Abstract
Leucine-rich Repeat Containing protein 10 (LRRC10) has recently been identified as a cardiac-specific factor in mice. However, the function of this factor remains to be elucidated. In this study, we investigated the developmental roles of Lrrc10 using zebrafish as an animal model. Knockdown of Lrrc10 in zebrafish embryos (morphants) using morpholinos caused severe cardiac morphogenic defects including a cardiac looping failure accompanied by a large pericardial edema, and embryonic lethality between day 6 and 7 post fertilization. The Lrrc10 morphants exhibited cardiac functional defects as evidenced by a decrease in ejection fraction and cardiac output. Further investigations into the underlying mechanisms of the cardiac defects revealed that the number of cardiomyocyte was reduced in the morphants. Expression of two cardiac genes was deregulated in the morphants including an increase in atrial natriuretic factor, a hallmark for cardiac hypertrophy and failure, and a decrease in cardiac myosin light chain 2, an essential protein for cardiac contractility in zebrafish. Moreover, a reduced fluorescence intensity from NADH in the morphant heart was observed in live zebrafish embryos as compared to control. Taken together, the present study demonstrates that Lrrc10 is necessary for normal cardiac development and cardiac function in zebrafish embryos, which will enhance our understanding of congenital heart defects and heart disease.
Keywords: Leucine-rich Repeat Containing protein 10, Heart development, Cardiac function, Zebrafish embryos, Cardiac looping defects, Pericardial edema
Introduction
Identification of molecular pathways involved in normal heart development will lead to the discovery of the genetic basis for human congenital heart disease as well as adult cardiac disease. Therefore, a better understanding of molecular mechanisms underlying normal cardiac development and function is a major focus of modern cardiovascular research (for reviews, see Chien, 2000; Gruber and Epstein, 2004; Heideman et al., 2005; Olson, 2004; Olson and Schneider, 2003; Srivastava and Olson, 2000). Cardiac development is a complex biological process requiring the integration of cell specification, differentiation, and morphogenesis. Many factors have been implicated in this process on the basis of their spatial and temporal expression patterns or their phenotypic effects when they are functionally inactivated in various animal models. These include diverse families of transcription factors, including Nkx2.5 (Lyons et al., 1995; Schott et al., 1998), GATA4 (Kuo et al., 1997; Molkentin et al., 1997), Tbx5, and Tbx20 (Bruneau et al., 2001; Krause et al., 2004; Stennard et al., 2005), and Jumonji/Jarid2, (Jung et al., 2005; Lee et al., 2000), and Fog-1 (Walton et al., 2006). In addition, secreted factors with their receptors (Bujak and Frangogiannis, 2006; Schneider et al., 2003), and cytoplasmic and contractile proteins (Bendig et al., 2006; Berdougo et al., 2003; Rottbauer et al., 2006) have been shown to be critical for cardiac development. However, the exact molecular mechanisms of embryonic cardiac development remain to be elucidated.
Therefore, we reasoned that there are yet unidentified cardiac factors that play critical roles in normal heart development as well as in maintaining normal cardiac function in adults. We have identified a mouse gene encoding Leucine-rich Repeat Containing protein 10 (LRRC10) as a cardiac-specific gene by employing in silico approaches (Passier et al., 2000; Wang et al., 2001), which exhibits dynamic expression patterns during development (Kim et al., in press). While we were characterizing this factor in zebrafish, LRRC10 was reported as a cardiac-restricted (HRLRRP) or cardiac-specific factor (SERDIN1) in mice (Adameyko et al., 2005; Nakane et al., 2004). Herein, we will refer to this gene as LRRC10 in accordance with the NCBI nomenclature. However, the developmental and molecular function of LRRC10 remains completely unknown.
Leucine-rich repeat (LRR) motifs are present in an increasing number of proteins with diverse functions such as enzyme inhibition, cell growth, cell adhesion, signal transduction, regulation of gene expression, apoptosis signaling, and development (Kobe and Kajava, 2001). LRR motifs are present in over 2000 proteins from viruses to eukaryotes. LRRs are 20–29 residue sequence motifs and the repeat number ranges from 2 to 45. The primary function of these motifs is believed to provide a structural framework for protein–protein interactions. Mouse and human LRRC10 contain seven LRR motifs and do not contain any N-terminal or C-terminal domains that are often present in other LRRC proteins (Fig. 1B). These additional domains seem to confer specific functions to LRRC proteins such as a kinase domain or a membrane spanning domain (Buchanan and Gay, 1996). Therefore, the LRRC10 gene product represents a unique member of the LRRC protein family, which contains only the leucine-rich repeat motif. The LRRC10 gene is conserved in most vertebrates including mouse, rat, human, chicken, and frog, suggesting an important function in an evolutionally conserved manner. Therefore, we set out to identify the roles of Lrrc10 in cardiac development using zebrafish (Danio rerio) as a vertebrate model.
Fig. 1.
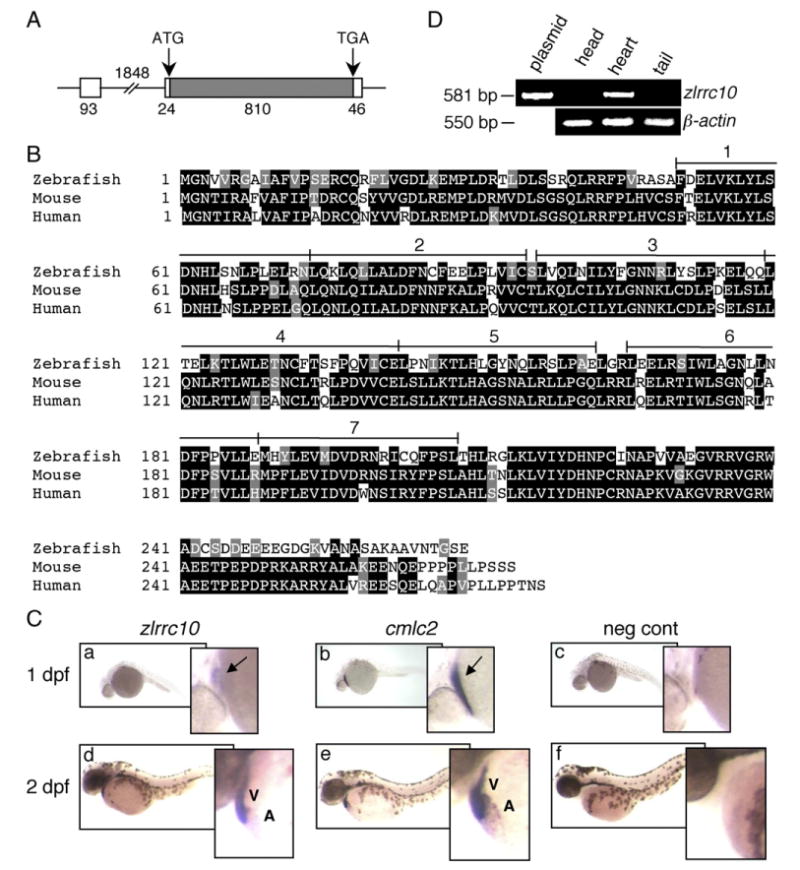
Cardiac-specific expression of zlrrc10. (A) The genomic structure of zlrrc10. The full-length zlrrc10 cDNA consists of two exons. The size of exons, intron and the 5′ and 3′ UTR regions are shown in bp. The amino acid coding regions are represented by the hatched box. (B) An alignment of zebrafish, mouse and human LRRC10 amino acid sequences. Identical residues between all three species are shaded black, and the residues with similar characteristics are shaded gray. The conserved seven LRR motifs are numbered. (C) Cardiac-specific expression of lrrc10 in zebrafish embryos. Zebrafish embryos at 1 dpf (a–c) and 2 dpf (d–f) were subjected to whole-mount in situ hybridization using digoxigenin labeled zlrrc10 antisense cRNA probes (a, d). cmlc2 antisense cRNA probe (b and e) was used for positive control to visualize the heart. Negative control was hybridized with sense zlrrc10 probe (c and f). Arrows indicate the linear heart region that expresses zlrrc10 and cmlc2. A and V indicate the atrium and ventricle, respectively. (D) Cardiac-specific expression of zlrrc10 in adult zebrafish. RT-PCR analysis was performed using total RNA isolated from three different parts of adult zebrafish as indicated. The zlrrc10 PCR products of 581 bp were visualized by agarose gel electrophoresis. The plasmid pCRII-TOPO-zlrrc10 was used as positive control for the amplification reactions. RT-PCR of β-actin was performed as a loading control.
The zebrafish has become a popular model for the study of cardiovascular development. It presents a prototypic vertebrate heart with only a single atrium and ventricle. The molecular mechanisms governing its patterning appear to be similar to those in more complex hearts of higher vertebrates (Fishman and Chien, 1997; Weinstein and Fishman, 1996). Assessing early embryonic heart morphology and function is facilitated in zebrafish embryos because they are transparent and not dependent on intact cardiovascular function during the first 6–7 days of development (Pelster and Burggren, 1996). The heart is the first definitive organ to develop and become functional as survival depends on its proper function. However, there is a period when the heart is already functional, but not yet essential in the early developmental stages. In the zebrafish embryos, this period actually lasts for several days because of the small size and relatively low metabolism. This enables analysis of mutants with compromised or no cardiac function for a considerable period of time (Stainier and Fishman, 1992; Stainier et al., 1996). In contrast a similar phenotype in the mouse would result in early embryonic lethality.
In this study, we describe the novel developmental roles of Lrrc10 in early cardiac development and function in zebrafish. The remarkable cardiac-specific expression of lrrc10 was observed in zebrafish. To identify the developmental roles of Lrrc10, Lrrc10 expression was reduced using lrrc10-specific antisense morpholinos. Knockdown of Lrrc10 caused early morphological defects such as cardiac looping defects accompanied by a large pericardial edema, suggesting cardiac insufficiency. In addition, defects in cardiac function were observed as evidenced by decreased ejection fraction and cardiac output. Further investigations into the underlying mechanisms of the cardiac defects indicated that the number of cardiomyocytes was reduced in the morphants. In addition, expression of important cardiac genes was deregulated in the morphants such as atrial natriuretic factor, a hallmark for cardiac failure, and cardiac myosin light chain 2, an essential protein for cardiac contractility. In addition, a reduced amount of NADH in the morphant heart was observed in live zebrafish embryos as compared to control, suggesting a decrease in energy reserve due to Lrrc10 reduction. The results presented here indicate that Lrrc10 is essential for early cardiac development and function in zebrafish embryos. Therefore, this study will provide a critical foundation to further study the biological functions of Lrrc10 in cardiomyocytes, which will enhance our understanding of the mechanisms that lead to normal cardiac development and function.
Materials and methods
Zebrafish maintenance
A cmlc2:dsRed2-nuc line of transgenic zebrafish (AB background) that expresses red fluorescent protein (RFP) specifically in the cardiomyocyte nucleus was used to determine the number of cardiac myocytes. A transgenic cmlc2::GFP zebrafish line (AB background) that marks cardiomyocytes with Green Fluorescence Protein (GFP) was used to visualize the abnormal contractions as described below (see Movies S1 and S2). The wild type AB strain of zebrafish was used for all other experiments. Pooled embryos from group matings were used in all experiments to control for variability between different clutches of embryos. All fish were bred and embryos were raised according to the procedures as described elsewhere (Westerfield, 1995). When applicable, fish were anesthetized with 1.67 mg/ml tricaine (Sigma).
Cloning of a full-length zebrafish lrrc10 cDNA
To identify novel cardiac-specific genes that are exclusively expressed in the heart, we performed a virtual subtractive screen (in silico approach) as described previously (Adameyko et al., 2005; Nakane et al., 2004; Passier et al., 2000; Wang et al., 2001). Mouse Expression Sequence Tag (EST) databases from embryonic day (E)13 heart were searched using the NCBI database. By subtracting genes that are expressed in other organs, we identified a list of putative cardiac-specific genes. These genes were subsequently subjected to in situ hybridization and Northern blot analyses to examine the expression patterns. From this expression screening, a mouse gene encoding Leucine-rich Repeat Containing protein 10 (LRRC10) was identified as a cardiac-specific gene (Kim et al., in press).
Zebrafish lrrc10 (zlrrc10) was cloned by RT-PCR using gene specific primers based on the reported zlrrc10 cDNA sequence, forward (F) 5′-ACAGGAGCATCAAACACCA and reverse (R) 5′-ACAAAAGTCACTAATTCACTC (Genbank nucleotide accession number; GI57222256). Total RNA was isolated from the adult zebrafish heart with Trizol Reagent (Invitrogen) and first-strand cDNA was synthesized from 1 μg total RNA by reverse transcriptase (SuperScriptIII, Invitrogen) with oligo (dT) primers followed by PCR. The profile for PCR reaction was 94 °C for 5 min, followed by 30 cycles of denaturation 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 1 min, then 7 min at 72 °C for the final extension. The full-length PCR product of 973 bp consisting of the entire coding sequence, 5′ and 3′ untranslated region was subcloned into pCRII-TOPO vector (Invitrogen) followed by sequencing. The nucleotide sequence of the cloned zlrrc10 is identical to the reported sequence.
To detect zlrrc10 in adult tissues (Fig. 1D), RT-PCR was performed as above using total RNA isolated from three different parts of adult zebrafish followed by PCR amplification with the following primers: zlrrc10, F 5′-CACGGCCACCAGTTTTC, R 5′-GGCGGGTAAACTCCGTAG; β-actin, F 5′-CACGAGACCACCTTCAACT, R 5′-CATTGTGAGGAGGGCAAAG.
Antisense morpholino experiments
Two independent antisense morpholinos (MOs) targeted against the translational start site of zlrrc10 were synthesized (Gene Tools). MOs were 3′ labeled with FITC to monitor for uniform oligonucleotide distribution in injected embryos. The morpholino sequences used were as follows: zlrrc10-MO1 (MO1, target − 11 to + 14), 5′-ACAACAT TTCCCATCTTCTTGGCCC; zlrrc10-MO1–5 bp mismatched control morpholino (CMO-5m), 5′-ACAAGAT TTGCCATGTTGTTGGGCC, mismatched base pairs indicated in bold; zlrrc10-MO2 (MO2, target −32 to −8), 5′-GCCCAAAATCATGGTCTCAGGTAGC.
The specificity of zlrrc10 morpholinos was confirmed by using a standard scrambled control morpholino (CMO) 5′-CCTCTTACCTCAGTTACAATTTATA (Gene Tools) and CMO-5m morpholino. The ability of MOs to specifically block translation of its cognate mRNA was analyzed using an in vitro transcription and translation assay (TNT kit; Promega). Briefly, in vitro transcription and translation of pCRII-TOPO-zlrrc10 was performed using T7 RNA polymerase. Reactions were performed according to manufacturer's protocol, except that to a 12.5 μl reaction was added 75 ng of template DNA and 0.75 μl of [35S]-methionine, in the presence or absence of various final concentrations of MO1, CMO, CMO-5m, or MO2 as indicated. After 90 min incubation at 30 °C radioactive translation products were resolved by 10% polyacrylamide gel electrophoresis followed by autoradiography.
The microinjection (Narishige IM300 Microinjector) of newly fertilized eggs was preformed at the 1–2 cell stages with approximately 2 nl of the appropriate morpholino solution (0.15 mM of zlrrc10-MO1, CMO or CMO-5m) (Antkiewicz et al., 2006). For MO2 injections, we used 0.75 mM of zlrrc10-MO2 or CMO. 1× Danieau's solution (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5 mM N-(2-hydroxyethyl)-piperazine-N'-2-ethane-sulfonic acid, pH 7.6) was used to dilute all morpholino stocks to the concentration appropriate for injection. The morpholino solution was injected approximately at the border between the cell and yolk, with a needle positioned such that it was piercing the cell. Embryos were screened under fluorescent light to determine injection success and even distribution of morpholino within the embryo at approximately 2 h postinjection.
Whole-mount in situ hybridization, immunostaining, and histology
To study the expression of zlrrc10, anf and cmlc2 mRNA in zebrafish embryos, pCRII-TOPO-zlrrc10 and pCRII-TOPO-anf plasmid were cloned. cmlc2 was a kind gift from Dr. Randall Peterson. To clone the partial anf cDNA, RT-PCR was performed using total RNA from the zebrafish adult heart with the following pairs of oligonucleotide primer sequences: anf: F 5′-ACACGTTGAGCAGACACAGC, R 5′-TGTTAACAAATTAAGCCGTATTGT.
Whole-mount in situ hybridization of zebrafish embryos was carried out by using digoxigenin-11-UTP-labeled probes (Roche Diagnostics Corp) as previously described (Yelon et al., 1999). The antisense probes for full-length zlrrc10, anf, and cmlc2 were synthesized by linearizing their respective plasmids with NotI and using Sp6 RNA polymerase. The sense probe for zlrrc10 was synthesized for negative control by linearizing the pCRII-TOPO-zlrrc10 plasmid with HindIII and using T7 RNA polymerase. To quantitatively analyze the differences of gene expression, semi-quantitative RT-PCR was performed. Briefly, following first-strand cDNA synthesis, PCR amplifications were performed with 2% of the first-strand reaction, 1 unit of Taq polymerase (Denville Scientific Inc, Metuchen, NJ), and 25 pmol of the appropriate primers in a reaction volume of 25 μl. The amplification was done at 94 °C for 1 min followed by 24 cycles of 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s. A final extension at 72 °C for 7 min was performed. Gene-specific primers for cmlc2 are F 5′-ACCGGGATGGAGTTATCA, and R 5′-CTCCTGTGGCATTAGGG. The authenticity of the amplified products was determined by size analysis on agarose gels.
Immunostaining of zebrafish embryos was performed as previously described (Berdougo et al., 2003) using the monoclonal antibody MF20 that recognizes the myosin heavy chain (MHC). Briefly, embryos fixed in 4% paraformaldehyde were dehydrated in a methanol series. For staining, rehydrated embryos were permeabilized by digestion in collagnase (1 mg/ml) for 30–40 min. Permeabilized larvae were blocked in 10% normal calf serum in PBS with 0.1% Tween-20 for 1 h. After overnight incubation with the MF20 antibody, embryos were incubated with a secondary antibody (Alexa-546 conjugated goat anti-mouse, Molecular Probes, OR, USA) for 5 h at room temperature and visualized by epifluorescence microscopy.
Histological experiments were performed by standard hematoxylin and eosin staining (H&E) as previously described (Lee et al., 2000). Zebrafish embryos fixed in 4% paraformaldehyde were serially dehydrated to 100% ethanol for paraffin embedding. Paraffin embedded embryos were sectioned at 10 μm thickness and stained with H&E to examine histology of the heart.
Red blood cell (RBC) perfusion rate and analyses of the number of cardiac myocytes
As an index of regional blood flow, RBC perfusion rate was measured in one readily identifiable vessel, the intersegmental blood vessel (ISV) in the tail as described elsewhere (Trumpp et al., 2001). Embryos were mounted in 3% methyl cellulose (Sigma) and high-speed video time-lapse recordings (125 frames per second) were used to image RBCs passing through an ISV of the mounted zebrafish in a 3 s period. These recordings were captured using a Motion Scope camera (Redlake) and a Nikon TE300 inverted microscope with a 10× lens. The total number of RBCs passing through an ISV during the entire recording was counted.
Cardiac myocytes were counted at 3 and 4 days post fertilization (dpf) in cmlc2::dsRed2-nuc transgenic zebrafish (Antkiewicz et al., 2005; Mably et al., 2003) using the method described by Antkiewicz et al. (2005). Briefly, embryos were anesthetized and flat-mounted on a glass microscope slide under a coverslip in Lebovitz's L15 cell culture media (Invitrogen). Epifluorescence images were captured using the Nikon TE300 inverted microscope (20× lens) and a Princeton Instruments Micromax charge-coupled device camera. Total number of visible cardiac myocyte nuclei per heart was counted.
Cardiac functional analyses
End-diastolic volume (EDV) and end-systolic volume (ESV), as well as stroke volume and cardiac output were determined at 3, 4, and 5 dpf from time-lapse recordings (Motion Scope camera, 250 frames/s) of a lateral view of a beating zebrafish heart as described previously (Antkiewicz et al., 2006; Carney et al., 2006). Briefly, frames representative of the ventricle in end-systole and end-diastole were used to calculate volume of the ventricle for both states according to the method of discs (Coucelo et al., 2000). This method assumes that ventricular volume can be represented as the sum of the volumes of individual discs into which the ventricle can be sliced. The two-dimensional image of the ventricle at end-diastole and end-systole was divided into slices of 10 μm thickness and their volume was assessed using the MetaMorph Imaging System (Universal Imaging Corporation). Time-lapse recordings of the heart were also used to calculate heart rate (HR), which was determined from the number of frames in which three consecutive heart contractions occurred. Stroke volume (SV) was calculated from the approximated EDV and ESV: SV=EDV–ESV; cardiac output (CO)=SV×HR.
Multiphoton fluorescence imaging
NADH is an intrinsically fluorescent molecule, which allows one to monitor NADH fluorescence to dynamically interpret the metabolic activity in vivo (Chance et al., 1962; Pappajohn et al., 1972). NADH images of the heart in live zebrafish embryos were collected with a purpose-built multiphoton imaging system (LOCI at the University of Wisconsin, Madison) (Bird et al., 2004; Bird et al., 2005). Briefly, a titanium sapphire laser (Coherent, Mira) tuned to wavelength at 780 nm excitation was the two-photon excitation source. The excitation light was sent to and the emitted light was collected from the sample through an inverted microscope (Nikon, TE2000U) with a 40× objective (Nikon, PlanApo NA 1.4) coupled to a laboratory-developed laser scanning system. Synchronized fluorescence data collection on a pixel-by-pixel basis was achieved using the x and y laser scanning signals generated for the laser scanning electronics. Acquisition was done with WiscScan, a laboratory-developed acquisition package (Bird et al., 2004).
Statistical analyses
One-way analysis of variance followed by the Fisher least significant difference test was used to determine statistical significance. The assumption of unequal variances for all data sets was checked using Levene's test. Results are presented as mean±standard error of the mean (SEM), and asterisks indicate a significant difference at p<0.05, or 0.01 as indicated. All analyses were performed using the Statistica 7.0 software package. In all cases, the observer was blinded to the treatment group until measurements or scoring was completed.
Results
Examination of zlrrc10 expression patterns
Information on the detailed expression patterns of a gene provides an important basis for understanding the biological roles of the gene product. Due to the remarkable cardiac-specific expression of LRRC10 in mouse (Adameyko et al., 2005; Nakane et al., 2004) (Kim et al., in press) and human (data not shown, 2007), we set out to determine whether Lrrc10 plays critical roles in cardiac development in the zebrafish embryo. It presents a prototypic vertebrate heart because the molecular mechanisms regulating cardiac development in zebrafish appear to be similar to those in more complex hearts of higher vertebrates (Weinstein and Fishman, 1996; Fishman and Chien, 1997).
In preparation for investigating the developmental roles of Lrrc10, we first examined the expression patterns of zlrrc10 in zebrafish embryos by whole-mount in situ hybridization using an antisense zlrrc10 probe (Fig. 1). The full-length cDNA consisting of the coding sequence and 5′ and 3′ untranslated regions of zlrrc10 was cloned by RT-PCR as described in Materials and methods section. The full-length zlrrc10 cDNA is encoded by two exons as depicted in Fig. 1A whereas the mouse LRRC10 is encoded by one exon. The coding sequence of zLrrc10 contains 270 amino acids, which is highly homologous to the mouse and human LRRC10, suggesting the conserved function of this gene product (Fig. 1B). The primary amino acid sequence of zLrrc10 is 62% or 64% identical to the mouse or human LRRC10, respectively. If the similar amino acids are included, the homology is 73% or 76% to the mouse or human LRRC10, respectively. The zLrrc10 consists of seven LRR motifs without any known functional domain as in mouse and human LRRC10.
To perform whole-mount in situ hybridization, zebrafish cardiac myosin light chain 2 (cmlc2) was used as a positive cardiac marker gene to visualize the developing heart (Fig. 1C, b and e). It has been demonstrated that cmlc2 is expressed both in the embryonic ventricle and atrium of zebrafish (Rottbauer et al., 2006; Yelon et al., 1999). lrrc10 in zebrafish was detected specifically, but weakly in the linear heart tube of 1 dpf embryos followed by a marked increase in the heart of 2 dpf embryos (Fig. 1C, a and d). zlrrc10 expression appears to be higher in the ventricle than the atrium, but it should be noted that the atrium contains fewer cardiomyocytes than the ventricle. zlrrc10 was not detected in any other tissues or organs in developing embryos. The negative control using a sense-zlrrc10 probe did not show any staining (Fig. 1C, c and f), indicating specificity of the antisense zlrrc10 probe. The cardiac-specific expression continues in adult zebrafish as indicated by RT-PCR assays. The zlrrc10 transcripts were detected only in the heart, but not in the head or tail region of adult zebrafish (Fig. 1D). These results demonstrate the cardiac-specific expression of lrrc10 in zebrafish from 1 dpf to adult.
Examination of morpholinos
To determine whether Lrrc10 plays critical roles in cardiac development, we performed antisense morpholino (MO) experiments during early zebrafish development. Zebrafish are well suited for gene function studies using MOs due to their rapid external development, transparent embryos, and the ease of delivery of the intervening MOs. We designed morpholinos against the translation initiation site in zlrrc10 mRNA to reduce Lrrc10 protein level in zebrafish embryos. The ability of morpholinos to block translation of zlrrc10 was examined using an in vitro translation reaction in the absence or presence of morpholino (Fig. 2A). In vitro translated and [35S]-methionine labeled Lrrc10 was detected as a single band at the expected size of 31 kDa (indicated by +). MO1 completely blocked translation of zlrrc10 at 0.26 μM whereas the control morpholino (CMO) did not inhibit translation even at a higher concentration of 1.3 μM. MOs display sequence-specific inhibition over a wide concentration range: 3–3000 nM in cell-free translation systems (Sumanas and Larson, 2002). To further confirm the specificity of MO1, we tested MO1 containing five mismatched base pairs (CMO-5m) as an additional specific negative control. CMO-5m did not block translation of zlrrc10, indicating the specific effect of MO1 on blocking translation of zlrrc10.
Fig. 2.

Effects of zlrrc10-MO1 on the zebrafish heart morphology and blood flow. (A) The effect of zlrrc10-MOs on blocking translation of zlrrc10. zlrrc10 cDNA yielded a [35S]-methionine labeled 31 kDa protein by in vitro transcription and translation reaction (+). Negative control (−) was incubated without template plasmid. To determine the effect of zlrrc10-MOs, zlrrc10 cDNA was in vitro translated in the presence of MO1 or MO2. To confirm the specificity of zlrrc10 MOs, two control morpholinos, CMO or CMO-5m were used. The final concentrations of morpholinos in the reaction are indicated. (B) Knockdown of Lrrc10 caused heart defects. CMO (a and d) and MO1 (b, c, e, and f) injected zebrafish were imaged at 3 dpf. d–f are high-power images of a–c, respectively, which show heart looping defects of the morphants (e and f). The arrowhead indicates a large pericardial edema. Blue and red indicate the ventricle and atrium, respectively. A, atrium; V, ventricle. (C) Reduced blood flow in the morphants. Zebrafish embryos were injected with zlrrc10-MO1 (hatched bar) or CMO (open bar). RBC perfusion rates in an intersegmental vessel were determined between 2 and 5 dpf as described in Materials and methods section. An asterisk indicates a significant difference in values between the test CMO and the MO1 at p<0.05.
It is important to use stringent negative controls for antisense experiments. While MOs can exhibit specific inhibition over a wide concentration range, non-specific interactions still happen, especially at higher doses. One likely reason for the possible non-specific effects is mistargeting. While random sequences of MOs are widely used as a control for general toxicity of MOs, and embryo handling, it does not address MO mistargeting, which is the likely cause of non-specific phenotypes. It becomes necessary to confirm the specificity of novel phenotypes either by RNA rescue or by using another MO of different sequence targeted against the same gene. RNA rescue is often complicated due to RNA overexpression artifacts and the inability to mimic the endogenous spatial and temporal gene expression pattern that is often critical for the proper function of the gene. This is the case for rescue experiments for Lrrc10 knockdown embryos because zlrrc10 is a cardiac-specific gene. However, the mistargeting issue is well addressed when two MOs of different sequence against the same gene produce the same phenotype. Therefore, we synthesized a different zlrrc10 morpholino, MO2, which also blocked the translation of zlrrc10 in vitro, but to a much lesser extent than MO1 (Fig. 2A). MO2 blocked the translation with about 90% efficiency at 3 μM while CMO did not (data not shown). Having confirmed the specific activity of zlrrc10 morpholinos in blocking translation in vitro, we generated the Lrrc10 knockdown embryos to examine the developmental roles of Lrrc10.
Knockdown of Lrrc10 caused cardiac morphological defects
To investigate the role of Lrrc10 in zebrafish development, we injected each morpholino into one-cell stage zebrafish embryos as described in the Materials and methods section and allowed them to develop for 6–7 dpf. The zlrrc10-MO1 knockdown embryos (morphants) did not show gross abnormalities at 1 dpf (data not shown). In normal zebrafish embryos, by 26 h post fertilization (hpf), the heart tube is formed and elongated to form an asymmetrically located tube, which then undergoes looping (Heideman et al., 2005). At about 36 hpf, the ventricular cells move to the right of the atrium in a process known as rightward looping, which lasts until approximately 48 hpf. Initially peristaltic waves of contraction in the simple heart drive circulation, beginning approximately 24 hpf. After 48 hpf, much of the basic architecture of the heart has formed, and a process of maturation continues in which the myocardial walls thicken and become trabeculated and actual valve leaflets form.
However, starting at 2 dpf, the zlrrc10 morphants exhibited gross abnormalities including a large pericardial edema and cardiac looping defects. The representative images of control (a and d) and the morphants (b, c, e and f) at 3 dpf are shown in Fig. 2B. The zlrrc10 morphants showed a large pericardial edema that was readily apparent (indicated by arrow Fig. 2B, b and c) that is absent in control (Fig. 2B, a), suggesting cardiac insufficiency. We did not observe any significant general toxicity in the control embryos (Fig. 2B, a). Because the heart has undergone looping in the control embryos, the ventricle and atrium of the control embryos are overlapped and not distinguishable by the lateral view (Fig. 2B, d). In contrast, the zlrrc10 morphants exhibited prominent looping defects at higher magnification as shown by two representative morphants (Fig. 2B, e and f). Most zlrrc10 morphants showed a complete looping failure, resulting in the linear heart with a stretched string-like atrium (Fig. 2B, e) or a round atrium (Fig. 2B, f). Some zlrrc10 morphants showed a partially looped heart (see Fig. 3G).
Fig. 3.
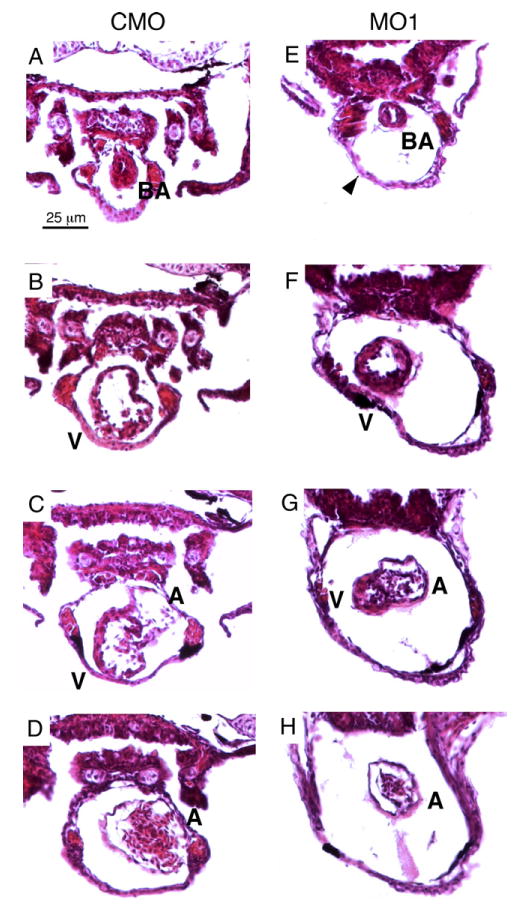
Histological analysis reveals cardiac defects in the zlrrc10 morphants. Zebrafish embryos injected with CMO (A–D) and MO1 (E–H) were fixed at 4 dpf and paraffin embedded. The embryos were sectioned (10 μm) and stained with H&E. Serial transverse sections are shown from the anterior end at the top of the figure and progressing to the posterior end at the bottom. Arrowhead indicates a large pericardial edema; BA, bulbus arteriosus; A, atrium; V, ventricle.
We analyzed the morphants daily and the results from four independent microinjection experiments are summarized in Table 1. The looping defect persisted and pericardial edema became progressively larger, suggesting severe cardiac insufficiency. By 4 dpf, about 90% of morphants showed looping defects with a large pericardial edema. The morphology of the ventricle in the morphants became abnormal by 4 dpf, as shown by it being compact and small compared to that of control embryos (see Fig. 3).
Table 1.
Phenotype analyses of embryos injected with zlrrc10 or control morpholinos
| Looping defect (%) | Pericardial edema (%) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MO1 | n | CMO | n | CMO-5m | n | MO2 | n | MO1 | n | CMO | n | CMO-5m | n | MO2 | n | |
| 2 dpf | 82.6±2.4 | 125 | 3.4±1.8 | 145 | 6.1±1.5 | 82 | 77.2±1.0 | 45 | 51.7±6.2 | 125 | 2.5±1.0 | 145 | 3.6±1.0 | 82 | 38.6±0.6 | 45 |
| 3 dpf | 89.5±3.7 | 124 | 2.1±1.2 | 114 | 9.0±2.8 | 66 | 78.8±3.8 | 43 | 80.6±4.9 | 124 | 2.6±1.0 | 114 | 7.5±1.3 | 66 | 66.3±3.8 | 43 |
| 4 dpf | 91.9±3.7 | 107 | 1.9±1.2 | 91 | 9.9±0.8 | 50 | 78.2±8.1 | 32 | 88.4±7.1 | 107 | 1.9±1.2 | 91 | 7.7±1.5 | 50 | 23.6±3.6 | 32 |
| 5 dpf | 97.7±1.6 | 80 | 2.2±1.6 | 77 | 6.9±202 | 43 | 80.2±10.2 | 31 | 99.4±0.6 | 80 | 3.2±1.9 | 77 | 9.3±0.2 | 43 | 17.1±2.9 | 31 |
Four or two independent microinjection experiments were performed for MO1 or MO2, respectively. Data are shown as mean±SEM. n, the number of embryos.
The tubular hearts in the morphants were able to contract and drive the circulation, but not as effectively as the control heart as evidenced by markedly slower moving blood cells in the ISVs (Fig. 2C). The number of blood cells passing through ISV in the tail caudal to the yolk was significantly decreased in the morphants. As early as 36 hpf, heart contractions appeared visibly abnormal or incomplete in zlrrc10 morphants, and this continued to be abnormal during development (Supplemental data provided by a movie using a transgenic zebrafish line cmlc2::GFP). In contrast, vigorous contractions were observed in control embryos. These results led us to analyze the cardiac function as described below (Fig. 4). Although the morphology of the heart was abnormal and cardiac function appeared defective, the morphants survived until 6–7 dpf when viability in the morphant started to deteriorate, as indicated by either no heart beat or a very weak heart beat.
Fig. 4.

Reduction of zLrrc10 expression causes the defects in heart function. (A) Zebrafish embryos were injected with MO1 or CMO as indicated. To determine EDV and ESV, time-lapse recording was used to follow ventricular movement from a lateral view in embryos at 3–5 dpf. (B) Stroke volume was calculated by subtracting ESV from EDV. (C) Ejection fraction (%)=(EDV −ESV)/EDV × 100. (D) Heart rate was calculated from the number of frames captured during three consecutive, complete ventricular contractions in the time-lapse recordings. (E) Cardiac output was calculated as the product of stroke volume and heart rate. Values are mean±standard error of the mean with n= 10. An asterisk indicates a significant difference in values between the morphants and control at p<0.05. (F) Representative fluorescence intensity images of the embryonic hearts are shown in vivo in lateral views with the head to the right using multiphoton microscopy. All images of the control and morphant embryos were obtained at 3 dpf under identical experimental conditions at 780 nm excitation. (G) Relative amounts of NADH were calculated when the control value was set at 100%. n=4.
To confirm that the effect of MO1 on cardiac development is caused by knockdown of Lrrc10, rather than non-specific effect due to mistargeting, zlrrc10-MO2 that is targeted at a different site was tested. As summarized in Table 1, MO2 injection also caused cardiac looping defects and pericardial edema in the morphants, which is the same phenotype by MO1 injection. Cardiac looping defects persisted during the course of examination, which were observed in about 80% of the MO2 morphants (Table 1). A large pericardial edema was observed in up to 66% of the MO2 morphants at 3 dpf followed by recovery at 4 and 5 dpf, suggesting cardiac function was improved as the morphants developed. It should be noted that MO2 blocks translation much less efficiently than MO1 (Fig. 2A). Moreover, it is highly likely that the effect of MO2 is reduced because the concentration of MO is diluted as embryos develop and the level of zLrrc10 expression increases as the heart develops (Fig. 1C, a and b; data not shown, 2007). In fact, it is believed that MOs are most efficient during the first 2 or 3 days of development, possibly due to dilution by ongoing cell divisions and by elimination from the embryo body due to metabolism and excretion of the MO (Sumanas and Larson, 2002). Nevertheless, it is interesting that the MO2 morphants with abnormal cardiac morphologies at 4–5 dpf seem to have improved cardiac function. We have not observed any defects in other regions of the morphants injected with MO1 or MO2, indicating the cardiac-specific effects of zlrrc10 MOs. In addition, neither scrambled control morpholino (CMO) nor CMO-5m caused any significant morphological defects, which indicates the specific effects of zlrrc10 MOs on Lrrc10 knockdown. Therefore, these results demonstrate that Lrrc10 is a critical factor for early cardiac development and suggest cardiac functional defects in the zlrrc10 morphants.
Histology of the heart is abnormal in the zlrrc10 morphant
The zebrafish embryos were sectioned and stained with H&E to analyze histology of the heart at 3 (results not shown) and 4 dpf (Fig. 3). By 48 hpf, the beginning of valve formation is evident as endocardial cells at the AV junction become distinct in morphology and gene expression, as they begin to form valve cushions. Blood flow is a critical factor in valve formation, and blocked circulation prevents proper valve development (Bartman et al., 2004; Hove et al., 2003). After 48 hpf, the zebrafish heart consists of a smooth-walled tube partitioned into all four segments with the definite structure of sinus venosus, atrium, ventricle, and bulbus arteriosus (Grimes et al., 2006; Hu et al., 2000). Each part is externally identified by a constriction between the segments. The myocardium is about one cell thickness in the atrium, but the ventricle has two to three cell layers. The heart is lined with endocardium, separated from the myocardium by a layer of cardiac jelly in the atrium. The ventricle and sinus venosus do not have a cardiac jelly layer. There are no valves separating the segments at this stage.
At 4 dpf, the zlrrc10 MO1 morphants showed similar abnormalities to the 3 dpf morphants. The position and morphology of the bulbus arteriosus in the morphants seemed normal (compare Figs. 3A and E). Two to three layers of cardiomyocytes were observed in the ventricles of both the morphants (Figs. 3F and G) and control (Figs. 3B and C). However, prominent morphological defects were observed in the ventricle of the zlrrc10 morphant, which was small and compact compared to the control. The AV valve that is forming seemed normal at the AV canal that was normally constricted (compare Figs. 3C and G). The looping defect in the zlrrc10 morphant was clearly evident as shown in Fig. 3G, where the atrium was located laterally as opposed to dorsally in the control. This particular embryo has undergone a partial looping process since the atrium was observed with the ventricle at the same plane of section. A cross-section of the stretched, string-like atrium was striking in the zlrrc10 morphant as compared to the control (compare Figs. 3D and H). In both control and the morphant, the atrial wall consisted of one myocardial layer and the endocardial layer with space for cardiac jelly in between. The impressive pericardial edema (indicated by an arrowhead) was evident in all transverse sections of the morphant heart. These results clearly demonstrate cardiac looping defects and suggest possible defective cardiac chamber growth when Lrrc10 expression is reduced.
Knockdown of zLrrc10 resulted in cardiac functional defects
In addition to defective cardiac morphogenesis, the heart of zlrrc10 morphants exhibited abnormal contraction. Vigorous peristaltic contraction waves traversing the heart tube were usually observed in wild type embryos by 36 hpf. In contrast, the heart of the morphant seemed to beat weakly. Since normal morphological development is critical for normal cardiac function, we investigated the cardiac function as described in Materials and methods. End-diastolic volume (EDV) and end-systolic volume (ESV) were measured for the beating heart (Fig. 4A). EDV was decreased in the zlrrc10 morphants during the period of 3–5 dpf as compared to the control heart. The decrease in EDV was prominent in the morphants at 5 dpf. In contrast, ESV in the morphant does not seem to differ from that of control. Accordingly, stroke volume and ejection fraction were significantly reduced in the morphants, especially at 5 dpf as compared to control during the course of study (Figs. 4B and C). Heart rate was also significantly decreased in the morphants compared to control (Fig. 4D). As a consequence of the reduced stroke volume and heart rate, cardiac output was markedly reduced (Fig. 4E), which led to embryonic lethality due to cardiac failure (no contraction) by 6–7 dpf.
Metabolic regulation is inextricably linked with cardiac function. This metabolism–function relationship is relevant to diseases that lead to cardiac hypertrophy and heart failure (Huss and Kelly, 2005). NADH is essential to generate ATP (Brandes and Bers, 1996) and the amount of ATP has been shown to be decreased in failing hearts because of decreased energy reserve (Nascimben et al., 2004). To determine whether the knockdown of zlrrc10 caused any perturbations in energy metabolism, fluorescence intensity images of the heart at 780 nm were collected in live zebrafish embryos using multiphoton microscopy, which reflects the amount of NADH (Fig. 4F). We have shown previously that the signal at 780 nm multiphoton excitation is dominated by the NADH signal (Skala et al., 2007). Interestingly, the fluorescence intensity in the morphant heart was reduced by 45% as compared to control (NIH Image J analysis), suggesting that energy reserve is likely reduced in the morphant heart (Fig. 4G).
These results so far demonstrate that reduced expression of Lrrc10 caused the decreased cardiac output during early development, in part due to malformation of the heart. These changes in the functional cardiac profile do not seem to correlate to adult cardiac hypertrophy in human or mice where there are no changes in ejection fraction. It should be noted that there was a marked increase in EDV in normal embryos from 4 to 5 dpf, which reflects the normal functional changes during development. The reduced EDV in the zlrrc10 morphant may be attributed to an inability to dilate after the contraction or to the defective cardiac chamber growth resulting in a small ventricle. Therefore, we investigated whether the number of cardiomyocytes is reduced in the morphant heart.
Myocardial cell number is reduced in the zlrrc10 morphants
Since the ventricle of the zlrrc10 morphants seemed smaller than control, the atrium showed a string-like shape at 3–4 dpf (Figs. 2 and 3), and EDV was reduced especially at 5 dpf (Fig. 4), we investigated whether chamber growth was defective in these morphants. We used a transgenic zebrafish line (cmlc2:: dsRed2-nuc) where cardiomyocytes are marked by the nuclear RFP to determine the effect of reduced Lrrc10 expression on the number of cardiomyocytes in the heart. MO1 or CMO was injected into cmlc2::dsRed2-nuc eggs after fertilization to count cardiomyocytes (Figs. 5A and B). In the normal developing heart, the number of cardiomyocytes per heart was significantly increased from 3 to 4 dpf, indicating normal developmental growth. In contrast, the total number of cardiomyocytes in the zlrrc10 morphant heart was not significantly increased. Instead, a 14.5% or 25.4% decrease in the total cardiomyocyte number was observed in the morphants at 3 or 4 dpf, respectively. These results suggest that cell proliferation is defective in the heart of the zlrrc10 morphant. However, it is not clear whether the reduced cell number results from very early defects in cells in the precardiac region (for example, reduced number of cardiac precursor cells) or from proliferation defects during cardiac development between 2 and 4 dpf. Although we have not observed a marked increase in apoptotic cells in the morphant heart (data not shown), we cannot exclude the possibility that slightly increased or aberrant apoptosis may have caused the reduced cardiomyocyte number or morphological defects in the morphant.
Fig. 5.

Knockdown of zLrrc10 causes a decrease in the number of cardiac myocytes in developing zebrafish embryos. Transgenic zebrafish embryos expressing RFP in the cardiomyocyte nuclei (cmlc2::dsRed2-nuc) were injected with CMO or MO1. At the indicated time points, fluorescence images were captured, and cells were counted in at least ten hearts per group. Embryos were grown in the presence of 0.003% phenylthiourea from 1 dpf to inhibit pigmentation, which is effective until 4 dpf. (A, B) Representative epifluorescence images of the CMO and MO1 injected cmlc2::dsRed2-nuc transgenic zebrafish hearts at 3 dpf. (C) The number of cardiomyocytes per heart in CMO and MO1 injected embryos. Values represent means±SEM. *p<0.01.
Cardiac gene expression is deregulated in the zlrrc10 morphants
The results so far demonstrate profound morphological and functional cardiac defects in Lrrc10 knockdown embryos. Next, to determine the underlying causes of these cardiac defects, we investigated alterations in gene expression of several important cardiac marker genes in the heart of morphants (Fig. 6). Atrial natriuretic factor (anf) gene expression in the zlrrc10 morphants was assessed because of its established responsiveness to various physiological and pathological stimuli including those causing hypertrophic or dilated cardiomyopathy in mammals (Rockman et al., 1994b). As in other species, the zebrafish anf gene is expressed in both the ventricle and atrium before becoming restricted to the atrium (Berdougo et al., 2003). ANF is known as a hallmark for cardiac hypertrophy because ANF expression is switched off in the normal adult heart, whereas it is re-expressed in hypertrophic and failing hearts. Whole-mount in situ hybridization of anf showed a marked increase in anf expression in both the ventricle and atrium of 2 day old morphants (Fig. 6A, b and d) as compared to control (Fig. 6A, a and c). These results suggest dysfunction of the heart in the zlrrc10 morphants as early as 2 dpf, when the heart has just finished an initial looping process. It is interesting that cardiac morphology of this particular zlrrc10 morphant seems normal at this stage of development. Therefore, these results suggest that induction of anf expression is very sensitive to altered cardiac function.
Fig. 6.
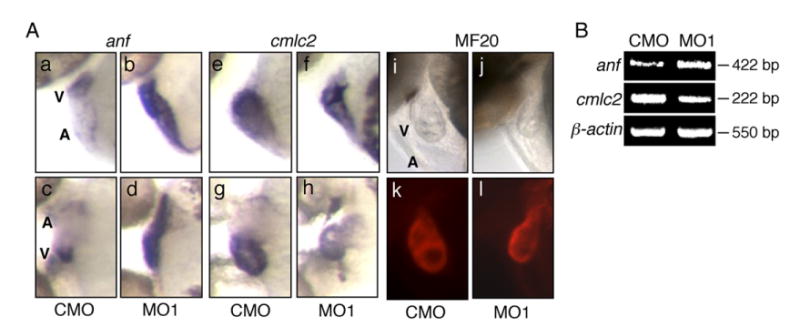
The expression levels of anf and cmlc2 were altered in the zlrrc10 morphants. (A) CMO (a, c, e, g, i and k) and MO1 (b, d, f, h, j and l) injected zebrafish embryos at 2 dpf were subjected to whole-mount in situ hybridization using digoxigenin labeled anf (a–d) and cmlc2 (e–h) antisense riboprobes. Images of whole-mount immunostaining using MF20 antibody are shown in k and l. i and j are the same fields as k and l, respectively. c, d, g and h are ventral views and others are lateral views, all with anterior to the left. Note the marked increase in anf expression in the zlrrc10 morphant (b and d) but the reduction of cmlc2 expression in the morphant (f and h). A ventral view of the morphant clearly shows the small ventricle and a looping defect (h). (B) For quantitative analyses of gene expression, RT-PCR was performed on anf and cmlc2. β-actin was used as a control.
We also examined expression of the gene encoding sarcomere proteins such as cmlc2 and myosin heavy chain (MHC) because genes encoding sarcomere components are often altered in response to pathologic conditions (Antos et al., 2002). cmlc2 has been shown to be expressed in embryonic heart and to have a critical role in thick-myofilament assembly and contractility in the zebrafish embryonic heart (Rottbauer et al., 2006; Yelon et al., 1999). In the morphant at 2 dpf, expression of cmlc2 was decreased in the ventricle and atrium as compared to control (in Fig. 6, compare e and g to f and h), which may be partly responsible for abnormal contractility and reduced cardiac output. Since there was only a 14.5% reduction in the cardiomyocyte number in the morphant at 3 dpf (see Fig. 5) and both the morphants and control have about two layers of myocytes in the ventricular wall (see Fig. 3), it is unlikely that the decreased expression of cmlc2 is entirely due to the reduced number of cardiomyocytes. Looping defects and the small ventricle were obvious in this morphant (Fig. 6A, f and h), which are consistent with the results shown in Fig. 3. To obtain quantitative expression data, semi-quantitative RT-PCR was performed (Fig. 6B). Expression of anf was up-regulated about 73%, but cmlc2 was down-regulated about 50% as compared to control (NIH Image J analysis), which correlates well with the in situ hybridization results.
Immunostaining of zebrafish embryos with MF20 antibody that recognizes sarcomere myosin heavy chain showed no detectable changes in the expression level between the zlrrc10 morphants and control (compare Fig. 6A, k and l). These data so far indicate that the zlrrc10 morphants exhibit increased expression of anf, a hallmark for cardiac hypertrophy and failure, but decreased expression of cmlc2.
Discussion
Lrrc10 is essential for early cardiac development
To advance our understanding of early cardiac development, we identified the cardiac-specific gene zlrrc10 and characterized its developmental roles in zebrafish in this study. We demonstrated for the first time that zlrrc10 plays critical roles in early cardiac development and cardiac function in the zebrafish embryo. Decreased expression of Lrrc10 in the zebrafish embryo resulted in a failure of the cardiac looping process and a significant reduction in cardiac output. The cardiac insufficiency is likely a cause for embryos dying at 6–7 day post fertilization when embryos require active transportation of nutrients for survival and growth. The zebrafish has become an attractive model for molecular biologists and geneticists, with most studies focused on the early embryonic stages (Alexander and Stainier, 1999; Chen et al., 1996; Weinstein and Fishman, 1996). Recently, information related to the morphology and function of the developing as well as mature zebrafish heart has been reported (Grimes et al., 2006; Hu et al., 2000; Hu et al., 2001), which provides a framework for structure–function analyses of the zebrafish cardiovascular system. The most striking phenotype observed in the zlrrc10 morphants is a cardiac looping failure accompanied by a large pericardial edema at 2 dpf, suggesting cardiac insufficiency at a very early developmental stage. The pericardial edema gets progressively larger, which indicates that cardiac insufficiency worsens as the zlrrc10 morphants develop.
It is often not clear whether morphogenic defects, a failure in the looping process in this case, cause cardiac malfunctions or vice versa since both looping defects and pericardiac edema were simultaneously observed at the very early developmental stage. It has been reported that proper cardiac contractility is necessary for normal cardiac morphogenesis. Hemodynamic parameters are known to be a determinant of myocardial growth, structure, and function in both the developing (Berdougo et al., 2003; Huang et al., 2003; Rottbauer et al., 2006) and adult heart and may be important in the etiology of heart failure (Hutchins et al., 1978; Rockman et al., 1994a). It has been postulated that normal flow and hemodynamics are also crucial determinants of normal cardiac morphogenesis and may play a causative role in congenital heart disease (Sedmera et al., 1998). In this regard, it is interesting that MO2, which is a much less efficient morpholino than MO1 in inhibiting translation of zlrrc10, caused looping defects that persisted during development. However, the pericardial edema began to be improved at 4–5 dpf, suggesting recovery of cardiac function. These results indicate that cardiac malfunctions in zlrrc10 morphants were not entirely due to morphogenic defects, but suggest an independent role of Lrrc10 in morphogenesis from cardiac function.
Underlying mechanisms of cardiac defects
Although we clearly demonstrated in this study that Lrrc10 is essential for early cardiac development and function, the molecular function of Lrrc10 remains to be elucidated. Due to lack of a functional domain in Lrrc10, it is difficult to hypothesize its exact molecular function from its deduced amino acid sequence. The leucine-rich repeat (LRR) motif is believed to function as a protein interaction domain. The zLrrc10 protein contains seven LRRs without any other functional domains as in mouse and human LRRC10. Therefore, it is plausible that LRRC10 regulates cellular processes by interacting with other factors that are critical for cardiac development and function. LRRC10 has been reported to be localized in the nucleus of immature cardiomyocytes, but in the cytoplasm of adult cardiomyocytes in mice (Adameyko et al., 2005). However, nuclear localization of LRRC10 in heart sections has been reported in adult mice (Nakane et al., 2004). This controversial but dynamic subcellular localization of LRRC10 suggests that it has critical and possibly changing roles as the heart matures. Therefore, it is necessary to further investigate the molecular function of LRRC10 as a nuclear or cytoplasmic factor. In this regard, the present study provides a solid foundation to further study the molecular function of LRRC10.
In an effort to determine what cellular changes occurred in the zlrrc10 morphants, we have performed candidate gene approaches (Fig. 6), which yielded two genes that are deregulated in the zlrrc10 morphants. anf expression was markedly increased both in the ventricle and atrium of the zlrrc10 morphant heart as compared to the control heart at 2 dpf. The ANF gene is expressed very early in embryonic development, at the stage when cells are committed to the cardiac phenotype. Throughout embryonic and fetal development, ANF expression characterizes both atrial and ventricular cells. However, ANF is switched off in the normal adult heart, but is re-expressed in hypertrophic and failing hearts. Therefore, ANF is a biomarker for cardiac hypertrophy. Likewise, the zebrafish anf gene is expressed in both the ventricle and atrium before becoming restricted to the atrium (Berdougo et al., 2003). Although cardiac hypertrophic responses have not been studied in the zebrafish embryonic heart, it is plausible that the zlrrc10 morphants are undergoing similar responses to cardiac hypertrophy/failure or cardiomyopathy as in the adult heart. Indeed, anf induction was reported in zebrafish wea mutant embryos, in which a loss of atrial function due to a mutation in atrial myosin heavy chain (aMHC) resulted in the small and compact ventricle (Berdougo et al., 2003).
Our candidate gene analyses indicate that cmlc2 in the heart is decreased in the zlrrc10 morphants as compared to the control heart. cmlc2 in zebrafish is expressed throughout the heart, unlike the chamber-specific pattern reported for regulatory MLCs in mouse and human. cmlc2 appears to be the only regulatory mlc gene expressed in the heart of the zebrafish (Rottbauer et al., 2006). It is well known that mutations in the cardiac regulatory myosin light chain-2 (cmlc2) gene cause hypertrophic cardiomyopathy. Recently, a mutation in zebrafish, tell tale heart (tel), which selectively perturbs contractility of the embryonic heart and gives rise to a defective morphology resembling looping defects, was identified as a mutation in the zebrafish cmlc2 gene (Rottbauer et al., 2006). Ultrastructural analysis of tel cardiomyocytes reveals complete absence of organized myofilaments, demonstrating in vivo that Cmlc2 is required for thick filament stabilization and contractility in the vertebrate heart. Therefore, it is possible that the reduced cmlc2 expression in the zlrrc10 morphants is in part responsible for reduced cardiac output leading to cardiac insufficiency.
It is interesting that the zlrrc10 morphants contain a reduced number of cardiomyocytes at 3 and 4 dpf as compared to the control heart. In the normal heart, the number of cardiomyocytes significantly increases from 3 to 4 dpf whereas it does not occur in the morphants. This reduced cardiomyocyte number in the zlrrc10 morphants may in part lead to the decreased EDV (Figs. 4 and 5), which in turn leads to cardiac insufficiency. It remains to be elucidated whether the decreased cardiomyocyte number in the morphants is primary or secondary to other cellular defects. Our current data cannot distinguish whether there are cardiomyocyte proliferation defects 3–4 dpf in the zlrrc10 morphant heart or this decrease in cardiomyocyte number is a result of a reduction in the number of cardiac precursor cells in early embryos. Although we have not detected any significant increases in apoptosis in the morphants (data not shown, 2007), there is a possibility that aberrant apoptosis may have caused the reduction in cell number. Apoptosis is a normal process of developing organs, which is also required for normal outflow tract development in chicken (Schaefer et al., 2004). The developing heart does not show a high number of apoptotic cells in the myocardium (data not shown, 2007), but apoptotic cells are detected in certain regions such as the apex of the heart, endocardial cushion area, and outflow tract during chicken or mouse development (Schaefer et al., 2004; Wikenheiser et al., 2006).
It is intriguing that total NADH amount in the morphant heart was decreased in live zebrafish embryos (Fig. 4F). There is a possibility that Lrrc10 directly regulates the metabolic pathways that produce NADH. It has been demonstrated that derangements in energy metabolism cause heart failure (Huss and Kelly, 2005). Alternatively, energy metabolism may have been altered as a consequence of cardiac malfunction/failure caused by Lrrc10 knockdown. NADH provides the electron transport chain with electrons necessary to build the proton motive force, ultimately regenerating ATP from ADP and Pi (Brandes and Bers, 1996). The amount of ATP has been reported to be decreased in failing hearts because of decreased energy reserve (Nascimben et al., 2004). In fact, heart failure of any cause is associated with a decline in the activity of the mitochondrial pathway (Huss and Kelly, 2005). We have recently demonstrated that LRRC10 in the cytoplasm is not localized in the mitochondria or at the sarcomeric Z-line, but is localized at a diad region where the sarcoplasmic reticulum is associated with the transverse tubule (Kim et al., in press). Therefore, zLrrc10 may not directly regulate the mitochondrial structure/function to regulate ATP production or be involved in sarcomeric organization. It is likely that Lrrc10 knockdown directly or indirectly caused reduced NADH amount and, therefore energy reserve in the form of ATP. The fluorescence intensity is mostly from NADH, but may include FAD (Bird et al., 2004; Bird et al., 2005), which is also a source of ATP generation. Although FAD can be detected with NADH in our measurement, the NADH amount produced by the glucose metabolism is five times higher than FAD that is mainly produced by fatty acid metabolism (Berg et al., 2002). Moreover, glucose is a preferred energy source in the embryonic heart (Huss and Kelly, 2005; Scheuer, 2004). In addition, spectral characterization of NADH and FAD at 780 nm using a spectrofluorometer (Fluorolog, Horiba) indicates that the primary signal is NADH (personal communication with Long Yan).
Altogether we have discovered that Lrrc10 is essential for normal cardiac development and cardiac function in the zebrafish embryo. Because early cardiac morphogenic defects accompanied by a pericardial edema were identified in the zlrrc10 morphants, we now know that a previously uncharacterized factor in zebrafish plays critical roles in heart development and function and could potentially be involved in human congenital heart defects and/or certain types of adult heart disease. Therefore, this study will provide the critical insights into the complex mechanisms of normal cardiac development, which will lead us to determine the molecular basis of congenital heart defects, and adult cardiac disease.
Supplementary Material
Appendix A. Supplementary data
Supplementary data associated with this article can be found,in the online version, at doi:10.1016/j.ydbio.2007.06.005.
Acknowledgments
We thank very much Drs. Junichi Sadoshima, Jeffery W. Walker, and Timothy J. Kamp for valuable discussions and Dr. Kyungmann Kim and Jihoon Kim for statistical analyses. We also thank Dorothy Nesbit for zebrafish maintenance and Matthew R. Mysliwiec for helpful comments on the manuscript. Supported by the National Institutes of Health (NIH) grant HL67050 (Y.L.), NIH grant EB000184 (K.E.), ES012716 from the National Institute of Environmental Health Sciences (NIEHS) (W.H. and R.E.P.) and NIH grant T32 ES07015 from the NIEHS (D.S.A.).
References
- Adameyko II, Mudry RE, Houston-Cummings NR, Veselov AP, Gregorio CC, Tevosian SG. Expression and regulation of mouse SERDIN1, a highly conserved cardiac-specific leucine-rich repeat protein. Dev Dyn. 2005;233:540–552. doi: 10.1002/dvdy.20368. [DOI] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Antkiewicz DS, Burns CG, Carney SA, Peterson RE, Heideman W. Heart malformation is an early response to TCDD in embryonic zebrafish. Toxicol Sci. 2005;84:368–377. doi: 10.1093/toxsci/kfi073. [DOI] [PubMed] [Google Scholar]
- Antkiewicz DS, Peterson RE, Heideman W. Blocking expression of AHR2 and ARNT1 in zebrafish larvae protects against cardiac toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2006;94:175–182. doi: 10.1093/toxsci/kfl093. [DOI] [PubMed] [Google Scholar]
- Antos CL, McKinsey TA, Frey N, Kutschke W, McAnally J, Shelton JM, Richardson JA, Hill JA, Olson EN. Activated glycogen synthase-3 beta suppresses cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2002;99:907–912. doi: 10.1073/pnas.231619298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman T, Walsh EC, Wen KK, McKane M, Ren J, Alexander J, Rubenstein PA, Stainier DY. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2004;2:E129. doi: 10.1371/journal.pbio.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, Just S, Trano N, Katus HA, Fishman MC, Rottbauer W. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 2006;20:2361–2372. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. Fifth. W.H. Freeman and Co; New York: 2002. 18.6. The Regulation of Cellular Respiration is Governed Primarily by the Need for ATP. [Google Scholar]
- Bird DK, Eliceiri KW, Fan CH, White JG. Simultaneous two-photon spectral and lifetime fluorescence microscopy. Appl Opt. 2004;43:5173–5182. doi: 10.1364/ao.43.005173. [DOI] [PubMed] [Google Scholar]
- Bird DK, Yan L, Vrotsos KM, Eliceiri KW, Vaughan EM, Keely PJ, White JG, Ramanujam N. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- Brandes R, Bers DM. Increased work in cardiac trabeculae causes decreased mitochondrial NADH fluorescence followed by slow recovery. Biophys J. 1996;71:1024–1035. doi: 10.1016/S0006-3495(96)79303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt–Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Buchanan SG, Gay NJ. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- Bujak M, Frangogiannis NG. The role of TGF-beta signaling in myocardial infarction and cardiac remodeling. Cardiovasc Res. 1996;74:184–195. doi: 10.1016/j.cardiores.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney SA, Chen J, Burns CG, Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol Pharmacol. 2006;70:549–561. doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- Chance B, Cohen P, Jobsis F, Schoener B. Intracellular oxidation–reduction states in vivo. Science. 1962;137:499–508. doi: 10.1126/science.137.3529.499. [DOI] [PubMed] [Google Scholar]
- Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Nusslein-Volhard C. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Chien KR. Genomic circuits and the integrative biology of cardiac diseases. Nature. 2000;407:227–232. doi: 10.1038/35025196. [DOI] [PubMed] [Google Scholar]
- Coucelo J, Joaquim N, Coucelo J. Calculation of volumes and systolic indices of heart ventricle from Halobatrachus didactylus: echocardiographic noninvasive method. J Exp Zool. 2000;286:585–595. doi: 10.1002/(sici)1097-010x(20000501)286:6<585::aid-jez5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Fishman MC, Chien KR. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- Grimes AC, Stadt HA, Shepherd IT, Kirby ML. Solving an enigma: arterial pole development in the zebrafish heart. Dev Biol. 2006;290:265–276. doi: 10.1016/j.ydbio.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Gruber PJ, Epstein JA. Development gone awry: congenital heart disease. Circ Res. 2004;94:273–283. doi: 10.1161/01.RES.0000116144.43797.3B. [DOI] [PubMed] [Google Scholar]
- Heideman W, Antkiewicz DS, Carney SA, Peterson RE. Zebrafish and cardiac toxicology. Cardiovasc Toxicol. 2005;5:203–214. doi: 10.1385/ct:5:2:203. [DOI] [PubMed] [Google Scholar]
- Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421:172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000;260:148–157. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Hu N, Yost HJ, Clark EB. Cardiac morphology and blood pressure in the adult zebrafish. Anat Rec. 2001;264:1–12. doi: 10.1002/ar.1111. [DOI] [PubMed] [Google Scholar]
- Huang C, Sheikh F, Hollander M, Cai C, Becker D, Chu PH, Evans S, Chen J. Embryonic atrial function is essential for mouse embryogenesis, cardiac morphogenesis and angiogenesis. Development. 2003;130:6111–6119. doi: 10.1242/dev.00831. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins GM, Bulkley BH, Moore GW, Piasio MA, Lohr FT. Shape of the human cardiac ventricles. Am J Cardiol. 1978;41:646–654. doi: 10.1016/0002-9149(78)90812-3. [DOI] [PubMed] [Google Scholar]
- Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn. 2005;232:21–32. doi: 10.1002/dvdy.20204. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim T, Micales BK, Lyons GE, Lee Y. Dynamic expression patterns of Leucine-rich Repeat Containing Protein 10 in the heart. Dev Dyn. doi: 10.1002/dvdy.21225. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273:106–120. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Nakane T, Satoh T, Inada Y, Nakayama J, Itoh F, Chiba S. Molecular cloning and expression of HRLRRP, a novel heart-restricted leucine-rich repeat protein. Biochem Biophys Res Commun. 2004;314:1086–1092. doi: 10.1016/j.bbrc.2003.12.202. [DOI] [PubMed] [Google Scholar]
- Nascimben L, Ingwall JS, Lorell BH, Pinz I, Schultz V, Tornheim K, Tian R. Mechanisms for increased glycolysis in the hypertrophied rat heart. Hypertension. 2004;44:662–667. doi: 10.1161/01.HYP.0000144292.69599.0c. [DOI] [PubMed] [Google Scholar]
- Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;10:467–474. doi: 10.1038/nm0504-467. [DOI] [PubMed] [Google Scholar]
- Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- Pappajohn DJ, Penneys R, Chance B. NADH spectrofluorometry of rat skin. J Appl Physiol. 1972;33:684–687. doi: 10.1152/jappl.1972.33.5.684. [DOI] [PubMed] [Google Scholar]
- Passier R, Richardson JA, Olson EN. Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech Dev. 2000;92:277–284. doi: 10.1016/s0925-4773(99)00330-5. [DOI] [PubMed] [Google Scholar]
- Pelster B, Burggren WW. Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio) Circ Res. 1996;79:358–362. doi: 10.1161/01.res.79.2.358. [DOI] [PubMed] [Google Scholar]
- Rockman HA, Ono S, Ross RS, Jones LR, Karimi M, Bhargava V, Ross J, Jr, Chien KR. Molecular and physiological alterations in murine ventricular dysfunction. Proc Natl Acad Sci U S A. 1994a;91:2694–2698. doi: 10.1073/pnas.91.7.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman HA, Wachhorst SP, Mao L, Ross J., Jr ANG II receptor blockade prevents ventricular hypertrophy and ANF gene expression with pressure overload in mice. Am J Physiol. 1994b;266:H2468–H2475. doi: 10.1152/ajpheart.1994.266.6.H2468. [DOI] [PubMed] [Google Scholar]
- Rottbauer W, Wessels G, Dahme T, Just S, Trano N, Hassel D, Burns CG, Katus HA, Fishman MC. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ Res. 2006;99:323–331. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- Schaefer KS, Doughman YQ, Fisher SA, Watanabe M. Dynamic patterns of apoptosis in the developing chicken heart. Dev Dyn. 2004;229:489–499. doi: 10.1002/dvdy.10463. [DOI] [PubMed] [Google Scholar]
- Scheuer J. Fueling the hypertrophied heart. Hypertension. 2004;44:623–624. doi: 10.1161/01.HYP.0000144295.29029.a2. [DOI] [PubMed] [Google Scholar]
- Schneider MD, Gaussin V, Lyons KM. Tempting fate: BMP signals for cardiac morphogenesis. Cytokine Growth Factor Rev. 2003;14:1–4. doi: 10.1016/s1359-6101(02)00053-9. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Sedmera D, Pexieder T, Hu N, Clark EB. A quantitative study of the ventricular myoarchitecture in the stage 21–29 chick embryo following decreased loading. Eur J Morphol. 1998;36:105–119. doi: 10.1076/ejom.36.2.105.4775. [DOI] [PubMed] [Google Scholar]
- Skala MC, Riching KM, Bird DK, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, Keely PJ, Ramanujam N. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J Biomed Opt. 2007;12:024014. doi: 10.1117/1.2717503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Fishman MC. Patterning the zebrafish heart tube: acquisition of anteroposterior polarity. Dev Biol. 1992;153:91–101. doi: 10.1016/0012-1606(92)90094-w. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Driever W, Fishman MC. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Sumanas S, Larson JD. Morpholino phosphorodiamidate oligonucleotides in zebrafish: a recipe for functional genomics? Brief Funct Genomic Proteomic. 2002;1:239–256. doi: 10.1093/bfgp/1.3.239. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Walton RZ, Bruce AE, Olivey HE, Najib K, Johnson V, Earley JU, Ho RK, Svensson EC. Fog1 is required for cardiac looping in zebrafish. Dev Biol. 2006;289:482–493. doi: 10.1016/j.ydbio.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Weinstein BM, Fishman MC. Cardiovascular morphogenesis in zebrafish. Cardiovasc Res 31 Spec No. 1996:E17–E24. [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. 1995. [Google Scholar]
- Wikenheiser J, Doughman YQ, Fisher SA, Watanabe M. Differential levels of tissue hypoxia in the developing chicken heart. Dev Dyn. 2006;235:115–123. doi: 10.1002/dvdy.20499. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


