux measurements to a detailed model of E. coli. We obtained highly consistent fits with 82 redundant measurements. The high accuracy and precision of our GC/MS data (<0.4 mol%), the small magnitude of residuals, and the large number of redundant measurements gave us very high degree of confidence in the calculated flux parameters. Metabolic fluxes were determined for 20 time points during the course of the fermentation, and as such we established for the first time detailed time profiles of intracellular fluxes. The estimated fluxes provided valuable insight into the physiology of PDO overproduction by E. coli, and furthermore confirmed the genotype of the E. coli strain. We related the D and G parameter values to the labeling history of the experiment. The G value was the fraction of labeled biomass in the sampled biomass pool, and the D value the biomass-averaged labeling of the precursor pool of labeled biomass. The predicted values for D and G parameters corresponded well with the estimated parameter values from MFA, which supports the validity of our modeling approach. Note that the estimated fluxes depend on the flux history of the sampled biomass, and thus should be seen as biomass-averaged fluxes. Iwatani et al. (Iwatani et al., 2007) measured the labeling of free intracellular and biomass amino acids to account for this effect, and Zamboni et al. (Zamboni et al., 2005) calculated mass isotopomer distributions for each time interval prior to flux estimation. An alternative and more rigorous method would be to perform fully dynamic flux analysis, i.e. where the fluxes, metabolite pools and isotopomer distributions are determined as a function of time. Developments of such techniques are ongoing and will be available in the near future. These tools will greatly facilitate global analysis of metabolic fluxes that are of industrial and medical importance.
Acknowledgments
We acknowledge the support of the DuPont-MIT Alliance and the NIH Metabolomics Roadmap Initiative DK070291.
APPENDIX A. Metabolic network model of E. coli
| Glycolysis | |
| v1 | G6P (abcdef) ↔ F6P (abcdef) |
| v2 | F6P (abcdef) + ATP → FBP (abcdef) |
| v3 | FBP (abcdef) ↔ DHAP (cba) + GAP (def) |
| v4 | DHAP (abc) ↔ GAP (abc) |
| v5 | GAP (abc) ↔ 3PG (abc) + ATP + NADH |
| v6 | 3PG (abc) ↔ PEP (abc) |
| v7 | PEP (abc) → Pyr (abc) + ATP |
| Pentose Phosphate Pathway | |
| v8 | G6P (abcdef) → 6PG (abcdef) + NADPH |
| v9 | 6PG (abcdef) → Ru5P (bcdef) + CO2 (a) + NADPH |
| v10 | Ru5P (abcde) ↔ X5P (abcde) |
| v11 | Ru5P (abcde) ↔ R5P (abcde) |
| v12 | X5P (abcde) ↔ TK-C2 (ab) + GAP (cde) |
| v13 | F6P (abcdef) ↔ TK-C2 (ab) + E4P (cdef) |
| v14 | S7P (abcdefg) ↔ TK-C2 (ab) + R5P (cdefg) |
| v15 | F6P (abcdef) ↔ TA-C3 (abc) + GAP (def) |
| v16 | S7P (abcdefg) ↔ TA-C3 (abc) + E4P (defg) |
| Entner-Doudoroff Pathway | |
| v17 | 6PG (abcdef) → KDPG (abcdef) |
| v18 | KDPG (abcdef) → Pyr (abc) + GAP (def) |
| TCA Cycle | |
| v19 | Pyr (abc) → AcCoA (bc) + CO2 (a) + NADH |
| v20 | OAC (abcd) + AcCoA (ef) → Cit (dcbfea) |
| v21 | Cit (abcdef) ↔ ICit (abcdef) |
| v22 | ICit (abcdef) ↔ AKG (abcde) + CO2 (f) + NADPH |
| v23 | AKG (abcde) → SucCoA (bcde) + CO2 (a) + NADH |
| v24 | SucCoA (abcd) ↔ Suc (½ abcd + ½ dcba) + ATP |
| v25 | Suc (½ abcd + ½ dcba) ↔ Fum (½ abcd + ½ dcba) + FADH2 |
| v26 | Fum (½ abcd + ½ dcba) ↔ Mal (abcd) |
| v27 | Mal (abcd) ↔ OAC (abcd) + NADH |
| Amphibolic Reactions | |
| v28 | Mal (abcd) → Pyr (abc) + CO2 (d) + NADPH |
| v29 | Mal (abcd) → Pyr (abc) + CO2 (d) + NADH |
| v30 | PEP (abc) + CO2 (d) → OAC (abcd) |
| v31 | OAC (abcd) + ATP → PEP (abc) + CO2 (d) |
| Acetic Acid Formation | |
| v32 | AcCoA (ab) ↔ Ac (ab) + ATP |
| PDO Biosynthesis | |
| v33 | DHAP (abc) + NADH ↔ Glyc3P (abc) |
| v34 | Glyc3P (abc) → Glyc (abc) |
| v35 | Glyc (abc) → HPA (abc) |
| v36 | HPA (abc) + NADPH → PDO (abc) |
| Amino Acid Biosynthesis | |
| v37 | AKG (abcde) + NADPH + NH3 → Glu (abcde) |
| v38 | Glu (abcde) + ATP + NH3 → Gln (abcde) |
| v39 | Glu (abcde) + ATP + 2 NADPH → Pro (abcde) |
| v40 | Glu (abcde) + CO2 (f) + Gln (ghijk) + Asp (lmno) + AcCoA (pq) + 5 ATP + NADPH → Arg (abcdef) + AKG (ghijk) + Fum (lmno) + Ac (pq) |
| v41 | OAC (abcd) + Glu (efghi) → Asp (abcd) + AKG (efghi) |
| v42 | Asp (abcd) + 2 ATP + NH3 → Asn (abcd) |
| v43 | Pyr (abc) + Glu (defgh) → Ala (abc) + AKG (defgh) |
| v44 | 3PG (abc) + Glu (defgh) → Ser (abc) + AKG (defgh) + NADH |
| v45 | Ser (abc) ↔ Gly (ab) + MEETHF (c) |
| v46 | Gly (ab) ↔ CO2 (a) + MEETHF (b) + NADH + NH3 |
| v47 | Thr (abcd) → Gly (ab) + AcCoA (cd) + NADH |
| v48 | Ser (abc) + AcCoA (de) + 3 ATP + 4 NADPH + SO4 → Cys (abc) + Ac (de) |
| v49 | Asp (abcd) + Pyr (efg) + Glu (hijkl) + SucCoA (mnop) + ATP + 2 NADPH → LL-DAP (abcdgfe) + AKG (hijkl) + Suc (½ mnop + ½ ponm) |
| v50 | LL-DAP (abcdefg) → Lys (abcdef) + CO2 (g) |
| v51 | Asp (abcd) + 2 ATP + 2 NADPH → Thr (abcd) |
| v52 | Asp (abcd) + METHF (e) + Cys (fgh) + SucCoA (ijkl) + ATP + 2 NADPH → Met (abcde) + Pyr (fgh) + Suc (½ ijkl + ½ lkji) + NH3 |
| v53 | Pyr (abc) + Pyr (def) + Glu (ghijk) + NADPH → Val (abcef) + CO2 (d) + AKG (ghijk) |
| v54 | AcCoA (ab) + Pyr (cde) + Pyr (fgh) + Glu (ijklm) + NADPH → Leu (abdghe) + CO2 (c) + CO2 (f) + AKG (ijklm) + NADH |
| v55 | Thr (abcd) + Pyr (efg) + Glu (hijkl) + NADPH → Ile (abfcdg) + CO2 (e) + AKG (hijkl) + NH3 |
| v56 | PEP (abc) + PEP (def) + E4P (ghij) + Glu (klmno) + ATP + NADPH → Phe (abcefghij) + CO2 (d) + AKG (klmno) |
| v57 | PEP (abc) + PEP (def) + E4P (ghij) + Glu (klmno) + ATP + NADPH → Tyr (abcefghij) + CO2 (d) + AKG (klmno) + NADH |
| v58 | Ser (abc) + R5P (defgh) + PEP (ijk) + E4P (lmno) + PEP (pqr) + Gln (stuvw) + 3 ATP + NADPH → Trp (abcedklmnoj) + CO2 (i) + GAP (fgh) + Pyr (pqr) + Glu (stuvw) |
| v59 | R5P (abcde) + FTHF (f) + Gln (ghijk) + Asp (lmno) + 5 ATP → His (edcbaf) + AKG (ghijk) + Fum (lmno) + 2 NADH |
| One-Carbon Metabolism | |
| v60 | MEETHF (a) + NADH → METHF (a) |
| v61 | MEETHF (a) → FTHF (a) + NADPH |
| Oxidative Phosphorylation | |
| v62 | NADH + ½ O2 → 3 ATP |
| v63 | FADH2 + ½ O2 → 2 ATP |
| Transhydrogenation | |
| v64 | NADH ↔ NADPH |
| ATP Hydrolysis | |
| v65 | ATP → ATP:ext |
| Transport | |
| v66 | Gluc.ext (abcdef) + ATP → G6P (abcdef) |
| v67 | Cit.ext (abcdef) → Cit (abcdef) |
| v68 | Glyc (abc) ↔ Glyc.ext (abc) |
| v69 | PDO (abc) → PDO.ext (abc) |
| v70 | Ac (ab) → Ac.ext (ab) |
| v71 | CO2 (a) → CO2.ext (a) |
| v72 | O2.ext → O2 |
| v73 | NH3.ext → NH3 |
| v74 | SO4.ext → SO4 |
| Biomass Formation | |
| v75 | 0.488 Ala + 0.281 Arg + 0.229 Asn + 0.229 Asp + 0.087 Cys + 0.250 Glu + 0.250 Gln + 0.582 Gly + 0.090 His + 0.276 Ile + 0.428 Leu + 0.326 Lys + 0.146 Met + 0.176 Phe + 0.210 Pro + 0.205 Ser + 0.241 Thr + 0.054 Trp + 0.131 Tyr + 0.402 Val + 0.205 G6P + 0.071 F6P + 0.754 R5P + 0.129 GAP + 0.619 3PG + 0.051 PEP + 0.083 Pyr + 2.510 AcCoA + 0.087 AKG + 0.340 OAC + 0.443 MEETHF + 33.247 ATP + 5.363 NADPH → 39.68 Biomass + 1.455 NADH |
Carbon atom transitions are represented using letter code. For each metabolite carbon atoms are identified using lower case letters to represent successive carbon atoms. The biomass formation reaction is based on precursor and cofactor requirements for E. coli as described by Neidhardt (Neidhardt, 1987).
APPENDIX B. Validation of carbon balance and degree of reduction balance
External fluxes were evaluated for gross errors by validating carbon balance and degree of reduction balance. The carbon balance is given by:
| (B1) |
The degree of reduction balance is given by:
| (B2) |
In the above equations vi is the net production rate, ci is the number of carbon atoms, and γi is the degree of reduction of metabolite i. The degree of reduction corresponds to the number of free electrons, where γi=0 for the following small molecules: H2O, H+, CO2, , , , . In general, the degree of reduction for any metabolite is calculated as follows:
| (B3) |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. 2006a doi: 10.1021/ac0708893. submitted to Anal Chem. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006b;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary Metabolite Units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunengraber H, Kelleher JK, Des RC. Applications of mass isotopomer analysis to nutrition research. Annu Rev Nutr. 1997;17:559–596. doi: 10.1146/annurev.nutr.17.1.559. [DOI] [PubMed] [Google Scholar]
- Dauner M, Sauer U. GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing. Biotechnol Prog. 2000;16:642–649. doi: 10.1021/bp000058h. [DOI] [PubMed] [Google Scholar]
- Des Rosiers C, Lloyd S, Comte B, Chatham JC. A critical perspective of the use of (13)C-isotopomer analysis by GCMS and NMR as applied to cardiac metabolism. Metab Eng. 2004;6:44–58. doi: 10.1016/j.ymben.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Drysch A, El MM, Mack C, Takors R, de Graaf AA, Sahm H. Production process monitoring by serial mapping of microbial carbon flux distributions using a novel Sensor Reactor approach: II--(13)C-labeling-based metabolic flux analysis and L-lysine production. Metab Eng. 2003;5:96–107. doi: 10.1016/s1096-7176(03)00005-3. [DOI] [PubMed] [Google Scholar]
- Fernandez CA, Des RC, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Iwatani S, Van DS, Shimbo K, Kubota K, Kageyama N, Iwahata D, Miyano H, Hirayama K, Usuda Y, Shimizu K, Matsui K. Determination of metabolic flux changes during fed-batch cultivation from measurements of intracellular amino acids by LC-MS/MS. J Biotechnol. 2007;128:93–111. doi: 10.1016/j.jbiotec.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Kelleher JK. Flux estimation using isotopic tracers: common ground for metabolic physiology and metabolic engineering. Metab Eng. 2001;3:100–110. doi: 10.1006/mben.2001.0185. [DOI] [PubMed] [Google Scholar]
- Kelleher JK, Masterson TM. Model equations for condensation biosynthesis using stable isotopes and radioisotopes. Am J Physiol. 1992;262:E118–E125. doi: 10.1152/ajpendo.1992.262.1.E118. [DOI] [PubMed] [Google Scholar]
- Klapa MI, Aon JC, Stephanopoulos G. Systematic quantification of complex metabolic flux networks using stable isotopes and mass spectrometry. Eur J Biochem. 2003;270:3525–3542. doi: 10.1046/j.1432-1033.2003.03732.x. [DOI] [PubMed] [Google Scholar]
- Lindenthal B, Aldaghlas TA, Holleran AL, Sudhop T, Berthold HK, von BK, Kelleher JK. Isotopomer spectral analysis of intermediates of cholesterol synthesis in human subjects and hepatic cells. Am J Physiol Endocrinol Metab. 2002;282:E1222–E1230. doi: 10.1152/ajpendo.00324.2001. [DOI] [PubMed] [Google Scholar]
- Nakamura CE, Whited GM. Metabolic engineering for the microbial production of 1,3-propanediol. Curr Opin Biotechnol. 2003;14:454–459. doi: 10.1016/j.copbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC. Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology. ASM Press; 1987. [Google Scholar]
- Schmidt K, Carlsen M, Nielsen J, Villadsen J. Modeling Isotopomer Distributions in Biochemical Networks Using Isotopomer Mapping Matrices. Biotechnol Bioeng. 1997:831–840. doi: 10.1002/(SICI)1097-0290(19970920)55:6<831::AID-BIT2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. Metabolic fluxes and metabolic engineering. Metab Eng. 1999;1:1–11. doi: 10.1006/mben.1998.0101. [DOI] [PubMed] [Google Scholar]
- Szyperski T. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur J Biochem. 1995;232:433–448. doi: 10.1111/j.1432-1033.1995.tb20829.x. [DOI] [PubMed] [Google Scholar]
- Wittmann C, Hans M, Heinzle E. In vivo analysis of intracellular amino acid labelings by GC/MS. Anal Biochem. 2002;307:379–382. doi: 10.1016/s0003-2697(02)00030-1. [DOI] [PubMed] [Google Scholar]
- Yoo H, Stephanopoulos G, Kelleher JK. Quantifying carbon sources for de novo lipogenesis in wild-type and IRS-1 knockout brown adipocytes. J Lipid Res. 2004;45:1324–1332. doi: 10.1194/jlr.M400031-JLR200. [DOI] [PubMed] [Google Scholar]
- Zamboni N, Fischer E, Muffler A, Wyss M, Hohmann HP, Sauer U. Transient expression and flux changes during a shift from high to low riboflavin production in continuous cultures of Bacillus subtilis. Biotechnol Bioeng. 2005;89:219–232. doi: 10.1002/bit.20338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.