Abstract
Strictosidine β-d-glucosidase (SG) follows strictosidine synthase (STR1) in the production of the reactive intermediate required for the formation of the large family of monoterpenoid indole alkaloids in plants. This family is composed of ∼2000 structurally diverse compounds. SG plays an important role in the plant cell by activating the glucoside strictosidine and allowing it to enter the multiple indole alkaloid pathways. Here, we report detailed three-dimensional information describing both native SG and the complex of its inactive mutant Glu207Gln with the substrate strictosidine, thus providing a structural characterization of substrate binding and identifying the amino acids that occupy the active site surface of the enzyme. Structural analysis and site-directed mutagenesis experiments demonstrate the essential role of Glu-207, Glu-416, His-161, and Trp-388 in catalysis. Comparison of the catalytic pocket of SG with that of other plant glucosidases demonstrates the structural importance of Trp-388. Compared with all other glucosidases of plant, bacterial, and archaeal origin, SG's residue Trp-388 is present in a unique structural conformation that is specific to the SG enzyme. In addition to STR1 and vinorine synthase, SG represents the third structural example of enzymes participating in the biosynthetic pathway of the Rauvolfia alkaloid ajmaline. The data presented here will contribute to deciphering the structure and reaction mechanism of other higher plant glucosidases.
INTRODUCTION
The large family of higher-plant monoterpenoid indole alkaloids is well defined at both the chemical and structural level. It consists of a large number of classes that, taken together, cover an enormous variety of skeletal types of fascinating complexity (Kisakürek et al., 1982). This alkaloid family was instrumental in boosting the development of mass spectrometry ∼50 years ago, helping it become one of the most powerful methods for structural elucidation of products of secondary metabolism (Hesse, 1974). Although broad biosynthetic knowledge is the prerequisite for metabolic engineering (Kutchan, 1995), the characterization of the enzymes responsible for the biosynthesis of these alkaloids is still somewhat preliminary, and details of the enzymatic formation have been obtained for only very few pathways. For instance, several enzymes were detected in cell suspensions of Catharanthus roseus G. Don, participating in the biosynthesis of ajmalicine and its isomers (Stöckigt, 1979; Treimer and Zenk, 1979). In addition, some enzymes involved in the formation of the alkaloid vindoline in plant seedlings and cell culture of C. roseus have been previously well characterized (De Luca et al., 1985; Fahn et al., 1985; St-Pierre et al., 1998; St-Pierre and De Luca, 2000). A major interest relating to the vindoline route has recently focused on the investigation of developmental regulation of biosynthesis in Catharanthus seedlings, its tissue-specific localization and comparison with undifferentiated systems, such as cell suspension cultures. Results demonstrated that an incomplete set of enzymes of vindoline formation are expressed in suspended cells, in sharp contrast with seedlings, and this explains the necessity of tissue and cellular differentiation (De Luca and St-Pierre, 2000). These findings are in agreement with the fact that vindoline has not been isolated from cultured Catharanthus cells, although it represents a typical alkaloid of Catharanthus leaves (Stöckigt and Soll, 1980). Since vindoline represents one of the ultimate biogenetic precursors of the dimeric anticancer alkaloids vinblastine and vincristine, the presence of the later alkaloids in any dedifferentiated Catharanthus cells has not been demonstrated. Therefore, a biotechnological application of Catharanthus cell suspensions for production of these highly valuable indole alkaloids has not been achieved so far. In contrast with the Catharanthus system, for alkaloids of the Indian medicinal plant Rauvolfia serpentina Bentham ex Kurz, all enzymes and stable intermediate alkaloids leading to the structurally complex alkaloid ajmaline, which exhibits nine chiral carbon atoms, have been detected in recent years from undifferentiated cell suspensions of Rauvolfia (Ruppert et al., 2005). In this case, alkaloid biosynthesis is therefore not necessarily dependent on the degree of cellular differentiation and tissue development. These results provided a comprehensive picture of the complete biosynthetic pathway and led to the development of the antiarrhythmic drug ajmaline. Including several branches of the ajmaline pathway, this represents one of the largest alkaloidal plant metabolomes known to date.
Starting from the early precursors tryptamine and secologanin, a variety of different enzymes are involved in the ajmaline pathway (Ruppert et al., 2005). Among these, two enzymes are of the most outstanding importance for biosynthesis of the entire alkaloid family. The first of these is strictosidine synthase (STR1; EC 4.3.3.2), which catalyzes the condensation of the above-mentioned precursors by a Pictet Spengler–type reaction, delivering the first biogenetic intermediate (strictosidine) of the whole alkaloid family (Stöckigt and Zenk, 1977). Heterologous expression of its cDNA (Kutchan et al., 1988) and its crystallization (Ma et al., 2004; Koepke et al., 2005) resulted in the recent elucidation of the three-dimensional structure of STR1 (Ma et al., 2006), providing both an insight into its specific catalytic mechanism as well as the structural basis for rational modulation of its substrate specificity.
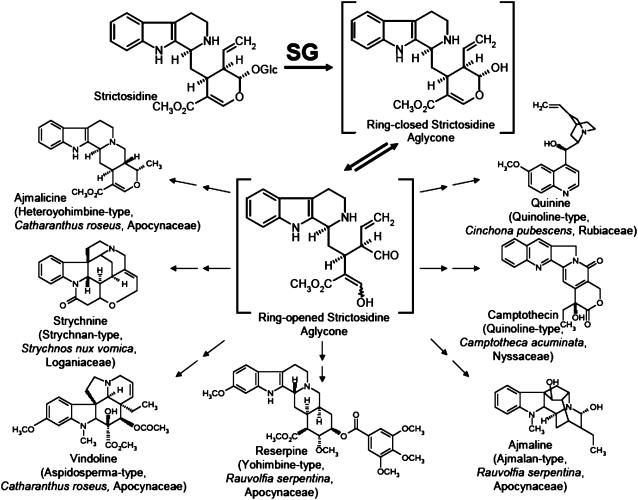
The second enzyme, the highly substrate-specific strictosidine β-d-glucosidase (SG; EC 3.2.1.105), activates the first pathway intermediate, strictosidine, by deglucosylation, forming a ring-opened, unstable, and reactive aglycone that remains to be isolated (Figure 1). The aglycone enters multiple pathways to the appropriate indole and quinoline alkaloid types in the three plant families Apocynaceae, Rubiaceae, and Loganiaceae (some of the structures are depicted in Figure 1), ultimately allowing plants to synthesize the enormous variety of ∼2000 monoterpenoid indole alkaloids. From a biosynthetic point of view, it is noteworthy that it is extremely rare for a glucoside such as the glucoalkaloid strictosidine to act as a precursor at the beginning of biosynthetic pathway and for it to become activated by deglucosylation. In natural product biosynthesis, glycosides usually appear at the end of pathways to convert end products into water-soluble compounds, which can easily be stored in the plant cell vacuole, as is the case for phenylpropanoids (Dixon and Paiva, 1995). Moreover, the deglucosylation process has been considered to potentially be part of a plant cell defense system in Catharanthus (Geerlings et al., 2000). Therefore, strictosidine glucosidase indeed plays an extraordinary role in indole alkaloid metabolism.
Figure 1.
The Key Role of the Deglucosylation of Strictosidine Catalyzed by SG in the Biosynthesis of Various Alkaloid Types Belonging to the Families of Monoterpenoide Indole and Quinoline Alkaloids.
Typical plant genera and species from which corresponding alkaloids were isolated are illustrated with the appropriate plant families.
Although SG has been previously described from Catharanthus and Rauvolfia (Hemscheidt and Zenk, 1980; Geerlings et al., 2000; Gerasimenko et al., 2002), details of the reaction mechanism of this enzyme and the reasons for its pronounced substrate specificity remain unknown. We recently developed an efficient overexpression and purification system for Rauvolfia strictosidine glucosidase in Escherichia coli, which was followed by crystallization and preliminary x-ray analysis (Barleben et al., 2005).
Here, we report the three-dimensional structure of native SG, its site-directed mutagenesis, and the structure of an inactive mutant in complex with the substrate strictosidine, thus defining the overall and active site architecture. The data deliver molecular details of the reaction center, providing insight into the formation of the enzyme-substrate complex and the catalytic mechanism of this central enzyme of indole alkaloid biosynthesis. This might help to gain a better understanding of the β-glucosidase enzyme superfamily, potentially allowing the rational design of mutant enzymes with altered substrate specificities to generate novel alkaloid libraries containing compounds with potential biological activities.
RESULTS AND DISCUSSION
Overall Structure of SG
Based on initial alignment studies, SG belongs unequivocally to the glycosyl hydrolase (GH) family 1 in the carbohydrate-active enzyme database (http://afmb.cnrs-mrs.fr/CAZY), which is based on overall amino acid sequences (Henrissat, 1991; Henrissat and Davies, 1997). GH family 1 is grouped together with 16 other families in clan GH-A, which is the biggest of the 13 clans of glycosidases. Although a number of three-dimensional structures already exist in family 1, only eight are from eukaryotic sources, six of which are of plant origin. These GHs are well known to be involved in plant defense mechanisms.
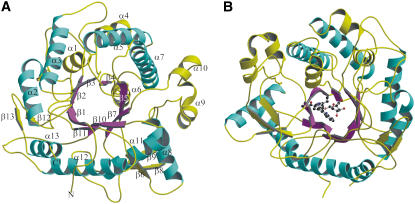
The overall fold of SG from Rauvolfia is shown in Figure 2A. SG has the expected (β/α)8 barrel fold of the large family 1 of glycosidases, which consists of ∼640 enzymes. The structure consists of 13 α-helices and 13 β-strands. The core of the structure contains eight parallel β-strands forming a β-barrel, and the barrel is surrounded by eight helices. Each β/α repeat is either connected by a loop or a combination of loops and helices or strands at the top of the barrel. The barrel hosts a binding site for the natural substrate strictosidine. The groove leading to this catalytic center is mainly formed by irregular loops between the secondary structures located on top of the enzyme (Figure 2B). The outer region of the structure also contains one main three-stranded, antiparallel β-sheet.
Figure 2.
Overall Structure of SG from R. serpentina, Illustrating Its (β/α)8 Fold, the Groove Leading to the Catalytic Center, and the Secondary Structure Elements.
(A) (β/α) repeats are illustrated in magenta and cyan, respectively. The extra helices and strands as well as connecting loops are shown in yellow. The secondary elements are labeled.
(B) (β/α)8 fold is shown in complex with the substrate strictosidine in ball-and-stick representation. The structure is rotated by 170° around the y axis with respect to (A). Color code is same as depicted in (A).
Site-Directed Mutagenesis and Reaction Mechanism
Sequence alignment of SG with glucosidases of various origins indicates complete conservation of amino acid residues Glu-207, Glu-416, and His-161. Based on earlier site-directed mutation experiments in the glucosidase family, Glu-207 is the proton donor that allows nucleophilic attack of Glu-416 at the anomeric carbon C-1 (McCarter and Withers, 1994). Both glutamic acids catalyze the concerted hydrolysis of the glucoside bond. In fact, mutations Glu207Gln, Glu207Asp, Glu416Gln, and Glu416Asp resulted in loss of enzyme activity, highlighting the importance of both glutamates for the deglucosylation of strictosidine (Table 1).
Table 1.
Kinetic Constants of SG Mutants
| Relative Activity (%) | Km (μM) | Kcat (s−1) | Kcat/Km (s−1 mM−1) | |
|---|---|---|---|---|
| Wild type | 100 | 90 | 9.8 | 106.4 |
| Phe373Thr | 100 | 2640 | 952.4 | 360.9 |
| Tyr481Phe | 90 | 220 | 27.0 | 121.4 |
| Gly386Ser | 10 | 370 | 3.4 | 9.3 |
| Trp388Ala | 1 | 720 | 0.3 | 0.4 |
| Glu207Asp | <0.1 | ND | ND | ND |
| Glu207Gln | <0.1 | ND | ND | ND |
| Glu416Asp | <0.1 | ND | ND | ND |
| Glu416Gln | <0.1 | ND | ND | ND |
| His161Asn | <0.1 | ND | ND | ND |
| His161Leu | <0.1 | ND | ND | ND |
For relative activity, the activity of the wild-type enzyme is set at 100. The detection limit was 0.1%. For mutants with detectable enzyme activity, further kinetic constants are given. ND, not detectable.
His-161 is also likely to play a critical role in the reaction catalyzed by SG because its replacement by Asn or Leu reduced enzyme activity to <0.1% (Table 1). His-161 has been shown to be an important residue in other glucosidases because it binds to the glucosidic part of the substrate and stabilizes the transition state during hydrolysis (Barrett et al., 1995). Based on these results, it was assumed that the Glu-207, Glu-416, and His-161 residues must be involved in the reaction mechanism encompassing strictosidine deglucosylation and should thus be located near the binding pocket or even within the catalytic center of SG. Two further mutants were prepared: Tyr481Phe and Gly386Ser. Whereas the first mutant still resulted in a high activity (90% relative activity), the larger substituent (Ser) at position 386 resulted in a 90% decrease in hydrolytic activity (Table 1). The explanation for these results became apparent upon analysis of the three-dimensional structure of native SG and the complex of an inactive mutant with strictosidine (see the next two Results sections).
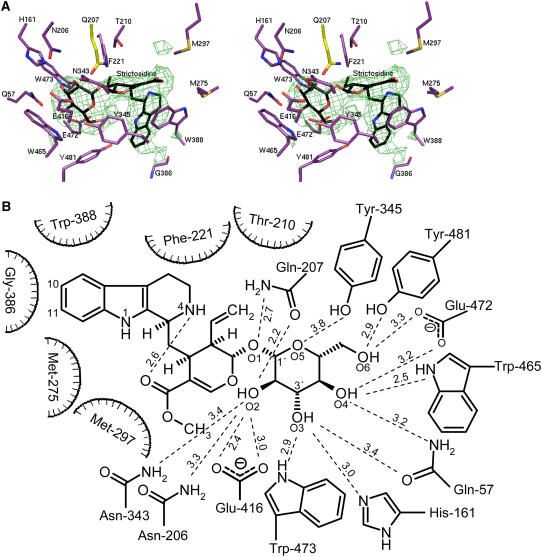
To gain additional information on the role of other individual amino acids of the binding pocket, the structure of the inactive mutant SG-Glu207Gln complexed with strictosidine was determined (Figures 3A and 3B).
Figure 3.
Structural Representation of SG: Ligand Complexes.
(A) Stereo view of strictosidine binding site of SG. The (Fo-Fc) SIGMAA-weighted electron density of strictosidine contoured at 4 σ is shown in green and strictosidine in black. In complex with the inactive mutant (Glu207Gln), residues within 4.1-Å distance from strictosidine are shown in violet, and Gln-207 is in yellow.
(B) Hydrogen binding network between the glucosidic part of strictosidine and residues in a distance ≤4.1 Å in the ligand structure of SG inactive mutant Glu207Gln.
Analysis of the Inactive SG-Glu207Gln Mutant in Complex with the Substrate Strictosidine and Architecture of the Catalytic Center
The Glu207Gln mutant showed no detectable catalytic activity with strictosidine compared with wild-type SG (Table 1), allowing soaking experiments with the natural substrate strictosidine to be performed. The structure of SG-Glu207Gln in complex with the strictosidine substrate was determined at 2.82 Å from tetragonal crystals soaked in ligand-containing solution (see Methods for details). The electron density was clearly visible for both the glycone and aglycone parts of the substrate molecule, allowing detailed characterization of the catalytic center of SG in the substrate binding state (Figures 3A and 3B). The binding pocket is located at the top of the barrel. The base of the pocket contains several charged residues, whereas the gate to the pocket is hydrophobic in nature. The glycone part is bound deep in the pocket, and the aglycone part points away toward the solvent, even though the aglycone part of the substrate is hydrophobic in nature. The aglycone part is surrounded by residues Phe-221, Trp-388, Gly-386, Met-275, Thr-210, and Met-297, the majority of which are hydrophobic (Figure 3B). Important residues for recognition of the aglycone unit seem to be Gly-386 and Trp-388. Gly-386 is in very close proximity to the indole system of strictosidine; therefore, larger residues decrease the enzyme activity, as demonstrated for the Gly386Ser mutant (Table 1). Trp-388 is discussed in the next paragraph. The glucose moiety interacts with a number of hydrophilic residues (Asn-206, Gln-207, Asn-343, Tyr-345, Glu-416, Trp-473, His-161, Gln-57, Trp-465, Glu-472, and Tyr-481; Figures 3A and 3B). As shown in Figures 3A and 3B, Gln-207 and Glu-416 of SG are located within the pocket near the sugar moiety, leaving a distance of 5.2 Å between their carboxyl carbons. This distance offers enough space for substrate entry and to place the glucosidic bond in an optimal position for hydrolysis. His-161 is not a catalytic residue, being located ∼5.8 Å away from the anomeric C-atom of the glucose moiety but forms a hydrogen bond to the O3 of the sugar moiety of strictosidine, helping to hold the substrate in the correct orientation for deglucosylation (Figures 3A and 3B). Tyr-481 corresponds to Tyr-473 in maize (Zea mays) β-glucosidase Zm Glu1. This Tyr was suggested to be important for aglycone recognition by the maize enzyme (Czjzek et al., 2000) and, moreover, is hydrogen bonded with the amide group of Trp-378 (Trp-388 in SG) (Verdoucq et al., 2003). Although Tyr-481 is positioned for a weak hydrogen bond with the glucose moiety, its major role is more likely related to its hydrophobic nature because the mutant Tyr481Phe still showed high enzyme activity (Table 1).
SG exhibits a pH optimum between 5.0 and 5.2, conditions in which the catalytic Glu-207 would not be expected to act as an acid. However, as suggested by White and Rose (1997), in all β-glycosyl hydrolases the pKa of the acidic residue Glu-207 in SG is expected to undergo significant changes of two to three pH units during the reaction. The high pKa of the proton donor Glu-207 might be due to electrostatic interactions with other carboxylate groups, as is the case for xylanases (Davoodi et al., 1995), or might be dependent upon a local pronounced hydrophobic environment (Keresztessy et al., 1994). Inspection of the binding pocket of SG around Glu-207 and Glu-416 indicates that both catalytic amino acids are shielded by a number of hydrophilic residues, which all are within a distance of 3.6 to 4.9 Å. In particular, the protonated Glu-207 is surrounded by the five residues Asn-273, Asn-343, Asn-206, Thr-210, and Trp-162, providing an overall hydrophilic pocket, whereas Glu-416 is only surrounded by Trp-465 and Tyr-345. All residues mentioned here, except Trp-162 and Asn-273, are within a 4.1 Å distance from strictosidine (Figures 3A and 3B).
Comparison of the Substrate Binding Site of SG with Other Plant Glucosidases
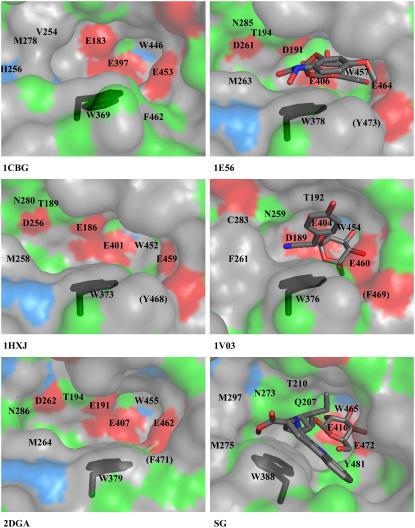
Here, we compare the binding pocket of SG-Glu207Gln in complex with strictosidine from R. serpentina with five glucosidases of plant origin, two of them as substrate complexes: cyanogenic β-glucosidase (CBG) (EC 3.2.1.21; Protein Data Bank [PDB] code 1CBG) from Trifolium repens; Zm Glu1 (EC 3.2.1.21; PDB code 1E1E; Zm Glu1-Glu191Asp in complex with DIMBOA-β-d-glucoside; PDB code 1E56) from Z. mays; Zmp60.1 (EC 3.2.1.21; PDB code 1HXJ) from Z. mays; Ta Glu1b (EC 3.2.1.21; PDB code 2DGA) from Triticum aestivum; and Dhurrinase1 (Dhr1) (EC 3.2.1.21; PDB code 1V02; Dhr1-Glu189Asp-dhurrin; PDB code 1V03) from Sorghum bicolor. The sequence identity and root mean square deviation of these plant glucosidases with reference to SG are in the range of 43 to 54% and 0.79 to 0.90 Å, respectively (see Supplemental Table 1 online). Ten of the eleven amino acids (Figure 3B) involved in the binding of the glucose moiety are conserved in the sequences of the glycosidases in family 1. In some glucosidases, only Tyr-481 is replaced by Phe (see Phe-462, Phe-469, and Phe-471 in 1CBG, 1V03, and 2DGA, respectively, in Figure 4; see Supplemental Figure 1 online). The root mean square deviation of the Cα of all 11 amino acids from the glucosidases with reference to SG ranges from 0.32 to 0.37 Å. In context with the acquired resolutions (1.8 to 2.5 Å), the different structures can be accepted as identical because the coordinate errors of the compared structures are in the range of 0.46 to 0.76 Å. With respect to the side chains, superimposed structures show a single difference at position SG-Glu472. As described by Verdoucq et al. (2004), the aglycone interactions are responsible for the orientation of the sugar in its binding pocket. Depending on the position of the aglycone, the sugar can rotate by 60°. The hydroxyl groups change their binding partners accordingly, only O6 of the glycone coordinates in both orientations with Glu-472. The side chain of Glu-472 adopts to the rotation as it is visible when the structures are superimposed (data not shown).
Figure 4.
Comparison of the Catalytic Center of Crystallized Plant Glucosyl Hydrolases of Family 1.
The conformation of Trp-388 is highlighted in black. Hydrophobic residues (Trp, Phe, Leu, Met, Cys, Ile, Ala, Gly, and Val), positively charged residues (His), negatively charged residues (Glu and Asp), and hydrophilic residues (Ser, Thr, Tyr, Pro, Gln, and Asn) are shown in gray, blue, red and green, respectively. 1CBG, cyanogenic β-glucosidase from T. repens; 1E56, Zm Glu1-Glu191Asp in complex with DIMBOA-β-d-glucoside from Z. mays; 1HXJ, Zmp60.1 from Z. mays; 1V03, Dhurrinase1-Glu189Asp in complex with dhurrin from S. bicolor; 2DGA, Ta Glu1b from T. aestivum; SG, strictosidine glucosidase-Glu207Gln in complex with strictosidine from R. serpentina. Equivalent residues are labeled (in 1CBG, there are nine instead of 10 residues displayed because the SG-corresponding Thr-210 is replaced by Gly, which is out of figure margins). Residues in brackets are screened from sight by other amino acids.
By contrast, the aglycone binding site is much more variable. The most striking difference in their active sites is observed for the residue Trp-388. This residue is conserved within the enzyme family 1, and its role in substrate recognition has been described previously (Czjzek et al., 2000). Figure 4 shows the conformation of Trp-388 from various glucosidases of plant origin. The Trp in all known plant glucosidases shows its χ angle ∼60° (which we term here plant glucosidase-typical conformation), but in SG, this conformation is changed to a χ angle of −180° and is named here as the SG-typical conformation. Such a change in orientation leads to more space within the catalytic pocket of SG, which may be required to accommodate the relative large substrate, strictosidine, compared with the smaller substrates acted upon by other β-glucosidases. More importantly, the residue Trp-388 forms a stacking interaction with strictosidine, as it does in other enzyme-substrate complexes (Czjzek et al., 2000). Moreover, based on this conformational change, the amino acids Gly-386, Met-275, Thr-210, and Met-297 become accessible to strictosidine (Figure 3B). Together with Phe-221 and Trp-388, which are also of importance in other β-glucosidases, all these amino acids form the substrate binding pocket of SG, as far as the hydrophobic part of strictosidine is concerned.
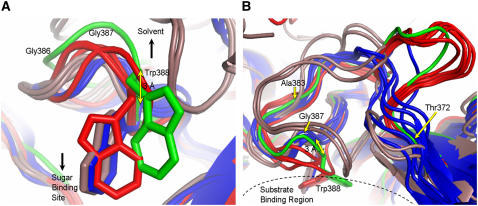
The reason why Trp-388 exists in an altered conformation in SG is not obvious. The different conformation of Trp-388 is not due to complex formation with strictosidine because, in the native structure of SG, Trp-388 is present in the same conformation as in the complex. To obtain a detailed insight, we performed a structure-based alignment of all the mentioned glucosidases (see Supplemental Figure 1 online). In SG, Trp-388 is in vicinity of the four amino acids Tyr-345, Thr-346, Phe-373, and Met-275. Analysis of these residues shows that Tyr-345 is part of the glycone recognition site and is completely conserved in plant glucosidases (Figures 3A and 3B; see Supplemental Figure 1 online). Thr-346 is also strictly conserved except in CBG. Phe-373 is found in CBG too, and even in the sequence of SG from C. roseus it is replaced by Ile-380, indicating that Phe-373 is of low significance for catalysis. When Phe-373 of R. serpentina SG was exchanged with Thr, the resulting mutant showed the same relative activity and a higher catalytic efficiency compared with the wild-type enzyme (Table 1). Met-275 is also present in both glucosidases from Z. mays. It is clear that all these residues have no direct or significant influence on the conformation of Trp-388, although they are located nearby. In the case of Dhr1, the SG-Met275 position is occupied by Phe, which is too close (2 Å) to support the Trp-388 conformation, typical of SG. In Zm Glu1, Zmp60.1, and Ta Glu1b, it is Arg-348, and in CBG, it is Tyr-348 (SG numbering; see Supplemental Figure 1 online), which might push the side chain of Trp by virtue of its size and hydrophilic nature to adopt the plant glucosidase-specific conformation. Perhaps these bulky residues either at position 275 or 348 or both positions may have an effect on the different conformation of Trp-388. The SG-typical conformation of Trp-388 remained an exception even if structural comparison was made with the other 15 glycosidases of family 1 (Figures 5A and 5B), despite those of thioglycoside-hydrolyzing enzymes (myrosinases), in which the appropriate Trp is absent. Structural comparison now indicates the presence of three different conformations of the Trp: (1) Trp points directly into the binding pocket (glycosidases from bacteria and archaea); (2) Trp points in the direction of the N terminus backbone (plant-specific conformation); and (3) Trp points in the direction of the C terminus backbone (SG-specific conformation). Figure 5B also illustrates structural differences in the region between Thr-372 and Trp-388, which depends on the origin of the enzymes. The conformation of the loop starting from Thr-372 to Trp-388 is variable. However, glycosidases from bacteria exhibit smaller loops (blue color). All plant-derived enzymes show significantly larger loops (red color, green for SG), with the major difference of the conformation of Trp-388. If additional examples of the SG-specific Trp-388 conformation are discovered in the future, β-glucosidases might then be structurally subdivided based on the new conformation of the Trp.
Figure 5.
Comparison of the Region around Trp-388 with Respect to Bacteria, Archaea, and Higher Plants.
(A) The three different conformations of Trp-388 in 16 β-glycosidases from bacteria (blue), archaea (brown), higher plants (red), and SG from Rauvolfia (green) described in this study. The location of the sugar binding site compared with the Trp side chains and direction to the enzyme surface are indicated by black arrows. Gly-386 and Gly-387 are labeled to indicate the direction to the N terminus, and the distance between Cα of Trp-388 in microbial and eukaryotic enzymes is illustrated by a yellow arrow. The sequence numbering is according to SG. The structures shown are as follows: 1NP2, β-glycosidase from Thermus nonproteolyticus HG102; 1UG6, β-glycosidase (TTHB087) from Thermus thermophilus HB8; 1GNX, β-glucosidase (Bgl3) from Streptomyces sp QM-B814; 1OD0, β-glucosidase A (BglA) from Thermotoga maritima MSB8; 1QOX, β-glucosidase (BglA) from Bacillus circulans subsp Alkalophilus; 1E4I, β-glucosidase A (BglA) from Paenibacillus polymyxa; 1CBG, cyanogenic β-glucosidase from T. repens; SG, strictosidine glucosidase from R. serpentina; 1E1E, Zm Glu1 from Z. mays; 1HXJ, Zmp60.1 from Z. mays; 1V02, Dhurrinase1 from S. bicolor; 2DGA, Ta Glu1b from T. aestivum; 1PBG, 6-P-β-galactosidase from Lactococcus lactis Z268; 1VFF, alkyl β-glycosidase (BGPh; PH0366) from Pyrococcus horikoshii OT3; 1GOW, β-glycosidase S (LacS;S-β-gly; SSO3019) from Sulfolobus solfataricus P2; 1QVB, β-glycosidase from Thermosphaera aggregans M11TL.
(B) To illustrate the different backbone orientations of the glycosidases, (A) was rotated by 90° around the x axis (colors are as in [A]). Near Thr-372 (yellow arrow), all the main chains diverge in three groups depending on their origin (bacteria, achaea, and higher plants). Archaea and bacteria backbones merge around residue Ala-383 (yellow arrow). Microbial and plant main chains merge after Trp-388. For better recognition, Gly-387 (yellow arrow) and the substrate binding region are marked. The sequence numbering is according to SG.
Functional Role of SG
It is generally accepted that one of the major functions of β-glucosidases in higher plants is defense related because of the formation of bitter or toxic small molecules, such as cyanide, isothiocyanates, or DIMBOA (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one) from their corresponding glucosides (Niemeyer, 1988; Poulton, 1990; Czjzek et al., 2001). A defense role was also discussed for the glucoside strictosidine and its deglucosylation product(s) generated by SG in C. roseus cells. Formed products have been proven to be active against several microorganisms leading to the conclusion that the compounds are involved in a plant protecting antimicrobial action (Luijendijk et al., 1996). Localization studies in Catharanthus cells indicated the storage of strictosidine to be in the vacuole, well separated from its specific glucosidase (SG) on the outside of the tonoplast (Stevens et al., 1993) or as later described in the endoplasmic reticulum (Luijendijk et al., 1998; Geerlings et al., 2000). SG has also been reported earlier to be a cytosolic enzyme (Deus-Neumann and Zenk, 1984). In addition, Rauvolfia SG lacks the signal peptide for translocation of the enzyme into the endoplasmic reticulum, making its localization in that compartment unlikely (Gerasimenko et al. 2002).
Regardless of its localization, SG and its substrate strictosidine, which shows the typical attributes of a classic defense-related glucoside, have been previously proposed to act as a damage-inducible biochemical defense system (Geerlings et al., 2000). Despite this, the function of SG is to generate the basic biosynthetic building block(s) for entering the multiple pathways illustrated in Figure 1 that are required for the synthesis of ∼2000 alkaloids. Apart from SG, there are only two additional examples in natural product biosynthesis in which the importance of glucosides and the appropriate glucosidases seem to be of synthetic relevance. Coniferin (coniferyl alcohol 4-O-β-d-glucoside) is discussed as the storage form of coniferyl alcohol, which is a biosynthetic precursor of one of the most stabilizing elements in higher plants, the lignin polymer (Tsuji et al., 2005). In addition to this, the glucosides deacetylipecoside and deacetylisoipecoside deliver the appropriate aglycone precursors for the biosynthesis of the relatively small family of the monoterpenoid isoquinoline alkaloids (De-Eknamkul et al., 2000). In contrast with SG, the three-dimensional structures of these enzymes are yet to be determined. In conclusion, strictosidine glucosidase may play a dual role in indole alkaloid metabolism, being of both defense- and synthesis-related importance.
Conclusion and Future Aspects of SG
Many attempts have been made in the past to increase the levels of alkaloid accumulation in in vitro plant organ systems (such as hairy roots) (Hughes et al., 2004) or in undifferentiated cell cultures (Endreß, 1994). Several strategies have been used to induce biosynthetic efficiency in plant cell suspensions, including application of growth regulators and exposure to various stress conditions, and of signaling molecules, such as methyl jasmonate (Blechert et al., 1995). Such approaches to enhance productivity might be successful for few selected natural products of special interest and high value. However, future demand of complex plant secondary metabolites of extreme structural diversity combined with variable biological properties will probably depend on a number of factors: detailed biosynthetic knowledge and rational molecular engineering of the enzymes and their synthetic application and likely by coexpression of appropriate biosynthetic genes in fast-growing cells, such as E. coli (Martin et al., 2003). Because of the decisive and multiple role of the aglycone of strictosidine described here, the enzyme SG may serve as one of the most attractive candidates for such an approach if its catalytic site and its substrate specificity can be broadly modulated.
Since the three-dimensional structure of the preceding enzyme in indole alkaloid biosynthesis, strictosidine synthase, has recently become available (Ma et al., 2006), initial attempts to engineer its substrate acceptance were successful (Loris et al., 2007). This result provides one of the basic requirements for future generation of large alkaloid libraries by structure-based enzyme design of STR1 and SG in combination with biomimetic approaches (Stöckigt and Ruppert, 1999).
METHODS
Overexpression, Purification, and Crystallization of Native SG
SG was obtained by transformation of Escherichia coli strain M15[pREP4] (Qiagen) using Rauvolfia serpentina SG cDNA (accession number AJ302044) as an SG-pQE-2 plasmid construct (Barleben et al., 2005). The bacterial culture was grown and the enzyme isolated and purified by nickel-nitrilotriacetic acid affinity chromatography as described (Barleben et al., 2005). The purity of SG was analyzed by SDS-PAGE and its concentration determined by the Bradford (1976) method. For crystallization, SG was concentrated to 4 mg/mL. Buffer containing 0.3 M (NH4)2SO4, 0.1 M NaOAc, pH 4.6, and 10% polyethylene glycol (PEG) 4000 was used as a precipitant. Crystals were grown by the hanging drop vapor diffusion method. Drops containing 2 μL of protein solution and 2 μL of precipitant buffer were equilibrated against 700 μL of precipitant buffer at 23°C, and crystals appeared in 1 to 3 d. Crystals of SG belonged to space group P4212 (Barleben et al., 2005).
Generation of SG Mutants by Site-Directed Mutagenesis
Mutagenesis was achieved using the QuickChange site-directed mutagenesis kit (Stratagene) and the SG-pQE-2 plasmid as template for PCR (Barleben et al., 2005). Using the following primer pairs, 10 different mutants (Glu207Asp, Glu207Gln, Glu416Asp, Glu416Gln, His161Asn, His161Leu, Phe373Thr, Gly386Ser, Trp388Ala, and Tyr481Phe) were generated (bases coding for substituted amino acids are underlined): Glu207Aspfor, 5′-GGACGACGTTCAATGATCCACATACCTTCGC-3′; Glu207Asprev, 5′-GCGAAGGTATGTGGATCATTGAACGTCGTCC-3′; Glu207Glnfor, 5′-GGACGACGTTCAATCAGCCACATACCTTCGC-3′; Glu207Glnrev, 5′-GCGAAGGTATGTGGCTGATTGAACGTCGTCC-3′; Glu416Aspfor, 5′-CCGGTGCTATATGTCACAGATAGTGGGATGG-3′ ; Glu416Asprev, 5′-CCATCCCACTATCTGTGACATATAGCACCGG-3′; Glu416Glnfor, 5′-CCGGTGCTATATGTCACACAGAGTGGGATGG-3′; Glu416Glnrev, 5′-CCATCCCACTCTGTGTGACATATAGCACCGG-3′; His161Asnfor, 5′-CCGTAACTCTCTTCAACTGGGATCTTCCTC-3′; His161Asnrev, 5′-GAGGAAGATCCCAGTTGAAGAGAGTTACGG-3′; His161Leufor, 5′-CCGTAACTCTCTTCAAATGGGATCTTCCTC-3′; His161Leurev, 5′-GAGGAAGATCCCATTTGAAGAGAGTTACGG-3′; Phe373Thrfor, 5′-GATCAAGTTACTAAGACTACTGAACGGAACCAAAAACCC-3′; Phe373Thrrev, 5′-GGGTTTTTGGTTCCGTTCAGTAGTCTTAGTAACTTGATC-3′; Gly386Serfor, 5′-CATTGGTCATGCGTTGTATAGCGGGTGGCAGCATGTCGTTC-3′; Gly386Serrev, 5′-GAACGACATGCTGCCACCCGCTATACAACGCATGACCAATG-3′; Trp388Alafor, 5′-CGTTGTATGGAGGGGCGCAGCATGTCGTTCCTTGG-3′; Trp388Alarev, 5′-CCAAGGAACGACATGCTGCGCCCCTCCATACAACG-3′; Tyr481Phefor, 5′-GGGTTATATATGTCGTTTTGGAATTATTCATGTTG-3′; Tyr481Pherev, 5′-CAACATGAATAATTCCAAAACGACATATATAACCC-3′. The resulting plasmids were transformed into E. coli TOP 10 for amplification, purified with the Nucleo Spin plasmid kit (Macherey-Nagel), and sequenced.
Enzyme Activity
Enzyme reactions were analyzed by HPLC as described by Gerasimenko et al. (2002). Mutants were tested by determination of the relative enzyme activity. In a total volume of 100 μL, 0.2 μg native SG, mutant or denatured SG, was incubated with strictosidine (90 μM) for 5 min at 30°C in presence of 100 mM KPi, pH 5.2; decreasing concentrations of substrates were compared in the assay. For mutants with a low activity, enzyme concentrations were increased to 10 μg SG, and incubation periods were prolonged to 50 min, leading to a detection limit of <0.1% relative activity. The Km values were determined in a similar way, with enzyme concentrations depending on the activity of the mutant from 0.05 to 5 μg and substrate concentration between 0.02 and 1 mM.
Crystallization of the Inactive Mutant SG-Glu207Gln and Preparation of Its Complex with the Substrate Strictosidine
Expression and purification of the SG-Glu207Gln mutant were performed in a similar way to wild-type SG, with optimal crystallization conditions found to be 0.35 M (NH4)2SO4, 0.1 M NaOAc, pH 4.6, and 11% PEG 4000 as precipitant buffer and 20°C for incubation temperature. Crystals of the SG-Glu207Gln mutant were transferred into soaking buffer [0.2 M (NH4)2SO4, 0.1 M NaOAc, pH 4.6, 9% PEG 4000, 5 mM strictosidine, and 2 mg/mL SG] for 4 h.
X-Ray Data Collection and Processing
Both native and strictosidine complex SG crystals were cryoprotected by addition of 20 to 25% glycerol to the precipitant buffer before flash-cooling in a stream of gaseous nitrogen at 100K. X-ray diffraction data from native crystals were collected on the BM14 beamline of the European Synchrotron Radiation Facility (ESRF) (Grenoble, France). A total of 240 images were measured with a 0.5° rotation per frame with a MARCCD 225-mm camera (Mar Research). Diffraction data from strictosidine-SG complex crystals were collected on the X13 beamline of the European Molecular Biology Laboratory at the DORIS storage ring of the Deutsches Elektronen-Synchrotron (Hamburg, Germany). A total of 1440 images were measured with 0.25° per frame with the MARCCD 165-mm camera. The beam properties coupled with a large mosaicity (0.3°) of the crystal, point spread function, and the small size of the detector (165 mm diameter) limit the attainable resolution for a 159 Å axis to 2.82 Å. Both data sets were indexed and processed with the program XDS and scaled employing XSCALE (Kabsch, 1993).
Structure Determination of Native SG, Model Building, and Refinement
The structure of native SG was determined by the method of molecular replacement using the program EPMR (Kissinger et al., 1999) in space group P4212. From packing considerations (Matthews, 1968), the content of one asymmetric unit of SG crystals was assumed to be two monomers. The structure of the homologous maize (Zea mays) enzyme (Czjzek et al., 2001) (PDB code 1E1E) was used, and its single monomer was used as a search model. Native data set (Table 2) was used for molecular replacement. Data in the range 10 to 4 Å were used for the rotation and translational function search. The best solution obtained for first monomer had a correlation coefficient of 0.26. The search for the second molecule was performed by fixing the first that yielded a solution with a correlation coefficient of 0.48. The output model from EPMR was subjected to rigid body refinement using CNS (Brünger et al., 1998), using the native data (Table 2) in the resolution range of 20 to 3.0 Å. A random set containing 2.4% of the total data was excluded from the refinement, and the agreement between calculated and observed structure factors corresponding to these reflections (Rfree) was used to monitor the progress of the refinement (Brünger, 1993). The crystallographic free R-factor (Rfree) was 40.4%. Data were then used in the refinement in the resolution range 20 to 2.48 Å. First, a round of positional and temperature factor refinement was performed, which lowered the R-factor to 29.3% and the Rfree to 34.4%. At this stage, sigmaA-weighted maps (Read, 1986) were calculated, and careful examination of the maps allowed corrections to be incorporated into the model. The graphics program XtalView/Xfit (McRee, 1999) was used for rebuilding of the model, which was refined using REFMAC5 (Murshudov et al., 1997; Winn et al., 2001). When the sequence of the first monomer was completely docked in the electron density, it was superimposed on the second monomer. The two monomers of the resultant model were then refined using the simulated annealing protocol of CNS. Manual building alternated with additional REFMAC cycles, which included bulk solvent correction, anisotropic scaling, and definition of each molecule as a TLS group in the modeling of anisotropy in the program REFMAC5. Following refinement, the root mean square deviation of all Cα atoms between the molecules in the asymmetric unit of SG is 0.32 Å. In the final rounds of refinement, 145 water molecules were included in the model. The refinement converged with good model parameters and R/Rfree factors of 21.1/24.2% (Table 2). The SG model exhibits all amino acids except the terminal residues 1 to 36 and 509 to 532, respectively, as well as five residues 355 to 359 being part of a loop at the surface of the enzyme. The refinement statistics for the native structure are shown in Table 2. Following refinement, the root mean square deviation of all Cα atoms between the molecules in the asymmetric unit of SG is 0.32 Å. The overall geometric quality of the model was assessed using PROCHECK (Laskowski et al., 1993). A total of 88.5% of the amino acid residues of SG were found in the most favorable, 10.5% in additionally allowed, and 1.0% in generously allowed regions of the Ramachandran plot, and no residues were in the disallowed region.
Table 2.
Data Collection and Refinement Statistics of SG
| Data Set | SG-Native | SG-Glu207Gln- Strictosidine Complex |
| Wavelength (Å) | 1.2828 | 0.8048 |
| Unit cell (Å) | a = b = 157.6 | a = b = 159.0 |
| c = 103.6 | c = 102.9 | |
| Space group | P4212 | P4212 |
| Total reflections | 441757 | 930353 |
| Unique reflections | 46616 | 32951 |
| Mosaicity | 0.22 | 0.32 |
| Resolution (Å) | 20–2.48 | 20–2.82 |
| Completeness (%)a | 99.7 (100) | 98.4 (99.1) |
| I/σ (I) | 14.6 (4.6) | 20.2 (7.6) |
| Rmerge (%)b | 9.1 (27.5) | 5.8 (11.0) |
| Refinement | ||
| Resolution (Å) | 20–2.48 | 20–2.82 |
| Rcryst/Rfree (%)c | 21.1/24.2 | 22.2/28.0 |
| Average B (Å) for protein | 28.0 | 22.0 |
| Average B (Å) for strictosidine | – | 34 |
| Number of atoms | ||
| Nonhydrogen | 7679 | 7607 |
| Water | 145 | 77 |
| Root mean square deviations | ||
| Bond (Å) | 0.013 | 0.014 |
| Angles (°) | 1.60 | 1.62 |
The values in parentheses correspond to the last resolution shell.
Rmerge = ΣhklΣi|Ii (hkl) − < I(hkl) >|/ΣhklΣi< Ii(hkl) >, where < I(hkl) > is the average intensity over symmetry equivalent reflections.
Rcryst (Rfree) = Σhkl‖Fo(hkl)| −|Fc(hkl)‖ /Σhkl|Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
Structure Determination of the SG-Glu207Gln-Strictosidine Complex
The soaked crystals were nonisomorphous with the native protein crystals. The structure of the complex was therefore solved using the molecular replacement protocol of Auto-Rickshaw: the automated crystal structure determination platform (Panjikar et al., 2005). The native structure was used as a search model. Within the pipeline, the program MOLREP (Vagin and Teplyakov, 1997) was used for molecular replacement. The rigid body, positional and B-factor refinement were performed using the program CNS. Refinement was further achieved using REFMAC5 in a similar manner to that used for the native structure. The root mean square deviation of all Cα atoms between the asymmetric unit of native enzyme and mutant Glu207Gln is 0.29 and 0.39 Å for chains A and B, respectively. The crystallographic information is summarized in Table 2.
Structural Analysis
Structure-based sequence alignment was performed using the program STAMP (Russell and Barton, 1992), and the sequence alignment was displayed with the program ESPRIPT (Gouet et al., 1999). The coordinate errors were calculated with the program SFTOOLS (Collaborative Computational Project, Number 4, 1994). The two-dimensional diagram (Figure 3B) was prepared using CHEMDRAW and edited using Microsoft PowerPoint. All figures were produced with the programs MOLSCRIPT (Kraulis, 1991), RASTER3D (Merrit and Murphy, 1994), and PyMol (DeLano, 2002).
Structural Data Deposition
The model coordinates and structural factor amplitudes have been deposited in the PDB for structures of the SG-Native (2JF7) and SG-E207Q-Strictosidine Complex (2JF6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Structure-Based Alignment of β-Glucosidases of Family 1 from Higher Plants.
Supplemental Table 1. Overall Comparison of Plant Glucosidases to SG.
Supplementary Material
Acknowledgments
Staff members of the EMBL X13 beamline at the DORIS storage ring (DESY, Hamburg, Germany) and BM14 beamline at ESRF (Grenoble, France) are kindly appreciated for their help. We also thank the Deutsche Forschungsgemeinschaft (Bonn, Bad-Godesberg, Germany), the Fonds der Chemischen Industrie (Frankfurt/Main, Germany), and the Bundesministerium für Bildung und Forschung (Bonn, Germany) for financial support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joachim Stöckigt (stoeckig@mail.uni-mainz.de).
Online version contains Web-only data.
References
- Barleben, L., Ma, X.Y., Koepke, J., Peng, G., Michel, H., and Stöckigt, J. (2005). Expression, purification, crystallization and preliminary X-ray analysis of strictosidine glucosidase, an enzyme initiating biosynthetic pathways to a unique diversity of indole alkaloid skeletons. Biochim. Biophys. Acta 1747 89–92. [DOI] [PubMed] [Google Scholar]
- Barrett, T., Suresh, C.G., Tolley, S.P., Dodson, E.J., and Hughes, M.A. (1995). The crystal structure of a cyanogenic beta-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure 3 951–960. [DOI] [PubMed] [Google Scholar]
- Blechert, S., Brodschelm, W., Holder, S., Kammerer, L., Kutchan, T.M., Mueller, M.J., Xia, Z.Q., and Zenk, M.H. (1995). The octadecanoic pathway: Signal molecules for the regulation of secondary pathways. Proc. Natl. Acad. Sci. USA 92 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. (1993). Assessment of phase accuracy by cross validation: The free R value. Methods and applications. Acta Crystallogr. D Biol. Crystallogr. 49 24–36. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Czjzek, M., Cicek, M., Zamboni, V., Bevan, D.R., Henrissat, B., and Esen, A. (2000). The mechanism of substrate (aglycone) specificity in beta-glucosidases is revealed by crystal structures of mutant maize beta-glucosidase-DIMBOA, -DIMBOAGlc, and dhurrin complexes. Proc. Natl. Acad. Sci. USA 97 13555–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czjzek, M., Cicek, M., Zamboni, V., Burmeister, W.P., Bevan, D.R., Henrissat, B., and Esen, A. (2001). Crystal structure of a monocotyledon (maize ZMGlu1) beta-glucosidase and a model of its complex with p-nitrophenyl beta-D-thioglucoside. Biochem. J. 354 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi, J., Wakarchuk, W.W., Campbell, R.L., Carey, P.R., and Surewicz, W.K. (1995). Abnormally high pka of an active-site glutamic acid residue in Bacillus circulans xylanase. The role of electrostatic interactions. Eur. J. Biochem. 232 839–843. [PubMed] [Google Scholar]
- De-Eknamkul, W., Suttipanta, N., and Kutchan, T.M. (2000). Purification and characterization of deacetylipecoside synthase from Alangium lamarckii Thw. Phytochemistry 55 177–181. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. (2002). The PyMOL Molecular Graphics System. (San Carlos, CA: DeLano Scientific).
- De Luca, V., Balsevich, J., and Kurz, W.G.W. (1985). Acetyl coenzyme A:deacetylvindoline O-acetyltransferase, a novel enzyme from Catharanthus. J. Plant Physiol. 121 417–428. [Google Scholar]
- De Luca, V., and St-Pierre, B. (2000). The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 4 168–173. [DOI] [PubMed] [Google Scholar]
- Deus-Neumann, B., and Zenk, M.H. (1984). A highly selective alkaloid uptake system in vacuoles of higher plants. Planta 162 250–260. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endreß, R. (1994). Plant cells as producers of secondary compounds. In Plant Cell Biotechnology, R. Endreß, ed (Berlin: Springer-Verlag), pp. 121–255.
- Fahn, W., Gundlach, H., Deus-Neumann, B., and Stöckigt, J. (1985). Late enzymes of the vindoline biosynthesis. Part I: Acetyl-CoA: 17-O-deacetylvindoline 17-O-acetyl-transferase. Plant Cell Rep. 4 333–336. [DOI] [PubMed] [Google Scholar]
- Geerlings, A., Ibañez Memelink, J., van der Heiden, R., and Verpoorte, R. (2000). Molecular cloning and analysis of strictosidine β-D-glucosidase, an enzyme in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. J. Biol. Chem. 275 3051–3056. [DOI] [PubMed] [Google Scholar]
- Gerasimenko, I., Sheludkov, Y., Ma, X.Y., and Stöckigt, J. (2002). Heterologous expression of a Rauvolfia cDNA encoding strictosidine glucosidase, a biosynthetic key to over 2000 monoterpenoid indole alkaloids. Eur. J. Biochem. 269 2204–2213. [DOI] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. (1999). ESPript: Multiple sequence alignments in PostScript. Bioinformatics 15 305–308. [DOI] [PubMed] [Google Scholar]
- Hemscheidt, T., and Zenk, M.H. (1980). Glucosidases involved in indole alkaloid biosynthesis of Catharanthus cell cultures. FEBS Lett. 110 187–191. [DOI] [PubMed] [Google Scholar]
- Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat, B., and Davies, G. (1997). Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7 637–644. [DOI] [PubMed] [Google Scholar]
- Hesse, M. (1974). Indolalkaloide, Part 1. Progress in Mass Spectrometry, Vol. 1, H. Budzikiewicz, ed (Weinheim, Germany: Verlag Chemie), p. IX.
- Hughes, E.H., Hong, S.B., Gibson, S.I., Shanks, J.V., and San, K.Y. (2004). Metabolic engineering of the indole pathway in Catharanthus roseus hairy roots and increased accumulation of tryptamine and serpentine. Metab. Eng. 6 268–276. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1993). Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26 795–800. [Google Scholar]
- Keresztessy, Z., Kiss, L., and Hughes, M.A. (1994). Investigation of the active site of the cyanogenic beta-D-glucosidase (linamarase) from Manihot esculenta Crantz (cassava). II. Identification of Glu-198 as an active site carboxylate group with acid catalytic function. Arch. Biochem. Biophys. 315 323–330. [DOI] [PubMed] [Google Scholar]
- Kisakürek, M.V., Leeuwenberg, A.J.M., and Hesse, M. (1982). A chemotaxonomic investigation of the plant families of apocynaceae, loganiaceae, and rubiaceae by their indole alkaloid content. In Alkaloids: Chemical and Biological Perspectives, Vol. 1, S.W. Pelletier, ed (New York: John Wiley & Sons), pp. 211–376.
- Kissinger, C.R., Gehlhaar, D.K., and Fogel, D.B. (1999). Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 55 484–491. [DOI] [PubMed] [Google Scholar]
- Koepke, J., Ma, X.Y., Fritzsch, G., Michel, H., and Stöckigt, J. (2005). Crystallization and preliminary X-ray analysis of strictosidine synthase and its complex with the substrate tryptamine. Acta Crystallogr. D Biol. Crystallogr. 61 690–693. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. (1991). Molscript: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Kutchan, T.M. (1995). Alkaloid biosynthesis – The basis for metabolic engineering of medicinal plants. Plant Cell 7 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchan, T.M., Hampp, N., Lottspeich, F., Beyreuther, K., and Zenk, M.H. (1988). The cDNA clone for strictosidine synthase from Rauvolfia serpentina. DNA sequence determination and expression in Escherichia coli. FEBS Lett. 237 40–44. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Loris, E., Panjikar, S., Ruppert, M., Barleben, L., Unger, M., Schüble, H., and Stöckigt, J. (2007). Structure-based engineering of strictosidine synthase: Auxiliary for alkaloid libraries. Chem. Biol., in press. [DOI] [PubMed]
- Luijendijk, T.J.C., Stevens, L.H., and Verpoorte, R. (1998). Purification and characterisation of strictosidine β-D-glucosidase from Catharanthus roseus cell suspension cultures. Plant Physiol. Biochem. 36 419–425. [Google Scholar]
- Luijendijk, T.J.C., van der Meijden, E., and Verpoorte, R. (1996). Involvement of strictosidine as a defensive chemical in Catharanthus roseus. J. Chem. Ecol. 22 1355–1366. [DOI] [PubMed] [Google Scholar]
- Ma, X., Koepke, J., Fritzsch, G., Diem, R., Kutchan, T.M., Michel, H., and Stöckigt, J. (2004). Crystallization and preliminary X-ray crystallographic analysis of strictosidine synthase from Rauvolfia – The first member of a novel enzyme family. Biochim. Biophys. Acta 1702 121–124. [DOI] [PubMed] [Google Scholar]
- Ma, X., Panjikar, S., Koepke, J., Loris, E., and Stöckigt, J. (2006). The structure of Rauvolfia serpentina strictosidine synthase is a novel six-bladed β-propeller fold in plant proteins. Plant Cell 18 907–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, V.J.J., Pitera, D.J., Withers, S.T., Newman, J.D., and Keasling, J.D. (2003). Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21 796–802. [DOI] [PubMed] [Google Scholar]
- Matthews, B.W. (1968). Solvent content of protein crystals. J. Mol. Biol. 33 491–497. [DOI] [PubMed] [Google Scholar]
- McCarter, J.D., and Withers, S.G. (1994). Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4 885–892. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. (1999). XtalView/Xfit - A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125 156–165. [DOI] [PubMed] [Google Scholar]
- Merrit, E.A., and Murphy, M.E. (1994). Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50 869–873. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53 240–255. [DOI] [PubMed] [Google Scholar]
- Niemeyer, H.M. (1988). Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defense chemicals in the gramineae. Phytochemistry 27 3349–3358. [Google Scholar]
- Poulton, J.E. (1990). Cyanogenesis in plants. Plant Physiol. 94 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjikar, S., Parthasarthy, V., Lamzin, V.S., Weiss, M.S., and Tucker, P.A. (2005). Auto-Rickshaw: An automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 61 449–457. [DOI] [PubMed] [Google Scholar]
- Read, R.J. (1986). Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42 140–149. [Google Scholar]
- Ruppert, M., Ma, X.Y., and Stöckigt, J. (2005). Alkaloid biosynthesis in Rauvolfia – cDNA cloning of major enzymes of the ajmaline pathway. Curr. Org. Chem. 9 1431–1444. [Google Scholar]
- Russell, R.B., and Barton, G.J. (1992). Multiple protein sequence alignment from tertiary structure comparison: Assignment of global and residue confidence levels. Proteins 14 309–323. [DOI] [PubMed] [Google Scholar]
- Stevens, L.H., Blom, T.J.M., and Verpoorte, R. (1993). Subcellular localization of tryptophan decarboxylase, strictosidine synthase and strictosidine glucosidase in suspension cultured cells of Catharanthus roseus and Tabernaemontana divaricata. Plant Cell Rep. 12 573–576. [DOI] [PubMed] [Google Scholar]
- Stöckigt, J. (1979). Enzymatic formation of intermediates in the biosynthesis of ajmaline: Strictosidine and cathenamine. Phytochemistry 18 965–971. [Google Scholar]
- Stöckigt, J., and Ruppert, M. (1999). Strictosidine – The biosynthetic key to monterpoid indole alkaloid. In Comprehensive Natural Products Chemistry: Amino Acids, Peptides, Porphyrins and Alkaloids, Vol. 4, D.H.R. Barton, K. Nakanishi, O. Meth-Cohn, and J.W. Kelly, eds (Amsterdam: Elsevier), pp. 109–139.
- Stöckigt, J., and Soll, H.J. (1980). Indole alkaloids from cell suspension cultures of Catharanthus roseus and C. ovalis. Planta Med. 40 22–30. [Google Scholar]
- Stöckigt, J., and Zenk, M.H. (1977). Strictosidine (isovincoside): The key intermediate in the biosynthesis of monoterpenoid indole alkaloids. J. Chem. Soc. Chem. Commun. 646–648.
- St-Pierre, B., and De Luca, V. (2000). Evolution of acyltransferase genes: Origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. In Recent Advances in Phytochemistry: Evolution of Metabolic Pathways, Vol. 34, J.T. Romeo, R. Ibrahim, L. Varin, and V. De Luca, eds (Amsterdam: Elsevier), pp. 285–316.
- St-Pierre, B., Laflamme, P., Alarco, A.-M., and De Luca, V. (1998). The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyltransfer. Plant J. 14 703–713. [DOI] [PubMed] [Google Scholar]
- Treimer, J.F., and Zenk, M.H. (1979). Purification and properties of strictosidine synthase, the key enzmye in indole alkaloid formation. Eur. J. Biochem. 101 225–233. [DOI] [PubMed] [Google Scholar]
- Tsuji, Y., Chen, F., Yasuda, S., and Fukushima, K. (2005). Unexpected behaviour of coniferin in lignin biosynthesis of Ginkgo biloba L. Planta 222 58–69. [DOI] [PubMed] [Google Scholar]
- Vagin, A., and Teplyakov, A. (1997). MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 30 1022–1025. [Google Scholar]
- Verdoucq, L., Czjzek, M., Moriniere, J., Bevan, D.R., and Esen, A. (2003). Mutational and structural analysis of aglycone specificity in maize and sorghum beta-glucosidases. J. Biol. Chem. 278 25055–25062. [DOI] [PubMed] [Google Scholar]
- Verdoucq, L., Moriniere, J., Bevan, D.R., Esen, A., Vasella, A., Henrissat, B., and Czjze, M. (2004). Structural determinants of substrate specificity in family 1 beta-glucosidases: Novel insights from the crystal structure of sorghum dhurrinase-1, a plant beta-glucosidase with strict specificity, in complex with its natural substrate. J. Biol. Chem. 279 31796–31803. [DOI] [PubMed] [Google Scholar]
- White, A., and Rose, D.R. (1997). Mechanism of catalysis by retaining beta-glucosyl hydrolases. Curr. Opin. Struct. Biol. 7 645–651. [DOI] [PubMed] [Google Scholar]
- Winn, M.D., Isupov, M.N., and Murshudov, G.N. (2001). Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57 122–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.