Abstract
The RPP5 (for recognition of Peronospora parasitica 5) locus in the Arabidopsis thaliana Columbia strain contains a cluster of paralogous disease Resistance (R) genes that play important roles in innate immunity. Among the R genes in this locus, RPP4 confers resistance to two races of the fungal pathogen Hyaloperonospora parasitica, while activation of SNC1 (for suppressor of npr1-1, constitutive 1) results in the resistance to another race of H. parasitica and to pathovars of the bacterial pathogen Pseudomonas syringae through the accumulation of salicylic acid (SA). Here, we demonstrate that other Columbia RPP5 locus R genes can be induced by transgenic overexpression of SNC1, which itself is regulated by a positive amplification loop involving SA accumulation. We also show that small RNA species that can target RPP5 locus R genes are produced in wild-type plants and that these R genes can be cosuppressed in transgenic plants overexpressing SNC1. Steady state expression levels of SNC1 increase in some mutants (dcl4-4, ago1-36, and upf1-5) defective in RNA silencing as well as in transgenic plants expressing the P1/Helper Component-Protease viral suppressor of RNA silencing. However, steady state levels of small RNA species do not change in mutants that upregulate SNC1. These data indicate many Columbia RPP5 locus R genes can be coordinately regulated both positively and negatively and suggest that the RPP5 locus is poised to respond to pathogens that disturb RNA silencing.
INTRODUCTION
Resistance (R) genes are used to recognize specific pathogens with cognate avirulence genes and initiate defense signaling that results in disease resistance (Dangl and Jones, 2001; Martin et al., 2003). The proteins encoded by the largest class of R genes carry a nucleotide binding site–leucine rich repeat (NBS-LRR) domain, and the N terminus shares similarity with either a coiled-coil domain or a Drosophila Toll/mammalian interleukin 1 receptor (TIR). The LRR domain seems to mediate specificity in pathogen recognition, while the N-terminal TIR or coiled-coil motif is likely to play a role in downstream signaling. Some R genes show a low steady state expression level in the absence of pathogens but can be induced by a positive feedback mechanism in which salicylic acid (SA) accumulation is required (Shirano et al., 2002; Xiao et al., 2003; Yang and Hua, 2004).
The Arabidopsis thaliana Columbia RPP5 (for recognition of Peronospora parasitica 5) locus is comprised of seven TIR-NBS-LRR class R genes, which are interspersed with three related sequences and two non-R genes (Noel et al., 1999) (see Supplemental Figure 1 online). All of the R genes and related sequences in the locus are more closely related to each other than any other R gene in the genome, consistent with the hypothesis that the locus was generated by local duplications and rearrangements (Baumgarten et al., 2003; Meyers et al., 2003). R genes found in this locus in the Columbia haplotype are highly diverged in nucleotide sequences from those in the Landsberg or Wassilewskija haplotype (Noel et al., 1999; Yang and Hua, 2004). Two R genes in the RPP5 locus, RPP4 and SNC1 (for suppressor of npr1-1, constitutive 1), have been demonstrated to be functional in disease resistance against bacterial and fungal pathogens (Stokes et al., 2002; van der Biezen et al., 2002; Zhang et al., 2003; Yang and Hua, 2004). The gain-of-function mutation snc1 is likely to increase SNC1 activity and thereby causes elevated disease resistance and dwarfism (Zhang et al., 2003). Two other mutations mapped to the RPP5 locus, cpr1 (for constitutive expresser of PR genes 1) and bal, display phenotypes similar to snc1 and exhibit a high degree of phenotypic instability after an exposure to ethyl methanesulfonate (Bowling et al., 1994; Stokes et al., 2002; Stokes and Richards, 2002). Even though the upregulation of SNC1 was reported in both mutants, neither the molecular nature of the mutations responsible for SNC1 activation nor the mechanism of phenotypic reversion has been characterized (Stokes et al., 2002; Yang and Hua, 2004).
As a first step to characterize bal and cpr1 mutants, we investigated how RPP5 locus R genes are regulated transcriptionally and posttranscriptionally. RNA silencing at the posttranscriptional level is conserved in diverse eukaryotic organisms and is mediated by small RNA (small interfering RNA [siRNA] and microRNA [miRNA]) that is complementary to target mRNA transcripts (Baulcombe, 2004). RNA silencing processing in plants involves (1) generation of double-stranded RNA (dsRNA), the formation of which may require RNA-dependent RNA polymerases (RDRs) (Dalmay et al., 2000; Mourrain et al., 2000; Xie et al., 2001, 2004); (2) production of 21- to 24-nucleotide small RNA by Dicer-like proteins (DCLs) (Park et al., 2002; Xie et al., 2004; Gasciolli et al., 2005; Yoshikawa et al., 2005); and (3) small RNA-guided slicing of target RNA by Argonaute family proteins (AGOs) (Baumberger and Baulcombe, 2005; Qi et al., 2005; Adenot et al., 2006; Qi et al., 2006).
The complexity of the RNA silencing machinery is exemplified by the model plant Arabidopsis, which has six RDRs, four DCLs, and 10 AGOs. Specialized but partially overlapping functions were observed among the four DCLs. DCL1 is almost exclusively responsible for the production of miRNA (Park et al., 2002; Rajagopalan et al., 2006). However, loss of one or more functions of DCL2, DCL3, and DCL4 can be partially substituted by other DCLs, although there is a preference or hierarchy in the use of specific DCLs for a given process (Gasciolli et al., 2005; Deleris et al., 2006; Fusaro et al., 2006; Henderson et al., 2006). The few functionally characterized AGOs and RDRs show diversified but still overlapping roles in RNA silencing (Fagard et al., 2000; Mourrain et al., 2000; Xie et al., 2001; Zilberman et al., 2003; Vazquez et al., 2004b; Baumberger and Baulcombe, 2005; Borsani et al., 2005; Katiyar-Agarwal et al., 2006; Qi et al., 2006; Kasschau et al., 2007). A wide range of processes in Arabidopsis, such as development, flowering time, stress tolerance, and resistance to bacterial and viral pathogens, are regulated by RNA silencing (Liu et al., 2004; Mallory et al., 2004a, 2004b; Vaucheret et al., 2004; Vazquez et al., 2004b; Borsani et al., 2005; Gasciolli et al., 2005; Deleris et al., 2006; Henderson et al., 2006; Katiyar-Agarwal et al., 2006).
Here, we report that several RPP5 locus R genes, including RPP4 and SNC1, are coordinately regulated by at least two different mechanisms. These R genes are positively regulated by transcriptional activation and negatively regulated by RNA silencing. We also found that SNC1 can be upregulated by mutations abrogating RNA silencing and overexpression of a viral RNA silencing suppressor. Our results suggest that the RPP5 locus, which is important for innate immunity, can be activated in response to challenges by plant pathogens that disturb RNA silencing.
RESULTS
Transgenic Overexpression of SNC1 Induces Two Distinct Morphological Syndromes Depending on Transgene Dosage
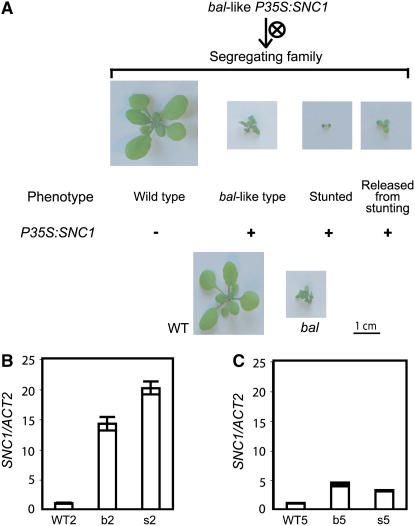
As reported previously, overexpression of a SNC1 genomic clone under the control of the strong 35S promoter can induce a bal-like morphology (twisted leaves and dwarf stature) (Stokes et al., 2002). Further characterization of transgenic plants revealed that the plants displaying bal-like phenotypes were hemizygous for the transgene and produced progeny with either wild-type, bal-like, or stunted (very small with unexpanded leaves) phenotypes in the absence of selection for the kanamycin resistance gene marker (Figure 1A). We found that progeny exhibiting wild-type phenotypes lacked the P35S:SNC1 transgene. By contrast, individuals with bal-like phenotypes carried the transgene, and their progeny contained plants with three distinctive phenotypes conforming to a 1:2:1 ratio (22 wild-type:48 bal-like:21 stunted plants). Stunted plants in segregating families also carried the P35S:SNC1 transgene, and they eventually developed a wild-type morphology after 3 weeks. This delayed normal growth syndrome bred true over three generations, suggesting that this syndrome is a consistent, reproducible phenotype of P35S:SNC1 homozygotes. Moreover, we observed a bal-like phenotype among all F1 progeny obtained from crosses between wild-type plants and P35S:SNC1 transgenic plants exhibiting the delayed normal growth syndrome.
Figure 1.
Transgenic Plants Overexpressing SNC1 (P35S:SNC1) Display Two Distinct Morphologies.
(A) Phenotypes of segregating progeny of bal-like transgenic plants overexpressing SNC1. The presence or absence of the transgene, determined by PCR using primers for the kanamycin resistance gene, is indicated by the + and − signs, respectively. Transgenic plants with the stunting phenotype eventually developed wild-type morphology after 3 weeks. bal, bal variant.
(B) SNC1 transcript level, relative to ACTIN2 (ACT2), determined by real-time RT-PCR using 2-week-old plants. WT2, wild-type plants; b2, bal-like transgenic plants; s2, stunted plants.
(C) Relative SNC1 transcript level determined by real-time RT-PCR using 5-week-old plants. WT5, wild-type plants; b5, bal-like transgenic plants; s5, plants released from stunting with wild-type morphology.
The genetic behavior described above suggested that the morphological consequences of SNC1 overexpression (bal-like versus stunted) depend on the dosage of the P35S:SNC1 transgene. Consistent with this explanation, the expression level of SNC1 was higher in homozygous stunted transgenic plants compared with hemizygous bal-like plants (Figure 1B). We suspected that the stunting and delayed normal growth syndrome, which we observed every generation in homozygous P35S:SNC1 transgenic plants, resulted from posttranscriptional gene silencing (PTGS), RNA silencing at the posttranscriptional level, triggered by overexpression of SNC1. Repeated onset of gene silencing in each generation is one feature of PTGS, and homozygosity-dependent PTGS has been observed for other Arabidopsis transgenes (Elmayan et al., 1998; Qin et al., 2003). Consistent with the hypothesis that the delayed normal growth in homozygous P35S:SNC1 plants was caused by PTGS of SNC1, we found that the steady state expression level of SNC1 in homozygous plants was lower compared with hemizygous plants when wild-type morphology was observed in homozygous plants (Figure 1C). We concluded that overexpression of SNC1 induces either bal-like or stunted phenotypes depending on the expression level of SNC1.
RPP5 Locus R Genes Are Coordinately Upregulated at the Transcriptional Level by a Positive Feedback Mechanism Mediated through SNC1
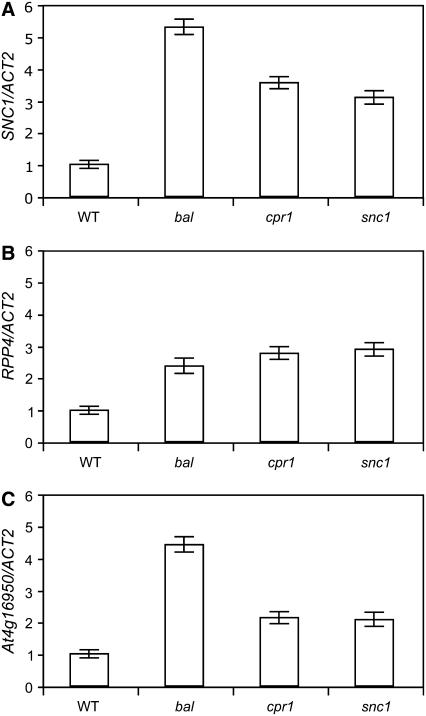
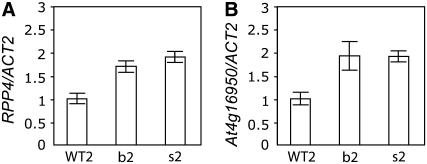
Transcriptional regulation of some R genes in Arabidopsis, including SNC1, is governed by a positive amplification loop requiring SA accumulation (Shirano et al., 2002; Xiao et al., 2003; Yang and Hua, 2004). Because all seven R genes located in the RPP5 locus, where SNC1 is located (see Supplemental Figure 1 online), appear to be generated by local tandem duplications (Baumgarten et al., 2003; Meyers et al., 2003; Leister, 2004), we suspected that other R genes in the locus might also be transcriptionally activated along with SNC1 using a conserved regulatory element in their promoters. Considering that RPP4 was shown to be functional in resistance against fungal pathogens and At4g16950 is the most similar R gene in Columbia haplotype to the RPP5 gene in Landsberg haplotype, we focused on RPP4 and At4g16950 (Parker et al., 1997; Noel et al., 1999; van der Biezen et al., 2002; Meyers et al., 2003). We found that expression levels of RPP4 and At4g16950 were higher in bal, cpr1, and snc1 mutants, all of which carry mutations mapping to the RPP5 locus and show constitutive upregulation of SNC1 (Bowling et al., 1994; Li et al., 2001; Stokes et al., 2002; Zhang et al., 2003), suggesting that RPP5 locus R genes are coordinately upregulated (Figures 2A to 2C). Using transgenic plants overexpressing SNC1, we directly tested whether expression levels of RPP5 locus R genes were transcriptionally induced by overexpression of SNC1 and found that expression levels of both RPP4 and At4g16950 were elevated in bal-like hemizygous and homozygous stunted P35S:SNC1 transgenic plants (Figures 3A and 3B). These data indicate that RPP4 and At4g16950, as well as SNC1, are regulated by a transcriptional amplification mechanism mediated by SNC1 activity.
Figure 2.
Steady State Expression Levels of RPP4 and At4g16950 Increase in Mutants That Upregulate SNC1.
(A) SNC1 transcript level, relative to ACT2, determined by real-time RT-PCR.
(B) and (C) Relative transcript levels of RPP4 and At4g16950 determined by real-time RT-PCR. Tissue for RNA isolation was collected from 2-week-old plants.
Figure 3.
Expression Levels of RPP4 and At4g16950 Correlate with SNC1 in P35S:SNC1 Transgenic Plants.
RPP4 and At4g16950 transcript levels, relative to ACT2, determined by real-time RT-PCR using 2-week-old plants. WT2, wild-type plants; b2, bal-like transgenic plants; s2, stunted plants.
RPP5 Locus R Genes Can Be Coordinately Downregulated by Cosuppression in Plants Overexpressing SNC1
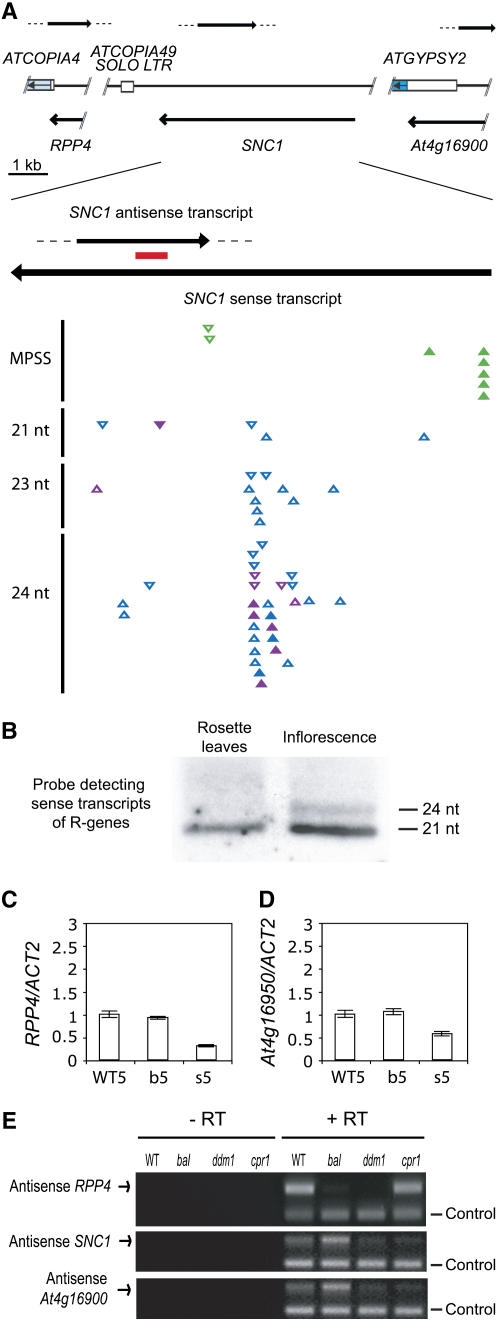
In recent years, many small RNA species in Arabidopsis have been identified from large-scale small RNA cDNA sequencing projects, and three research groups have independently reported diverse small RNA species originating from RPP5 locus R genes (Nakano et al., 2006; Rajagopalan et al., 2006; Kasschau et al., 2007). Although most of these small RNA species were identified only a single time and many of them showed perfect matches to more than one RPP5 locus R gene, including SNC1, some SNC1-specific small RNA were also isolated (Figure 4A). A collective data set revealed that small RNA species in various size classes are generated from both strands of SNC1 gene and that these small RNA species are predominantly 24 nucleotides and produced in the central part of the SNC1 gene, downstream of the region encoding the NBS domain.
Figure 4.
Small RNA Species Are Detected from Columbia RPP5 Locus R Genes.
(A) Small RNA species of various size are generated from SNC1. Each left-to-right arrow shows the positions of antisense transcripts in the RPP5 locus detected in (E). Below the antisense transcripts, the positions of a part of At Copia4 and At Gypsy2 LTRs, as well as the At Copia49 solo LTR, are indicated by rectangles. Arrows in the shaded rectangles represent the putative retrotransposon polyproteins. The coding regions of RPP4, SNC1, and At4g16900 sense transcripts are indicated by right-to-left arrows below the representation of transposon-related sequences. The red bar in the overlapping region of SNC1 antisense and sense transcripts shows the position of the strand-specific probe used in small RNA gel blot analysis ([B]; Figure 6). The 5′ and 3′ ends of the antisense transcripts, including that of SNC1, have not been determined and are therefore represented by dotted lines. The triangles show positions of all the reported small RNA signatures whose nucleotide sequences are perfect matches to those in SNC1. The small RNA species reported by Meyers/Green (MPSS), Carrington, and Bartel labs are colored in green, purple, and blue, respectively (Nakano et al., 2006; Rajagopalan et al., 2006; Kasschau et al., 2007), and are arranged by size class. SNC1-specific small RNA species are indicated by filled triangles, while small RNA species whose sequences are conserved among different RPP5 locus R genes, including RPP4 and At4g16950, are indicated by open triangles. Small RNA species originating from sense and antisense transcripts are represented by triangles pointing upward and downward, respectively. Small RNA species that were isolated multiple times are indicated by multiple triangles. Note that the small RNA species shown here are cataloged regardless of tissues or genotypes from which a given small RNA species was isolated. nt, nucleotides.
(B) RNA gel blot analysis detects small RNA species corresponding to the sense strand of the LRR region of R genes from the locus in rosette leaves and inflorescence tissue. The sizes of 21- and 24-nucleotide small RNA species were determined by subsequent hybridization to detect small RNA species of known size.
(C) and (D) Relative transcript levels of RPP4 and At4g16950 determined by real-time RT-PCR using 5-week-old plants. WT5, wild-type plants; b5, bal-like transgenic plants; s5, plants released from stunting with wild-type morphology.
(E) Antisense transcripts overlapping with RPP4, SNC1, and At4g16900 sense transcripts are detected by strand-specific RT-PCR. −RT and + RT, first-strand cDNA library constructed without and with reverse transcriptase, respectively. ddm1, ddm1-2 mutants that were inbred for two generations; control, internal loading control for multiplex RT-PCR, glyceraldehyde 3-phosphate dehydrogenase (GAPC).
We confirmed that 21- to 24-nucleotide small RNA species, the hallmark of RNA silencing, are generated from RPP5 locus R gene transcripts by performing a small RNA gel blot experiment. Discrete 21-nucleotide small RNA species were detected in vegetative tissues using a hybridization probe corresponding to the conserved LRR domain in RPP5 locus R genes (Figure 4B). In inflorescence tissue, 24-nucleotide small RNA species and 21-nucleotide species were observed, although the smaller size class was more abundant. The difference in the prevalence of the different size classes between the small RNA sequencing projects and our experiments may be due to the specific hybridization probe used in our small RNA gel blot experiment and the preferential use of inflorescence tissue in the small RNA sequencing projects.
We investigated the possibility that SNC1 and other RPP5 locus R genes are coordinately regulated by RNA silencing at the posttranscriptional level based on their nucleotide sequence similarity. We tested whether the nucleotide sequences of SNC1 and other RPP5 locus R genes are similar enough to be targeted by RNA silencing in a concerted manner, as was suggested by the presence of small RNA species that are perfect matches to more than one R gene in the RPP5 locus. We reasoned that expression levels of RPP4 and At4g16950 might also be reduced in homozygous P35S:SNC1 plants exhibiting wild-type morphology compared with those in bal-like hemizygotes, if RPP5 locus R genes can be coordinately regulated by RNA silencing. We found that expression levels of both RPP4 and At4g16950 are lower in wild type–like P35S:SNC1 homozygotes (released from stunting) than in bal-like hemizygotes at this developmental stage (Figures 4C and 4D). The expression level of RPP4 is comparable in wild-type plants and snc1 r1 mutants, which carry a null allele of SNC1 (Zhang et al., 2003) (see Supplemental Figure 2A online), suggesting that the reduction of RPP4 and At4g16950 expression in normal-looking homozygous transgenic plants is not caused by downregulation of SNC1. We concluded that RPP5 locus R genes can be coordinately suppressed by RNA silencing based on their nucleotide sequence similarity.
Antisense Transcripts Overlapping with RPP4, SNC1, and At4g16900 Are Produced in the RPP5 Locus
The generation and amplification of siRNA species, which can coordinately regulate homologous genes through RNA silencing, requires the formation of foldback structures or dsRNA (Vaucheret, 2006). Antisense transcripts originating from downstream transposons or genes have been implicated in the formation of dsRNA with sense transcripts from upstream genes (Aravin et al., 2001; Kashkush et al., 2003; Borsani et al., 2005; Katiyar-Agarwal et al., 2006). To identify the source of the small RNA species detected above, we focused on the genomic region around SNC1 where three retrotransposon sequences, At Copia4 (At4g16870), At Copia49 solo long terminal repeat (LTR), and At Gypsy2 (At4g16910) are located (Figure 4A). Signatures in the Arabidopsis massively parallel signature sequencing (MPSS) database suggested that antisense transcripts are made from RPP4, SNC1, or At4g16900 (Nakano et al., 2006). First, we determined the positions of the 3′ ends of RPP4, SNC1, and At4g16900 sense transcripts using rapid amplification of cDNA ends and obtained results consistent with those reported from 3′ end or full-length sequencing of cDNA clones from these R genes (Figure 4A) (van der Biezen et al., 2002; Castelli et al., 2004). The presence of antisense transcripts, which overlap at least 1 kb with sense transcripts, was validated using strand-specific RT-PCR for all three R genes (Figure 4E). Compared with wild-type plants, expression of the RPP4 antisense transcript did not show any significant change in the cpr1 mutant but showed a dramatic decrease in the bal variant. Antisense transcripts of RPP4 were almost abolished in aphenotypic ddm1-2 (for decrease in DNA methylation 1-2) mutants, suggesting that this change originated from the inbred ddm1-2 mutant background from which the bal variant was derived (Kakutani et al., 1996). By contrast, a slight increase in the levels of antisense transcripts of SNC1 and At4g16900 was observed in the bal variant compared with wild-type plants or the cpr1 mutant (Figure 4E). Therefore, it is unlikely that the downregulation of antisense transcripts is directly responsible for the upregulation of overlapping sense transcripts. Our results show that sense and antisense transcript pairs, which may form dsRNA and generate small RNA species, are produced from RPP5 locus R genes.
The Steady State Expression Level of SNC1 Is Elevated in Mutants Associated with Defects in RNA Silencing
Our findings that discrete small RNA species corresponding to the RPP5 locus R gene transcripts are produced and that RPP5 locus R genes can be cosuppressed led us to examine the effect of mutations affecting RNA silencing on SNC1 expression. Small RNA species, which are loaded into AGOs and used to direct sequence-specific cleavage of target transcripts, are produced by DCLs from RNA duplexes whose formation may depend on RDR activities. In Arabidopsis, three out of six RDRs, all four DCLs, and four out of 10 AGOs have been characterized for their function in RNA silencing (Vaucheret, 2006; Zheng et al., 2007). Another protein that has been implicated in RNA silencing is UPF1, a conserved component in nonsense-mediated mRNA decay (NMD), which degrades mRNA transcripts with premature stop codons in eukaryotes (Behm-Ansmant and Izaurralde, 2006). The UPF1 ortholog in Caenorhabditis elegans is necessary for the persistence of RNA silencing (Domeier et al., 2000; Kim et al., 2005), and the Arabidopsis ortholog is required for RNA silencing triggered by an inverted repeat transgene (Arciga-Reyes et al., 2006). The steady state expression levels of SNC1 transcripts in wild-type plants and various mutants were compared using quantitative real-time RT-PCR. As expected for pairs of overlapping sense and antisense transcripts, which can form dsRNA over a 1-kb region without RDR activity, we found that RDR1, RDR2, and RDR6 activity was dispensable for the full suppression of SNC1 (Figure 5A). We cannot rule out the possibility that other RDRs, which were not tested here, contribute to the generation of dsRNA in the RPP5 locus, but we note that the MPSS mRNA signatures, which originally suggested the presence of antisense transcripts in the locus, were generated using polyadenylated RNA, most likely produced by RNA polymerase II.
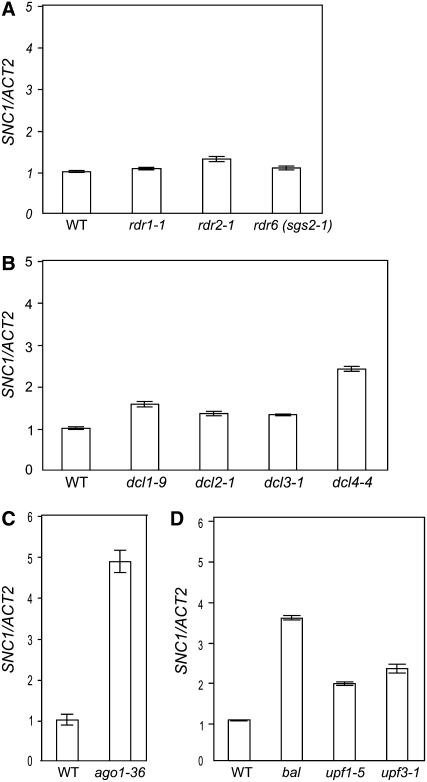
Figure 5.
Steady State Expression Levels of SNC1 Are Elevated in Mutants Affecting RNA Silencing.
(A) and (B) SNC1 transcript levels, relative to ACT2, determined by real-time RT-PCR using plants homozygous for various rdr mutations and four dcl mutations, respectively.
(C) Relative SNC1 transcript levels in ago1-36 mutants compared with wild-type siblings plants.
(D) Relative SNC1 transcript levels compared among wild-type control plants, the bal variant, and upf1-5, and upf3-1 mutants. Plants were grown in soil except for samples used in (C), where plants were grown using in vitro culture conditions. Tissue for RNA isolation was collected from 2-week-old plants.
We found that the steady state expression level of SNC1 transcript was elevated in some mutants defective in RNA silencing, for example, the dcl4-4 mutant (Dunoyer et al., 2005) (Figure 5B). DCL4 is responsible for the generation of 21-nucleotide siRNA species in the trans-acting siRNA pathway as well as those produced from inverted repeat transgenes and viruses (Dunoyer et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Deleris et al., 2006; Fusaro et al., 2006). DCL4 was also reported to be responsible for the production of two miRNA species in Arabidopsis (Rajagopalan et al., 2006). No significant changes in SNC1 expression were observed in other dcl mutants compared with wild-type plants (Figure 5B). Among the four AGOs with a demonstrated role in RNA silencing, AGO1 physically associates with 21-nucleotide small RNA species almost exclusively (Baumberger and Baulcombe, 2005; Qi et al., 2006). We found that the steady state expression level of SNC1 was elevated in the ago1-36 mutant (Baumberger and Baulcombe, 2005) (Figure 5C). However, we did not observe any significant change in SNC1 expression level in rpd2a-2 rpd2b-1, dcl3, or rdr2 mutants, all of which specifically affect the accumulation of 24-nucleotide small RNA (Figures 5A and 5B; see Supplemental Figure 2B online) (Onodera et al., 2005; Kasschau et al., 2007). In addition, increases in the steady state expression levels of SNC1 were observed in hypomorphic upf1-5 and upf3-1 mutants, both of which are compromised for NMD (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006) (Figure 5D). The activation of SNC1 in the NMD-defective mutants might be related to the fact that alternatively spliced transcripts with premature stop codons in retained introns are produced for SNC1, thereby making SNC1 a direct target of NMD (see Supplemental Figure 3 online).
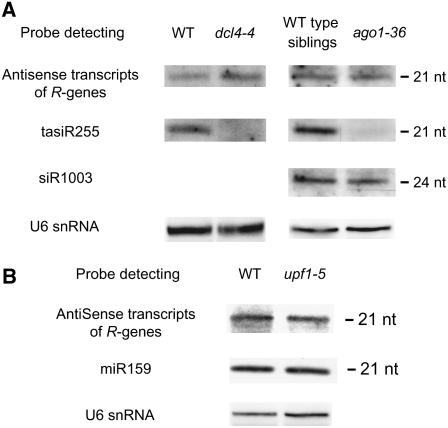
Next, we examined small RNA species generated from the RPP5 locus R genes in dcl4-4, ago1-36, upf1-5, and upf3-1 mutants. Specifically, we monitored the abundance of 21-nucleotide antisense small RNA species corresponding to the LRR region. DCL4 and AGO1 are involved in the biogenesis and use of 21-nucleotide small RNA. The LRR region was chosen because antisense small RNA species from this conserved region are expected to negatively regulate multiple R genes in the RPP5 locus. As expected, the accumulation of tasiR255 is abolished in dcl4 and ago1 mutants (Gasciolli et al., 2005; Adenot et al., 2006). By contrast, we did not observe any significant decrease in the levels of small RNA species corresponding to antisense transcripts of RPP5 locus R genes in any of the mutants that showed increase in steady state expression of SNC1 (Figure 6; see Supplemental Figure 4 online).
Figure 6.
Accumulation of Small RNA Species Originating from Antisense Transcripts of RPP5 Locus R Genes Does Not Significantly Decrease in Mutants with an Elevated SNC1 Expression Level.
Small RNA gel blot analysis was used to detect the RNA species indicated on the left. Results shown here were obtained from a single membrane by sequential hybridization after an extensive washing. The sizes of 21- and 24-nucleotide small RNA species were determined by subsequent hybridization to detect small RNA species of known size. Note that wild-type siblings and ago1-36 mutants were grown under in vitro culture conditions. WT siblings, siblings of ago1-36 with wild-type morphology (i.e., wild-type plants and heterozygous ago1-36 mutants). Tissue for RNA isolation was collected from 2-week-old plants. Similar results were found in at least two independent biological replicates of these hybridization experiments.
DISCUSSION
Here, we report two mechanisms that can coordinately regulate many paralogous R genes in the RPP5 locus. Many RPP5 locus R genes are positively regulated at the transcriptional level by a feedback amplification loop involving SNC1, and they can be cosuppressed when SNC1 is expressed over a certain threshold. In addition, we show that the steady state expression level of SNC1, which can induce the activation of other RPP5 locus R genes, is elevated in dcl4-4 and ago1-36 mutants.
RPP5 Locus R Genes Are Coordinately Upregulated by a Positive Feedback Mechanism Mediated by SNC1
It was originally proposed that SNC1 is regulated by a positive feedback mechanism through SA accumulation (Yang and Hua, 2004). We confirmed and extended these findings by demonstrating that RPP4 and At4g16950, two additional R genes in the locus, are also transcriptionally induced by overexpression of SNC1 (Figure 3). Our results are consistent with the analysis of data from 128 publicly available microarray experiments that identified Copia4 (At4g16870), RPP4 (At4g16860), At4g16880, and SNC1 (At4g16890) as one group of 226 highly coexpressed neighboring genes in Arabidopsis (Zhan et al., 2006). Our data also show that another R gene in the locus, At4g16950, is coregulated with RPP4 and SNC1 when SNC1 is overexpressed. Even though we propose that RPP4 and At4g16950 are directly activated through SA accumulation resulting from SNC1 activation, we cannot rule out the possibility that coordinated upregulation of RPP4, SNC1, and At4g16950 resulted indirectly from transcriptional activation of SNC1, which could change the local chromatin structure of the locus. Activation of a number of related R genes that function in defense signaling pathways via SA accumulation may provide plants a broader spectrum of resistance to closely related pathogens.
SNC1 and Other RPP5 Locus R Genes Can Be Cosuppressed
Beginning with the identification of small RNA species showing perfect matches to many R genes in the RPP5 locus, we showed that several R genes in the locus can be cosuppressed (Figure 4). Overexpression of SNC1, which constitutively activates defense signaling, reduces fitness (Heidel et al., 2004; van Hulten et al., 2006). RNA silencing might play an important role in minimizing the fitness cost associated with excessive SNC1 expression. It is possible that RNA silencing machinery is used to quickly remove SNC1 transcripts when SNC1 expression level reaches a certain threshold level that cannot be tolerated or after activation by pathogen challenge. Alternatively, RNA silencing may allow the evolution of RPP5 locus R genes while minimizing the possible deleterious effect caused by the recombination or multiplication of these R genes, as suggested for other rapidly expanding gene families in Arabidopsis (Howell et al., 2007).
SNC1 Is Activated When DCL4 and AGO1 Are Compromised
Our results show that SNC1, overexpression of which induces upregulation of other RPP5 locus R genes (Figure 3), is activated when two components in RNA silencing (DCL4 and AGO1) are compromised (Figures 5B and 5C). DCL4 is responsible for the production of 21-nucleotide small RNA species from various sources (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Deleris et al., 2006; Fusaro et al., 2006), and AGO1 is almost exclusively associated with 21-nucleotide small RNA species (Baumberger and Baulcombe, 2005; Qi et al., 2006). We detected convergent, overlapping sense and antisense transcripts along with 21-nucleotide small RNA species corresponding to several R genes in the RPP5 locus. Moreover, the genetic requirement of DCL4 and AGO1 for suppression of SNC1 expression is consistent with our detection of 21-nucleotide small RNA species from these R genes. The simplest model predicts that the abundance of 21-nucleotide RNA species corresponding to the antisense strand of the R genes would decrease in dcl4-4 and ago1-36 mutants. However, we did not observe an inverse correlation between the abundance of these small RNA species and their putative targets in either dcl4-4 or ago1-36 mutants (Figure 6). One possibility is that the probe used in our gel blot hybridization experiments did not detect the functional small RNA species responsible for downregulating R gene transcripts. The probe was chosen to recognize the conserved LRR region corresponding to the antisense 21-nucleotide small RNA species (Figure 4B), which are the best candidates for coordinate regulation of RPP5 locus R genes. An alternative hypothesis is that the 21-nucleotide small RNA species detected in Figure 6 are responsible for targeting R gene turnover and are only transiently reduced in the dcl4-4 and ago1-36 mutants. The result in Figure 6 may reflect a steady state equilibrium where a wild-type level of small RNA species, which are produced and stabilized by other DCLs and AGOs, is now balanced against a higher concentration of R gene transcript targets. A third possibility is that the activation of the RPP5 locus R genes in dcl4-4 and ago1-36 mutant backgrounds is indirect, caused by an RNA silencing pathway defect affecting one or more upstream components that transcriptionally activate SNC1 (Yang and Hua, 2004).
Whether SNC1 is directly or indirectly activated by release of RNA silencing, our results show that SNC1 can be upregulated if some components in RNA silencing are functionally compromised. Therefore, RNA silencing may function as a sensor to detect any interference in RNA silencing machinery by pathogens, leading to the initiation of defense responses that are mediated through the activation of SNC1 and other RPP5 locus R genes. Consistent with this hypothesis, an increase in steady state expression level of SNC1 was observed from transgenic plants overexpressing a viral suppressor of RNA silencing suppressor, P1/Helper Component-Protease (P1/HC-Pro) (see Supplemental Figure 5 online). Furthermore, DCL4, which is required for the full suppression of SNC1, is the primary DCL involved in viral defense and is a target of p38, another viral RNA silencing suppressor (Deleris et al., 2006). Several RNA silencing pathways, which require different combinations of RDRs, DCLs, AGOs, and other components, have been identified in Arabidopsis (Brodersen and Voinnet, 2006; Vaucheret, 2006). However, the genetic requirement of DCL4 and AGO1 for the suppression of SNC1 is different from the requirements defined for other pathways regulating endogenous targets. For example, DCL4 is required for the production of 21-nucleotide trans-acting siRNA, but this pathway also requires the concerted action of DCL1, RDR6 (SGS2), and SGS3 to generate dsRNA (Gasciolli et al., 2005). Our results show that loss of DCL1, RDR6, or SGS3 does not affect steady state expression level of SNC1 significantly (Figures 5A and 5B; see Supplemental Figure 6A online). Instead, the genetic requirement to suppress SNC1 expression is similar to that expected for the RNA silencing pathway targeting inverted repeat transgenes (IR-PTGS pathway), which has been widely used to knock down specific genes in transgenic plants (Wesley et al., 2001). The IR-PTGS pathway requires DCL4 and AGO1 (Dunoyer et al., 2005, 2007), but RDR6 and HEN1 are dispensable (Boutet et al., 2003; Li et al., 2005), as is the case for the suppression of SNC1 (Figure 5A; see Supplemental Figure 6B online). The overlap of genetic requirements between the IR-PTGS pathway and suppression of SNC1 suggests that SNC1 itself or an upstream component in the signaling cascade that results in the activation of SNC1 is one natural target of the IR-PTGS pathway.
Accumulating evidence shows that RNA silencing is used to regulate endogenous genes more extensively than previously thought. For example, posttranscriptional control of the chalcone synthase gene family has been reported for naturally generated variants of soybean (Glycine max), maize (Zea mays), and petunia (Petunia hybrida) (Senda et al., 2004; Della Vedova et al., 2005; Koseki et al., 2005). Many clustered paralogous genes in plant genomes might generate inverted repeat transcripts or antisense transcripts, thereby providing an opportunity for coordinated suppression of these genes by RNA silencing. Our results show that RNA silencing can be used to restrict the fitness cost associated with the constitutive activation of defense signaling involving RPP5 locus R genes, which are controlled by a feedback amplification loop. In addition, our results demonstrate that defense mechanisms can be activated in response to pathogen attack that disturbs the RNA silencing machinery.
METHODS
Plants and Growth Conditions
All plant genotypes used in this study were in the Columbia background and inbred a few generations except hypomorphic dcl1-9 mutation that causes female sterility. Our dcl1-9 strain carrying the Columbia RPP5 locus was generated by crossing a Landsberg strain carrying dcl1-9 (Jacobsen et al., 1999) to wild-type plants in the Columbia background and subsequent segregation. We previously described bal and cpr1 as well as the ddm1-2 mutant (Kakutani et al., 1996; Stokes et al., 2002; Stokes and Richards, 2002). rdr1-1, rdr2-1 (Xie et al., 2004), rdr6 (sgs2-1) (Elmayan et al., 1998), dcl2-1, dcl3-1 (Xie et al., 2004), dcl4-4 (Dunoyer et al., 2005), snc1 r1 (Zhang et al., 2003), hen1-5 (Vazquez et al., 2004a), rpd2a-2 rpd2b-1 mutants (Onodera et al., 2005), and transgenic plants overexpressing the P1/HC-Pro viral suppressor (Chapman et al., 2004; Mlotshwa et al., 2005) were kindly provided by James C. Carrington (dcl2-1, dcl3-1, rdr1-1, rdr2-1, and P1/HC-Pro), Xin Li (snc1 r1), Craig S. Pikaard (rpd2a rpd2b), Vicki B. Vance (P1/HC-Pro), Hervé Vaucheret (rdr6 and hen1-5), and Olivier Voinnet (dcl4-4). In addition, upf1-5 (Arciga-Reyes et al., 2006), upf3-1 (Hori and Watanabe, 2005), and sgs3-13 (Peragine et al., 2004) mutants were obtained from the ABRC (Alonso et al., 2003).
All plants except ago1 and sibling plants were grown on soil in growth chambers under long-day conditions (16 h light) as described previously (Stokes et al., 2002). Homozygous ago1-36 mutants (Baumberger and Baulcombe, 2005) and segregating siblings were grown on solid media (half-strength Murashige and Skoog basal salt mixture [Sigma-Aldrich], 1% sucrose, and 4% Agarasel [Sigma-Aldrich]) in growth chambers under long-day conditions (16 h light).
Nucleic Acid Isolation
Total RNA and low molecular weight–enriched RNA were isolated from aerial parts of either 2- or 5-week-old plants using TRIzol reagent (Invitrogen) and the mirVana miRNA isolation kit (Ambion), respectively, following the manufacturers' protocols. Genomic DNA for genotyping was isolated by the urea lysis miniprep protocol (Cocciolone and Cone, 1993).
Expression Analysis
To analyze the expression level of genes by real-time RT-PCR, first-strand cDNA libraries were constructed with total RNA samples using the SMART RACE cDNA amplification kit (Clontech), after treating total RNA with RQ1 RNase-free DNase I (Promega). Antisense strand-specific libraries were constructed with GAPCR and balANTI primers, instead of the 3′ RACE CDS primer A in the kit. Nucleotide sequences of the primers and Taqman probes used in our experiments are listed in Supplemental Table 1 online. For quantitative real-time PCR, the Applied Biosystems 7500 Real Time PCR system and Taqman MGB probe were used. Triplicate PCR reactions were used to generate error bars in every experiment, and at least two biologically independent experiments were performed for every case to confirm the results. For multiplex RT-PCR, the ratio between GAPC primers with or without dideoxy modification was empirically determined (Kerschen et al., 2004).
Small RNA gel blot analysis was performed as previously described with minor modifications (Onodera et al., 2005). In brief, a denaturing 15% (v/v) acrylamide gel was used to separate low-molecular RNA, and PefectHyb Plus hybridization buffer (Sigma-Aldrich) was used. Strand-specific probes were made with the mirVana miRNA probe construction kit (Ambion) using oligonucleotides or PCR products with T7 polymerase binding sites. After hybridization at 40°C, membranes were washed as follows at the same temperature: 15 min in 2× SSC and 0.1% SDS, 15 min in 1× SSC and 0.1% SDS, and 15 min in 0.5× SSC and 0.1% SDS. Nucleotide sequences for oligonucleotides or primers used in probe construction are in Supplemental Table 1 online.
Accession Numbers
The following loci were studied in this article: At4g16860 (RPP4), At4g16870 (Copia4), At4g16890 (SNC1), At4g16900, At4g16910 (Gypsy2), and At4g16950.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Organization of R Genes Located in RPP5 Locus of the Columbia Haplotype.
Supplemental Figure 2. The Relative Quantity of RPP4 in the snc1 r1 Mutant and That of SNC1 in the rpd2a-2 rpd2b-1 Double Mutant Are Not Changed Compared with Wild-Type Plants.
Supplemental Figure 3. SNC1 Transcripts Are Alternatively Spliced to Produce Transcripts with or without Introns.
Supplemental Figure 4. No Significant Difference in the Accumulation of Small RNA Species Originating from Antisense Transcripts of RPP5 Locus R Genes Was Detected between Wild-Type Plants and upf3-1 Homozygotes.
Supplemental Figure 5. Expression Levels of SNC1 and RPP4 Are Elevated in Transgenic Plants Overexpressing a Viral Suppressor of RNA Silencing, P1/HC-Pro.
Supplemental Figure 6. The Relative Quantity of SNC1 Is Comparable in the sgs3-13 or hen1-5 Mutant Compared with Wild-Type Plants.
Supplemental Table 1. Nucleotide Sequences for Oligonucleotides or Primers Used.
Supplementary Material
Acknowledgments
We thank Douglas Chalker, Barbara Kunkel, Craig Pikaard, and members of the Richards lab for helpful comments on the manuscript and Mike Dyer and the greenhouse staff for plant care. Seeds were generous gifts from James Carrington, Xin Li, Craig Pikaard, Vicki Vance, Hervé Vaucheret, and Olivier Voinnet. Additional genetic material was obtained from the ABRC. This work was supported by grants from the National Science Foundation to E.J.R. (MCB-0321990 and MCB-0548597). Additional support was provided by the Danforth Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eric J. Richards (richards@wustl.edu).
Online version contains Web-only data.
References
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11 1017–1027. [DOI] [PubMed] [Google Scholar]
- Arciga-Reyes, L., Wootton, L., Kieffer, M., and Davies, B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47 480–489. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., and Baulcombe, D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten, A., Cannon, S., Spangler, R., and May, G. (2003). Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant, I., and Izaurralde, E. (2006). Quality control of gene expression: A stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev. 20 391–398. [DOI] [PubMed] [Google Scholar]
- Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R., and Zhu, J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet, S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. (2003). Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Castelli, V., et al. (2004). Whole genome sequence comparisons and “full-length” cDNA sequences: A combined approach to evaluate and improve Arabidopsis genome annotation. Genome Res. 14 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, E.J., Prokhnevsky, A.I., Gopinath, K., Dolja, V.V., and Carrington, J.C. (2004). Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone, S.M., and Cone, K.C. (1993). Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543–553. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K.D., Carrington, J.C., and Voinnet, O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313 68–71. [DOI] [PubMed] [Google Scholar]
- Della Vedova, C.B., Lorbiecke, R., Kirsch, H., Schulte, M.B., Scheets, K., Borchert, L.M., Scheffler, B.E., Wienand, U., Cone, K.C., and Birchler, J.A. (2005). The dominant inhibitory chalcone synthase allele C2-Idf (inhibitor diffuse) from Zea mays (L.) acts via an endogenous RNA silencing mechanism. Genetics 170 1989–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeier, M.E., Morse, D.P., Knight, S.W., Portereiko, M., Bass, B.L., and Mango, S.E. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289 1928–1931. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Himber, C., Ruiz-Ferrer, V., Alioua, A., and Voinnet, O. (2007). Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 39 848–856. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Himber, C., and Voinnet, O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37 1356–1360. [DOI] [PubMed] [Google Scholar]
- Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro, A.F., Matthew, L., Smith, N.A., Curtin, S.J., Dedic-Hagan, J., Ellacott, G.A., Watson, J.M., Wang, M.B., Brosnan, C., Carroll, B.J., and Waterhouse, P.M. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli, V., Mallory, A.C., Bartel, D.P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15 1494–1500. [DOI] [PubMed] [Google Scholar]
- Heidel, A.J., Clarke, J.D., Antonovics, J., and Dong, X. (2004). Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., and Jacobsen, S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 38 721–725. [DOI] [PubMed] [Google Scholar]
- Hori, K., and Watanabe, Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43 530–540. [DOI] [PubMed] [Google Scholar]
- Howell, M.D., Fahlgren, N., Chapman, E.J., Cumbie, J.S., Sullivan, C.M., Givan, S.A., Kasschau, K.D., and Carrington, J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126 5231–5243. [DOI] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J.A., Flowers, S.K., Munakata, K., and Richards, E.J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M., and Levy, A.A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33 102–106. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Fahlgren, N., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., and Carrington, J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A., Jr., Zhu, J.K., Staskawicz, B.J., and Jin, H. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 103 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschen, A., Napoli, C.A., Jorgensen, R.A., and Muller, A.E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566 223–228. [DOI] [PubMed] [Google Scholar]
- Kim, J.K., Gabel, H.W., Kamath, R.S., Tewari, M., Pasquinelli, A., Rual, J.F., Kennedy, S., Dybbs, M., Bertin, N., Kaplan, J.M., Vidal, M., and Ruvkun, G. (2005). Functional genomic analysis of RNA interference in C. elegans. Science 308 1164–1167. [DOI] [PubMed] [Google Scholar]
- Koseki, M., Goto, K., Masuta, C., and Kanazawa, A. (2005). The star-type color pattern in Petunia hybrida ‘red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol. 46 1879–1883. [DOI] [PubMed] [Google Scholar]
- Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 20 116–122. [DOI] [PubMed] [Google Scholar]
- Li, J., Yang, Z., Yu, B., Liu, J., and Chen, X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Clarke, J.D., Zhang, Y., and Dong, X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14 1131–1139. [DOI] [PubMed] [Google Scholar]
- Liu, J., He, Y., Amasino, R., and Chen, X. (2004). siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 18 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004. a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004. b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G.B., Bogdanove, A.J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54 23–61. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S., Schauer, S.E., Smith, T.H., Mallory, A.C., Herr, J.M., Jr., Roth, B., Merchant, D.S., Ray, A., Bowman, L.H., and Vance, V.B. (2005). Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Nakano, M., Nobuta, K., Vemaraju, K., Tej, S.S., Skogen, J.W., and Meyers, B.C. (2006). Plant MPSS databases: Signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 34 D731–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel, L., Moores, T.L., van Der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11 2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Onodera, Y., Haag, J.R., Ream, T., Nunes, P.C., Pontes, O., and Pikaard, C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 613–622. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabo, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Denli, A.M., and Hannon, G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428. [DOI] [PubMed] [Google Scholar]
- Qi, Y., He, X., Wang, X.J., Kohany, O., Jurka, J., and Hannon, G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443 1008–1012. [DOI] [PubMed] [Google Scholar]
- Qin, H., Dong, Y., and von Arnim, A.G. (2003). Epigenetic interactions between Arabidopsis transgenes: Characterization in light of transgene integration sites. Plant Mol. Biol. 52 217–231. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda, M., Masuta, C., Ohnishi, S., Goto, K., Kasai, A., Sano, T., Hong, J.S., and MacFarlane, S. (2004). Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.L., Kunkel, B.N., and Richards, E.J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.L., and Richards, E.J. (2002). Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc. Natl. Acad. Sci. USA 99 7792–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29 439–451. [DOI] [PubMed] [Google Scholar]
- van Hulten, M., Pelser, M., van Loon, L.C., Pieterse, C.M., and Ton, J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crete, P., and Vaucheret, H. (2004. a). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14 346–351. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004. b). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16 69–79. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Brown, S., Patrick, E., Brearley, C., and Turner, J.G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Wilken, A., and Carrington, J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Fan, B., Chen, C., and Chen, Z. (2001). An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA 98 6516–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., and Hua, J. (2004). A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, S., Horrocks, J., and Lukens, L.N. (2006). Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J. 45 347–357. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., Zhu, J., Kapoor, A., and Zhu, J.K. (2007). Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.