Abstract
Rationale: Replication of gene-disease associations has become a requirement in complex trait genetics.
Objectives: In studies of childhood asthma from two different ethnic groups, we attempted to replicate associations with five potential asthma susceptibility genes previously identified by positional cloning.
Methods: We analyzed two family-based samples ascertained through an asthmatic proband: 497 European-American children from the Childhood Asthma Management Program and 439 Hispanic children from the Central Valley of Costa Rica. We genotyped 98 linkage disequilibrium–tagging single-nucleotide polymorphisms (SNPs) in five genes: ADAM33, DPP10, GPR154 (HUGO name: NPSR1), HLA-G, and the PHF11 locus (includes genes SETDB2 and RCBTB1). SNPs were tested for association with asthma and two intermediate phenotypes: airway hyperresponsiveness and total serum immunoglobulin E levels.
Measurements and Main Results: Despite differing ancestries, linkage disequilibrium patterns were similar in both cohorts. Of the five evaluated genes, SNP-level replication was found only for GPR154 (NPSR1). In this gene, three SNPs were associated with asthma in both cohorts, although the opposite alleles were associated in either study. Weak evidence for locus-level replication with asthma was found in the PHF11 locus, although there was no overlap in the associated SNP across the two cohorts. No consistent associations were observed for the three other genes.
Conclusions: These results provide some further support for the role of genetic variation in GPR154 (NPSR1) and PHF11 in asthma susceptibility and also highlight the challenges of replicating genetic associations in complex traits such as asthma, even for genes identified by linkage analysis.
Keywords: bronchial hyperreactivity, immunoglobulin E, linkage disequilibrium, NPSR1, single-nucleotide polymorphism
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Asthma candidate genes have been identified by positional cloning, although these results have not been consistently replicated, even in the initial reports.
What This Study Adds to The Field
This study provides further support for the role of GPR154 in asthma susceptibility and the first independent replication for the PHF11 locus.
Replication of the results of a gene–disease association study in independent samples has emerged as a standard for demonstrating the relevance of a candidate gene for a complex trait (1–3). Although there are now many examples of reproducible associations in which a specific genetic variant is consistently associated with a specific phenotype, these are greatly outnumbered by studies in which true replication is claimed, yet the associations observed do not exactly replicate the initial report, as evidenced by differences in the particular variants or phenotypes studied. Replication of the association with the specific polymorphism is the strongest evidence; however, positive association with other variants at the locus may still point to the importance of the particular candidate gene. Phenotypic heterogeneity (e.g., childhood vs. adult asthma) is an issue in many complex diseases and may be especially problematic in diseases such as asthma that lack an explicit diagnostic test (4). Different population origins and linkage disequilibrium (LD) patterns may be an additional cause of nonreplication.
Asthma is the most common obstructive lung disease, affecting 12% of children in the United States (5) and accounting for substantial morbidity in children and adults worldwide. Family studies strongly support an important genetic contribution to asthma susceptibility (6). Genomewide linkage studies have identified no fewer than 20 loci with evidence suggestive of linkage, although few of these linkages have met stringent criteria for significance, even in well-powered samples (7). These data suggest that asthma is a complex polygenic disease, with multiple genes of modest effect interacting with each other and with environmental factors. Starting with the identification of ADAM metallopeptidase domain 33 (ADAM33) on chromosome 20p by Van Eerdewegh and colleagues (8), six asthma susceptibility genes have been identified by positional cloning, including PHD finger protein 11 (PHF11), a locus on chromosome 13 that includes two additional genes: regulator of chromosome condensation (RCC1) and BTB (POZ) domain–containing protein 1 (RCBTB1) and SET domain, bifurcated 2 (SETDB2) (9), dipeptidyl-peptidase 10 (DPP10, on chromosome 2q) (10), neuropeptide S receptor 1 (NPSR1, previously GPR154 or GPRA, on chromosome 7p) (11), HLA-G histocompatibility antigen, class I, G (HLA-G, on chromosome 6p) (12), and cytoplasmic FMR1-interacting protein 2 (CYFIP2, on chromosome 5q) (13).
Because linkage analyses are typically powered to detect risk loci with large effects, disease genes identified through linkage might be expected to exert a larger effect on the phenotype, and therefore be more amenable to replication. For the positionally cloned asthma genes presented in Table 1, replication at the marker level was not always observed, even in the initial reports. In follow-up studies (Table 2), stringent replication at the level of the single-nucleotide polymorphism (SNP) or haplotype described in the first report has not been common. Given the lack of definitive functional data for these genes, it remains unclear which of these findings are valid. We hypothesized that the genetic associations for positionally cloned asthma genes would not all be consistently replicated across additional populations. To test this hypothesis, we examined the first five of the six reported positional asthma genes (ADAM33, PHF11, DPP10, GPR154 [NPSR1], and HLA-G) in two family-based samples ascertained through an asthmatic proband: a cohort of non-Hispanic white North American children from the Childhood Asthma Management Program (CAMP) and a cohort of Hispanic children from the Central Valley of Costa Rica. Results from this study have been previously reported as an abstract (14).
TABLE 1.
ORIGINAL REPORTS OF ASTHMA SUSCEPTIBILITY GENES IDENTIFIED BY POSITIONAL CLONING
| No. of SNPs Typed | Association Results
|
Level of Replication
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene (Ref.) | Population | Sample Size | Design | Phenotype | Single Marker | Haplotype | Independent Population | Phenotype | Marker | |
| ADAM33 (8) | U.S. and U.K. | 130 cases, 217 controls | CC | Asthma | 135 | 6 SNPs | Yes | |||
| 460 families | FBTA | Asthma | 5 | 5 SNPs | Yes | No | N/A | N/A | ||
| PHF11 (9) | Initial population | |||||||||
| Australia | 80 families, 364 subjects | FBTA | IgE | 54 | 24 SNPs | Yes | ||||
| Replication populations | ||||||||||
| Australia | 150 families | FBTA | IgE | 6 | NR | Yes | Yes | Yes | Yes | |
| U.K. | 87 families | FBTA | IgE | 6 | NR | Yes | Yes | Yes | Yes | |
| U.K. | 150 families | FBTA | IgE | 6 | NR | Yes | Yes | Yes | Yes | |
| U.K. | 130, adult severe asthma; 49, child severe asthma; 92, mild asthma; 28, blood donor controls | CC | Asthma | NR | 2 SNPs | NR | Yes | No | Yes | |
| DPP10 (10) | Initial population | |||||||||
| Australia and U.K. | 244 families, 1,112 subjects (239 asthmatic offspring) | FBTA | Asthma | 82 SNPs, 1 STR | 10 SNPs, 1 STR | Yes | ||||
| IgE | 13 SNPs | NR | ||||||||
| Replication populations | ||||||||||
| Germany | 118 cases, 139 controls | CC | Asthma | 2 SNPs, 1 STR | 1 STR | Yes | Yes | Yes | Yes | |
| U.K. | 179, severe asthma; 92, mild asthma; 304 controls | CC | Asthma | 1 SNP, 1 STR | NR | Yes | Yes | No | Yes | |
| GPR154 (11) | Initial population | |||||||||
| Kainuu, Finland | 106 families, 361 subjects | FBTA | IgE | 51 | 43 SNPs | Yes | ||||
| Replication populations | ||||||||||
| Quebec, Canada | 193 trios | FBTA | Asthma | 22 | NR | Yes | Yes | No | No | |
| North Karelia, Finland | 31 trios | FBTA | IgE | 29 | NR | Yes | Yes | Yes | No | |
| HLA-G (12) | Initial populations | |||||||||
| U.S. (Chicago) | 35 families, 138 subjects | FBTA | Asthma | 59 | 9 SNPs | Yes | ||||
| U.S. (Chicago) | 46 trios | FBTA | Asthma | 59 | 2 SNPs | Yes | ||||
| Replication populations | ||||||||||
| U.S. (Hutterite) | 156 cases, 434 controls | CC | AHR | 21 | 3 SNPs | NR | Yes | No | No | |
| Netherlands | 200 families | FBTA | Atopy | 6 | 1 SNP | NR | Yes | No | No | |
| AHR | NS | NR | Yes | No | No | |||||
Definition of abbreviations: AHR = airway hyperresponsiveness; CC = case–control; FBTA = family-based test of association; N/A = not applicable; NR = not reported; NS = not significant; SNP = single-nucleotide polymorphism.
TABLE 2.
PEER-REVIEWED PUBLISHED REPLICATION STUDIES FOR POSITIONALLY CLONED ASTHMA SUSCEPTIBILITY GENES IN THE ENGLISH LANGUAGE LITERATURE
| Study: First Author (Ref.) | No. SNPs Typed | Associations with Asthma*
|
Additional Phenotypes | Association Results | Level of Replication
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | Sample Size | Design | Single Marker | Haplotype | Phenotype | Marker | ||||||||||||||
| ADAM33 | ||||||||||||||||||||
| Lind (47) | Puerto Rican | 318 trios | FBTA | 6 | NS | NS | IgE | NS | No | No | ||||||||||
| Mexican | 265 trios | FBTA | 6 | NS | NS | IgE | NS | No | No | |||||||||||
| Howard (48) | Dutch | 153 cases, 124 controls | CC | 8 | ST+7, V4 | Yes | IgE | NS | Yes | Yes | ||||||||||
| African American | 160 cases, 256 controls | CC | 8 | S2 | Yes | IgE | NS | Yes | No | |||||||||||
| US white | 219 cases, 225 controls | CC | 8 | ST+7, T1, T2 | Yes | IgE | NS | Yes | Yes | |||||||||||
| US Hispanic | 112 cases, 126 controls | CC | 8 | S2, T2 | NS | IgE | NS | Yes | No | |||||||||||
| Werner (30) | German white | 171 families, 732 subjects | FBTA | 15 | F+1, ST+4, ST+5 | Yes | IgE | F+1, ST+5 | Yes | Yes | ||||||||||
| AHR | F+1, S2 | |||||||||||||||||||
| German white | 48 cases, 499 controls | CC | 15 | ST+7 | Yes | IgE | ST+7 | Yes | Yes | |||||||||||
| AHR | ST+5 | |||||||||||||||||||
| Lee (31) | Korean | 326 cases, 151 controls | CC | 5 | NS | NS | AHR | T1 | No | No | ||||||||||
| Raby (22) | US white | 474 families | FBTA | 17 | NS | NS | IgE, AHR | NS | No | No | ||||||||||
| African American | 66 families | FBTA | 17 | NS | NS | IgE, AHR | NS | No | No | |||||||||||
| US Hispanic | 47 families | FBTA | 17 | T1, T+1 | Yes | IgE | T1, T+1 | Yes | No | |||||||||||
| AHR | NS | |||||||||||||||||||
| Blakey (49) | Iceland | 348 cases, 262 controls | CC | 13 | NS | NR | No | No | ||||||||||||
| UK | 60 families, 240 subjects | FBTA | 13 | NS | NR | No | No | |||||||||||||
| Simpson (50) | UK | 470 children | Cohort | 17 | NS | NS | FEV1 | F+1, M+1, T1, T2 | No | No | ||||||||||
| Kedda (51) | Australian white | 612 cases, 473 controls | CC | 10 | NS | Yes | No | No | ||||||||||||
| Schedel (52) | German white | 624 cases, 1,248 controls | CC | 10 | NS | Yes | AHR | NS | No | No | ||||||||||
| German white | 824 children | Cohort | 10 | NS | NS | AHR | NS | No | No | |||||||||||
| Hirota (53) | Japanese | 504 cases, 651 controls | CC | 14 | S2, T1, T2, V-3 | Yes | IgE | NS | Yes | No | ||||||||||
| Wang (54) | Chinese | 296 cases, 270 controls | CC | 3 | NS | NS | No | No | ||||||||||||
| Noguchi (55) | Japanese | 155 families, 538 subjects | FBTA | 23 | S+1, ST+4, T2 | NS | IgE | NS | Yes | Yes | ||||||||||
| GPR154 | ||||||||||||||||||||
| Shin (56) | Korean | 439 cases, 374 controls | CC | 1 | NS | — | IgE, AHR | NS | No | No | ||||||||||
| Melen (57) | Western European | 265 cases, 2,848 controls | CC† | 7 | NR | H5 | Allergic sensitization | H5, H6 | Yes | Yes | ||||||||||
| Swedish | 176 cases, 624 controls | CC | 7 | NR | NS | Allergic sensitization | NS | No | No | |||||||||||
| Kormann (32) | German white | 351 cases, 1,769 controls | CC | 6 | 546333, 585883 | H4 | Yes | Yes | ||||||||||||
| AHR | NS | |||||||||||||||||||
| German white | 112 cases, 1,737 controls | CC | 6 | IgE | 546333, 585883 | |||||||||||||||
| Feng (33) | Chinese | 451 cases, 232 controls | CC | 7 | NR | NS | AHR | rs324981, rs324987 | No | No | ||||||||||
| Chinese | 264 cases, 241 controls | CC | 1 | AHR | NS | |||||||||||||||
| Malerba (58) | Italian | 211 families, 927 subjects | FBTA | 7 | NS | H5, H6 | IgE | 546333, 585883, H4 | Yes | Yes | ||||||||||
Included here are all published studies in the English literature that test for genetic association with asthma or a related phenotype and include at least one population with a minimum of 100 cases and 100 controls or 100 families. Studies that do not include reports of association testing with asthma are not included. An association is claimed if reported with a p value less than 0.05 in the primary report. Marker replication is claimed only if the marker associated in the replication study is that reported in the initial study. Combination phenotypes (i.e., asthma + AHR) are not considered. For ADAM33, the six markers associated with asthma in the combined population in reference (8) were Q-1, S1, ST+4, ST+7, V-1, and V4. For GPR154, the primary replication is for haplotypes H2, H4, H5, and H7, and for SNP522363, which differentiates these risk haplotypes from three nonrisk haplotypes (H1, H3, and H6).
SNPs associated with asthma are listed.
Population-based case–control study.
METHODS
See the online supplement for detailed methods.
CAMP Genetics Ancillary Study
CAMP is a multicenter North American clinical trial designed to investigate the long-term effects of inhaled antiinflammatory medications in children with mild to moderate asthma (15, 16). This analysis includes nuclear families of 497 non-Hispanic white children. A diagnosis of asthma was based on methacholine hyperresponsiveness (provocative concentration of methacholine causing a 20% fall in FEV1 [PC20] ⩽ 12.5 mg/ml) and one or more of the following criteria for at least 6 months in the year before recruitment: (1) asthma symptoms at least two times per week, (2) at least two uses per week of an inhaled bronchodilator, and (3) use of daily asthma medication (15). The Institutional Review Board of the Brigham and Women's Hospital (Boston, MA), as well as those of the other CAMP study centers, approved this study. Informed assent and consent were obtained from the study participants and their parents to collect DNA for genetic studies.
Costa Rica Study
Schoolchildren aged 6–14 years with asthma were recruited through 95 schools in the Central Valley of Costa Rica; subject enrollment and phenotyping protocols have been previously described (17). The Central Valley is a relatively genetically isolated population (18), with extensive genealogical records that can be used to trace ancestry back to approximately 4,000 founding individuals in the 1697 census (19). Inclusion criteria included physician-diagnosed asthma, at least two respiratory symptoms (cough, wheeze, or dyspnea) or asthma attacks in the previous year, and a high probability of having at least six great-grandparents born in the Central Valley of Costa Rica. Parents provided written informed consent for themselves and for their children, who also gave written assent. The study was approved by the Institutional Review Boards of Brigham and Women's Hospital and the Hospital Nacional de Niños (San José, Costa Rica).
SNP Selection and Genotyping
Using CEPH (Centre d'Etude du Polymorphisme Humain) genotype data from the International HapMap project (Utah residents with northern and western European ancestry) (20), we applied an LD tagging algorithm to capture common variation (r2 > 0.8, minor allele frequency > 0.1) across the genes studied (21). Results for 16 SNPs in ADAM33 have been previously published for CAMP (22). SNPs were genotyped in a highly multiplexed allele-specific hybridization assay with a BeadStation 500G (Illumina, San Diego, CA). Mendelian transmission was tested with PedCheck (23), and inconsistent SNPs and individuals were removed from analysis. See Table E1 in the online supplement for a list of SNPs successfully genotyped in both cohorts.
Statistical Analysis
Pairwise LD was expressed as both D′ and r2, calculated using Haploview (24). To ensure phenotype comparability to the CAMP enrollment criteria, a strict definition of asthma was used in the Costa Rica study, which included methacholine hyperresponsiveness (provocative dose of methacholine causing a 20% fall in FEV1 [PD20] ⩽ 16.81 μmol) or bronchodilator responsiveness plus the recruitment criteria above. In addition to asthma, two intermediate phenotypes were analyzed: (1) airway hyperresponsiveness (AHR), measured as log10-transformed dose–response slope to methacholine (25); and (2) log10-transformed total serum immunoglobulin E levels (26). Haplotype blocks were defined using the Gabriel algorithm (27), and haplotype-tagging SNPs were identified in Haploview. Family-based association testing and power calculations were performed with PBAT software (28). Additional statistical analyses were performed in SAS version 9.1 (SAS Institute, Cary, NC) and in R. Because these genes have all been associated with asthma phenotypes in prior studies, we used P < 0.05 in both samples to define statistical significance in the setting of multiple tests, instead of an adjusted P value (29).
RESULTS
Study Participants
Characteristics of children with asthma from Costa Rica and non-Hispanic white CAMP participants are shown in Table 3. Participant ages were similar in both studies, which had a predominance of boys consistent with the demographics of childhood asthma. The majority of children in both cohorts were atopic and the distributions of total serum IgE levels were similar. AHR, as measured by methacholine challenge, was also similar, with the vast majority of children in both studies demonstrating a high degree of responsiveness. However, in CAMP PC20 distributions were right censored at 12.5 mg/ml as a result of enrollment criteria.
TABLE 3.
CHARACTERISTICS OF CHILDREN WITH ASTHMA IN COSTA RICA STUDY AND IN CHILDHOOD ASTHMA MANAGEMENT PROGRAM STUDY
| Characteristic | Costa Rica | CAMP |
|---|---|---|
| Number of children | 439 | 497 |
| Age, yr, mean (range) | 9.1 (6.0–14.2) | 8.8 (5.2–13.2) |
| Sex, no. (%) | ||
| Female | 163 (37.1) | 190 (38.2) |
| Male | 276 (62.9) | 307 (61.8) |
| Methacholine PD20 (Costa Rica; μmol) or PC20 (CAMP; mg/ml), median (interquartile range)* | 1.34 (0.71–3.58) | 1.06 (0.48–2.78) |
| Total serum IgE level (IU/ml), median (interquartile range)† | 413 (117–967) | 399 (157–1,068) |
| At least one positive skin test, no. (%) | 375 (85.8) | 429 (86.3) |
| Atopic, no. (%)‡ | 396 (90.2) | 453 (91.2) |
Definition of abbreviations: CAMP = Childhood Asthma Management Program; PC20 = provocative concentration of methacholine causing a 20% fall in FEV1; PD20 = provocative dose of methacholine causing a 20% fall in FEV1.
Costa Rica, n = 350; CAMP, n = 495.
CAMP, n = 490.
Atopic is defined as having at least one positive skin-prick test or a total serum IgE level of at least 100 IU/ml.
Genotyping Quality and Linkage Disequilibrium
Ninety-eight SNPs mapping to the five positionally cloned asthma candidates were genotyped in both study populations (see Table E1). Genotype distributions for all SNPs in both cohorts were consistent with Hardy-Weinberg equilibrium (at a threshold P < 0.01), and parent–child genotype inconsistencies were rare: five in Costa Rica and three instances in the new CAMP data (quality control for ADAM33 SNPs has been previously reported [22]). Genotyping completion rates averaged 99.4% in Costa Rica and 97.5% in CAMP.
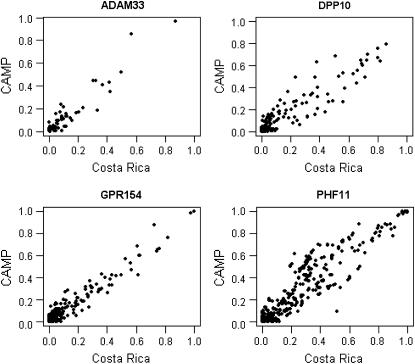
The linear correlations in the plots of pairwise r2 values (four of five genes studied are presented in Figure 1) in the CAMP and Costa Rica cohorts for the candidate gene SNPs suggest that despite the known differences in ancestry between our two populations, regional LD at these five loci is similar. In general, there was strong similarity in LD patterns between the two populations, particularly at ADAM33 and GPR154 (NPSR1). r2 Variability was slightly greater in DPP10 and PHF11, particularly for SNP pairs with intermediate degrees of LD (r2 = 0.3–0.7). Only two SNPs were genotyped in HLA-G, so the r2 plot is not presented; for the two HLA-G SNPs, r2 = 0.31 in CAMP and r2 = 0.41 in Costa Rica. Only a small proportion of pairwise comparisons demonstrated r2 ⩾ 0.8, because most SNPs were selected to tag LD at this threshold.
Figure 1.
Correlation of pairwise r2 values between all pairs of single-nucleotide polymorphisms (SNPs) in four of five studied asthma genes in the Costa Rica and Childhood Asthma Management Program (CAMP) studies. Because only two SNPs were genotyped, data for HLA-G are not shown.
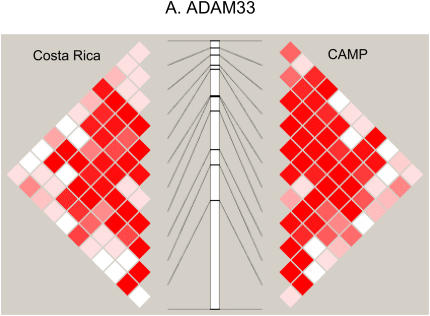
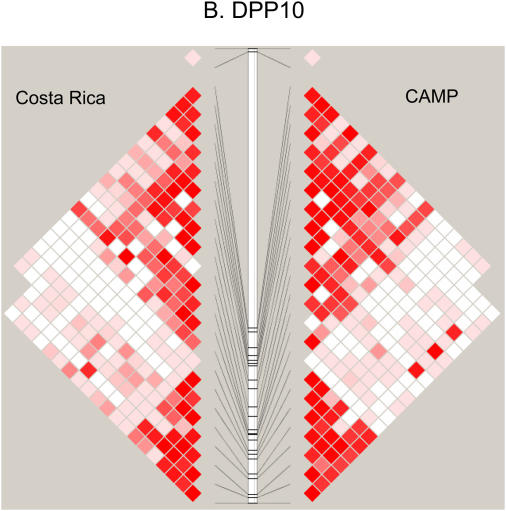
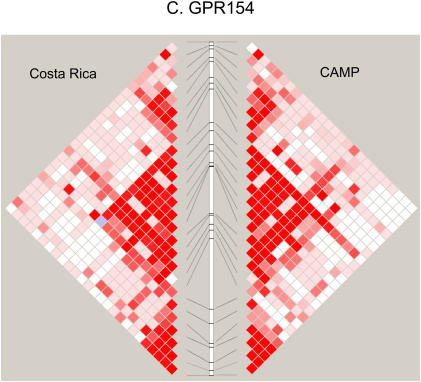
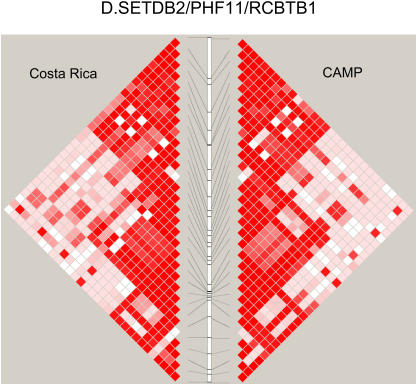
Figure 2 compares the LD patterns in the populations by plotting pairwise D′ in physical order. In the regions encompassing DPP10, GPR154 (NPSR1), PHF11, and HLA-G (data not shown), side-by-side comparison of the LD plots demonstrates that the LD patterns appear strikingly similar, suggesting shared ancestral mutation and recombination events at these loci in these two populations. Interestingly, at the ADAM33 locus, LD extends further in CAMP than in the relatively isolated population from Costa Rica. In general, the LD and haplotype patterns (data not shown) observed are similar at all five loci.
Figure 2.
(A–D) Comparison of linkage disequilibrium (LD) patterns in four of five studied asthma genes across the Costa Rica and Childhood Asthma Management Program (CAMP) studies. LD is measured as D′, with darker red colors indicating higher values. Because only two SNPs were genotyped, data for HLA-G are not shown.
Family-based Association Analysis
Family-based tests of association for asthma with P ⩽ 0.05 are presented in Table 4. Of the five candidate genes evaluated, GPR154 (NPSR1) was the only one that demonstrated association with asthma in both populations, with three associated SNPs. The most significant associations were observed with SNP rs1379928 (P = 0.003 and 0.0006 in Costa Rica and CAMP, respectively), which is located 5′ to the risk haplotype described in the initial report (11). However, the direction of these SNP effects are opposite in the two samples, with all three SNPs showing undertransmission of the minor alleles in Costa Rica and overtransmission in CAMP. Of the four other candidate genes tested, only PHF11 demonstrated evidence of association with asthma, with different polymorphisms in each cohort demonstrating allelic transmission distortion (P values, 0.03–0.05). SNP rs9316454 in PHF11 showed a trend toward overtransmission of the minor allele in both cohorts. However, the strength of this association was weak and not statistically significant in either cohort separately. No associations with asthma were observed in either cohort for variants in ADAM33, DPP10, or HLA-G.
TABLE 4.
GENETIC ASSOCIATIONS WITH ASTHMA IN COSTA RICA AND CAMP STUDIES
| Costa Rica
|
CAMP
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | MAF | Families* | P Value† | T:U Ratio‡ | MAF | Families* | P Value† | T:U Ratio‡ |
| GPR154 | rs2609234 | 0.12 | 160 | 0.03 | 0.71 | 0.18 | 202 | 0.001 | 1.50 |
| GPR154 | rs1006392 | 0.33 | 235 | 0.03 | 1.32 | 0.47 | 342 | 0.6 | 1.06 |
| GPR154 | rs714588 | 0.45 | 284 | 0.03 | 0.80 | 0.44 | 343 | 0.01 | 1.25 |
| GPR154 | rs1379928 | 0.14 | 176 | 0.003 | 0.62 | 0.20 | 211 | 0.0006 | 1.52 |
| GPR154 | rs963218 | 0.48 | 278 | 0.04 | 0.77 | 0.44 | 339 | 0.1 | 1.14 |
| GPR154 | rs2609215 | 0.09 | 105 | 0.5 | 0.83 | 0.10 | 128 | 0.01 | 1.48 |
| GPR154 | rs323917 | 0.04 | 56 | 0.8 | 0.92 | 0.07 | 86 | 0.02 | 1.61 |
| PHF11 | rs9316454 | 0.46 | 254 | 0.05 | 1.23 | 0.46 | 336 | 0.07 | 1.19 |
| PHF11 | rs7332573 | 0.07 | 84 | 0.4 | 1.21 | 0.07 | 109 | 0.04 | 0.68 |
| PHF11 | rs3829366 | 0.43 | 258 | 0.2 | 0.84 | 0.47 | 342 | 0.05 | 0.84 |
| PHF11 | rs3186013 | 0.43 | 259 | 0.2 | 0.83 | 0.47 | 338 | 0.03 | 0.82 |
| PHF11 | rs11148151 | 0.24 | 211 | 0.7 | 1.06 | 0.12 | 190 | 0.04 | 0.76 |
| PHF11 | rs7981396 | 0.24 | 213 | 0.7 | 1.07 | 0.12 | 191 | 0.03 | 0.75 |
Definition of abbreviation: MAF = minor allele frequency.
SNPs with P ⩽ 0.05 in either cohort are listed. Boldface signifies SNPs with stringent replication in both populations. No SNPs in ADAM33, DPP10, or HLA-G (see Table E1 for list) were significant in either cohort.
Number of informative families.
P value from family-based association test.
T:U ratio, transmitted:untransmitted ratio from transmission disequilibrium test.
In previous reports, associations with AHR have been observed with SNPs in ADAM33 (30, 31), GPR154 (NPSR1) (30–33), and HLA-G (12). In our cohorts, associations with AHR were observed with SNPs in DPP10, GPR154 (NPSR1), and PHF11 (Table 5), although none of these SNP associations overlap in both cohorts. Using PD20 (Costa Rica) or PC20 (CAMP) to measure AHR, the associations with GPR154 rs323917 were strengthened, with P < 0.05 in each cohort (Costa Rica, P = 0.006; CAMP, P = 0.03); the minor allele led to increased AHR in each study. Significant linkages and subsequent associations with total serum IgE levels were the initial findings that resulted in the identification of DPP10, GPR154 (NPSR1), and PHF11 as asthma susceptibility genes, and associations with IgE have also been reported with ADAM33. However, in our cohort there was evidence supporting an association with total serum IgE levels for only one SNP in GPR154 (NPSR1) in the Costa Rica cohort, and one SNP in PHF11 in CAMP (see Table E2).
TABLE 5.
GENETIC ASSOCIATIONS WITH AIRWAY HYPERRESPONSIVENESS IN COSTA RICA AND CAMP STUDIES
| Costa Rica
|
CAMP
|
||||||
|---|---|---|---|---|---|---|---|
| Gene | SNP | MAF | Families* | P Value | MAF | Families* | P Value |
| DPP10 | rs843385 | 0.05 | 66 | 0.04 | 0.09 | 130 | 0.9 |
| DPP10 | rs7576583 | 0.06 | 71 | 0.7 | 0.10 | 132 | 0.04 |
| DPP10 | rs272071 | 0.23 | 207 | 0.4 | 0.28 | 302 | 0.05 |
| GPR154 | rs323917 | 0.04 | 55 | 0.02 | 0.07 | 86 | 0.06 |
| GPR154 | rs10278663 | 0.14 | 144 | 0.007 | 0.16 | 220 | 0.2 |
| PHF11 | rs2057413 | 0.20 | 200 | 0.4 | 0.30 | 304 | 0.03 |
| PHF11 | rs2031532 | 0.23 | 215 | 1.0 | 0.35 | 326 | 0.05 |
| PHF11 | rs2274278 | 0.15 | 173 | 0.09 | 0.17 | 221 | 0.04 |
AHR was measured as the log-transformed dose–response slope to methacholine. SNPs with p ⩽ 0.05 in either cohort are listed. No SNPs in ADAM33 or HLA-G (see Table E1 for list) were significant in either cohort.
Number of informative families.
Family-based analysis of haplotypes within blocks did not show any consistent association with asthma, AHR, or IgE levels across the two cohorts, nor did analysis of SNPs in GPR154 (NPSR1) defining the risk haplotypes in the initial report (data not shown) (11).
DISCUSSION
The publication of the first genomewide linkage analysis to asthma-related traits generated interest in the potential of positional cloning to identify genetic variants that underlie asthma pathogenesis (34). Using this approach, six asthma candidate genes have been identified, further intensifying hopes that such findings would translate to clinical applications. However, follow-up studies (Tables 1 and 2) for several of these genes have yielded inconsistent results, dampening enthusiasm for the initial findings. Multiple reasons have been proposed to explain nonreplication in genetic association studies, including small effect sizes and subsequent lack of power; differences in population ascertainment, phenotype definitions, and trait distributions; differences in LD patterns; and population stratification (35, 36). In the present study, we set out to reproduce associations for five of these genes in two family-based cohorts by eliminating many of these potential problems in replication analyses. In both studies, enrollment criteria, phenotyping protocols, and phenotype definitions were similar. As a consequence, the age, sex, and trait distributions were comparable. The family-based design eliminated the potential effects of population stratification. Despite these strategies, replication results were largely negative, with the exception of GPR154 (NPSR1), where three SNPs replicated across both cohorts. Although the effects of these three SNPs on asthma phenotypes were mostly opposite across the two samples, SNP rs323917 was consistently associated with increased AHR. Lin and coworkers have demonstrated that “flip–flop” associations may be due to different LD patterns between populations or failure to consider gene–gene and/or gene–environment interactions (37).
Gene-level replication was noted for the PHF11 locus, yet the associated SNPs were different in the two samples. Although our results are the first to provide any evidence of independent replication of this locus, the inconsistency in the SNPs associated and the lack of association with serum IgE levels (the quantitative trait that was initially used to map the gene) raise doubts about the significance of the findings. For the remaining three genes, no consistent evidence of association was observed in either cohort. Taken together, these results provide additional support for the relevance of GPR154 (NPSR1) (and perhaps the PHF11 locus) in asthma pathogenesis, but also illustrate the substantial challenges in the replication of genetic associations in asthma, especially among candidate genes identified by linkage analysis and subsequent positional cloning. Genes identified in this manner are likely to be novel and therefore have an unknown relationship to asthma pathobiology. Replication may be easier if the gene is known to be associated with asthma, as these genes may have a known biological function. We have successfully replicated associations with IL-13, a known asthma candidate gene, using the same study design and populations that we have used for the positionally cloned genes in the present study (38).
Inadequate statistical power due to small genetic effects and inadequate sample size is perhaps the most common reason for lack of replication but is an unlikely explanation for our findings. The CAMP and Costa Rican cohorts are each larger than any of those used in the original reports of association; a larger sample size is necessary for a replication study, because the initial effect estimate is often inflated (35). For a disease state such as asthma, with 5% prevalence in the general population, we had 80% power to detect an odds ratio of 1.66 for association with an SNP with 10% minor allele frequency and an odds ratio of 1.50 for an SNP with 20% MAF in the Costa Rica trios; we had power to detect even lower odds ratios in the CAMP study, given the larger number of subjects. For a quantitative trait, there was 80% power to find an association with a 10% MAF SNP that explained 1.2% of the trait variation in Costa Rica. Lower heritability was detectable for more common SNPs and for SNPs in CAMP. We had ample power to detect association in both cohorts for all of these genes, including those for which no association was detected in either cohort.
Among the most important differences between our cohorts is their distinct ancestral histories and resultant genetic architecture. Whereas the children in the CAMP study represent a relatively heterogeneous sample of non-Hispanic whites, those from Costa Rica are descended from the relatively genetically isolated population of mixed Spanish and Amerindian ancestry that populated the Central Valley (39). Although genome-wide comparisons suggest that LD in this population extends over larger distances than in more heterogeneous populations of European ancestry (18), it is likely that regional similarities exist. In fact, LD was largely similar across the five loci studied, although differences were evident, particularly for PHF11 and DPP10. Although GPR154 (NPSR1) had the most similar LD structure across our two samples, the observed “flip–flop” associations may imply specific LD differences with the (untyped) functional locus (see Figure E1 in the online supplement). Regional differences in LD may also explain the lack of replication for the other loci. To date, functional effects have not been demonstrated for the asthma-associated variants in any of the five positional candidate genes. The SNPs evaluated here are unlikely to be causal, but rather in LD with the unidentified functional variants. Replication efforts using these markers should be considered as indirect tests of replication. In this context, even slight differences in LD patterns between populations can affect genotype–phenotype associations, making stringent replication more difficult to achieve. Although several studies have demonstrated portability of LD-tagging SNPs derived from Western European–American genotype data to populations of Spanish (40), Amerindian (41), and U.S. Hispanic origin (42), our data illustrate that this is not always the case, and that the role of genetic heterogeneity should be assessed on a gene-by-gene basis (40–42).
In mapping complex genetic traits in human populations, independent corroboration of findings is among the most important criteria for their validation and acceptance and is now a requirement for publication in many leading journals. In the field of asthma, it has been difficult to meet this rigorous standard, with only the initial report of PHF11 by Zhang and coworkers (9) demonstrating internally consistent replication with respect to the phenotype, marker, and direction of effect in at least one other population. The inherent problems of precise replication for complex traits have been detailed (2, 43, 44). The central assumptions underlying the replication standard are that each cohort is representative of the population from which it is sampled and that all cohorts are largely homogeneous with respect to the collection of genetic and environmental factors that interact, leading to the clinical phenotypes. Although this assumption may hold for monogenic or oligogenic traits with few environmental determinants, this seems unrealistic in complex traits such as asthma, hypertension, and heart disease, in which diverse epistatic and environmental factors play a central role. In these circumstances, only a fraction of risk alleles would be expected to replicate across studies, namely those with high allele frequency whose effects are not heavily influenced by interactions with environmental factors or other genetic loci. Although consistent replication of association provides strong evidence of a gene's importance, the lack of replication does not necessarily render the initial associations invalid. Studies in a single ethnic group may demonstrate the relevance of a candidate gene in that group only, whereas replication in multiple ethnic groups may determine the generalizability of findings across populations in different environments or with different patterns of epistasis.
In our study of five genes and three asthma phenotypes, one must consider the possibility of spurious results due to multiple testing. However, in the statistical genetics literature, there is no clear consensus regarding the optimal methodology to adjust for multiple testing (45). Moreover, most of the available methods are more appropriate for detecting novel associations than for confirming previously reported candidate genes, especially genes identified through positional cloning, which may be expected to have a higher probability of true association (46). Instead of ap-plying a correction factor (e.g., Bonferroni or false discovery rate), we have relied on replication in two samples to protect against spurious results. The use of P < 0.05 to confirm previously iden-tified candidate genes has been applied in a genomewide association study of type 2 diabetes (29). Even with this liberal threshold, we did not find strong evidence of replication. To limit the num-ber of tests, we considered only the three asthma phenotypes most commonly reported in previous studies (Tables 1 and 2) and did not examine gene–gene and gene–environment interactions. Examination of additional phenotypes and consideration of interaction effects are possible avenues for future studies.
In summary, we have evaluated five asthma susceptibility genes identified by positional cloning for evidence of association in two childhood asthma cohorts, and provide evidence of replication for GPR154 (NPSR1), although the associated alleles were mostly opposite across the two study samples. Our data and prior studies suggest that common genetic variation in GPR154 (NPSR1) influences asthma phenotypes in populations of differing ethnicity. Although the functional variants for this gene have not been conclusively identified, further investigations into the precise role of GPR154 (NPSR1) in asthma are warranted. In addition, our work highlights the challenges of replicating genetic associations in complex traits such as asthma, especially for genes identified by positional cloning. Innovative strategies, such as integration of gene expression data and consideration of epistatic interaction in gene–gene networks, are likely to increase the statistical power of genetic association studies and thus may aid in the future identification of novel asthma candidate genes.
Supplementary Material
Acknowledgments
The authors thank the participating families from the Genetic Epidemiology of Asthma in Costa Rica Study and from the CAMP Genetics Ancillary Study for their enthusiastic cooperation. We also acknowledge the CAMP investigators and research team, supported by the NHLBI, for collection of CAMP Genetic Ancillary Study data, and the members of the field team in Costa Rica. The authors also thank Ankur Patel for assistance with genotyping. All work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women's Hospital under appropriate CAMP policies and human subjects protections.
Supported by NIH grants HL66289, HL04370, and HL074193 (Genetic Epidemiology of Asthma in Costa Rica Study) and by NHLBI grant N01-HR-16049 (CAMP Genetics Ancillary Study). Additional support for this research came from grants U01HL065899, P50HL67664, and T32HL07427. C.P.H. and B.A.R. are recipients of Mentored Clinical Scientist Development Awards (K08 HL080242 and HL074193, respectively); C.P.H. is also supported by a grant from the Alpha-1 Foundation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200704-592OC on August 16, 2007
Conflict of Interest Statement: C.P.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.A.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.E.S.-Q. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.J.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.A. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.L.-S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.S.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.J.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.T.W. was a consultant for Pfizer from 2002 to 2003, Schering Plough from 1999 to 2000, Variagenics in 2002, Genome Therapeutics in 2003, and Merck Frost in 2002. S.T.W. received a $500,000 grant for genomic equipment from Roche Pharmaceuticals from 2002 to 2003 and was a consultant for Roche Pharmaceuticals in 2000, but did not receive any financial compensation for this consultancy. He received $10,000 from 2005 to 2006 for serving as an advisor and chair of the advisory board to the TENOR Study for Genentech, and a $900,065 grant from AstraZeneca from 1997 to 2003 for the Asthma Policy Modeling Study. S.T.W. was a coinvestigator on a grant from Boehringer Ingelheim to investigate a COPD natural history model from 2003. J.C.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Freely associating [editorial]. Nat Genet 1999;22:1–2. [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn JN, Altshuler D. Once and again: issues surrounding replication in genetic association studies. J Clin Endocrinol Metab 2002;87:4438–4441. [DOI] [PubMed] [Google Scholar]
- 3.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet 2001;29:306–309. [DOI] [PubMed] [Google Scholar]
- 4.Lemanske RF Jr, Busse WW. Asthma. JAMA 1997;278:1855–1873. [PubMed] [Google Scholar]
- 5.Bloom B, Dey AN. Summary health statistics for US children: National Health Interview Survey, 2004. Vital Health Stat 10 2006;227:1–85. [PubMed] [Google Scholar]
- 6.Los H, Koppelman GH, Postma DS. The importance of genetic influences in asthma. Eur Respir J 1999;14:1210–1227. [DOI] [PubMed] [Google Scholar]
- 7.Howard TM, Celedon JC. Linkage analyses of asthma and its intermediate phenotypes. In: Postma DS, Weiss ST, editors. Genetics of asthma and COPD. New York: Taylor & Francis; 2006. pp. 199–210.
- 8.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 2002;418:426–430. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, Edser P, Bhattacharyya S, Dunham A, Adcock IM, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet 2003;34:181–186. [DOI] [PubMed] [Google Scholar]
- 10.Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, Bhattacharyya S, Tinsley J, Zhang Y, Holt R, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet 2003;35:258–263. [DOI] [PubMed] [Google Scholar]
- 11.Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, Makela S, Rehn M, Pirskanen A, Rautanen A, et al. Characterization of a common susceptibility locus for asthma-related traits. Science 2004;304:300–304. [DOI] [PubMed] [Google Scholar]
- 12.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Kuldanek S, Donfack J, Kogut P, Patel NM, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet 2005;76:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi E, Yokouchi Y, Zhang J, Shibuya K, Shibuya A, Bannai M, Tokunaga K, Doi H, Tamari M, Shimizu M, et al. Positional identification of an asthma susceptibility gene on human chromosome 5q33. Am J Respir Crit Care Med 2005;172:183–188. [DOI] [PubMed] [Google Scholar]
- 14.Hersh CP, Raby BA, Soto-Quiros ME, Avila L, Weiss ST, Celedon JC. Comprehensive replication of positionally cloned asthma susceptibility genes in two distinct populations [abstract]. Am J Respir Crit Care Med 2007;175:A458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- 16.Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med 2000;343:1054–1063. [DOI] [PubMed] [Google Scholar]
- 17.Hunninghake GM, Soto-Quiros ME, Avila L, Ly NP, Liang C, Sylvia JS, Klanderman BJ, Silverman EK, Celedon JC. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol 2007;119:654–661. [DOI] [PubMed] [Google Scholar]
- 18.Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha JA, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet 2006;38:556–560. [DOI] [PubMed] [Google Scholar]
- 19.Melendez C. Conquistadores y pobladores: origenes historico-sociales de los Costarricenses. San Jose, Costa Rica: Editorial Universidad Estatal a Distancia; 1982.
- 20.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 21.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005;37:1217–1223. [DOI] [PubMed] [Google Scholar]
- 22.Raby BA, Silverman EK, Kwiatkowski DJ, Lange C, Lazarus R, Weiss ST. ADAM33 polymorphisms and phenotype associations in childhood asthma. J Allergy Clin Immunol 2004;113:1071–1078. [DOI] [PubMed] [Google Scholar]
- 23.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor G, Sparrow D, Taylor D, Segal M, Weiss S. Analysis of dose–response curves to methacholine: an approach suitable for population studies. Am Rev Respir Dis 1987;136:1412–1417. [DOI] [PubMed] [Google Scholar]
- 26.Barbee RA, Halonen M, Lebowitz M, Burrows B. Distribution of IgE in a community population sample: correlations with age, sex, and allergen skin test reactivity. J Allergy Clin Immunol 1981;68:106–111. [DOI] [PubMed] [Google Scholar]
- 27.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–2229. [DOI] [PubMed] [Google Scholar]
- 28.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885. [DOI] [PubMed] [Google Scholar]
- 30.Werner M, Herbon N, Gohlke H, Altmuller J, Knapp M, Heinrich J, Wjst M. Asthma is associated with single-nucleotide polymorphisms in ADAM33. Clin Exp Allergy 2004;34:26–31. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Park HS, Park SW, Jang AS, Uh ST, Rhim T, Park CS, Hong SJ, Holgate ST, Holloway JW, et al. ADAM33 polymorphism: association with bronchial hyper-responsiveness in Korean asthmatics. Clin Exp Allergy 2004;34:860–865. [DOI] [PubMed] [Google Scholar]
- 32.Kormann MS, Carr D, Klopp N, Illig T, Leupold W, Fritzsch C, Weiland SK, von Mutius E, Kabesch M. G-protein–coupled receptor polymorphisms are associated with asthma in a large German population. Am J Respir Crit Care Med 2005;171:1358–1362. [DOI] [PubMed] [Google Scholar]
- 33.Feng Y, Hong X, Wang L, Jiang S, Chen C, Wang B, Yang J, Fang Z, Zang T, Xu X. G protein–coupled receptor 154 gene polymorphism is associated with airway hyperresponsiveness to methacholine in a Chinese population. J Allergy Clin Immunol 2006;117:612–617. [DOI] [PubMed] [Google Scholar]
- 34.Daniels SE, Bhattacharrya S, James A, Leaves NI, Young A, Hill MR, Faux JA, Ryan GF, le Souef PN, Lathrop GM, et al. A genome-wide search for quantitative trait loci underlying asthma. Nature 1996;383:247–250. [DOI] [PubMed] [Google Scholar]
- 35.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet 2003;361:865–872. [DOI] [PubMed] [Google Scholar]
- 36.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med 2002;4:45–61. [DOI] [PubMed] [Google Scholar]
- 37.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip–flop phenomenon. Am J Hum Genet 2007;80:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunninghake GM, Soto-Quiros ME, Avila L, Su J, Murphy A, Demeo DL, Ly NP, Liang C, Sylvia JS, Klanderman BJ, et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol 2007;120:84–90. [DOI] [PubMed] [Google Scholar]
- 39.Carvajal-Carmona LG, Ophoff R, Service S, Hartiala J, Molina J, Leon P, Ospina J, Bedoya G, Freimer N, Ruiz-Linares A. Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet 2003;112:534–541. [DOI] [PubMed] [Google Scholar]
- 40.Ribas G, Gonzalez-Neira A, Salas A, Milne RL, Vega A, Carracedo B, Gonzalez E, Barroso E, Fernandez LP, Yankilevich P, et al. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet 2006;118:669–679. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez-Neira A, Ke X, Lao O, Calafell F, Navarro A, Comas D, Cann H, Bumpstead S, Ghori J, Hunt S, et al. The portability of tagSNPs across populations: a worldwide survey. Genome Res 2006;16:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet 2006. [DOI] [PubMed]
- 43.Page GP, George V, Go RC, Page PZ, Allison DB. “Are we there yet?”: Deciding when one has demonstrated specific genetic causation in complex diseases and quantitative traits. Am J Hum Genet 2003;73:711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan SG. Regression to the truth: replication of association in pharmacogenetic studies. Pharmacogenomics 2003;4:201–207. [DOI] [PubMed] [Google Scholar]
- 45.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet 2006;7:781–791. [DOI] [PubMed] [Google Scholar]
- 46.Roeder K, Bacanu SA, Wasserman L, Devlin B. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet 2006;78:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lind DL, Choudhry S, Ung N, Ziv E, Avila PC, Salari K, Ha C, Lovins EG, Coyle NE, Nazario S, et al. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med 2003;168:1312–1316. [DOI] [PubMed] [Google Scholar]
- 48.Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, Xu J, Bleecker ER, Meyers DA. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol 2003;112:717–722. [DOI] [PubMed] [Google Scholar]
- 49.Blakey J, Halapi E, Bjornsdottir US, Wheatley A, Kristinsson S, Upmanyu R, Stefansson K, Hakonarson H, Hall IP. Contribution of ADAM33 polymorphisms to the population risk of asthma. Thorax 2005;60:274–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson A, Maniatis N, Jury F, Cakebread JA, Lowe LA, Holgate ST, Woodcock A, Ollier WE, Collins A, Custovic A, et al. Polymorphisms in a disintegrin and metalloprotease 33 (ADAM33) predict impaired early-life lung function. Am J Respir Crit Care Med 2005;172:55–60. [DOI] [PubMed] [Google Scholar]
- 51.Kedda MA, Duffy DL, Bradley B, O'Hehir RE, Thompson PJ. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet 2006;14:1027–1036. [DOI] [PubMed] [Google Scholar]
- 52.Schedel M, Depner M, Schoen C, Weiland SK, Vogelberg C, Niggemann B, Lau S, Illig T, Klopp N, Wahn U, et al. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res 2006;7:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirota T, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, Shirakawa T, Doi S, Fujita K, Suzuki Y, et al. Association between ADAM33 polymorphisms and adult asthma in the Japanese population. Clin Exp Allergy 2006;36:884–891. [DOI] [PubMed] [Google Scholar]
- 54.Wang P, Liu QJ, Li JS, Li HC, Wei CH, Guo CH, Gong YQ. Lack of association between ADAM33 gene and asthma in a Chinese population. Int J Immunogenet 2006;33:303–306. [DOI] [PubMed] [Google Scholar]
- 55.Noguchi E, Ohtsuki Y, Tokunaga K, Yamaoka-Sageshima M, Ichikawa K, Aoki T, Shibasaki M, Arinami T. ADAM33 polymorphisms are associated with asthma susceptibility in a Japanese population. Clin Exp Allergy 2006;36:602–608. [DOI] [PubMed] [Google Scholar]
- 56.Shin HD, Park KS, Park CS. Lack of association of GPRA (G protein–coupled receptor for asthma susceptibility) haplotypes with high serum IgE or asthma in a Korean population. J Allergy Clin Immunol 2004;114:1226–1227. [DOI] [PubMed] [Google Scholar]
- 57.Melen E, Bruce S, Doekes G, Kabesch M, Laitinen T, Lauener R, Lindgren CM, Riedler J, Scheynius A, van Hage-Hamsten M, et al. Haplotypes of G protein–coupled receptor 154 are associated with childhood allergy and asthma. Am J Respir Crit Care Med 2005;171:1089–1095. [DOI] [PubMed] [Google Scholar]
- 58.Malerba G, Lindgren CM, Xumerle L, Kiviluoma P, Trabetti E, Laitinen T, Galavotti R, Pescollderungg L, Boner AL, Kere J, et al. Chromosome 7p linkage and GPR154 gene association in Italian families with allergic asthma. Clin Exp Allergy 2007;37:83–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.