Abstract
It is not clear how transcription factors bound at distal enhancer and proximal promoter sequences cooperate to stimulate transcription in vivo. To distinguish between different models for the action of enhancer elements, we have directly measured DNA binding of the Drosophila activator zeste by in vivo UV crosslinking. Experiments in Drosophila embryos show that binding of zeste protein to either the proximal promoter of the Ultrabithorax (Ubx) gene or to a Ubx enhancer element does not require the presence of the other element. However, significant transcription is observed only when both elements are present and bound by zeste. The results indicate that stimulation by an enhancer can occur by a mechanism other than increasing the occupancy of an activator to binding sites near the start site of transcription.
In eukaryotes, transcriptional activation by proteins bound to distant enhancer elements frequently requires transcription factors bound just upstream of the TATA box (1–4). These proteins stimulate transcription in a synergistic manner, since the activation by enhancer and promoter factors working in combination is much greater than the additive activities of the individual transcription factors. In vitro, some activator proteins bound at enhancer and proximal promoter elements have been shown to directly contact each other, looping out the intervening DNA, and increasing the binding of one or more of the interacting proteins to DNA (3, 5–7). Therefore, in principle, an enhancer might activate transcription solely by cooperatively increasing the binding of proteins to DNA sequences just upstream of the TATA box, so that these proximal promoter factors could then directly stimulate transcription (8). However, further in vitro studies have demonstrated that synergistic interactions between transcription factors bound to distantly separated sites are observed even when DNA binding sites for all of the activators are saturated (9). From these experiments, it has been argued that the mutual requirement for enhancer and proximal promoter factors may be due to the fact that significant activation requires simultaneous interactions between both sets of proteins and the general transcription factors, such as TFIID (9–13).
Therefore, an important question is, how do endogenous eukaryotic transcription factors behave in the organism? Are they present at sufficient concentrations to bind enhancer and promoter proximal regions independently, or is cooperative binding between the distant elements required to target the proteins to DNA? Here, we have experimentally addressed these questions by measuring the DNA binding of the endogenous zeste protein to a series of transgenic Ubx promoter constructs with an in vivo UV crosslinking technique (14–17).
MATERIALS AND METHODS
Analysis of Expression Patterns.
Expression of β-galactosidase from the Ubx promoter transgenes (18, 19) was examined as described (18). The transgenic embryos shown are 6 hr old and were stained for β-galactosidase activity in parallel for an identical time.
In Vivo UV Crosslinking.
In vivo UV crosslinking, immunoprecipitation of chromatin, and affinity purification of anti-zeste antibodies were performed as described (14–17). The Southern blots used to observe the proximal promoter were probed with a 1-kb StuI/EcoRI fragment from Ubx genomic clone 3102, whereas the Southern blots used to observe the BXD element were probed with a 1.6-kb HindIII/Asp718 fragment from pBS bxd, a subcloned BXD fragment from Ubx genomic clone 3105.
Quantitation of Crosslinking Results.
The crosslinking efficiency was determined by comparing the immunoprecipitation signals with their respective dilution series of total DNA in the immunoprecipitation reaction. These values were then standardized to the signal of the internal control restriction fragment derived from the endogenous Ubx gene. Signals were quantified using a Molecular Dynamics imaging densitometer and imagequant software. Two independent transformant lines for each contruct were examined, except for ssDpry, where a single transformant line was tested. The variation among the different transformants of the same constructs was less than 15%.
RESULTS AND DISCUSSION
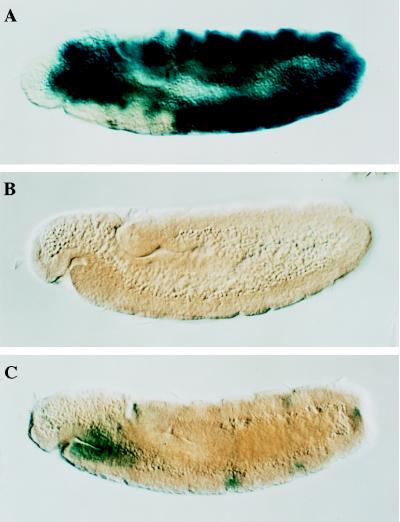
Differently mutated Ubx promoter constructs have previously been introduced into Drosophila by P-element-mediated transformation and their expression patterns analyzed (18, 19). Fig. 1 directly compares the relative levels of transcription from these promoter constructs, demonstrating that significant transcription from Ubx promoter constructs in 6- to 14-hr-old embryos requires the presence of both the BXD enhancer and zeste protein-binding sites at the proximal promoter. Strong transgene expression was observed in the Uβ construct, in which the proximal promoter and BXD enhancer were present (Fig. 1A). However, deletion of either the proximal promoter element (Uβ Δ−200/−31; Fig. 1B) or the BXD enhancer (ssDpry; Fig. 1C) dramatically reduced expression. Zeste protein appears to be a major activator of the Uβ expression pattern, since transcription of this transgene is reduced 30- to 50-fold in zeste mutant embryos (18).
Figure 1.
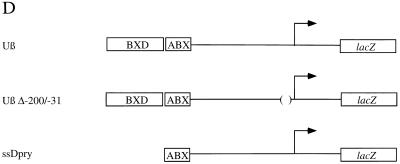
The distant BXD enhancer and a proximal promoter element of Ubx interact synergistically to drive high levels of expression in the Drosophila embryo. (A) Expression of the Uβ promoter construct that contains the BXD and ABX enhancer elements and the intact proximal promoter of Ubx (D). (B) Expression of promoter construct Uβ Δ−200/−31, which is identical to Uβ except that promoter elements between nucleotides −200 and −31 have been deleted (D). (C) Expression of ssDpry promoter construct, which is identical to Uβ, except that it lacks the BXD enhancer (D). (D) Schematic representations of the Ubx promoter constructs are diagrammed. Ubx promoter DNA between nucleotides −3.1 kb and +1 kb is drawn as a horizontal line, and the ABX and BXD distant regulatory elements and the Escherichia coli lacZ gene are boxed. The start site of transcription is denoted by an arrow. The extent of the deletion at the proximal promoter in Uβ Δ−200/−31 is also indicated. This deletion removes the five zeste DNA binding sites in this region.
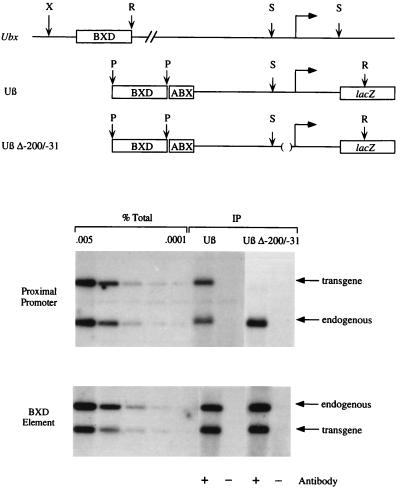
Because zeste protein binds to both the proximal promoter and the distant BXD enhancer (14, 20), we asked whether cooperative binding of zeste molecules to these widely separated sites is responsible for the observed synergistic interactions between the regulatory elements. In vivo UV crosslinking was used to compare the binding of zeste molecules to the BXD enhancer in the Uβ and Uβ Δ−200/−31 constructs. Fig. 2 indicates that the level of zeste protein crosslinking to the BXD enhancer in the Uβ Δ−200/−31 construct is the same as that to the BXD enhancer on the Uβ transgene (see also Table 1). Thus, deletion of the proximal high-affinity binding sites for zeste has no effect on the occupancy of sites in the BXD enhancer. It is not known whether the binding of zeste to the BXD enhancer is required for activation, but this result illustrates that at least one protein can access its binding sites in the distant element even when the enhancer has been functionally inactivated by deletion of the proximal promoter. Finally, as expected, zeste protein crosslinks to the proximal promoter of the Uβ transgene and the endogenous Ubx gene, but not to the proximal promoter of the Uβ Δ−200/−31 construct (Fig. 2).
Figure 2.
In vivo UV crosslinking of zeste protein to the Uβ and Uβ Δ−200/−31 transgenes. (Upper) The endogenous Ubx gene, and the Uβ and Uβ Δ−200/−31 transgenes are diagrammed using the same conventions employed in Fig. 1D. The positions of the relevant XhoI (X), EcoRV (R), StuI (S), and PstI (P) restriction sites are shown. (Lower) Autoradiograms of Southern blots, which show the results of immunoprecipitation experiments with crosslinked chromatin isolated from Uβ or Uβ Δ−200/−31 transformant embryos (18). Two independent transformant lines of each construct were examined with the same result. Chromatin digested with StuI and EcoRV (proximal promoter) or with XhoI, EcoRV, and PstI (BXD element) was immunoprecipitated (IP) with affinity-purified anti-zeste antibody (+ antibody) or with anti-IgG antibody (− antibody). The coprecipitating DNA was then analyzed by Southern blot analysis. To allow quantitation, 0.005%, 0.002%, 0.0005%, 0.0002%, and 0.0001% of the total DNA (% Total) in the immunoprecipitation reaction is also shown. The restriction fragments from the endogenous Ubx gene and the transgenes are indicated. Note that the fragment derived from the proximal promoter of the Uβ Δ−200/−31 transgene is 169 base pairs smaller than the proximal promoter fragment of the Uβ transgene.
Table 1.
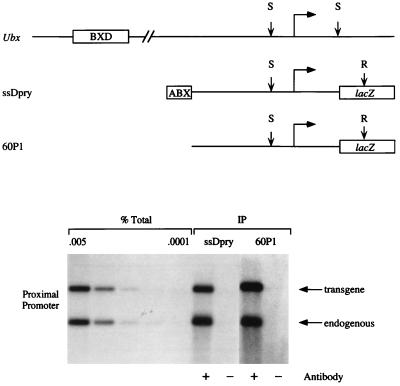
Relative levels of immunoprecipitation of restriction fragments from the various Ubx transgenic constructs. The numbers represent the in vivo crosslinking efficiency of zeste protein to the proximal promoter and BXD element in each transgenic construct, relative to the efficiency of crosslinking to the proximal promoter of the Uβ Δ−200/−31 transgene (see Materials and Methods for quantitation methods). No values for crosslinking to the BXD element in ssDpry or 60P1 transformants are given because the enhancer element is not present in these constructs.
| Regulatory region | Uβ | Uβ Δ−200/−31 | ssDpry | 60P1 |
|---|---|---|---|---|
| Proximal promoter | 20 | 1 | 15 | 25 |
| BXD element | 40 | 30 | — | — |
We next examined whether zeste protein can still bind to the proximal promoter region of two different Ubx promoter constructs that lack either the BXD enhancer (ssDpry) or both the BXD and ABX enhancer (60P1) (Fig. 3). In both cases, zeste crosslinked to the proximal promoter at levels virtually identical to that observed on the Uβ promoter construct (Table 1). Therefore, neither zeste nor any other protein bound to the BXD enhancer acts by increasing the binding of zeste to the Ubx proximal promoter in embryos. Previous in vitro experiments indicate that zeste molecules can form dimers and higher-order oligomers. Furthermore, these molecules appear to form a bridge between widely separated DNA sites and bind distant DNA sites cooperatively (20, 23). Thus, zeste molecules bound to the enhancer and proximal promoter could directly contact each other and loop out the intervening DNA. Because our data indicate that this cannot be acting to increase the binding of zeste to DNA in vivo, the transcriptional synergy between the proximal promoter and enhancer may instead result from simultaneous interactions between the proteins bound to these regulatory elements and the general transcription factor machinery. This supports the conclusions of earlier in vitro experiments with other transcription factors (9–13).
Figure 3.
In vivo UV crosslinking of zeste protein to the ssDpry and 60P1 transgenes. (Upper) Endogenous Ubx gene, and ssDpry and 60P1 transgenes. (Lower) Autoradiograms of Southern blots from in vivo crosslinking and immunoprecipitation experiments with chromatin isolated from ssDpry and 60P1 transformant embryos (21, 22). The conventions are described in the legend to Fig. 2. One ssDpry line and two independent 60P1 lines have been examined.
The results presented here contrast with those obtained in other in vivo studies of two other eukaryotic enhancers. These enhancers appear to function, at least in part, by increasing the binding of other transcription factors to sequences near the start site of transcription (24–26). Instead, our data are more similar to that of a well-characterized prokaryotic enhancer, which acts not by recruiting RNA polymerase to the promoter, but instead by activating an already bound polymerase (27, 28). Given the complexity and diversity of promoter architectures and the differences in the concentrations of various transcription factors in cells, it is not surprising that multiple mechanisms for enhancer action appear to exist in vivo.
Acknowledgments
We thank Mariann Bienz and Jeff Simon for generously providing fly stocks and Trevor Williams and Sankar Ghosh for helpful comments on this manuscript. This work was funded by grants to M.D.B. from the National Institutes of Health and the Pew Charitable Trust.
References
- 1.Treisman R, Maniatis T. Nature (London) 1985;315:72–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Vigneron M, Matthes H, Wildeman A, Zenke M, Chambon P. Nature (London) 1986;319:121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- 3.Li R, Knight J D, Jackson S P, Tjian R, Botchan M R. Cell. 1991;65:493–505. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- 4.Ptashne M. A Genetic Switch. Cambridge, MA: Blackwell Scientific Publications; 1992. [Google Scholar]
- 5.Schleif R. Science. 1988;240:127–128. doi: 10.1126/science.3353710. [DOI] [PubMed] [Google Scholar]
- 6.Knight J D, Li R, Botchan M. Proc Natl Acad Sci USA. 1991;88:3204–3208. doi: 10.1073/pnas.88.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mastrangelo I A, Courey A J, Wall J S, Jackson S P, Hough P V C. Proc Natl Acad Sci USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ptashne M. Nature (London) 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y-S, Carey M, Ptashne M, Green M R. Nature (London) 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 10.Carey M, Lin Y-S, Green M, Ptashne M. Nature (London) 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 11.Chi T, Lieberman P, Ellwood K, Carey M. Nature (London) 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 12.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 13.Chi T, Carey M. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 14.Walter J, Dever C A, Biggin M D. Genes Dev. 1994;8:1678–1692. doi: 10.1101/gad.8.14.1678. [DOI] [PubMed] [Google Scholar]
- 15.Walter J, Biggin M D. Proc Natl Acad Sci USA. 1996;93:2680–2685. doi: 10.1073/pnas.93.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laney J D, Biggin M D. Development. 1996;122:2303–2311. doi: 10.1242/dev.122.7.2303. [DOI] [PubMed] [Google Scholar]
- 17.Gilmour D S, Lis J T. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laney J D, Biggin M D. Genes Dev. 1992;6:1531–1541. doi: 10.1101/gad.6.8.1531. [DOI] [PubMed] [Google Scholar]
- 19.Müller J, Bienz M. EMBO J. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson M, Pirrotta V. EMBO J. 1988;7:3907–3915. doi: 10.1002/j.1460-2075.1988.tb03277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bienz M, Saari G, Tremml G, Müller J, Züst B, Lawrence P A. Cell. 1988;53:567–576. doi: 10.1016/0092-8674(88)90573-9. [DOI] [PubMed] [Google Scholar]
- 22.Simon J, Peifer M, Bender W, O’Connor M. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J D, Pirrotta V. EMBO J. 1993;12:2061–2073. doi: 10.1002/j.1460-2075.1993.tb05855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordingley M G, Riegel A T, Hager G L. Cell. 1987;48:261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein T, Forrester W C, Qiu R-G, Grosschedl R. Genes Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 26.Archer T K, Lefebvre P, Wolford R G, Hager G L. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 27.Sasse-Dwight S, Gralla J D. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]