Abstract
As an accompanying manuscript to the release of the honey bee genome, we report the entire sequence of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) ribosomal RNA (rRNA)-encoding gene sequences (rDNA) and related internally and externally transcribed spacer regions of Apis mellifera (Insecta: Hymenoptera: Apocrita). Additionally, we predict secondary structures for the mature rRNA molecules based on comparative sequence analyses with other arthropod taxa and reference to recently published crystal structures of the ribosome. In general, the structures of honey bee rRNAs are in agreement with previously predicted rRNA models from other arthropods in core regions of the rRNA, with little additional expansion in non-conserved regions. Our multiple sequence alignments are made available on several public databases and provide a preliminary establishment of a global structural model of all rRNAs from the insects. Additionally, we provide conserved stretches of sequences flanking the rDNA cistrons that comprise the externally transcribed spacer regions (ETS) and part of the intergenic spacer region (IGS), including several repetitive motifs. Finally, we report the occurrence of retrotransposition in the nuclear large subunit rDNA, as R2 elements are present in the usual insertion points found in other arthropods. Interestingly, functional R1 elements usually present in the genomes of insects were not detected in the honey bee rRNA genes. The reverse transcriptase products of the R2 elements are deduced from their putative open reading frames and structurally aligned with those from another hymenopteran insect, the jewel wasp Nasonia (Pteromalidae). Stretches of conserved amino acids shared between Apis and Nasonia are illustrated and serve as potential sites for primer design, as target amplicons within these R2 elements may serve as novel phylogenetic markers for Hymenoptera. Given the impending completion of the sequencing of the Nasonia genome, we expect our report eventually to shed light on the evolution of the hymenopteran genome within higher insects, particularly regarding the relative maintenance of conserved rDNA genes, related variable spacer regions and retrotransposable elements.
Keywords: honey bee, Apis mellifera, ribosomal RNA, rRNA, rDNA, 18S, 5.8S, 28S, 5S, 16S, 12S, ITS-1, ITS-2, ETS, IGS, secondary structure, alignment, retrotransposition, R1, R2, R element, reverse transcriptase
Introduction
Ribosomal RNA (rRNA)-encoding genes (rDNA) and related genetic elements have been well studied for over six decades (see Noller, 2005 for a recent review), with interests ranging from pharmaceutical and biochemical investigations to comparative biological studies garnering a wealth of information on the structural, functional and evolutionary characteristics of these molecules. Phylogenetic studies, in particular, have propagated a large number of rRNA gene sequences on public genetic databases, as the organismal universality and typically high gene copy number per cell facilitate gene amplification and sequencing (e.g. Woese, 1987; Woese et al., 1990; Winker & Woese, 1991). Within rRNA genes, varying evolutionary rates of base substitution can be localized to conserved core rRNA sequences (e.g. Gutell et al., 1985; Van de Peer et al., 1993; Vawter & Brown, 1993; Sullivan et al., 1995; Cannone et al., 2002; Misof et al., 2002; Baele et al., 2006) as well as rapidly evolving variable regions and expansion segments (e.g. Hassouna et al., 1984; Gerbi, 1985, 1996; Gorski et al., 1987; Neefs & De Wachter, 1990; Hancock, 1995; Schnare et al., 1996). Rates of substitution are even faster in transcribed spacer regions and non-coding spacer regions, which can reveal high levels of divergence even when compared across closely related species (e.g. Long & Dawid, 1980; Hillis & Dixon, 1991; Osorio et al., 2005), as well as within individual genomes (e.g. Gruendler et al., 1991; Lanadu et al., 1992; Lakshmikumaran & Negi, 1994; Rocheford, 1994). These conserved and variable regions of rRNA genes have not only been useful for recovering phylogenetic relationships (e.g. Woese et al., 1990; Friedrich & Tautz, 1997a,b), but also for understanding the structural and functional constraints of both the coding and the non-coding organizational sequences within rRNA genes (see Noller, 2005).
Recent crystal structures of the ribosome (Cate et al., 1999; Ban et al., 2000; Schluenzen et al., 2000; Wimberly et al., 2000; Spahn et al., 2001; Yusupov et al., 2001) have verified many of the long-standing structures of the rRNAs that were predicted by comparative sequence analysis (Woese et al., 1980; Noller & Woese, 1981; Noller et al., 1981; Brimacombe et al., 1983; Gutell et al., 1985, 1986; Woese & Pace, 1993) or substantiated with site-directed or random mutagenesis (e.g. Musters et al., 1989, 1991; Sweeney & Yao, 1989; Green et al., 1990). Still, these three-dimensional models have yielded further information on the organization of the small and large subunits (SSU and LSU, respectively) with the numerous ribosomal proteins comprising the mature ribosome. In particular, new information on enzymatic function (e.g. Nissen et al., 2000), non-canonical base pairing (e.g. Leontis & Westhof, 2003; Lee & Gutell, 2004; Lescoute et al., 2005), tertiary interactions via coaxial stacking (e.g. Elgavish et al., 2001; Ogle et al., 2001), RNA turn motifs (e.g. Gutell et al., 2000; Klein et al., 2001), rRNA–protein interactions (e.g. Brodersen et al., 2002; Hoang et al., 2004; Klein et al., 2004a; Noller, 2004), metal ions and rRNA–rRNA packing interactions (Klein et al., 2004b), lone pair triloops (Lee et al., 2003) and dynamism (Carter et al., 2001; Harms et al., 2001; Noller & Baucom, 2001) has been elicited from observations of tertiary models not easily obtained from comparative sequence analysis. It is essential that these characteristics be incorporated into multiple sequence alignments to improve the assignment of structurally and functionally homologous bases, as well as to verify that generated sequences are indeed functional rRNA genes and not paralogues and/or pseudogenes (e.g. Buckler et al., 1997; Muir et al., 2001; Bailey et al., 2003; Márquez et al., 2003) or artefacts of the sequencing process (see Clark & Whittam, 1992; States, 1992; Hickson et al., 1996; Gillespie et al., 2005a).
In this study, we predict entire secondary and some tertiary structures of the nuclear (nl) and mitochondrial (mt) SSU and LSU rRNAs from the honey bee, Apis mellifera, as the recent compilation of its genome (The Honey Bee Genome Sequencing Consortium, 2006) has provided sequences spanning the entire rDNA region. Wherever possible, we include information from published crystal structures of the ribosome and chemical probing studies to annotate and update diagrams of arthropod nl and mt rRNA structure. These diagrams (and related multiple sequence alignments) will be valuable for related studies on the phylogeny of arthropods, as well as the evolution of the structure and function of eukaryotic and organellar rRNAs.
With the completion of the honey bee genome comes the opportunity to begin analysing the intragenomic variation associated with repetitive sequence motifs, particularly those associated with multicopy genes. Here, we make an attempt to characterize the sequences associated with the externally transcribed spacer regions (ETS) and intergenic spacers (IGS) that are conserved across all rDNA cistrons in the honey bee genome. We describe several repeat regions that occur in the 5′- and 3′-ends of the IGS regions. Our preliminary mapping of these complete ETS sequences and partial IGS sequences will be useful for future studies that attempt to span the further repetitive motifs of the honey bee rRNA genes that occur in the central portion of the IGS, including promoter and enhancer regions.
As a result of the compilation of all rRNA genes in the honey bee genome, we were able to assess the degree of rRNA gene inactivation by the insertion of retrotransposable elements. Two types of retrotransposable elements, R1 and R2, can disrupt the complete transcription of arthropod rRNA genes by inserting into conserved sequences of the nl LSU rDNA (e.g. Long & Dawid, 1979; Jamrich & Miller, 1984; Jakubczak et al., 1990, 1991; Burke et al., 1993, 1999; Eickbush, 2002). Our analysis of R1 and R2 element retrotransposition in the honey bee rRNA genes is somewhat consistent with other studies on arthropod retrotransposition, and we provide here the predicted open reading frame (ORF) of an R2 element retrotransposase (RT).
Results and discussion
Predicted rRNA secondary structures
Our results are presented within the typical organization and pattern of transcription of eukaryotic rRNA genes. A schematic diagram illustrates the composition of typical arthropod rRNA genes within the nuclear and mitochondrial genomes (Fig. 1). Within the honey bee genome, the nl rRNA gene arrays have been identified as occurring on two different chromosomes (Beye & Moritz, 1993) subsequently localized to linkage groups 6 and 12 (Aquino-Perez, unpublished data), while the mt rRNA genes are organized in the ancestral arthropod position (see Boore et al., 1995; Boore, 1999) within the mitochondrial genome (Crozier & Crozier, 1993). In insects, pioneering structural studies of rRNA molecules were conducted on flies (Diptera) (Clary & Wolstenholme, 1985, 1987; Gutell & Fox, 1988; Hancock & Dover, 1988; Hancock et al., 1988; Tautz et al., 1988) and one beetle (Coleoptera) (Hendriks et al., 1988a). While these studies predicted structures mostly from comparisons with other early published complete rDNA sequences from bacteria (Glotz & Brimacombe, 1980; Woese et al., 1980; Noller & Woese, 1981; Noller et al., 1981; Stiegler et al., 1981; Zwieb et al., 1981), yeast (Veldman et al., 1981), frog (Ware et al., 1983; Clark et al., 1984; Dunon-Bluteau & Brun, 1986), rodent (Bibb et al., 1981; Hassouna et al., 1984; Michot et al., 1984; Gutell et al., 1985), cow (Gutell et al., 1985), human (Gorski et al., 1987) and other mammals (Hixson & Brown, 1986; Wool, 1986), they were lacking a larger collection of arthropod sequences to make thorough comparisons of most of the variable regions outside of the core rRNA [although see the early structural model of the mt LSU rRNA of the bird spider (Hendriks et al., 1988b)]. Thus, the following sections review subsequent structural studies that were performed with comparisons across several to many arthropod/insect rDNA sequences.
Figure 1.
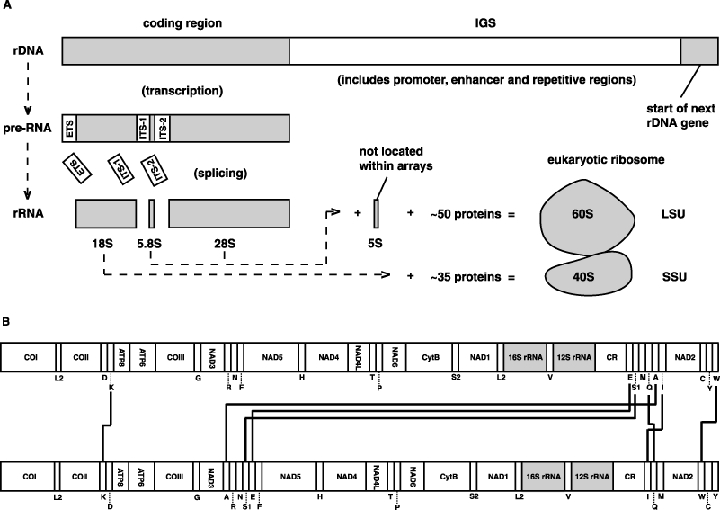
Organization of the rRNA genes within the honey bee genome. (A) Typical organization of the nuclear rRNA genes of eukaryotes. IGS, intergenic spacer; SSU, small subunit; LSU, large subunit. (B) Position of the rRNA genes (shaded) within the honey bee mitochondrial genome (top) and the ancestral arthropod mitochondrial genome (bottom). Rearrangements in the honey bee genome are depicted with solid lines. See Clary & Wolstenholme (1985, 1987) and Crozier & Crozier (1993) for information on honey bee mitochondrial genes.
Nuclear SSU rRNA
This model (Fig. 2A) is in concordance with the conserved 16S (prokaryote) and 16S-like (eukaryote) structures of the SSU rRNA from the literature (Woese et al., 1980; Noller & Woese, 1981; Stiegler et al., 1981; Gutell et al., 1985; Huysmans & De Wachter, 1986; Dams et al., 1988; Neefs et al., 1990, 1991, 1993; De Rijk et al., 1992; Gutell, 1994, 1996; Van de Peer et al., 1994, 1996, 1997, 1998, 1999; De Rijk et al., 1998; Wuyts et al., 2004) and is based on an updated model of Drosophila melanogaster (Gutell, 1993, 1994; Van de Peer et al., 2000; Cannone et al., 2002). Aside from D. melanogaster, structural models of five insect taxa have been published, including Tenebrio molitor (Coleoptera) (Hendriks et al., 1988a), Acyrthosiphon pisum (Heteroptera) (Kwon et al., 1991), peloridiid Hemiptera (Ouvrard et al., 2000), Loricera foveata (Coleoptera) (Van de Peer et al., 2000) and ichneumonoid Hymenoptera (Gillespie et al., 2005c). Our model is based loosely on these earlier models, and relies more heavily on the structure predictions from three recent studies wherein the 18S rRNA secondary structure was predicted across most insect orders (Kjer, 2004; Gillespie et al., 2005a; Yoshizawa & Johnson, 2005) as well as evidence from SSU rRNA crystal structures (Schluenzen et al., 2000; Wimberly et al., 2000; Yusupov et al., 2001). For referencing purposes, our new model follows the convention (variable region nomenclature, helix numbering, etc.) put forth by Gillespie et al. (2005a). The SSU rRNA contains 65 helices that are conserved across arthropods (Fig. 2A).
Figure 2.
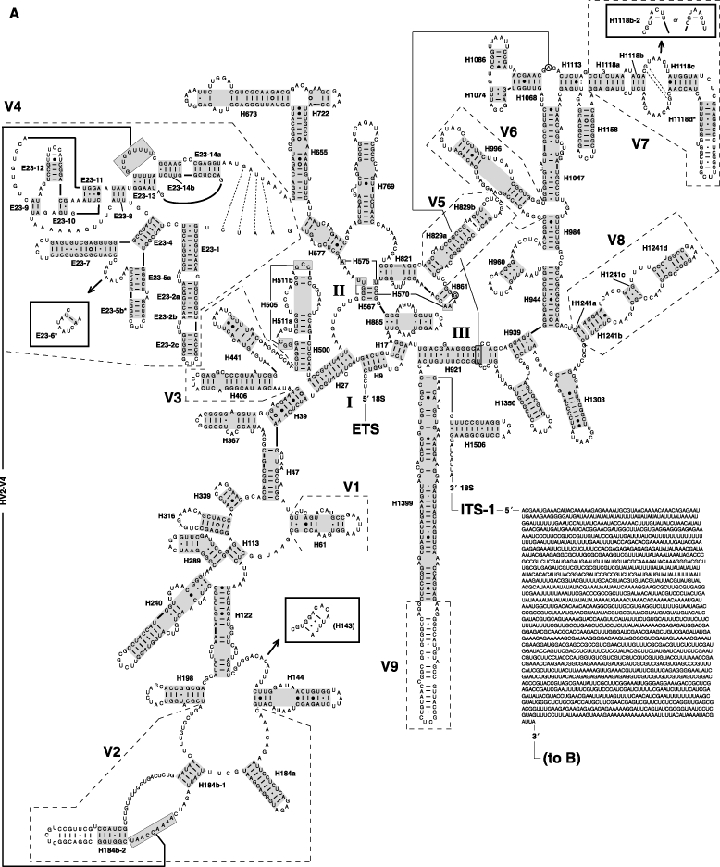
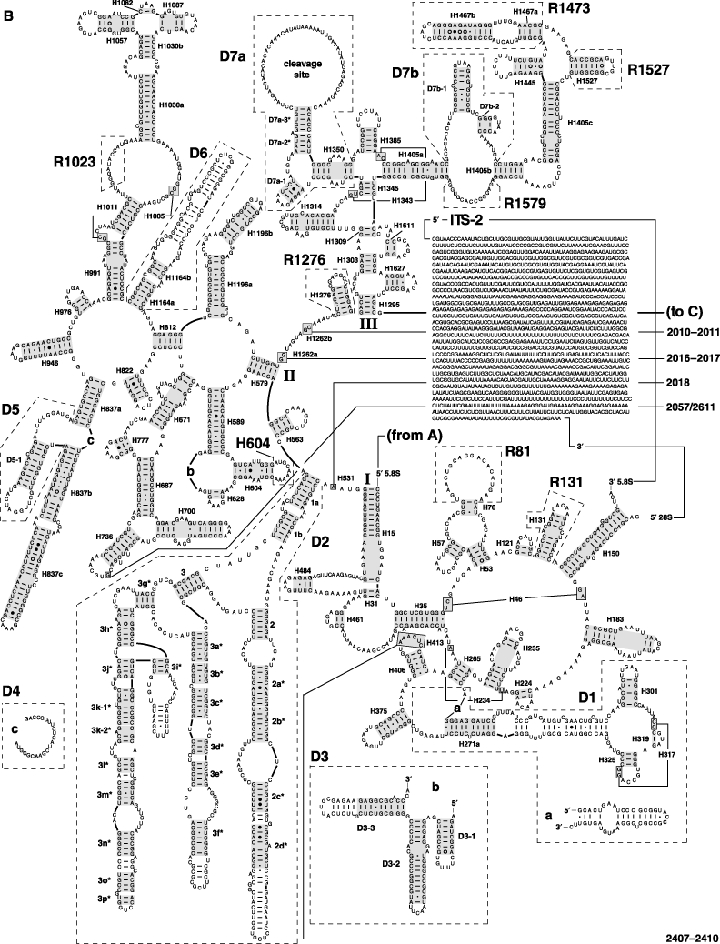
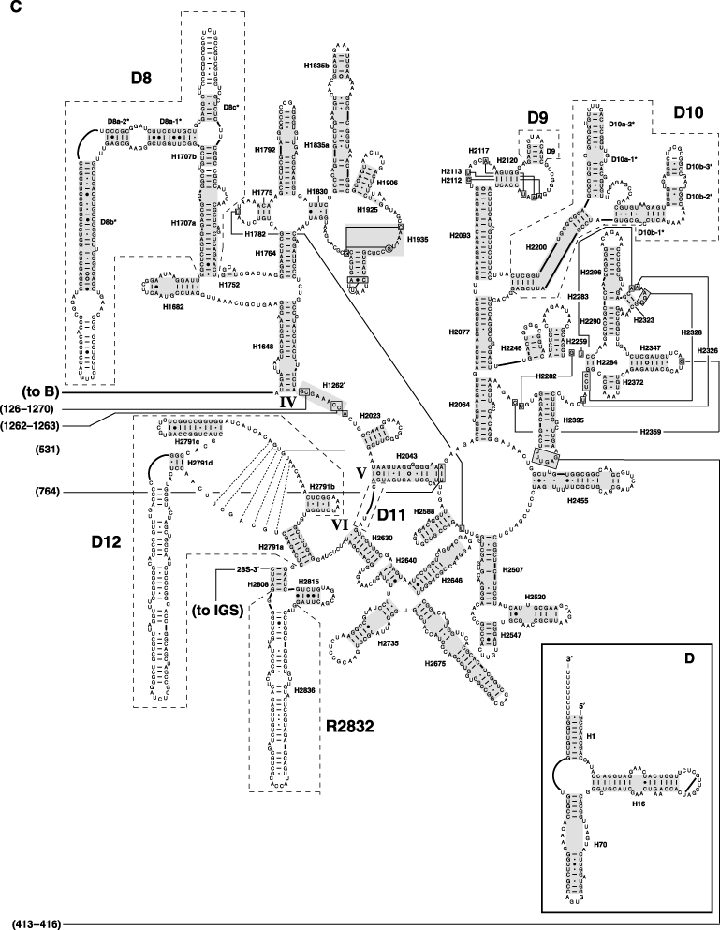
The secondary structure model of the nuclear rRNA (18S + 5.8S + 28S + 5S) from the honey bee, Apis mellifera. Variable regions are enclosed within dashed boxes and the naming follows either Schnare et al. (1996), Van de Peer et al. (1999) or Gillespie et al. (2005a,b,c). Helix numbering follows the system of Cannone et al. (2002), except for variable region 4 (V4) for which the notation of Wuyts et al. (2000) and Gillespie et al. (2005a) is used. Helices aligned across all sampled panarthropods are boxed in grey. Tertiary interactions (where there is strong comparative support) and base triples are shown connected by continuous lines. Base pairing is indicated as follows: standard canonical pairs by lines (C-G, G-C, A-U, U-A); wobble G·U pairs by dots (G·U); A·G pairs by open circles (A○G); other non-canonical pairs by filled circles (e.g. C•A). (A) Domains I–III of SSU rRNA (18S). Regions with alternative structures are boxed. (B) Domains I–III of LSU rRNA (5.8S + 28S). (C) Domains IV-VI of LSU rRNA (28S). (D) 5S rRNA. Diagrams were generated using the program XRNA (Weiser, B. & Noller, H., University of California at Santa Cruz) with manual adjustment.
The predicted structure of most regions of the 18S rRNA from honey bee are consistent with structures of the major lineages of Arthropoda, and differences between the nl SSU rRNA model of Gillespie et al. (2005a) and that presented here are illustrated on the jRNA website (see Experimental procedures). Variable region four (V4) (Neefs & De Wachter, 1990; Van de Peer et al., 1999) is shown with refinements to the double pseudoknot model of Wuyts et al. (2000), such that their proposed base triple is removed in place of the recently predicted tertiary interaction between the hairpin loop of the second pseudoknot and an unpaired sequence in variable region 2 (V2) (Alkemar & Nygård, 2003, 2004). Our alignment across arthropod V4 sequences spanning most orders shows strong support for the internal seven base pairs of this helix (HV2–V4); however, base pairs 1 and 9 are minimally supported with covariation evidence across arthropods (CRW Site bp frequency tables; Gillespie et al., 2005a).
Accurate structural models of arthropod V4 are of particular interest because this region of 18S rRNA is the most commonly sequenced marker for higher level arthropod phylogeny estimation (reviewed in Kjer, 2004). Still, multiple structural predictions exist for this large variable region of SSU rRNA. For instance, Ouvrard et al. (2000) proposed a secondary structure model for insect V4 based on the comparative analysis of 22 insect sequences. That same year, Wuyts et al. (2000) presented a refined model with the 3′-half of this variable region consisting of two pseudoknots. One of the pseudoknots was previously predicted (Neefs & De Wachter, 1990), while the other, a rare type of pseudoknot similar to that found in plant viruses (van Batenburg et al., 2000), was newly reported. Within these two contrasting models of the 3′-half of arthropod V4, 32 nucleotides form different base pairs (Fig. 3A). Six nucleotides form the same base pairs but within different structural contexts (grey dashed lines in Fig. 3A). Interestingly, across most eukaryotes this region alternates the number of base pairs within helices E23-13 and E23-14 (Wuyts et al., 2000). Our analysis of honey bee (and most arthropod) 18S rRNA sequences is consistent with an expanded helix E23-13 (6–11 bp) and a contracted helix E23-14 (11–3 bp) (Fig. 3B). A base pair frequency table (available at the jRNA website) suggests that the model of Wuyts et al. (2000), including HV2–V4 (Alkemar & Nygård, 2003, 2004), is more probable based on more covariation at the positions in the proposed base pairs; however, a minimal amount of compensatory base change evidence supports the Ouvrard et al. (2000) model such that a dynamic nature of this region of nl SSU rRNA cannot be disproved. These findings are consistent with a recent comparison of the Wuyts et al. (2000) and Alkemar & Nygård (2003, 2004) models in the V2 region across most insect lineages (Gillespie et al., 2005a; see table 3).
Figure 3.
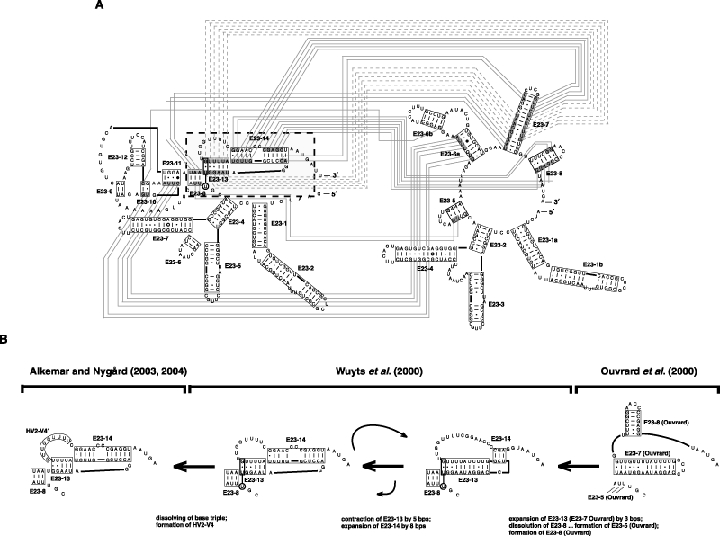
An illustration of several proposed secondary structural models for variable region V4 (V4) of arthropod 18S rRNA. (A) A comparison of the Wuyts et al. (2000) model (left) with the Ouvrard et al. (2000) model. Nucleotides in the 3′-half of V4 that are base paired in both models are shaded. Shaded nucleotides forming different base pairs in each model are connected with dashed lines. Shaded nucleotides forming the same base pairs in both models are connected with solid lines. The region within the dashed box is illustrated in (B). (B) An example of a possible dynamic relationship between three proposed models for the 18S○V4. See Fig. 2 legend for explanations of base pair symbols, helix numbering and reference for software used to construct structure diagrams.
Of the remaining regions of insect 18S rRNA, domain I (including variable regions V1–V3) is sequenced less frequently than domain III (including variable regions V6–V9) (Cannone et al., 2002). Variable regions V1, V3, V6 and V8 (and V5 of domain II) are considerably smaller in size than V4 and yield little phylogenetic information (Gillespie et al., 2005a). The V2 region is difficult to align with confidence due to length variation. Studies on arthropod V7 have divulged a region characteristic of extraordinary variation in the length of helices (Crease & Colbourne, 1998; Crease & Taylor, 1998; McTaggart & Crease, 2005). Sequence alignments of V7 across divergent insect taxa contain many variable positions, yet structure is necessary to guide the alignment. This is because, while only one helix is present in nearly all non-holometabolous orders, as well as in Amphiesmenoptera (Lepidoptera + Trichoptera) (Gillespie et al., 2005a), the remaining seven (minus Thysanoptera) holometabolous orders contain a second helix within V7. Additionally, this smaller less-conserved helix, H1118b-2 (Gillespie et al., 2005a), can be present or absent within these orders, with its base pairs often containing minimal comparative support. Finally, sometimes it is difficult to predict base pairing in the V9 at the end of the penultimate helix because many of the published sequences are missing the extreme 3′-end of the 18S rRNA (Cannone et al., 2002).
Nuclear LSU rRNA
This model is in concordance with the conserved 23S (prokaryote) and 23S-like (eukaryote) structures of the LSU rRNA from the literature (Noller et al., 1981; Wool, 1986; Gutell & Fox, 1988; Gutell et al., 1990, 1992, 1993; Larsen, 1992; De Rijk et al., 1994, 1997, 2000; Gutell, 1996; Schnare et al., 1996; Wuyts et al., 2001, 2002, 2004). With existing predicted structure models for Aedes albopictus (Kjer et al., 1994), D. melanogaster (Schnare et al., 1996; others therein), Acyrthosiphon pisum (Amako et al., 1996), Tenebrio sp. (Gillespie et al., 2004) and ichneumonoid Hymenoptera (Gillespie et al., 2005b), this is the sixth predicted structure of a complete or nearly complete insect nl LSU rRNA (Fig. 2B,C). To date, complete comparative models (alignments) for insect 28S rRNA have not been published, so we present our model following the nearly complete model of ichneumonoid Hymenoptera (Gillespie et al., 2005b) with adherence to LSU rRNA crystal structures (Cate et al., 1999; Ban et al., 2000; Spahn et al., 2001; Yusupov et al., 2001). Eukaryotic nl LSU rRNA is interrupted by an internally transcribed spacer region (ITS-2) in the terminal loop of helix H150 that separates the LSU into 5.8S and 28S rRNAs (Veldman et al., 1981; Michot et al., 1982). Flies (Diptera) have a second ITS in the nl LSU that occurs within the terminal loop of helix H131 (Tautz et al., 1988). The A. mellifera LSU rRNA (minus 5S rRNA) structure model contains 143 helices (133 in the 28S rRNA and five in the 5.8S rRNA, with five helices comprised of both 28S and 5.8S rRNAs) that are conserved across arthropods (Fig. 2B,C).
Thirteen regions previously described as extremely variable or expansion segments (Clark et al., 1984) of the 28S rRNA sequence are difficult to align across the arthropods (e.g. Hwang et al., 1998). These regions of honey bee rRNA have the typical variation seen across insect orders and families. Regarding Hymenoptera, the structures for two of the largest of these regions, expansion segments D2 and D3, were recently predicted in studies of 349 ichneumonoid wasps (Gillespie et al., 2005c; Wharton et al., 2006) and are comparable to the model predicted across 527 chalcidoid wasps (Gillespie et al., 2005b) and 60 evanioid wasps (Deans et al., 2006). Two other studies have used secondary structure as an alignment guide for phylogeny estimation of hymenopteran D2 sequences but did not produce global structural models (Belshaw & Quicke, 1997, 2002). While expansion segment D2 is phylogenetically informative within orders and families of arthropods (for a comparison of our model with Coleoptera see Gillespie et al., 2004), the alignment of these sequences across highly divergent taxa (i.e. class and phylum levels) is extraordinarily difficult (see Schnare et al., 1996). However, the expansion segment D3 is conserved enough in its secondary structure to be of practical use in arthropod phylogeny estimation (e.g. Whiting et al., 1997; Wheeler et al., 2001; Hovmöller et al., 2002), although its phylogenetic signal is weak when analysed without other data partitions (see Kjer, 2004).
As expansion segments D2 and D3 of the 28S rRNA can be amplified in one PCR reaction, and because they usually contain characteristic variation in sequence length and base composition that is informative across family, genus and species levels, this region of nl LSU rRNA has become one of the most common phylogenetic markers (e.g. Gillespie et al., 2003; Schulmeister, 2003). Thus, the majority of published insect nl LSU rRNA sequences are comprised of expansion segments D2 and D3 and related core sequences (Cannone et al., 2002). Some insect phylogenetic studies have generated sequences for expansion segments D4 to D10 (e.g. Hwang et al., 1998; Belshaw & Quicke, 2002; Wiegmann et al., 2003); however, in-depth structural predictions with reference to previous models were not provided. This lack of structural information from these studies, coupled with limited sequences covering the majority of the 3′-half of the 28S rRNA, makes it difficult to propose models that are thoroughly supported with base pair covariation. Therefore, our proposed structures for this region of the nl LSU rRNA, particularly expansion segments D7, D10, D12 and R2832, should not be considered well supported. Our labs are currently generating more sequences from the 3′-half of the 28S rRNA from a wide range of insect taxa that should improve these preliminary structure predications.
Sequences beyond the D3 in domain II include expansion segments D4–D6, which are relatively small for candidate phylogenetic markers (Fig. 2B). Earlier predicted structures for these regions in insects (Hancock et al., 1988; Kjer et al., 1994; Hwang et al., 1998) have recently been modified based on a more thorough taxon sampling (Gillespie et al., 2005c). In domain IV, a notable expansion segment, D7a, has been identified as the region of metazoan nl LSU rRNA wherein an internally processed cleavage site termed the ‘hidden break’ occurs. As originally characterized in the dipteran Sciara coprophila (Ware et al., 1985), this cleavage site separates the 28S rRNA into α and β fragments of roughly equal size (Applebaum et al., 1966; Balazas & Agosin, 1968; Greenberg, 1969; Ishikawa & Newburgh, 1972; Ishikawa, 1977; de Lanversin & Jacq, 1989). Aside from pea aphid (Acyrthosiphon pisum) 28S rRNA (Ogino et al., 1990); which does not separate into α and β fragments, most insects contain this hidden break (e.g. Shine & Dalgarno, 1973; Park & Fallon, 1990), which was speculated to occur near an unpaired 5′-UAAU-3′ sequence within expansion segment D7 (Ware et al., 1985). Despite this prevalent pattern, the proposed cleavage signal is absent in many hymenopteran 28S rDNA sequences (Gillespie et al., 2005c), including the honey bee. However, it is probable that this region of the D7a contains a cleavage site in the 28S rRNA, given that the loop formed by helix D7a-3 is extraordinarily variable in sequence length and base composition and contains no detectable conserved secondary structural elements across arthropods (e.g. Gillespie et al., 2005c) or other metazoan taxa studied (e.g. van Keulen et al., 1991; Zarlenga & Dame, 1992). Also, some insect D7a sequences contain repeated microsatellite motifs (Gillespie et al., 2005c) that are indicative of slippage events during replication of the rRNA gene array (Levinson & Gutman, 1987; Hancock & Dover, 1988), further suggesting that this portion of the 28S rRNA is non-functional and likely cleaved out of the mature rRNA molecule. This observation is consistent with site-directed mutagenesis studies on the yeast Saccharomyces cerevisiae, which show that the D7a variable region of the 26S LSU rRNA is dispensable (Musters et al., 1991). Recently, Basile-Borgia et al. (2005) provided evidence for the possible involvement of a highly conserved eukaryotic processing machinery responsible for cleavage of the α and β fragments in LSU rRNA, as microinjected Sciara coprophila (fungus fly) rDNA into Xenopus laevis oocytes resulted in transcription and pre28S rRNA fragmentation. Still, without a characterized processing machinery, an autocatalytic nature of the cleavage of cytoplasmic LSU rRNA cannot be ruled out.
Nearly half of domain IV of arthropod 28S rRNA is comprised of expansion segment D8 (Fig. 2C). Site-directed mutagenesis studies have implicated the D8 region with ribosome function (Sweeney et al., 1994), and it is likely that small nucleolar RNA E2 interacts with D8 in the human LSU rRNA (Rimoldi et al., 1993). Gillespie et al. (2005c) have characterized the structure of expansion segment D8 across aculeate Hymenoptera, and our analyses of more diverse taxa support their predicted structure. Domain V of arthropod 28S rRNA contains expansion segments D9, which is small and highly conserved in many Hymenoptera, including the honey bee, and D10, which is larger and highly variable in sequence composition but conserved in secondary structure (Gillespie et al., 2005c). As more sequences accumulate on genetic databases, it is likely that expansion segment D10 will become a more commonly used marker for phylogeny estimation, especially because it can be amplified in some arthropod taxa with all of expansion segment D12 (Gillespie, unpublished data).
Expansion segment D11 is positioned between domains V and VI in cytoplasmic LSU rRNA (Fig. 4C) and is typically short and without base pairing (Schnare et al., 1996). The remaining variable regions of arthropod 28S rRNA are expansion segments D12 and R2832, for which little information on base pairing can be derived given the paucity of published arthropod sequences spanning these regions (Cannone et al., 2002). We found minimal compensatory base change support for the four helices within expansion segment D12, and the remainder of the structured bases within this region should be considered preliminary at best. Similarly, despite having a complete 28S rRNA sequence for the honey bee, our prediction of variable region R2832 is supported minimally because most ‘complete’ published arthropod rDNA sequences are nearly complete, having the 3′-primer designed from the D. melanogaster sequence (it is no coincidence that the majority of published arthropod sequences containing complete or nearly complete R2832 sequences are flies). The difficulty in sequencing the entire 3′-half of the 28S rDNA is a consequence of the extreme variability within the IGS region that immediately flanks the last nucleotides of the 28S rDNA (see below).
Figure 4.
The secondary structure model of the mitochondrial rRNA (12S + 16S) from the honey bee, Apis mellifera. Differences between our sequence and previously published A. mellifera sequences (U65190 and U65191) are in bold, with insertions (dark arrows), deletions (open arrows) and substitutions (parentheses) shown. Helices aligned across all sampled panarthropods are boxed in grey. Tertiary interactions (where there is strong comparative support) and base triples are shown connected by continuous lines. (A) SSU rRNA (12S). Misaligned sequences 1 and 2 (discussed in text) are within dashed boxes and connected to redrawn structures from Hickson et al. (1996) and Page (2000) with dashed lines. (B) LSU rRNA (16S). See Fig. 2 legend for explanations of base pair symbols, helix numbering and reference for software used to construct structure diagrams.
The nuclear-encoded cytoplasmic large subunit of the ribosome contains a 5S rRNA in Archaea, Bacteria and Eukaryota, and while chloroplast and plant mitochondria also have a 5S rRNA in their large ribosomal subunit, the mitochondria in fungi, animals and many protists do not (Bullerwell et al., 2003). Our predicted structure of the nuclear-encoded honey bee 5S rRNA (Fig. 2D) is similar to other published structures of arthropod 5S rRNA (e.g. Barciszewska et al., 1995; Paques et al., 1995), and we confirmed our model with a recent interpretation (Szymanski et al., 2002) of the crystal structure of the LSU rRNA of the halophile archaea Haloarcula marismortui (Ban et al., 2000). We have no supporting evidence that this 5S rRNA, or an additional 5S rRNA encoded in the nucleus, is imported into mitochondria; however, evidence suggests that nuclear-encoded 5S rRNA is a constituent of mt ribosomes in several eukaryotes, including trypanostomatids (Mahapatra et al., 1994; Tan et al., 2002), yeast (Entelis et al., 2002) and mammals (Yoshionari et al., 1994; Magalhães et al., 1998; Entelis et al., 2001).
Mitochondrial SSU rRNA
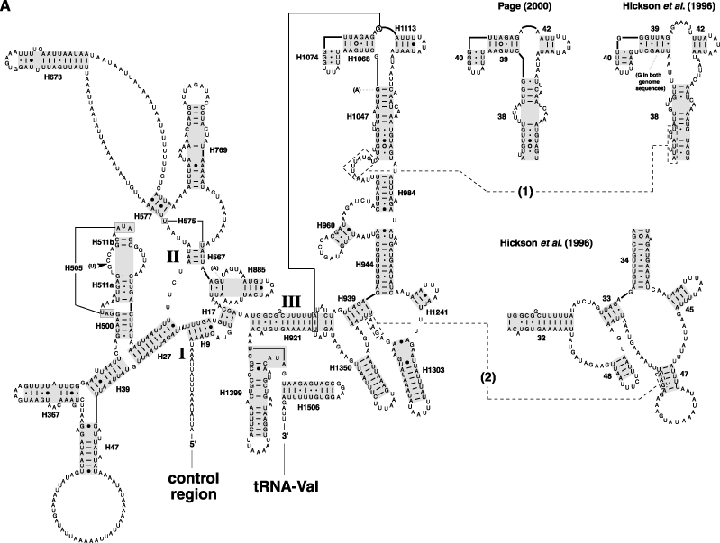
In most insects, the 12S rRNA gene is flanked on the 5′-side by the non-coding portion of the mitochondrial genome that contains the control region and on the 3′-side by the valine transfer RNA (tRNA-Val). The honey bee 12S rRNA gene follows this conserved arrangement of the rRNA genes, despite seven gene rearrangements within the mitochondrial genome (Crozier & Crozier, 1993). Our structural model (Fig. 4A) follows the Drosophila virilis model of Cannone et al. (2002) with minor modifications made based on an alignment of 83 neopteran 12S rRNA sequences. Our numbering system follows the convention established at the CRW Site (see Experimental procedures). There are 30 helices within insect 12S rRNA that are supported by compensatory base change evidence across our alignment.
Other than Drosophila spp. (Clary & Wolstenholme, 1985, 1987), the first structural models for insect 12S rRNA were based on comparisons with other mt SSU rRNA models, such as mouse (Bibb et al., 1981), frog (Dunon-Bluteau & Brun, 1986), primate (Dunon-Bluteau & Brun, 1986; Hixson & Brown, 1986) and sea anemone (Pont-Kingdon et al., 1994). Initially used for large-scale phylogenetic analyses (e.g. Pace et al., 1986; Field et al., 1988), it soon after became clear that the phylogenetic utility of SSU mt rRNA genes was limited to lower level divergences (Thomas et al., 1989; Simon et al., 1990). Thus, a plethora of lower level studies within Insecta utilizing 12S rDNA sequences emerged. Some sequences were collected in various structural compilations (Dams et al., 1988; Neefs et al., 1993; Gutell, 1994), and together with more in-depth models on birds (Cooper, 1994), vertebrate classes (Hickson, 1993), rodents (Sullivan et al., 1995) and insects (Simon et al., 1990, 1994; Simon, 1991), Hickson et al. (1996) presented a refined model of animal 12S rRNA that differed from previous models in that several variable regions were adjusted from the global SSU rRNA model to fit a custom animal model [results also reached by Kjer (1995, 1997) in his analysis of amphibians]. Of particular note, the honey bee 12S rRNA sequence confounded the alignment of helix H1047 [helix 38 of Hickson et al. (1996)] due to its high AT bias. This resulted in the proposal for a radical structure for this helix in honey bee. Analysing the secondary structure of 225 insect 12S rRNA sequences using maximum weighted matching, Page (2000) predicted a structure for domain III of honey bee 12S rRNA that was in more agreement with earlier insect models (e.g. Gutell, 1994). Our predicted model for domain III of honey bee 12S rRNA is more consistent with the model of Page (2000) than that of Hickson et al. (1996), and we have identified two regions within the alignment of Hickson et al. (1996) that were misaligned, causing the majority of domain III to be predicted inaccurately (Fig. 4A, boxed sequences 1 and 2). The second misaligned sequence (5′-GAA-3′) was noted by Page (2000) as being misaligned, prompting a follow-up note about the dangers of relying on conserved motifs to anchor alignments (Page, 2001). Our realignment of the first sequence (5′-AUUUAUGU-3′) further suggests that caution should be used when anchoring variable regions with thermodynamic evidence rather than support from crystal structures, as helix H1047 contains several non-canonical base pairs in its proximal region across many arthropod taxa (Cannone et al., 2002). As with Page (2001), we note that, despite its misalignment in Hickson et al. (1996), the honey bee 12S rDNA sequence likely did not affect the analysis by Hickson et al. (2000), wherein their structural alignment was used to evaluate the performance of a suite of automated alignment programs.
Despite the continued use of 12S rDNA sequences, few recent studies have presented structural predictions. The predicted structures for lice (Phthiraptera) (Page et al., 2002), Trirhabda leaf beetles (Coleoptera: Chrysomelidae) (Swigonova & Kjer, 2004), species in the taxon Arthropleona (Collembola) (Carapelli et al., 2004) and burnet moths (Lepidoptera) (Niehuis et al., 2006) all slightly differ from our model in that they are refined for the lower taxonomic levels analysed in each study. This latter study is interesting because, while nearly all previous studies of 12S rRNA structure and evolution have focused primarily on domain III, Niehuis et al. (2006) predict a structure of the entire 12S rRNA molecule, providing primers to amplify the less common sequence domains I and II. Hopefully more studies will utilize this information, as well as our 12S rRNA structural diagram and related alignments, to compile more sequences of domains I and II from diverse taxa to improve our knowledge of this seldom-sequenced region of arthropod 12S rRNA (Cannone et al., 2002).
Mitochondrial LSU rRNA
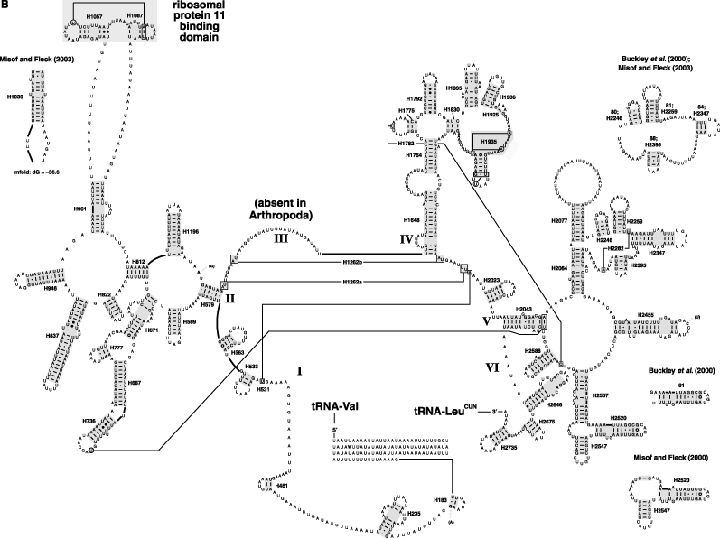
In most insects, the 16S rRNA gene is flanked on the 5′-side by the tRNA-Val gene, and on the 3′-side by the tRNA-LeuCUN gene. Like the 12S rRNA gene, the honey bee 16S rRNA gene follows this conserved arrangement (Fig. 1). Our secondary structure model (Fig. 4B) follows the D. melanogaster model of Cannone et al. (2002) with minor modifications made based on an alignment of 244 neopteran 16S rRNA sequences. Our numbering system follows the convention established at the CRW Site (see Experimental procedures). There are 49 helices within insect 16S rRNA that are supported by compensatory base changes across our alignment. Despite the presence of a complete sequence for nearly 20 years (Vlasak et al., 1987), we believe that this is the first published secondary structural model for honey bee mitochondrial 16S rRNA.
Since the earliest structural predictions for arthropod 16S rRNA, namely those from Drosophila yakuba (Clary & Wolstenholme, 1985), Locusta spp. (Uhlenbusch et al., 1987), D. melanogaster (Gutell & Fox, 1988) and gypsy moth, Lymantria dispar (Davis et al., 1994), more refined models have been proposed based on larger comparisons across divergent taxa, e.g. Deltocephalus-like leafhoppers (Fang et al., 1993), families of Hymenoptera (Dowton & Austin, 1994), families of Dermaptera (Kambhampati et al., 1996), families of Orthoptera (Flook & Rowell, 1997) and groups of spiders (Smith & Bond, 2003). These studies have undoubtedly prompted refinements to older structural diagrams present on the CRW Site and other rRNA secondary structure databases.
Buckley et al. (2000) analysed 16S rRNA secondary structure from over 400 taxa representing 13 insect orders. We assume that honey bee was included in their study; however, no secondary structure diagram was included for honey bee, and no honey bee sequence was present in their condensed alignment (Buckley et al., 2000). Aside from minor deviances, the authors noted several major differences between their predicted insect model and previous structures proposed by Gutell (unpublished then, now compiled on the CRW Site) and De Rijk et al. (1997). Specifically, helices 84 and 91 (our H2347 and H2520) were proposed to form different base pairs within their models than in the older models, and these structures are drawn as alternatives in our 16S rRNA structural diagram (Fig. 4B). Our search across 244 neopteran 16S rDNA sequences did not yield a high amount of compensatory substitutions supporting these alternative structures (data not shown).
Consistent with most structural and phylogenetic studies on 12S rRNA (see above), nearly all studies on 16S rRNA have analysed only a portion of the 3′-half of the molecule, representing conserved domains IV and V (Fig. 4B). Recently, however, Misof & Fleck (2003) analysed domains I, II, IV and V of 16S rRNA across major groups of dragonflies and damselflies (Odonata), predicting a nearly complete model for this large insect group (domain III is absent in arthropods and domain VI is rarely sequenced due to the position of the universal 3′-primer). Much of the odonate model is consistent with our predicted 16S rRNA structure presented here, with the major differences resulting from additional base pairs proposed in regions left unpaired in our model, for example the region in domain II that interacts with ribosomal protein 11 (Fig. 4B; discussed below), and helices H1–H3 in domain I proposed by Misof & Fleck (2003). Additionally, as with Buckley et al. (2000), helices H2347 and H2520 of our model are different in the Misof & Fleck (2003) model. Interestingly, many of the regions in the odonate model that differ from our model, likely by being more odonate-based, do not reflect phylogenetic information consistent with expected relationships within Odonata (Misof & Fleck, 2003; see Hypsa, 2006).
Mitochondrial rRNA structural conservation.
Arthropod mt rRNA has a characteristic structure (Fig. 4) that may be correlated with genome reduction in mitochondria (e.g. Lang et al., 1997) such that many features common to rRNA from all domains of life are absent in the organellar rRNA structure. A lack of recombination in the mitochondria (e.g. Lynch, 1995) likely contributes to a high level of non-canonical base pairs or unstructured regions that are highly conserved in non-organellar rRNAs of other organisms. The highly oxidative nature of the mitochondria is probably responsible for mutational loads not seen in most regions of the nucleus (e.g. Martin & Palumbi, 1993), particularly in nuclear coding sequences. Rates of evolution within the mitochondrial genome are typically much faster than genes within the nucleus (e.g. Crozier et al., 1989; Dowton & Austin, 1995; Sullivan et al., 1995; Lynch, 1997; Page et al., 1998; Lin & Danforth, 2004), a possible consequence of the lack of recombination and proofreading, and high oxidative environment. Studies showing gene rearrangement in arthropod mtDNA are growing (e.g. Boore et al., 1995; Black & Roehrdanz, 1998; Campbell & Barker, 1998, 1999; Dowton & Austin, 1999; Masta, 2000; Dowton & Campbell, 2001; Shao et al., 2001a,b; Shao & Barker, 2003; Masta & Boore, 2004; Covacin et al., 2006), and it has recently been demonstrated that mt genes in arthropods show a positive correlation between high rates of nucleotide substitution and gene rearrangement (Shao et al., 2003). While once considered stable in their position within the arthropod mitochondrial genome, evidence for rearrangements of the rRNA genes is growing (e.g. Masta, 2000; Shao & Barker, 2003; Covacin et al., 2006). It is interesting that, despite some of the highest levels of rates of substitution and AT bias (Crozier et al., 1989; Crozier & Crozier, 1993), the honey bee mt rRNA genes are arranged in the conserved ancestral manner of most other arthropods (Fig. 1B).
The paucity of crystal structures of mt rRNAs, coupled with the difficulty of aligning their variable regions due to a high AT bias, does not permit us to evaluate accurately the regions of mt rRNAs that are truncated or deleted relative to the cytoplasmic rRNAs. Ideally, significant differences from our conserved structural model, i.e. the louse 12S rRNA model (Page et al., 2002) or the odonate 16S rRNA model (Misof & Fleck, 2003), should be evaluated across all published arthropod mt rRNA sequences to determine the base pairings with the strongest positional covariation. Unfortunately, this difficult task is beyond the scope of our current investigation. However, we stress that regions of insect mt rRNA that differ across diverse taxa in both base composition and predicted secondary structures, may in fact be conserved in tertiary interactions within the ribosome. For example, the ribosomal protein 11 binding-domain within domain II of LSU rRNA (Fig. 4B) and associated ribosomal protein 11 (L11) is one of three structural domains that are proximal on the 50S subunit, forming the GTPase-associated centre (Wimberly et al., 1999). Recent evidence suggests that the structure of the LSU rRNA stabilized by the C-terminal domain of L11 is necessary to alleviate elongation factor G (EF-G) binding in the post-translocation state of the ribosome (Bowen et al., 2005). In arthropod 16S rRNA sequences between the L11 binding-domain and helix H991, which are shorter and closer to the core rRNA due to the loss of pseudoknot H1005 and helices H1011 and H1030, nucleotides must remain unpaired and in a ‘stretched’ conformation in order for the L11–L11 binding domain to proximally interact with two other structural domains in the GTPase-associated centre of the LSU rRNA. Thus, base pairing in these regions, even if supported by compensatory base change evidence (which by chance alone would outperform unpaired sites; see mfold structure in Fig. 4B), would be deleterious to ribosome assembly and function, as recently suggested for mammalian 16S rRNA (Mears et al., 2006). Given this, we doubt the formation of secondary structure in this region of 16S rRNA, although, as in the case of V4 of 18S rRNA (Fig. 3), it cannot be ruled out that base pairing in these regions may be temporary, ultimately dissolving during the tertiary interactions formed before and during translation. However, unlike the V4 region of 18S rRNA, transitionally less optimal structures are so far not supported across all arthropod sequences, and hence cannot support a hypothetical dynamic nature of this region of LSU rRNA.
Taxon-specific structures in mt rRNA variable regions would seemingly imply changes to associated ribosomal proteins and other ribosome cofactors, such that all of these molecules would coevolve to maintain the structural integrity and function of the mt ribosome. This is unlikely as ribosomal proteins are usually highly conserved across deep levels of divergence (e.g. Graybeal, 1994; Koonin et al., 2004; Stuart & Berry, 2004). Studies characterizing the coevolution of mitochondrial ribosomal proteins and their variable rRNA counterparts are needed to determine, if any, the secondary structures that are homologous across insects.
Characterization of IGS- and ETS-rDNA boundaries
Despite the growing number of sequenced eukaryotic genomes, few studies have predicted full sequences spanning the entire IGS, especially in arthropods. Usually, only those studies that sequence clones report complete IGS sequences, as intergenomic variation compounds other simpler methods. For genome sequencing studies, two main problems confound the accurate assemblage of IGS sequences: variation in subrepeat number, and the presence of multiple subrepeats that are not contiguous (i.e. separated by conserved or other repetitive sequences). This second problem is critical because if the variation within subrepeats is high then multiple non-contiguous subrepeats can be spuriously assembled in the wrong locale of the IGS, ultimately compounding the rebuilding of the accurate IGS sequence. Methods for accommodating these problems are in great need, as IGS regions contain the rDNA promoter region, as well as enhancer sequences that facilitate transcription of arthropod rRNA genes (e.g. Hayward & Glover, 1988). These regions are also useful for species identification in arthropods (e.g. McLain et al., 1989; Pepera et al., 1998; Mukha et al., 2000; Whang et al., 2002; Ohnishi & Yamamoto, 2004) and assessing the fitness and the evolutionary ecology of organisms (e.g. Cluster et al., 1987; Weider et al., 2005).
Relative to coding regions of the rRNA genes, complete IGS sequences from arthropods have only been characterized for a few taxa, and many of these are dipterans: D. melanogaster (Simeone et al., 1985; Tautz et al., 1987; Hayward & Glover, 1988; Tautz et al., 1988), D. orena, D. virilis, D. hydei (Tautz et al., 1987), Aedes albopictus (Baldridge & Fallon, 1992), A. aegypti (Wu & Fallon, 1998), Anopheles sinensis (Whang et al., 2002), the tstetse fly, Glossina sp. (Glossinidae) (Cross & Dover, 1987a,b), the black fly, Simulium sanctipauli (Simuliidae) (Morales-Hojas et al., 2002) and several species of Chironomus midges (Chironomidae) (Degelmann et al., 1979; Israelewski & Schmidt, 1982; Israelewski, 1983). Other published arthropod complete IGS sequences are from the German cockroach Blattella germanica (Blattaria: Blattellidae) (Mukha et al., 2002), the bulldog ant, Myrmecia croslandi (Hymenoptera: Formicidae) (Ohnishi & Yamamoto, 2004) and the crustaceans Daphnia pulex (Cladocera) (Crease, 1993) and Tigriopus californicus (Copepoda) (Burton et al., 2005). Much of the IGS and ETS have also been characterized for the pea aphid, Acyrthosiphon pisum (Kwon & Ishikawa, 1992) and the silkmoth, Bombyx mori (Fujiwara & Ishikawa, 1987). In agreement with complete IGS sequences from other eukaryotes, including the nematode Caenorhabditis elegans (Ellis et al., 1986), several diverse plants (Gruendler et al., 1991; Lanadu et al., 1992; Lakshmikumaran & Negi, 1994; Rocheford, 1994; Chiang et al., 1998; Fernández et al., 2000; Macas et al., 2003; Hsieh et al., 2004) and trypanostomatids (Schnare et al., 2000; Orlando et al., 2002; Boucher et al., 2004), IGS sequences contain one to several ‘types’ of repeat regions that are variable within species and difficult to compare across even closely related species (e.g. Murtif & Rae, 1985). Because these repetitive regions are associated with the rDNA transcription promoter, it is no surprise that RNA polymerase I complexes of one species fail to transcribe the rRNA genes of another (e.g. Dover & Flavell, 1984; Dover & Tautz, 1986).
IGS
We predicted the sequence and structure of the conserved regions flanking the 3′-end of the 28S rDNA (473 nts of the IGS) and the 5′-end of the 18S rDNA (2042 nts of the IGS + ETS) mostly from the repeat reads of assembly 3 (Fig. 5). While a distinct boundary between the IGS and ETS was not determined, we treat the description of the ETS separately (next section). Immediately flanking the boundary of the 28S rDNA and the IGS we predict a helix of eight base pairs that can also form in the bulldog ant (S1 in Fig. 5). Interestingly, Ohnishi & Yamamoto (2004) predicted a compound helical region in this same position in bulldog ant consisting of over 639 nts. While not experimentally proven, it is possible that this region of the IGS fosters a helical element; however, structural prediction in honey bee yielded nothing substantially significant other than helix S1 (data not shown). The remaining nts of the IGS flanking the 3′-end of the 28S rDNA contain six conserved repetitive (CRp) regions, three variable repetitive (VRp) regions, one A-rich region, one T-rich region and a subrepetitive (SRp) region (Fig. 5). This first SRp region, SRp1, is estimated to repeat from four to 11 times in the honey bee genome, but could likely exceed this range. Due to the variability in sequence length and base composition of SRp1, we could not accurately predict sequences flanking the 3′-end (dashed line in Fig. 5).
Figure 5.
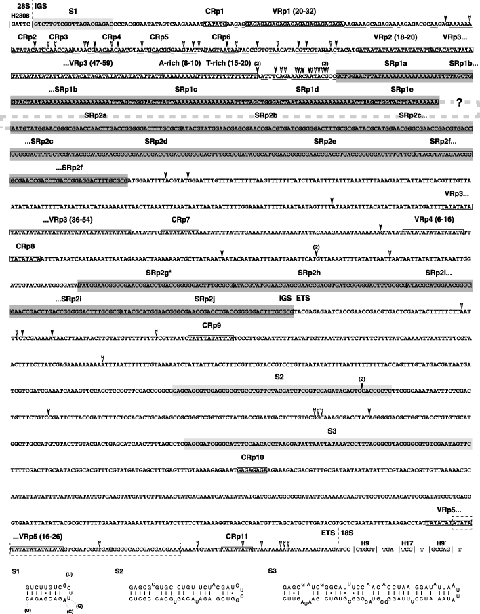
Regions of the IGS and ETS flanking the 3′-half of the 28S rDNA and the 5′-half of the 18S rDNA in the honey bee. Diagram depicts a consensus of assembled contigs, with infrequent insertions (filled arrows) and deletions (open arrows) shown in conserved regions of the alignment, as well as in conserved repetitive (CRp) regions. Length variation within variable repetitive (VRp) regions is given as ranges. Variation in sequence length and base composition is not provided for sub-repetitive (SRp) regions. All repetitive regions, as well as single-base length variable regions, are boxed and contiguous (SRp) regions are darkly shaded. The region of the IGS for which sequence identity was not possible due to intragenomic heterogeniety is depicted with a dashed line linking two large (SRp) regions. A putative promoter sequence, as predicted with the Neural Network Promoter Prediction tool (Reese, 2001) at the Berkeley Drosophila Genome project (http://www.fruitfly.org/seq_tools/promoter.html) is within a dashed box, with the bolded nucleotide depicting the predicted transcription start site. Conserved rRNA helices flanking the IGS (H2808) and ETS (H9) are within vertical bars (|). Regions with putative secondary structures (S1–S3) are lightly shaded, with structural diagrams shown below the alignment. The structure for S1 is shown with variable bases in a related hymenopteran, Myrmecia croslandi, shown in parentheses. Alignments are available at the jRNA website.
As with the region of the IGS flanking the 3′-end of the 28S rDNA, the region of the IGS adjacent to the ETS cannot be predicted beyond a SRp region (Fig. 5). This second SRp region, SRp2, is more complicated than SRp1 because it is repeated at least twice within conserved stretches of sequence. SRp2 is present from three to six times (and likely more), and is then interrupted by a conserved sequence containing two CRp regions and two VRp regions (Fig. 5). SRp2 then repeats again from two to four times before flanking a conserved stretch of 1113 nts that is likely comprised mostly of the ETS. It is difficult to predict the composition and structure of the internal region of the IGS flanked by SRp1 and SRp2 for two reasons. First, the number of SRp regions vary in arthropods, ranging from one in the pea aphid (Kwon & Ishikawa, 1992) to three in D. melanogaster (Tautz et al., 1988). Thus, although we identified two distinct SRp regions in honey bee IGS, a third (or more) SRp region may exist in the central portion of the spacer. It is interesting, however, that the bulldog ant has two SRp regions, and this may be a characteristic of hymenopteran IGS sequences. Second, the composition of the central portion of the IGS in arthropods is variable, in that it can be comprised entirely of repetitive regions (e.g. bulldog ant) or alternating conserved and repetitive regions (e.g. A. sinensis). While we have not reported a complete sequence for the IGS from honey bee, we predict that our characterization of the conserved sequences flanking two SRp regions will allow for primer design and sequencing of the remaining internal sequence.
ETS
Three CRp regions and one VRp region occur within a highly conserved stretch of nts that contains the rDNA gene promoter and probable transcription enhancer elements. Spacer and gene promoters have been determined for several arthropods (listed in Crease, 1993), with a consensus sequence of 5′-TA-TATANGRRRR-3′ accounting for the variation across diverse taxa. We did not find a sequence within the putative ETS region of honey bee that matched this motif; however, using the Neural Network Promoter Prediction tool (Reese, 2001) at the Berkeley Drosophila Genome project (http://www.fruitfly.org/seq_tools/promoter.html), a promoter sequence 5′-ATATATATATATATATATAGTCGATCGGTGAGGGGGCACCGACGACGAAA-3′ was predicted with a score of 1.0 (Fig. 5). A second promoter sequence 5′-ATTATATATATATATATATATATATATAGTCGATCGGTGAGGGGGCACCG-3′, shifted nine nts upstream from the first predicted promoter, was predicted with a score of 0.93. The bold nucleotide in each sequence is the predicted transcription initiation site (TIS), and the predicted TIS in the more optimal promoter is shaded in Fig. 5. Despite high scores, these promoter sequences are likely too close to the start of the 18S rDNA, especially when compared to the location of promoters in other arthropod ETS sequences. These putative promoter sequences and TIS should be taken with caution, as it will be necessary to determine the true promoter and TIS with S1 nuclease mapping.
Despite the high level of variation across ETS regions from different taxa, some characteristics are conserved and may allude to functional constraints not easily identifiable from primary sequence comparison. For instance, the GC content within the putative ETS region of honey bee is 35.3% and similar to other holometabolous insects; e.g. B. mori (36.7%) (Fujiwara & Ishikawa, 1987), D. melanogaster (24%) and D. orena (23.7%) (Tautz et al., 1987) and tsetse fly, Glossina morsitans (28%) (Cross & Dover, 1987a). The GC content of a non-holometabolous insect, the pea aphid, is 69% (Kwon & Ishikawa, 1992), indicating a possible correlation between base composition in the ETS and phylogeny. Helical elements have been predicted within the ETS region in other eukaryotes (Fernández et al., 2000; Schnare et al., 2000; Orlando et al., 2002; Boucher et al., 2004) and likely have an important role in the regulation of transcription and processing of rRNA. Using secondary structure, these regions can be aligned across closely related species (Fernández et al., 2000; Schnare et al., 2000; Orlando et al., 2002), providing information on putative rRNA processing sites (Schnare et al., 2000). Within the honey bee ETS region we calculated two putative helices using mfold (Mathews et al., 1999; Zuker, 2003) that warrant further validation from closely related hymenopteran taxa (Fig. 5). We were unable to identify these helices in the bulldog ant; however, given the variability present in other ETS helical elements, e.g. in legumes (Fernández et al., 2000) or trypanostomatids (Schnare et al., 2000), the bulldog ant and honey bee may have very divergent structures involved in the processing of the ETS region.
Retrotransposition in 28S rDNA sequences
R2-element insertion sites
We report the presence of three distinct R2 elements inserted into the 28S rDNA at the typical target sites described for other arthropods (e.g. Long & Dawid, 1979; Jamrich & Miller, 1984; Jakubczak et al., 1990, 1991; Burke et al., 1993, 1999; Eickbush, 2002). In general, most arthropod R2 elements insert between nucleotides 1928 and 1929 of the lonepair triloop (LPTR) H1925, however, R2 elements have also been shown to insert in helices H1835a and H1906 (Jakubczak et al., 1991). It now appears that only the initial nick in the target 28S rDNA sequence is conserved across arthropods, with the cleavage of the second strand highly variable such that the 5′-junctions of the R2 element and 28S rDNA are highly variable in sequence length and base composition, even within species (Burke et al., 1999). Indeed, our findings suggest at least two types of R2 element insert within nucleotides 1928 and 1929 of LPTR H1925 (Fig. 6A, B). Type 1 R2 elements are more conserved at their 5′-junction with the 28S rDNA and have a putative 5′-untranslated region (5′-UTR) of approximately 335 nts (Fig. 6E). In contrast, type 2 R2 elements are extraordinarily variable in their junction with the 28S rDNA and have a putative 5′-UTR of approximately 1039 nts (Fig. 6D,E). The 3′-region of this larger 5′-UTR from type 2 R2 elements is nearly identical with the entire 5′-UTR of type 1 R2 elements (Fig. 6E), suggesting that the additional nucleotides in the 5′-half of the 5′-UTR of type 2 R2 elements are not likely contributing to the structure and function of the conserved 5′-UTR. The ultimate 3′-nucleotides of the 5′-UTR in all type 1 and type 2 R2 elements from honey bee flank a predicted start codon of the putative ORF of the RT protein (Figs 6E and 7A).
Figure 6.
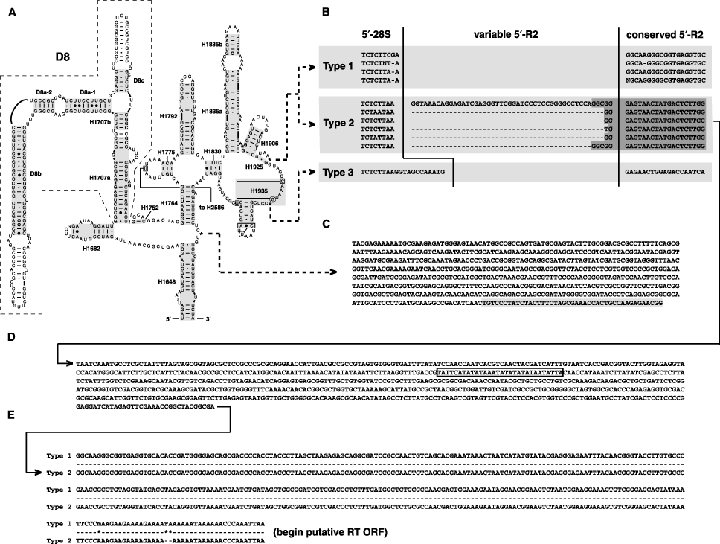
R1 and R2 element insertion sites in honey bee 28S rDNA sequences, and variable R2 element 5′-UTRs. (A) Predicted secondary structure of domain IV of honey bee 28S rRNA, with asterisks depicting R1 and R2 element insertion sites. (B) Variable 5′-junction of R2 element insertion sites. Shaded region in type II elements depict conserved regions flanking a highly variable junction. (C) Three-prime junction of partial putative R1 element in honey bee 28S rDNA. Shaded sequence depicts 28S rDNA. Note: no 5′-junction was recovered for this partial R1 element (see text). (D) Conserved 758 nts in the 5′-UTR of type II R2 elements. Boxed sequence contains variation across unassembled reads. (E) Conserved 335 nts of the 5′-UTR of type I and type II R2 elements. See Fig. 2 legend for explanations of base pair symbols, helix numbering and reference for software used to construct structure diagrams.
Figure 7.
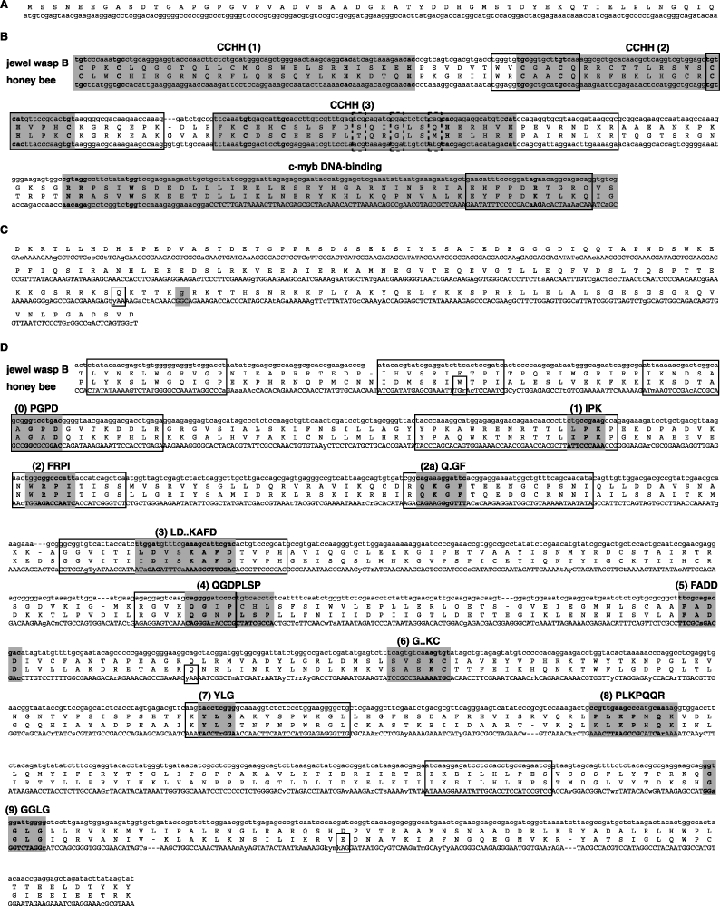
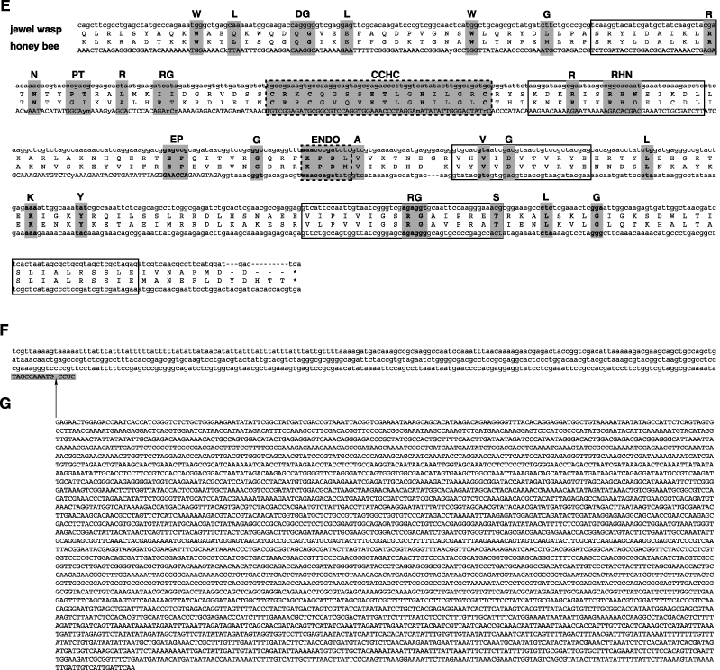
Characteristics of the honey bee R2 element ORF, 3′-UTR and potential imprecise insertion site. Bold amino acid residues are conserved across 80% or more arthropod R2 elements (Burke et al., 1999). Boxed aligned regions depict highly conserved sequence across parasitic Hymenoptera. Single-boxed residues depict codons with IUPAC ambiguity codes with the alternative nucleotide causing a stop codon. (A) Non-conserved 5′ sequence of the ORF with the putative initiation codon Met. (B) Comparison of jewel wasp B sequence (Nasonia sp., Pteromalidae) (GenBank accession no. AF090145) to honey bee in the conserved amino-terminal domains. Shaded regions depict conserved motifs of DNA-binding proteins, with dashed boxes showing the three residues believed to interact with the α-helical region of DNA. CCHH, Cys-His motifs; c-myb, proto-oncogene protein. (C) Non-conserved sequence flanked by the conserved amino-terminal motifs and the 5′-end of the RT domain. Note: the jewel wasp B sequence contains 255 amino acid residues compared to the 178 of the honey bee. The shaded glycine residue was recovered in only one unassembled read. (D) Comparison of jewel wasp sp. B and honey bee in the highly conserved reverse transcriptase (RT) domain, including the fingers/palm and thumb motifs. The 11 shaded regions depict motifs conserved in the RTs of all retroelements (Xiong & Eickbush, 1990; Burke et al., 1999). (E) Comparison of jewel wasp B and honey bee in the conserved carboxyl-terminal domains. The DNA-binding motif CCHC and the KPDI sequence (ENDO) are shaded and within dashed boxes, representing the endonculease domain. Other shaded motifs depict conserved residues in arthropods (Burke et al., 1999). (F) Predicted 3′-UTR. The shaded sequence is the 3′-junction with the 28S rDNA (see Fig. 5), with the space depicting the potential imprecise insertion site of R2 element type 3. (G) Consensus sequence of R2 element type 3, which seemingly inserts 12 nucleotides downstream from the typical R2 insertion site. No ORF has been predicted for this element (see text).
We also discovered a third type of R2 element in honey bee 28S rDNA. Type 3 R2 elements were found to insert in an unusual position in the 28S rDNA, helix H1935 (Fig. 6A,B), and did not reveal a typical ORF of other arthropod R2 elements (Fig. 6G). This, coupled with a lack of a 3′-junction with the 28S rDNA, and the fact that we only detected a few copies of type 3 R2 elements, led us to doubt that this position in honey bee 28S rDNA fosters the insertion of R elements. It could also be that this particular region of 28S rDNA was erroneously assembled in the building of the genome. Still, as the number of arthropod genome sequences continue to grow, it is likely that new insertion sites in 28S rDNA will be discovered, especially within this region of domain IV, given that it is one of the most interrupted regions of rDNA across all domains of life (Jackson et al., 2002).
Predicted R2-element ORF and 3′-UTR
Given that ORFs for the RT of R2 elements have been predicted for a variety of arthropods (e.g. Burke et al., 1999; Kojima & Fujiwara, 2004, 2005; Bunikis & Barbour, 2005), we aligned and predicted the domain structures for honey bee R2-RT sequences by comparison with another hymenopteran insect, the jewel wasp Nasonia sp. (Chalcidoidea: Pteromalidae) (Fig. 7B,D,E). Nasonia sp. is remarkable because it contains two different types of R2 elements: one that is comparable to most other arthropod R2 elements (type A), and a second that differs mainly in the presence of two additional zinc-finger domains in the amino-terminal region (type B) (Burke et al., 1999). Only one other arthropod, the horseshoe crab Limulus polyphemus, has a type B R2 element (Burke et al., 1999). Interestingly, not only does our analysis of the honey bee genome identify the presence of a type B R2 element similar to horseshoe crab and Nasonia sp. type B elements (a finding consistent with Kojima & Fujiwara, 2005) (Fig. 7B), it also suggests that the honey bee does not have the more common type A R2 element. Within a phylogenetic context of Hymenoptera (e.g. Dowton & Austin, 1994), wherein Chalcidoidea is derived in regards to a more primitive Apidae, this implies that Nasonia sp. has retained both A and B type R2 elements, whereas honey bee has lost the more typical A type but retained the less frequent B type, which is likely a more primitive parasite of arthropod genomes (Kojima & Fujiwara, 2005). Two types of R2 elements have been detected in the cicada killer, Sphecius speciosus (Sphecoidea; Sphecidae); however, the limited portion of the published sequence cannot lead us to determine if this insect contains type A or B R2 elements (or both). In response to this, we have identified several regions within the ORF alignment of Nasonia sp. B and honey bee R2 elements that are highly conserved for the design of PCR and sequencing primers (Fig. 7). We are currently using primers from these regions to determine the presence or absence of type B R2 elements across hymenopteran families. Given that R2 elements typically evolve at rates similar to their rDNA hosts (Eickbush & Eickbush, 1995; Eickbush et al., 1995), we expect our investigation to yield a reasonable estimate of the evolution of R2 elements across Hymenoptera, and the degree of vertical and horizontal transmission of these mobile elements.
The remaining conserved domains of the honey bee R2 element ORF, namely the RT domain with fingers/palm and thumb motifs (Fig. 7D) and the carboxyl-terminal domains that include the CCHC DNA-binding motif and endonuclease domain (Fig. 7E), are highly conserved across honey bee and jewel wasp. The sequence between the ORF stop codon (UGA) and the 3′-junction of the 28S rDNA, or 3′-UTR, is more conserved than the 5′-UTR and comprises 504 nts. Thus, it is similar in size to the Nasonia spp. 3′-UTR, which is 594 nts (Burke et al., 1999). While predicted secondary structures exist for R2 element-3′-UTRs from several arthropods, including the lepidopterans Bombyx mori (silkmoth), Samia cynthia (Ailanthus silkmoth), Callosamia promethea (Promethea silkmoth), Coscinocera hercules (Hercules moth) and Saturnia pyri (great peacock moth), the earwig (Forficula auricularia) and several Drosophila spp. (Mathews et al., 1997; Ruschak et al., 2004), no secondary structures have been predicted for hymenopteran insects. This is likely due to the large size of the hymenopteran R2 element-3′-UTR as compared to other arthropods, but also due to a lack of more sequences to facilitate the comparative method for predicting structure. Even using the Nasonia spp. (A and B) for comparative analysis, our attempts at predicting a secondary structure for honey bee R2 element-3′-UTR were futile.
While non-LTR retrotransposable elements are likely highly abundant in arthropod genomes (Perez-Gonzalez & Eickbush, 2001), most copies are likely defective elements that drift with their inactivated rRNA gene hosts (e.g. Jamrich & Miller, 1984; Weiner et al., 1986; Luan & Eickbush, 1996; Sassaman et al., 1997). Subject to the process of concerted evolution or biased gene conversion of rDNA arrays (Bigot et al., 1992; Jakubczak et al., 1992), non-functional R2 elements within inactivated rRNA genes are rapidly eliminated from arthropod genomes (Perez-Gonzalez & Eickbush, 2001, 2002). Despite the likely non-functional putative type 3 R2 element described above, we found limited evidence for non-functional R2 elements within the rRNA genes of the honey bee. This is probably an artefact of our method for constructing the R2 element sequences from conserved flanking regions of 28S rDNA, as the majority of non-functional R2 elements would likely reside within heavily mutated 28S rDNA sequences, which would not have been sampled in our procedure. Of the conserved R2 elements with putatively functional ORFs, we identified four nucleotide positions that contain variation in excess of single nucleotide polymorphisms (SNPs). All four cases pertain to either first or third codon positions of predicted amino acids, with the state in less frequency resulting in a stop codon (Fig. 7C,D). These variable positions likely indicate the presence of some non-functional R2 elements that have yet to be pruned out of the genome by concerted evolution of rRNA genes.
Lack of functional R1 elements in the honey bee genome
R1 elements are found in arthropod 28S rRNA genes in a conserved insertion site 74 nts downstream of the R2 insertion site (Roiha et al., 1981) (Fig. 6A). Our search for R1 elements within the honey bee genome did not recover any functional or complete sequences. Only truncated copies of putative R1 elements were assembled at the 3′-junction of the 28S rDNA (Fig. 6C). It is surprising that the honey bee does not contain complete and functional R1 elements, given that other hymenopteran insects have been purported to harbour them (Jakubczak et al., 1991; Bigot et al., 1992; Varricchio et al., 1994). According to Jakubczak et al. (1991), Nasonia sp., S. speciosus and the carpenter bee, Xylocopa sp., all contain R1 elements, although ORFs have not been determined for any of these species, and only the 3′-junctions with the 28S rDNA were actually sequenced. Similarly, Bigot et al. (1992) detected the presence of R1 elements in 12 Hymenoptera, including the honey bee; however, the distinction between functional and truncated copies could not be determined by Southern blot analysis. The results of their RFLP analysis of the R1 insertion site (Fig. 3, Bigot et al., 1992) showed a small amount of DNA in honey bee as compared to other sampled Hymenoptera, a finding consistent with our detection of only truncated R1 elements. Indeed, 5′-truncated R1 elements of sizes 0.5 and 1 kb long are known from the 28S rRNA genes of D. melanogaster (Jakubczak et al., 1990, 1991). In agreement, our truncated consensus R1 element is 577 nts long, and thus could reflect the remnant of once functional R1 elements in the genome of the honey bee that have since been pruned by rRNA gene homogenization. Alternatively, R1 elements could occur in such low frequency in the honey bee genome that no functional copies were included in its assembly.
Conclusion
Our analysis of the rRNA genes from the honey bee suggests that the functional rRNA-coding regions are structurally conserved and homogeneous throughout the nuclear and mitochondrial genomes. Some regions of mt rRNAs that are variable in sequence length and base composition do not contain secondary structures that are conserved across Insecta. Studies evaluating these variable regions within the tertiary structure of the ribsome, including rRNA-rRNA and rRNA–ribosomal protein interactions, are needed to determine their structural and functional significance. Our preliminary characterization of the IGS and ETS regions linking nl rRNA genes in the honey bee suggests that these highly variable sequences are relatively similar to other holometabolous insects in organization, repetitive nature and base composition. Like most other arthropods studied, honey bee rRNA genes are subject to parasitism by retrotransposable elements, although they lack both the most common type of R2 element and functional R1 elements. In Hymenoptera, it has been hypothesized that the intriguing haplo-diploid system, in which males come from parthenogenic eggs (n) and females come from fertilized eggs (2n), is correlated with a low level of genetic variability, relative to other arthropod genomes (Grauer, 1985; Woods & Guttman, 1987; Bigot et al., 1992). The characteristics of the honey bee rRNA genes we present here cannot contest this claim.
Experimental procedures
rRNA secondary structure prediction
The assembled honey bee rDNA sequences were integrated into the arthropod rRNA models (nl 18S, 5.8S, 28S, 5S rRNAs; mt 12S and 16S rRNAs) predicted and compiled at the Comparative RNA website (CRW Site) (http://www.rna.icmb.utexas.edu) and the jRNA website (http://hymenoptera.tamu.edu/rna). Helix numbering follows the E. coli system available at the CRW Site. Information pertaining to the alignment of RNA sequences using secondary structure models, including covariation analysis, thermodynamic algorithms, and ambiguously aligned regions, is available at both websites. Secondary structure model diagrams were generated with the program XRNA (developed by B. Weiser and H. Noller, University of Santa Cruz, CA). Base pair frequency tables and structural diagrams are available at the CRW Site. Differences between our previous arthropod rRNA structures and those presented here are illustrated at the jRNA website.
IGS sequence comparison
Conserved rDNA sequences flanking the 3′-terminal end of the 28S rRNA and 5′-terminal end of the 18S rRNA were used for Blast (Altschul et al., 1990) searches at the Honey Bee Genome Database (http://racerx00.tamu.edu/bee_resources.html). All options in assembly 3 were explored, including unassigned groups (bin 0); however, almost without exception, all Blast results were within the repeat reads of assembly 3. Default Blast settings were used, except that we did not filter for low complexity. Results were viewed as master-slave with identities and displayed with 500 descriptions and alignments. We used master-slave to identify reads used in subsequent Blast searches to extend the sequence. Only sequence differences repeated four or more times were reported. Unique and rare SNP differences were not reported. Sequences were then aligned manually in SeAl v2.0a11 (Rambaut, 1996). Only one alignment was made for the 5′-end of the IGS, while three alignments were made for the 3′-end of the IGS and ETS regions. Alignments are available at the jRNA website.
R1 and R2 element prediction rDNA sequences spanning the conserved insertion sites for R1 and R2 elements in arthropods were compiled using the Blast strategy discussed above. Results containing non-rDNA sequences inserted either at the 5′- or 3′-end of the rDNA insertion site were exported to SeAl for manual alignment. Further Blast searches were performed using conserved regions of aligned sequences, allowing us to ‘walk’ across the R elements from both the 5′- and 3′-ends. Upon completion of the R2 elements, we translated the consensus sequence in six frames to determine the ORF of RT protein. This allowed for the identification of putative start and stop codons. Nucleotide and amino acid sequences of honey bee R2 element were aligned with the R2-B element of the jewel wasp, Nasonia sp. (GenBank accession no. AF090145). Alignment was performed manually with reference to the published structure of RT proteins from arthropods (Burke et al., 1999).
Supplementary material
The following material is available for this article online:
S1. Base pair frequency table comparing the models of Wuyts et al. (2000) and Ouvrard et al. (2000) for variable region 4 (V4) of arthropod 18S rRNA.
S2. Alignments of the IGS and ETS regions of unassembled honey bee rDNA sequences.
S3. Alignments of R1 and R2 retrotransposable elements of the honey bee.
This material is available from the Comparative RNA website, http://www.rna.icmb.utexas.edu, and the jRNA website http://hymenoptera.tamu.edu.
Acknowledgments
We are grateful to Thomas Eickbush and Deborah Stage (Rochester University) for helpful insight on retrotransposable elements in arthropods, to Teresa Crease (University of Guelph) for helpful discussions on IGS regions, and to Karl Kjer (Rutgers University) for his profundity on insect rRNA secondary structure. J.J.G. especially thanks Bob Wharton at Texas A & M University (NSF DEB 0328920) for support throughout the completion of this project. This manuscript was greatly improved by the thorough critique of an anonymous reviewer. Our work was partially funded by a grant from the National Institute of Health to R.R.G. (GM67317). ‘Where is the wisdom we have lost in knowledge? Where is the knowledge we have lost in information?’– T. S. Eliot.
Note added in proof
Carapelli et al. (2006) recently predicted the secondary structure of the mt 16S rRNA for the strepsipteran Xenos vesparum. There are significant differences between their model and the model presented here, particularly in domain V of mt LSU rRNA.
Ref:
Carapelli A, Vannini L, Nardi F, Boore JL, Beani L, Dallai R, Frati F. The mitochondrial genome of the entomophagous endoparasite Xenos vesparum (Insecta: Strepsiptera) Gene. 2006;376:248–259. doi: 10.1016/j.gene.2006.04.005.
References
- Alkemar G, Nygård O. A possible tertiary rRNA interaction between expansion segments ES3 and ES6 in eukaryotic 40S ribosomal subunits. RNA. 2003;9:20–24. doi: 10.1261/rna.2108203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkemar G, Nygård O. Secondary structure of two regions in expansion segments ES3 and ES6 with the potential of forming a tertiary interaction in eukaryotic 40S ribosomal subunits. RNA. 2004;10:403–411. doi: 10.1261/rna.5135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amako D, Kwon OY, Ishikawa H. Nucleotide sequence and presumed secondary structure of the 28S rRNA of pea aphid: implication for diversification of insect rRNA. J Mol Evol. 1996;206:469–475. doi: 10.1007/BF02337519. [DOI] [PubMed] [Google Scholar]
- Applebaum SW, Ebstein RP, Wyatt GR. Dissociation of ribosomal ribonucleic acid from silkmoth pupae by heat and dimethylsulfoxide: evidence for specific cleavage points. J Mol Biol. 1966;21:29–41. doi: 10.1016/0022-2836(66)90077-5. [DOI] [PubMed] [Google Scholar]
- Baele G, Raes J, Van de Peer Y, Vansteelandt S. An improved statistical method for detecting heterotachy in nucleotide sequences. Mol Biol Evol. 2006;23:1397–1405. doi: 10.1093/molbev/msl006. [DOI] [PubMed] [Google Scholar]
- Bailey CD, Carr TG, Harris SA, Hughes CE. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol Phylogenet Evol. 2003;29:435–455. doi: 10.1016/j.ympev.2003.08.021. [DOI] [PubMed] [Google Scholar]
- Balazas I, Agosin M. Isolation and characterization of ribonucleic acid from Musca domestica (L.) Comp Biochem Physiol. 1968;27:227–237. doi: 10.1016/0010-406x(68)90766-4. [DOI] [PubMed] [Google Scholar]
- Baldridge GD, Fallon AM. Primary structure of the ribosomal DNA intergenic spacer from the mosquito, Aedes albopictus. DNA Cell Biol. 1992;11:51–59. doi: 10.1089/dna.1992.11.51. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Barciszewska MZ, Gawronski A, Szymanski M, Erdmann VA. The primary structure of Harpalus rufipes 5S ribosomal RNA: a contribution for understanding insect evolution. Mol Biol Rep. 1995;21:165–167. doi: 10.1007/BF00997239. [DOI] [PubMed] [Google Scholar]
- Basile-Borgia AE, Dunbar DA, Ware VC. Heterologous rRNA gene expression: internal fragmentation of Sciara coprophila 28S rRNA within microinjected Xenopus laevis oocytes. Insect Mol Biol. 2005;14:523–536. doi: 10.1111/j.1365-2583.2005.00583.x. [DOI] [PubMed] [Google Scholar]
- van Batenburg FHD, Gultyaev AP, Pleij CWA, Ng J, Oliehoek J. PseudoBase: a database with RNA pseudoknots. Nucleic Acids Res. 2000;28:201–204. doi: 10.1093/nar/28.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Quicke DLJ. A molecular phylogeny of the Aphidiinae (Hymenoptera: Braconidae) Mol Phylogenet Evol. 1997;7:281–293. doi: 10.1006/mpev.1996.0400. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Quicke DLJ. Robustness of ancestral state estimates: Evolution of life history strategy in ichneumonoid parasitoids. Syst Biol. 2002;51:450–477. doi: 10.1080/10635150290069896. [DOI] [PubMed] [Google Scholar]
- Beye M, Moritz RFA. In situ hybridization of rDNA on chromosomes of the honeybee, Apis mellifera L. Experientia. 1993;49:337–338. [Google Scholar]
- Bibb MJ, van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bigot Y, Lutcher F, Hamelin M-H, Périquet G. The 28S ribosomal RNA-encoding gene of Hymenoptera: inserted sequences in the retrotransposon-rich regions. Gene. 1992;121:347–352. doi: 10.1016/0378-1119(92)90142-c. [DOI] [PubMed] [Google Scholar]
- Black WC, Roehrdanz RL. Mitochondrial gene order is not conserved in arthropods: prostriate and metastriate tick mitochondrial genomes. Mol Biol Evol. 1998;15:1772–1785. doi: 10.1093/oxfordjournals.molbev.a025903. [DOI] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Collins TM, Stanton D, Daehler LL, Brown WM. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- Boucher N, McNicoll F, Laverdière M, Rochette A, Chou M-N, Papadopoulou B. The ribosomal RNA gene promoter and adjacent cis-acting DNA sequences govern plasmid DNA partitioning and stable inheritance in the parasitic protozoan Leishmania. Nucleic Acids Res. 2004;32:2925–2936. doi: 10.1093/nar/gkh617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WS, Van Dyke N, Murgola EJ, Lodmell JS, Hill WE. Interaction of thiostrepton and elongation factor-G with the ribosomal protein L11-binding domain. J Biol Chem. 2005;280:2934–2943. doi: 10.1074/jbc.M407008200. [DOI] [PubMed] [Google Scholar]
- Brimacombe R, Maly P, Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16S RNA. J Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Buckler ESI, Ippolito A, Holtsford TP. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics. 1997;145:821–832. doi: 10.1093/genetics/145.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley TR, Simon C, Flook PK, Misof B. Secondary structure and conserved motifs of the frequently sequenced domains IV and V of the insect mitochondrial large subunit rRNA gene. Insect Mol Biol. 2000;9:565–580. doi: 10.1046/j.1365-2583.2000.00220.x. [DOI] [PubMed] [Google Scholar]
- Bullerwell CE, Schnare MN, Gray MW. Discovery and characterization of Acanthamoeba castellanii mitochondrial 5S rRNA. RNA. 2003;9:287–292. doi: 10.1261/rna.2170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis J, Barbour AG. Ticks have R2 retrotransposons but not the consensus transposon target site of other arthropods. Insect Mol Biol. 2005;14:465–474. doi: 10.1111/j.1365-2583.2005.00577.x. [DOI] [PubMed] [Google Scholar]
- Burke WD, Eickbush DG, Xiong Y, Jakubczak JL, Eickbush TH. Sequence relationship of retrotransposable elements R1 and R2 within and between divergent insect species. Mol Biol Evol. 1993;10:163–185. doi: 10.1093/oxfordjournals.molbev.a039990. [DOI] [PubMed] [Google Scholar]
- Burke WD, Malik HS, Jones JP, Eickbush TH. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol Biol Evol. 1999;16:502–511. doi: 10.1093/oxfordjournals.molbev.a026132. [DOI] [PubMed] [Google Scholar]
- Burton RS, Metz EC, Flowers JM, Willett CS. Unusual structure of ribosomal DNA in the copepod Tigriopus californicus: intergenic spacer sequences lack internal subrepeats. Gene. 2005;344:105–113. doi: 10.1016/j.gene.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Campbell NJH, Barker SC. An unprecedented major rearrangement in an arthropod mitochondrial genome. Mol Biol Evol. 1998;15:1786–1787. doi: 10.1093/oxfordjournals.molbev.a025904. [DOI] [PubMed] [Google Scholar]
- Campbell NJH, Barker SC. The novel mitochondrial gene arrangement of the cattle tick, Boophilus microplus: Fivefold tandem repetition of a coding region. Mol Biol Evol. 1999;16:732–740. doi: 10.1093/oxfordjournals.molbev.a026158. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, et al. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:15. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapelli AF, Soto-Adames N, Simon C, Frati F, Nardi F, Dallai R. Secondary structure, high variability and conserved motifs for domain III of 12S rRNA in the Arthropleona (Hexapoda: Collembola) Insect Mol Biol. 2004;13:659–670. doi: 10.1111/j.0962-1075.2004.00528.x. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- Chiang T-Y, Chiang Y-C, Chou C-H. Complete nucleotide sequence of the intergenic spacer between 25S and 17S rDNA in Miscanthus sinensis var. glaber. Bot Bull Acad Sin. 1998;39:241–244. [Google Scholar]
- Clark AG, Whittam TS. Sequencing errors and molecular evolutionary analysis. Mol Biol Evol. 1992;9:744–752. doi: 10.1093/oxfordjournals.molbev.a040756. [DOI] [PubMed] [Google Scholar]
- Clark CG, Tague BW, Ware VC, Gerbi SA. Xenopus laevis 28S ribosomal RNA: a secondary structural model and its evolutionary and functional implications. Nucleic Acids Res. 1984;12:6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. The ribosomal RNA genes of Drosophila DNA. Nucleic Acids Res. 1985;13:4029–4045. doi: 10.1093/nar/13.11.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. Drosophila mitochondrial DNA: conserved sequences in the A+T-rich region and supporting evidence for a secondary structure model of the small ribosomal RNA. J Mol Evol. 1987;25:116–125. doi: 10.1007/BF02101753. [DOI] [PubMed] [Google Scholar]
- Cluster PD, Marinkovic D, Allard RW, Ayala FJ. Correlations between development rates, enzyme activities, ribosomal DNA spacer-length phenotypes, and adaptation in Drosophila melanogaster. Proc Natl Acad Sci USA. 1987;84:610–614. doi: 10.1073/pnas.84.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. PhD Thesis. New Zealand: Victoria University of Wellington; 1994. Molecular evolutionary studies of New Zealand birds. [Google Scholar]
- Covacin C, Shao R, Cameron S, Barker SC. Extraordinary number of gene rearrangements in the mitochondrial genomes of lice (Phthiraptera: Insecta) Insect Mol Biol. 2006;15:63–68. doi: 10.1111/j.1365-2583.2005.00608.x. [DOI] [PubMed] [Google Scholar]
- Crease TJ. Sequence of the intergenic spacer between the 28S and 18S rRNA-encoding genes of the crustacean, Daphnia pulex. Gene. 1993;134:245–249. doi: 10.1016/0378-1119(93)90101-8. [DOI] [PubMed] [Google Scholar]
- Crease TJ, Colbourne JK. The unusually long small-subunit ribosomal RNA of the crustacean, Daphnia pulex: sequence and predicted secondary structure. J Mol Evol. 1998;46:307–313. doi: 10.1007/pl00006307. [DOI] [PubMed] [Google Scholar]
- Crease TJ, Taylor DJ. The origin and evolution of variable-region helices in V4 and V7 of the small-subunit ribosomal RNA of branchiopod crustaceans. Mol Biol Evol. 1998;15:1430–1446. doi: 10.1093/oxfordjournals.molbev.a025871. [DOI] [PubMed] [Google Scholar]
- Cross NC, Dover GA. Tsetse fly rDNA: an analysis of structure and sequence. Nucleic Acids Res. 1987a;15:15–30. doi: 10.1093/nar/15.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross NC, Dover GA. A novel arrangement of sequence elements surrounding the rDNA promoter and its spacer duplications in tsetse species. J Mol Biol. 1987b;195:63–74. doi: 10.1016/0022-2836(87)90327-5. [DOI] [PubMed] [Google Scholar]
- Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier RH, Crozier YC, Mackinlay AG. The CO-I and CO-II region of honeybee mitochondrial DNA: evidence for variation in insect mitochondrial evolutionary rates. Mol Biol Evol. 1989;4:399–411. doi: 10.1093/oxfordjournals.molbev.a040553. [DOI] [PubMed] [Google Scholar]
- Dams E, Hendriks L, Van de Peer Y, Neefs J-M, Smits G, Vandenbempt I, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988;16(Suppl.):r87–173. doi: 10.1093/nar/16.suppl.r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Kelly TJ, Masler EP, Thyagaraja BS, Rote CA, Imberski RB. Complete base sequences for the mitochondrial large subunit ribosomal RNA of the gypsy moth Lymantria dispar (L.) Insect Mol Biol. 1994;3:219–228. doi: 10.1111/j.1365-2583.1994.tb00170.x. [DOI] [PubMed] [Google Scholar]
- De Rijk P, Neefs J-M, Van de Peer Y, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1992;20(Suppl.):2075–2089. doi: 10.1093/nar/20.suppl.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk P, Van de Peer Y, Chapelle S, De Wachter R. Database on the structure of large ribosomal subunit RNA. Nucleic Acids Res. 1994;22:3495–3501. doi: 10.1093/nar/22.17.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk P, Van de Peer Y, De Wachter R. Database on the structure of the large ribosomal RNA. Nucleic Acids Res. 1997;25:117–123. doi: 10.1093/nar/25.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk P, Caers A, Van de Peer Y, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1998;26:183–186. [PMC free article] [PubMed] [Google Scholar]
- De Rijk P, Wuyts J, Van de Peer Y, Winkelmans T, De Wachter R. The European large subunit ribosomal RNA database. Nucleic Acids Res. 2000;28:177–178. doi: 10.1093/nar/28.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AR, Gillespie JJ, Yoder MJ. An evaluation of ensign wasp classification (Hymenoptera: Evaniidae) based on molecular data and insights from ribosomal RNA secondary structure. Syst Entomol. 2006;31:517–528. [Google Scholar]
- Degelmann A, Royer H-D, Hollenberg CP. The organization of the ribosomal RNA genes of Chironomus tentans and some closely related species. Chromosoma. 1979;71:263–281. doi: 10.1007/BF00287136. [DOI] [PubMed] [Google Scholar]
- Dover GA, Flavell RB. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984;38:622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Dover GA, Tautz D. Conservation and divergence in multigene families: alternatives to selection and drift. Philos Trans Roy Soc Lond B Biol Sci. 1986;312:275–289. doi: 10.1098/rstb.1986.0007. [DOI] [PubMed] [Google Scholar]
- Dowton M, Austin AD. Molecular phylogeny of the insect order Hymenoptera: Apocritan relationships. Proc Natl Acad Sci USA. 1994;91:9911–9915. doi: 10.1073/pnas.91.21.9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowton M, Austin AD. Increased genetic diversity in mitochondrial genes is correlated with the evolution of parasitism in the Hymenoptera. J Mol Evol. 1995;41:958–965. doi: 10.1007/BF00173176. [DOI] [PubMed] [Google Scholar]
- Dowton M, Austin AD. Evolutionary dynamics of a mitochondrial rearrangement ‘hotspot’ in the Hymenoptera. Mol Biol Evol. 1999;16:298–309. doi: 10.1093/oxfordjournals.molbev.a026111. [DOI] [PubMed] [Google Scholar]
- Dowton M, Campbell NJH. Intramitochondrial recombination: is it why some mitochondrial genes sleep around? Trends Ecol Evol. 2001;16:269–271. doi: 10.1016/s0169-5347(01)02182-6. [DOI] [PubMed] [Google Scholar]
- Dunon-Bluteau D, Brun G. The secondary structures of the Xenopus laevis and human mitochondrial small ribosomal subunit RNA are similar. FEBS Lett. 1986;198:333–337. doi: 10.1016/0014-5793(86)80431-8. [DOI] [PubMed] [Google Scholar]
- Eickbush TH. R2 and related site-specific non-long terminal repeat retrotransposons. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 813–835. [Google Scholar]
- Eickbush DG, Eickbush TH. Vertical transmission of the retrotransposable elements R1 and R2 during the evolution of the Drosophila melanogaster species subgroup. Genetics. 1995;139:671–684. doi: 10.1093/genetics/139.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush DG, Lathe WD, Francino MP, Eickbush TH. R1 and R2 retrotransposable elements of Drosophila evolve at rates similar to those of nuclear genes. Genetics. 1995;139:685–695. doi: 10.1093/genetics/139.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgavish T, Cannone JJ, Lee JC, Harvey SC, Gutell RR. AA.AG@Helix.Ends: A:A and A:G base-pairs at the ends of 16S and 23S rRNA helices. J Mol Biol. 2001;310:735–753. doi: 10.1006/jmbi.2001.4807. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Sulston JE, Coulson AR. The rDNA of C. elegans: sequence structure. Nucleic Acids Res. 1986;14:2345–2364. doi: 10.1093/nar/14.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis NS, Kolesnikova OA, Dogan S, Martin RP, Tarassov IA. 5S rRNA and tRNA import into human mitochondria: comparison of in vitro requirements. J Biol Chem. 2001;276:45642–45653. doi: 10.1074/jbc.M103906200. [DOI] [PubMed] [Google Scholar]
- Entelis N, Kolesnikova O, Kazakova H, Brandina I, Kamenski P, Martin RP, Tarassov I. Import of nuclear encoded RNAs into yeast and human mitochondria: experimental approaches and possible biomedical applications. Genet Eng. 2002;24:191–213. doi: 10.1007/978-1-4615-0721-5_9. [DOI] [PubMed] [Google Scholar]
- Fang Q, Black WCIV, Blocker HD, Whitcomb RF. A phylogeny of New World Deltocephalus-like leafhopper genera based on mitochondrial 16S ribosomal DNA sequences. Mol Phylogenet Evol. 1993;2:119–131. doi: 10.1006/mpev.1993.1012. [DOI] [PubMed] [Google Scholar]
- Fernández M, Polanco C, Ruiz ML, Pérez de la Vega M. A comparative study of the structure of the rDNA intergenic spacer of Lens culinaris Medik. & other legume species. Genome. 2000;43:597–603. doi: 10.1139/g00-022. [DOI] [PubMed] [Google Scholar]
- Field KG, Olsen GJ, Lane DJ, Giovannoni SJ, Ghiselin MT, Raff EC, Pace NR, Raff RA. Molecular phylogeny of the animal kingdom. Science. 1988;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- Flook PK, Rowell CHF. The effectiveness of mitochondrial rRNA gene sequences for the reconstruction of the phylogeny of an insect order, Orthoptera. Mol Phylogenet Evol. 1997;8:177–192. doi: 10.1006/mpev.1997.0425. [DOI] [PubMed] [Google Scholar]
- Friedrich M, Tautz D. An episodic change of rDNA nucleotide substitution rate has occurred at the time of the emergence of the insect order Diptera. Mol Biol Evol. 1997a;14:644–653. doi: 10.1093/oxfordjournals.molbev.a025804. [DOI] [PubMed] [Google Scholar]
- Friedrich M, Tautz D. Evolution and phylogeny of the Diptera: a molecular phylogenetic analysis using 28S rDNA sequences. Syst Biol. 1997b;46:674–698. doi: 10.1093/sysbio/46.4.674. [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Ishikawa H. Structure of the Bombyx mori rDNA: Initiation site for its transcription. Nucleic Acids Res. 1987;15:1245–1258. doi: 10.1093/nar/15.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA. Evolution of ribosomal DNA. In: MacIntyre RJ, editor. Molecular Evolutionary Genetics. New York, NY: Plenum; 1985. pp. 419–517. [Google Scholar]
- Gerbi SA. Expansion segments: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Synthesis. Boca Raton, FL: CRC Press; 1996. pp. 71–87. [Google Scholar]
- Gillespie JJ, Kjer KM, Duckett CN, Tallamy DW. Convergent evolution of cucurbitacin feeding in spatially isolated rootworm taxa (Coleoptera: Chrysomelidae; Galerucinae, Luperini) Mol Phylogenet Evol. 2003;29:161–175. doi: 10.1016/s1055-7903(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Cannone JJ, Gutell RR, Cognato AI. A secondary structural model of the 28S rRNA expansion segments D2 and D3 from rootworms and related leaf beetles (Coleoptera: Chrysomelidae; Galerucinae) Insect Mol Biol. 2004;13:495–518. doi: 10.1111/j.0962-1075.2004.00509.x. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, McKenna CH, Yoder MJ, Gutell RR, Johnston JS, Kathirithamby J, Cognato AI. Assessing the odd secondary structural properties of nuclear small subunit ribosomal RNA sequences (18S) of the twisted-wing parasites (Insecta: Strepsiptera) Insect Mol Biol. 2005a;14:625–643. doi: 10.1111/j.1365-2583.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Munro JB, Heraty JM, Yoder MJ, Owen AK, Carmichael AE. A secondary structural model of the 28S rRNA expansion segments D2 and D3 for chalcidoid wasps (Hymenoptera: Chalcidoidea) Mol Biol Evol. 2005b;22:1593–1608. doi: 10.1093/molbev/msi152. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Yoder MJ, Wharton RA. Predicted secondary structures for 28S and 18S rRNA from Ichneumonoidea (Insecta: Hymenoptera: Apocrita): impact on sequence alignment and phylogeny estimation. J Mol Evol. 2005c;61:114–137. doi: 10.1007/s00239-004-0246-x. [DOI] [PubMed] [Google Scholar]
- Glotz C, Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980;8:2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JL, Gonzalez KL, Schmicuez RD. The secondary structure of human 28S rRNA: The structure and evolution of a mosaic rRNA gene. J Mol Evol. 1987;24:236–251. doi: 10.1007/BF02111237. [DOI] [PubMed] [Google Scholar]
- Grauer G. Gene diversity in Hymenoptera. Evolution. 1985;39:190–199. doi: 10.1111/j.1558-5646.1985.tb04091.x. [DOI] [PubMed] [Google Scholar]
- Graybeal A. Evaluating the phylogenetic utility of genes: a search for genes informative about deep divergences among vertebrates. Syst Biol. 1994;43:174–193. [Google Scholar]
- Green R, Ellington AD, Szostak JW. In vitro genetic analysis of the Tetrahymena self-splicing intron. Nature. 1990;347:406–408. doi: 10.1038/347406a0. [DOI] [PubMed] [Google Scholar]
- Greenberg JR. Synthesis and properties of ribosomal RNA in Drosophila. J Mol Biol. 1969;46:85–98. doi: 10.1016/0022-2836(69)90059-x. [DOI] [PubMed] [Google Scholar]
- Gruendler P, Unfried I, Pascher K, Schweizer D. rDNA intergenic region from Arabidopsis thaliana: structural analysis, infraspecific variation and functional implications. J Mol Biol. 1991;221:1209–1222. doi: 10.1016/0022-2836(91)90929-z. [DOI] [PubMed] [Google Scholar]
- Gutell RR. Collection of Small Subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993;21:3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR. Collection of Small Subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR. Comparative sequence analysis and the structure of 16S and 23S rRNA. In: Dahlberg AE, Zimmerman RA, editors. Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Synthesis. Boca Raton, Florida: CRC Press; 1996. pp. 111–128. [Google Scholar]
- Gutell RR, Fox GE. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res. 1988;16S:r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Noller HF, Woese CR. Higher order structure in ribosomal RNA. EMBO J. 1986;5:1111–1113. doi: 10.1002/j.1460-2075.1986.tb04330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Schnare MN, Gray MW. A compilation of large subunit (23S-like) ribosomal RNA sequences presented in a secondary structure format. Nucleic Acids Res. 1990;18S:2319–2330. doi: 10.1093/nar/18.suppl.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Schnare MN, Gray MW. A compilation of large subunit (23S- and 23S-like) ribosomal RNA structures. Nucleic Acids Res. 1992;20S:2095–2109. doi: 10.1093/nar/20.suppl.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Gray MW, Schnare MN. A compilation of large subunit (23S- and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993;21S:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Cannone JJ, Du Shang ZY, Serra MJ. A story: unpaired adenosine bases in ribosomal RNAs. J Mol Biol. 2000;304:335–354. doi: 10.1006/jmbi.2000.4172. [DOI] [PubMed] [Google Scholar]
- Hancock JM. The contribution of DNA slippage to eukaryotic nuclear 18S rRNA evolution. J Mol Evol. 1995;40:629–639. doi: 10.1007/BF00160511. [DOI] [PubMed] [Google Scholar]
- Hancock JM, Dover GA. Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol Biol Evol. 1988;5:377–392. doi: 10.1093/oxfordjournals.molbev.a040505. [DOI] [PubMed] [Google Scholar]
- Hancock JM, Tautz D, Dover GA. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Mol Biol Evol. 1988;5:393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Hassouna N, Michot B, Bachellerie J-P. The complete nucleotide sequence of mouse 28S rRNA gene: Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12:3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward DC, Glover DM. Analysis of the Drosophila rDNA promoter by transient expression. Nucleic Acids Res. 1988;25:4253–4268. doi: 10.1093/nar/16.10.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks L, de Baere R, van Broeckhoven C, De Wachter R. Primary and secondary structure of the 18S ribosomal RNA of the insect species Tenebrio molitor. FEBS Lett. 1988a;232:115–120. doi: 10.1016/0014-5793(88)80398-3. [DOI] [PubMed] [Google Scholar]
- Hendriks L, van Broeckhoven C, Vandenberghe A, Van de Peer Y, De Wachter R. Primary and secondary structure of the 18S ribosomal RNA of the bird spider Eurypelma californica and evolutionary relationships among eukaryotic phyla. Eur J Biochem. 1988b;177:15–20. doi: 10.1111/j.1432-1033.1988.tb14339.x. [DOI] [PubMed] [Google Scholar]
- Hickson RE. PhD Thesis. New Zealand: Massey University; 1993. Evolutionary tails from the South Pacific. Being a wondrous and exciting account of investigations into the histories of New Zealand skinks, and in which it is shown that things are not what they seem. [Google Scholar]
- Hickson RE, Simon C, Cooper A, Spicer GS, Sullivan J, Penny D. Conserved sequence motifs, alignment, and secondary structure for the third domain of animal 12S rRNA. Mol Biol Evol. 1996;13:150–169. doi: 10.1093/oxfordjournals.molbev.a025552. [DOI] [PubMed] [Google Scholar]
- Hickson RE, Simon C, Perrey SW. The performance of several multiple-sequence alignment programs in relation to secondary-structure features for an rRNA sequence. Mol Biol Evol. 2000;17:530–539. doi: 10.1093/oxfordjournals.molbev.a026333. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Quart Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Hixson JE, Brown WM. A comparison of the small ribosomal RNA genes from the mitochondrial DNA of the great apes and humans: sequence, structure, evolution, and phylogenetic implications. Mol Biol Evol. 1986;3:1–18. doi: 10.1093/oxfordjournals.molbev.a040379. [DOI] [PubMed] [Google Scholar]
- Hoang L, Fredrick K, Noller HF. Creating ribosomes with an all-RNA 30S subunit P site. Proc Natl Acad Sci USA. 2004;101:12439–12443. doi: 10.1073/pnas.0405227101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovmöller R, Pape T, Kallersjö M. The Paleoptera problem: basal pterygote phylogeny inferred from 18S and 28S rDNA sequences. Cladistics. 2002;18:313–323. doi: 10.1111/j.1096-0031.2002.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Hsieh H-M, Hou R-J, Chen K-F, Tsai L-C, Liu S-W, Liu K-L, Linacre A, Lee JC-I. Establishing the rDNA IGS structure of Cannabis sativa. J Forensic Sci. 2004;49:1–4. [PubMed] [Google Scholar]
- Huysmans E, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1986;14(Suppl.):r73–r118. doi: 10.1093/nar/14.suppl.r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang UI, Kim W, Tautz D, Friedrich M. Molecular phylogenetics at the Felsenstein zone: approaching the Strepsiptera problem using 5.8S and 28S rDNA sequences. Mol Phylogenet Evol. 1998;9:470–480. doi: 10.1006/mpev.1998.0518. [DOI] [PubMed] [Google Scholar]
- Hypsa V. Parasite histories and novel phylogenetic tools: alternative approaches to inferring parasite evolution from molecular markers. Int J Parasitol. 2006;36:141–155. doi: 10.1016/j.ijpara.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Evolution of ribosomal RNA. Comp Biochem Physiol B. 1977;58:1–7. doi: 10.1016/0305-0491(77)90116-x. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Newburgh RW. Studies of thermal conversion of 28S RNA of Galleria mellonella (L.) to an 18S product. J Mol Biol. 1972;64:135–144. doi: 10.1016/0022-2836(72)90325-7. [DOI] [PubMed] [Google Scholar]
- Israelewski N. Structure and function of an AT-rich, interspersed repetitive sequence from Chironomus thummi: solenoidal DNA, 142 bp palindrome-frame and homologies with the sequence for site-specific recombination of bacterial transposons. Nucleic Acids Res. 1983;11:6985–6996. doi: 10.1093/nar/11.20.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelewski N, Schmidt ER. Spacer size heterogeneity in ribosomal DNA of Chironomus thummi is due to a 120 bp repeat homologous to a predominantly centromeric repeated sequence. Nucleic Acids Res. 1982;10:7689–7700. doi: 10.1093/nar/10.23.7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, Cannone JJ, Lee JC, Gutell RR, Woodson SA. Distribution of rRNA introns in the three-dimensional structure of the ribosome. J Mol Biol. 2002;323:35–52. doi: 10.1016/s0022-2836(02)00895-1. [DOI] [PubMed] [Google Scholar]
- Jakubczak JL, Xiong Y, Eickbush TH. Type I (R1) and type II(R2): ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol. 1990;212:37–52. doi: 10.1016/0022-2836(90)90303-4. [DOI] [PubMed] [Google Scholar]
- Jakubczak JL, Burke WD, Eickbush TH. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc Natl Acad Sci USA. 1991;88:3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak JL, Zenni MK, Woodruff RC, Eickbush TH. Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics. 1992;131:129–142. doi: 10.1093/genetics/131.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamrich M, Miller OL., Jr The rare transcripts of interrupted rRNA genes in Drosophila melanogaster are processed or degraded during synthesis. EMBO J. 1984;3:1541–1545. doi: 10.1002/j.1460-2075.1984.tb02008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhampati S, Kjer KM, Thorne BL. Phylogenetic relationships among termite families based on DNA sequence of mitochondrial LSU ribosomal RNA gene. Insect Mol Biol. 1996;5:229–238. doi: 10.1111/j.1365-2583.1996.tb00097.x. [DOI] [PubMed] [Google Scholar]
- van Keulen H, Mertz PM, LoVerde T, Shi H, Rekosh DM. Characterization of a 54-nucleotide gap region in the 28S rRNA gene of Schistosoma mansoni. Mol Biochem Parasitol. 1991;45:205–214. doi: 10.1016/0166-6851(91)90087-m. [DOI] [PubMed] [Google Scholar]
- Kjer KM. Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs. Mol Phylogenet Evol. 1995;4:314–330. doi: 10.1006/mpev.1995.1028. [DOI] [PubMed] [Google Scholar]
- Kjer KM. An alignment template for amphibian 12S rRNA, domain III: conserved primary and secondary structural motifs. J Herpetol. 1997;31:599–604. [Google Scholar]
- Kjer KM. Aligned 18S and insect phylogeny. Syst Biol. 2004;53:506–514. doi: 10.1080/10635150490445922. [DOI] [PubMed] [Google Scholar]
- Kjer KM, Baldridge GD, Fallon AM. Mosquito large subunit ribosomal RNA: simultaneous alignment of primary and secondary structure. Biochim Biophys Acta. 1994;1217:147–155. doi: 10.1016/0167-4781(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004a;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004b;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima KK, Fujiwara H. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol. 2004;21:207–217. doi: 10.1093/molbev/msg235. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Fujiwara H. Long-term inheritance of the 28S rDNA-specific retrotransposon R2. Mol Biol Evol. 2005;22:2157–2165. doi: 10.1093/molbev/msi210. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7.1–R7.28. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O-Y, Ishikawa H. Unique structure in the intergenic and 5′ external transcribed spacer of the ribosomal RNA gene from the pea aphid Acyrthosiphon pisum. Eur J Biochem. 1992;206:935–940. doi: 10.1111/j.1432-1033.1992.tb17003.x. [DOI] [PubMed] [Google Scholar]
- Kwon OY, Ogino K, Ishikawa H. The longest 18S ribosomal RNA ever known. Eur J Biochem. 1991;202:827–833. doi: 10.1111/j.1432-1033.1991.tb16439.x. [DOI] [PubMed] [Google Scholar]
- Lakshmikumaran M, Negi MS. Structural analysis of two length variants of the rDNA intergenic spacer from Eruca sativa. Plant Mol Biol. 1994;24:915–927. doi: 10.1007/BF00014445. [DOI] [PubMed] [Google Scholar]
- Lanadu C, du Montcel HT, Jolivot MP, Glaszmann JC, de Leon DG. Variation of ribosomal gene spacer length among wild and cultivated banana. Hereditas. 1992;68:147–156. doi: 10.1038/hdy.1992.23. [DOI] [PubMed] [Google Scholar]
- Lang BF, Burger G, O’Kelly CJ, Cedergren R, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:454–455. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- de Lanversin G, Jacq B. Sequence and secondary structure of the central domain of Drosophila 26S rRNA: a universal model for the central domain of the large rRNA containing the region in which the central break may happen. J Mol Evol. 1989;28:403–417. doi: 10.1007/BF02603076. [DOI] [PubMed] [Google Scholar]
- Larsen N. Higher order interactions in 23S rRNA. Proc Natl Acad Sci USA. 1992;89:5044–5048. doi: 10.1073/pnas.89.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Gutell RR. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J Mol Biol. 2004;344:1225–1249. doi: 10.1016/j.jmb.2004.09.072. [DOI] [PubMed] [Google Scholar]
- Lee JC, Cannone JJ, Gutell RR. The lonepair triloop: a new motif in RNA structure. J Mol Biol. 2003;325:65–83. doi: 10.1016/s0022-2836(02)01106-3. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lin C-P, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogenet Evol. 2004;30:686–702. doi: 10.1016/S1055-7903(03)00241-0. [DOI] [PubMed] [Google Scholar]
- Long EO, Dawid IB. Restriction analysis of spacers in ribosomal DNA of Drosophila melanogaster. Nucleic Acids Res. 1979;7:205–215. doi: 10.1093/nar/7.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol Cell Biol. 1996;16:4726–4734. doi: 10.1128/mcb.16.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in transfer RNAs: molecular evidence for Muller's ratchet in mitochondrial genomes. Mol Biol Evol. 1995;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 1997;14:914–925. doi: 10.1093/oxfordjournals.molbev.a025834. [DOI] [PubMed] [Google Scholar]
- Macas J, Navrátilová A, Mézáros T. Sequence subfamilies of satellite repeats related to rDNA intergenic spacer are differentially amplified on Vicia sativa chromosomes. Chromosoma. 2003;112:152–158. doi: 10.1007/s00412-003-0255-3. [DOI] [PubMed] [Google Scholar]
- Magalhães PJ, Andreu AL, Schon EA. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol Biol Cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Ghosh T, Adhya S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 1994;22:3381–3386. doi: 10.1093/nar/22.16.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez LM, Miller DJ, MacKenzie JB, van Oppen MJH. Pseudogenes contribute to the extreme diversity of nuclear ribosomal DNA in the hard coral Acropora. Mol Biol Evol. 2003;20:1077–1086. doi: 10.1093/molbev/msg122. [DOI] [PubMed] [Google Scholar]
- Martin AP, Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masta SE. Mitochondrial sequence evolution in spiders: Intraspecific variation in tRNAs lacking the TΨC arm. Mol Biol Evol. 2000;17:1091–1100. doi: 10.1093/oxfordjournals.molbev.a026390. [DOI] [PubMed] [Google Scholar]
- Masta SE, Boore JL. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol. 2004;21:893–902. doi: 10.1093/molbev/msh096. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Banerjee AR, Luan DD, Eickbush TH, Turner DH. Secondary structure model of the RNA recognized by the reverse transcriptase from the R2 retrotransposable element. RNA. 1997;3:1–16. [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McLain DK, Collins FH, Brandling-Bennet AD, Were JBO. Microgeographic variation in rDNA intergenic spacers of Anopheles gambiae in Western Kenya. Heredity. 1989;62:257–264. doi: 10.1038/hdy.1989.36. [DOI] [PubMed] [Google Scholar]
- McTaggart S, Crease T. Selection on the structural stability of a ribosomal RNA expansion segment in Daphnia obtusa. Mol Biol Evol. 2005;22:1309–1319. doi: 10.1093/molbev/msi119. [DOI] [PubMed] [Google Scholar]
- Mears JA, Sharma MR, Gutell RR, McCook AS, Richardson PE, Caulfield TR, Agrawal RK, Harvey SC. A structural model for the large subunit of the mammalian mitochondrial ribosome. J Mol Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B, Bachellerie J-P, Raynal F. Sequence and secondary structure of mouse 28S rRNA 5′-terminal domain: organisation 5 8S−28S rRNA complex. Nucleic Acids Res. 1982;10:5273–5283. doi: 10.1093/nar/10.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B, Hassouna N, Bachellerie J-P. Secondary structure of mouse 28S rRNA and general model for the folding of the large rRNA in eukaryotes. Nucleic Acids Res. 1984;12:4259–4279. doi: 10.1093/nar/12.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Fleck G. Comparative analysis of mt LSU rRNA secondary structures of Odonates: structural variability and phylogenetic signal. Insect Mol Biol. 2003;12:535–547. doi: 10.1046/j.1365-2583.2003.00432.x. [DOI] [PubMed] [Google Scholar]
- Misof B, Anderson CL, Buckley TR, Erpenbeck D, Rickert A, Misof KJ. An empirical analysis of mt 16S rRNA covarion-like evolution in insects: site-specific rate variation is clustered and frequently detected. J Mol Evol. 2002;554:460–469. doi: 10.1007/s00239-002-2341-1. [DOI] [PubMed] [Google Scholar]
- Morales-Hojas R, Post RJ, Wilson MD, Cheke RA. Completion of the sequence of the nuclear ribosomal DNA subunit of Simulium sanctipauli, with descriptions of the 18S, 28S genes and the IGS. Med Vet Entomol. 2002;16:386–394. doi: 10.1046/j.1365-2915.2002.00392.x. [DOI] [PubMed] [Google Scholar]
- Muir G, Fleming CC, Schlötterer C. Three divergent rDNA clusters predate the species divergence in Quercus petrea (Matt.) Liebl. & Quercus robur L. Mol Biol Evol. 2001;18:112–119. doi: 10.1093/oxfordjournals.molbev.a003785. [DOI] [PubMed] [Google Scholar]
- Mukha DV, Sidorenko AP, Lazebnaya IV, Wiegmann BM, Schal C. Analysis of intraspecies polymorphism in the ribosomal DNA cluster of the cockroach Blattella germanica. Insect Mol Biol. 2000;9:217–222. doi: 10.1046/j.1365-2583.2000.00175.x. [DOI] [PubMed] [Google Scholar]
- Mukha DV, Wiegmann BM, Schal C. Saltatory changes in the structure of the ribosomal DNA external transcribed spacer during the evolution of cockroaches of genus Blattella. Doklady Biol Sci. 2002;387:549–552. doi: 10.1023/a:1021745709410. [DOI] [PubMed] [Google Scholar]
- Murtif VL, Rae PM. In vivo transcription of rDNA spacers in Drosophila. Nucleic Acids Res. 1985;13:3221–3239. doi: 10.1093/nar/13.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Venema J, van der Linden G, van Heerikhuizen H, Klootwijk J, Planta RJ. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989;9:551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Goncalves PM, Boon K, Raué HA, van Heerikhuizen H, Planta RJ. The conserved GTPase center and variable region V9 from Saccharomyces cerevisae 26 rRNA can be replaced by their equivalents from other prokaryotes or eukaryotes without detectable loss of ribosomal function. Proc Natl Acad Sci USA. 1991;88:1469–1473. doi: 10.1073/pnas.88.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J-M, De Wachter R. A proposal for the secondary structure of a variable area of eukaryotic small ribosomal subunit RNA involving the existence of a pseudoknot. Nucleic Acids Res. 1990;18:5695–5704. doi: 10.1093/nar/18.19.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J-M, Van de Peer Y, Hendriks L, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990;18(Suppl.):2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J-M, Van de Peer Y, De Rijk P, Goris A, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1991;19:1987–2015. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J-M, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Naumann CM, Misof B. Identification of evolutionary conserved structural elements in the mt SSU rRNA of Zygaenoidea (Lepidoptera): a comparative sequence analysis. Organ Division Evol. 2006;6:17–32. [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Noller HF. The driving force for molecular evolution of translation. RNA. 2004;10:1833–1837. doi: 10.1261/rna.7142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- Noller HF, Baucom A. Structure of the 70 S ribosome: implications for movement. Biochem Soc Trans. 2001;30:1159–1161. doi: 10.1042/bst0301159. [DOI] [PubMed] [Google Scholar]
- Noller HF, Woese CR. Secondary structure of 16S ribosomal RNA. Science. 1981;212:403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Noller HF, Kop J, Wheaton V, Brosius J, et al. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Edafujiwara H, Fujiwara H, Ishikawa H. What causes the aphid 28S ribosomal RNA to lack the hidden break? J Mol Evol. 1990;30:509–513. doi: 10.1007/BF02101106. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MT, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ohnishi H, Yamamoto M-T. The structure of a single unit of ribosomal RNA gene (rDNA) including intergenic subrepeats in the Australian bulldog ant Myrmecia croslandi (Hymenoptera: Formicidae) Zoo Sci. 2004;21:139–146. doi: 10.2108/zsj.21.139. [DOI] [PubMed] [Google Scholar]
- Orlando TC, Rubio MA.T, Sturm NR, Campbell DA, Floeter-Winter LM. Intergenic and external transcribed spacers of ribosomal RNA genes in lizard-infecting Leishmania: molecular structure and phylogenetic relationship to mammal-infecting Leishmania in the subgenus Leishmania (Leishmania) Mem Inst Oswaldo Cruz, Rio Janeiro. 2002;97:1–7. doi: 10.1590/s0074-02762002000500020. [DOI] [PubMed] [Google Scholar]
- Osorio CR, Collins MD, Romalde JL, Toranzo AE. Variation in 16S−23S rRNA intergenic spacer regions in Photobacterium damselae: a mosaic-like structure. Appl Environ Microbiol. 2005;71:636–645. doi: 10.1128/AEM.71.2.636-645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouvrard D, Campbell BC, Bourgoin T, Chan KL. 18S rRNA secondary structure and phylogenetic position of Peloridiidae (Insecta, Hemiptera) Mol Phylogenet Evol. 2000;16:403–417. doi: 10.1006/mpev.2000.0797. [DOI] [PubMed] [Google Scholar]
- Pace NR, Olsen GJ, Woese CR. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986;45:325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- Page RDM. Comparative analysis of insect mitochondrial small subunit ribosomal RNA using maximum weighted matching. Nucleic Acids Res. 2000;28:3839–3845. doi: 10.1093/nar/28.20.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. On the dangers of aligning RNA sequences using ‘conserved’ motifsTechnical reports in taxonomy. Available at: http://taxonomy.zoology.gla.ac.uk/publications/tech-reports/index.html.
- Page RD, Lee PL, Becher SA, Griffiths R, Clayton DH. A different tempo of mitochondrial DNA evolution in birds and their parasitic lice. Mol Phylogenet Evol. 1998;9:276–293. doi: 10.1006/mpev.1997.0458. [DOI] [PubMed] [Google Scholar]
- Page RD.M, Cruickshank R, Johnson KP. Louse (Insecta: Phthiraptera) mitochondrial 12S rRNA secondary structure is highly variable. Insect Mol Biol. 2002;11:361–369. doi: 10.1046/j.1365-2583.2002.00346.x. [DOI] [PubMed] [Google Scholar]
- Paques F, Samson ML, Jordan P, Wegnez M. Structural evolution of the Drosophila 5S ribosomal genes. J Mol Evol. 1995;41:615–621. doi: 10.1007/BF00175820. [DOI] [PubMed] [Google Scholar]
- Park Y-J, Fallon AM. Mosquito ribosomal RNA genes: characterization of gene structure and evidence for changes in copy number during development. Insect Biochem. 1990;20:1–11. [Google Scholar]
- Pepera OP, Cockburn AF, Mitchell SE, Conn J, Seawright JA. Species-specific repeat units in the intergenic spacer of the ribosomal RNA cistron of Anopheles aquasalis Curry. Am J Trop Med Hyg. 1998;59:673–678. doi: 10.4269/ajtmh.1998.59.673. [DOI] [PubMed] [Google Scholar]
- Perez-Gonzalez CE, Eickbush TH. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila. Genetics. 2001;158:1557–1567. doi: 10.1093/genetics/158.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez CE, Eickbush TH. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics. 2002;162:799–811. doi: 10.1093/genetics/162.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont-Kingdon GA, Beagley CT, Okimoto R, Wolstenholme DR. Mitochondrial DNA of the sea anemone, Metridum senile (Cnidaria): Prokaryote-like genes for tRNA (f-Met) and small-subunit ribosomal RNA, and standard genetic code specificities for AGR and ATA codons. J Mol Evol. 1994;39:387–399. doi: 10.1007/BF00160271. [DOI] [PubMed] [Google Scholar]
- Rambaut A. Se-Al: Sequence Alignment Editor. Available at http://evolve.zoo.ox.ac.uk/
- Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- Rimoldi OJ, Raghu B, Mag MK, Eliceiri GL. Three new small nucleolar RNAs that are psoralen cross-linked in vivo to unique regions of pre-rRNA. Mol Cell Biol. 1993;13:4382–4390. doi: 10.1128/mcb.13.7.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheford TR. Change in ribosomal DNA intergenic spacer-length composition in maize recurrent selection populations. 1. Analysis of BS13, BSSS, and BSCB1. Theor Appl Genet. 1994;88:541–547. doi: 10.1007/BF01240916. [DOI] [PubMed] [Google Scholar]
- Roiha H, Miller RM, Woods LC, Glover DM. Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature. 1981;290:749–753. doi: 10.1038/290749a0. [DOI] [PubMed] [Google Scholar]
- Ruschak AM, Mathews DH, Bibillo A, Spinelli SL, Childs JL, Eickbush TH, Turner DH. Secondary structure models of the 3′ untranslated regions of diverse R2 RNAs. RNA. 2004;10:978–987. doi: 10.1261/rna.5216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kinberland ML, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schnare MN, Collings JC, Spencer DF, Gray MW. The 28S−18S rDNA intergenic spacer from Crithidia fasciculata: repeated sequences, length heterogeneity, putative processing sites and potential interactions between U3 and small nucleolar RNA and the ribosomal precursor. Nucleic Acids Res. 2000;28:3452–3461. doi: 10.1093/nar/28.18.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare MN, Damberger SH, Gray MW, Gutell RR. Comprehensive comparison of structural characteristics in eukaryotic cytoplasmic large subunit (23S-like) ribosomal RNA. J Mol Biol. 1996;256:701–719. doi: 10.1006/jmbi.1996.0119. [DOI] [PubMed] [Google Scholar]
- Schulmeister S. Simultaneous analysis of basal Hymenoptera (Insecta): Introducing robust-choice sensitivity analysis. Biol J Linn Soc. 2003;79:245–275. [Google Scholar]
- Shao R, Barker SC. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- Shao R, Campbell NJH, Barker SC. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera) Mol Biol Evol. 2001a;18:858–865. doi: 10.1093/oxfordjournals.molbev.a003867. [DOI] [PubMed] [Google Scholar]
- Shao R, Campbell NJH, Schmidt ER, Barker SC. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol Biol Evol. 2001b;18:1828–1832. doi: 10.1093/oxfordjournals.molbev.a003970. [DOI] [PubMed] [Google Scholar]
- Shao R, Dowton M, Murrell A, Barker SC. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol. 2003;20:1612–1619. doi: 10.1093/molbev/msg176. [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. Occurrence of heat-dissociable ribosomal RNA in insects: presence of 3 polynucleotide chains in S26 RNA from cultured Aedes aegypti cells. J Mol Biol. 1973;75:57–72. doi: 10.1016/0022-2836(73)90528-7. [DOI] [PubMed] [Google Scholar]
- Simeone A, La Volpe A, Boncinelli E. Nucleotide sequence of a complete ribosomal spacer of D. melanogaster. Nucleic Acids Res. 1985;13:1089–1101. doi: 10.1093/nar/13.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C. Molecular systematics at the species boundary. In: Hewitt GM, Johnston AWB, Young JPW, editors. Exploiting Conserved and Variable Regions of the Mitochondrial Genome of Animals Via Direct Sequencing from Amplified DNA. Springer Verlag; 1991. pp. 33–71. Molecular Techniques in Taxonomy. NATO ASI Series, Vol. H57. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- Simon C, Paabo S, Kocher T, Wilson AC. Evolution of the mitochondrial ribosomal RNA in insects as shown by the polymerase chain reaction. In: Cleggs M, O’brien S, editors. Molecular Evolution. New York: Liss; 1990. pp. 235–244. UCLA Symposia on Molecular and Cellular Biology, New Series, Vol. 122. [Google Scholar]
- Smith SD, Bond JE. An analysis of the secondary structure of the mitochondrial large subunit rRNA gene (16S) in spiders and its implications for phylogenetic reconstruction. J Arach. 2003;31:44–54. [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- States DJ. Molecular sequence accuracy: analysing imperfect data. Trends Genet. 1992;8:52–55. doi: 10.1016/0168-9525(92)90349-9. [DOI] [PubMed] [Google Scholar]
- Stiegler P, Carbon P, Ebel JP, Ehresmann C. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem. 1981;120:487–495. doi: 10.1111/j.1432-1033.1981.tb05727.x. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Berry MW. An SVD-based comparison of nine whole eukaryotic genomes supports a coelomate rather than ecdysozoan lineage. BMC Bioinformatics. 2004;5:204. doi: 10.1186/1471-2105-5-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J, Holsinger KE, Simon C. Among-site rate variation and phylogenetic analysis of 12S rRNA in sigmodontine rodents. Mol Biol Evol. 1995;12:988–1001. doi: 10.1093/oxfordjournals.molbev.a040292. [DOI] [PubMed] [Google Scholar]
- Sweeney R, Yao M-C. Identifying functional regions of rRNA by insertion mutagenesis and complete gene replacement in Tetrahymena thermophila. EMBO J. 1989;8:933–938. doi: 10.1002/j.1460-2075.1989.tb03454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney R, Chen L, Yao M-C. An rRNA variable region has an evolutionary conserved essential role despite sequence divergence. Mol Cell Biol. 1994;14:4203–4215. doi: 10.1128/mcb.14.6.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z, Kjer KM. Phylogeny and host–plant association in the leaf beetle genus Trirhabda LeConte (Coleoptera: Chrysomelidae) Mol Phylogenet Evol. 2004;32:358–374. doi: 10.1016/j.ympev.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Szymanski JB. 5S ribosomal RNA database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan THP, Pach R, Crausaz A, Ivens A, Schneider A. tRNAs in Trypanosoma brucei: genomic organization, expression, and mitochondrial import. Mol Cell Biol. 2002;22:3707–3717. doi: 10.1128/MCB.22.11.3707-3716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Hancock JM, Webb DA, Tautz C, Dover GA. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol Biol Evol. 1988;5:366–376. doi: 10.1093/oxfordjournals.molbev.a040500. [DOI] [PubMed] [Google Scholar]
- Tautz D, Tautz C, Webb D, Dover GA. Evolutionary divergence of promoters and spacers in the rDNA family of four Drosophila species. J Mol Biol. 1987;195:525–542. doi: 10.1016/0022-2836(87)90181-1. [DOI] [PubMed] [Google Scholar]
- The Honey Bee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006 doi: 10.1038/nature05260. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WK, Maa J, Wilson AC. Shifting constraints on tRNA genes during mitochondrial DNA evolution in animals. New Biologist. 1989;1:93–100. [PubMed] [Google Scholar]
- Uhlenbusch I, McCracken A, Gellissen G. The gene for the large (16S) ribosomal RNA from the Locusta migratoria mitochondrial genome. Curr Genet. 1987;11:631–638. doi: 10.1007/BF00393927. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Neefs J-M, De Rijk P, De Wachter R. Reconstructing evolution from eukaryotic small-ribosomal-subunit RNA sequences: calibration of the molecular clock. J Mol Evol. 1993;37:221–232. doi: 10.1007/BF02407359. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, van den Broeck, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1994;22:3488–3392. doi: 10.1093/nar/22.17.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Nicolai S, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1996;24:86–91. doi: 10.1093/nar/24.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Jansen J, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1997;25:111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Caers A, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1998;26:179–182. [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Robbrecht E, de Hoog S, Caers A, De Rijk P, De Wachter R. Database on the structure of small subunit ribosomal RNA. Nucleic Acids Res. 1999;27:179–183. doi: 10.1093/nar/27.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, De Rijk P, Wuyts J, Winkelmans T, De Wachter R. The European Small Subunit Ribosomal RNA database. Nucleic Acids Res. 2000;28:175–176. doi: 10.1093/nar/28.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varricchio P, Gargiulo G, Graziani F, Manzi A, Pennacchio F, Digilio M, Tremblay E, Malva C. Characterization of Aphidius ervi (Hymenoptera: Braconidae) ribosomal genes and identification of site-specific insertion elements belonging to the non-LTR retrotransposon family. Insect Biochem Mol Biol. 1994;25:603–612. doi: 10.1016/0965-1748(94)00102-n. [DOI] [PubMed] [Google Scholar]
- Vawter L, Brown WM. Rates and patterns of base change in the SSU ribosomal RNA gene. Genetics. 1993;134:597–608. doi: 10.1093/genetics/134.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman GM, Klootwijk J, De Regt VCFH, Planta RJ, Branlant C, Krol A, Ebel J-P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak I, Burgschwaiger S, Kreil G. Nucleotide sequence of the large ribosomal RNA of honeybee mitochondria. Nucleic Acids Res. 1987;15:2388. doi: 10.1093/nar/15.5.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware VC, Tague BW, Clark CG, Gourse RL, Brand RC, Gerbi SA. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983;11:7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware VC, Renkawitz R, Gerbi SA. rRNA processing: removal of only nineteen bases at the gap between 28S alpha and 28S beta rRNAs in Sciara coprophila. Nucleic Acids Res. 1985;13:3581–3597. doi: 10.1093/nar/13.10.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider LJ, Elser JJ, Crease TJ, Mateos M, Cotner JB, Markow TA. The functional significance of ribosomal (r) DNA variation: Impacts on the evolutionary ecology of organisms. Annu Rev Ecol Evol Syst. 2005;36:219–242. [Google Scholar]
- Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Whang I-J, Jung J, Par J-K, Min G-S, Kim W. Intragenomic length variation of the ribosomal DNA intergenic spacer in a malaria vector, Anopheles sinensis. Mol Cells. 2002;14:158–162. [PubMed] [Google Scholar]
- Wharton RA, Yoder MJ, Gillespie JJ, Patton JC, Honeycutt RL. Relationships of Exodontiella, a non-alysiine, exodont member of the family Braconidae (Insecta, Hymenoptera) Zool Scripta. 2006;35:323–340. [Google Scholar]
- Wheeler WC, Whiting MF, Wheeler QD, Carpenter JC. The phylogeny of extant insect orders. Cladistics. 2001;17:113–169. doi: 10.1111/j.1096-0031.2001.tb00115.x. [DOI] [PubMed] [Google Scholar]
- Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC. The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol. 1997;46:1–68. doi: 10.1093/sysbio/46.1.1. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Yeates DK, Thorne JL, Kishino H. Time flies, a new molecular time-scale for brachyceran fly evolution without a clock. Syst Biol. 2003;52:745–756. [PubMed] [Google Scholar]
- Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Winker S, Woese CR. A definition of the domains Archaea, Bacteria and Eucarya, in terms of small subunit ribosomal RNA characteristics. Syst Appl Microbiol. 1991;14:305–310. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Pace NR. Probing RNA structure, function and history by comparative analysis. In: Gesteland RF, Atkins JF, editors. The RNA World. New York, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 91–117. [Google Scholar]
- Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell RR, Hogan JJ, Noller HF. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Archaea, Bacteria, Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods PE, Guttman SI. Genetic variation in Neodiprion (Hymneoptera: Symphyta: Diprionnidae) sawflies and a comment on low levels of genetic diversity within Hymenoptera. Ann Entomol Soc Am. 1987;80:590–599. [Google Scholar]
- Wool IG. Studies of the structure of eukaryotic (mammalian) ribosomes. In: Hardesty J, Kramer G, editors. Structure, Function and Genetics of Ribosomes. New York: Springer-Verlag; 1986. pp. 391–411. [Google Scholar]
- Wu CC, Fallon AM. Analysis of a ribosomal DNA intergenic spacer region from the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 1998;7:19–29. doi: 10.1046/j.1365-2583.1998.71194.x. [DOI] [PubMed] [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, De Wachter R. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 2000;28:4698–4708. doi: 10.1093/nar/28.23.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Winkelmans T, De Wachter R. The European large subunit ribosomal RNA database. Nucleic Acids Res. 2001;29:175–177. doi: 10.1093/nar/29.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Van de Peer Y, Winkelmans T, De Wachter R. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 2002;30:183–185. doi: 10.1093/nar/30.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Perriere G, Van de Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshionari S, Koike T, Yokogawa T, Nishikawa K, Ueda T, Miura K, Watanabe K. Existence of nuclear-encoded 5S rRNA in bovine mitochondria. FEBS Lett. 1994;338:137–142. doi: 10.1016/0014-5793(94)80351-x. [DOI] [PubMed] [Google Scholar]
- Yoshizawa K, Johnson KP. Aligned 18S for Zoraptera (Insecta): phylogenetic position and molecular evolution. Mol Phylogenet Evol. 2005;37:572–580. doi: 10.1016/j.ympev.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zarlenga DS, Dame JB. The identification and characterization of a break within the large subunit ribosomal RNA of Trichinella spiralis: comparison of gap sequences within the genus. Mol Biochem Parasitol. 1992;51:281–290. doi: 10.1016/0166-6851(92)90078-x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Glotz C, Brimacombe R. Secondary structure comparisons between SSU ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981;9:3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]


