Abstract
Rhizobium Nod factors (NFs) are specific lipochitooligosaccharides that activate host legume signaling pathways essential for initiating the nitrogen-fixing symbiotic association. This study describes the characterization of cis-regulatory elements and trans-interacting factors that regulate NF-dependent and epidermis-specific gene transcription in Medicago truncatula. Detailed analysis of the Mt ENOD11 promoter using deletion, mutation, and gain-of-function constructs has led to the identification of an NF-responsive regulatory unit (the NF box) sufficient to direct NF-elicited expression in root hairs. NF box–mediated expression requires a major GCC-like motif, which is also essential for the binding of root hair–specific nuclear factors. Yeast one-hybrid screening has identified three closely related AP2/ERF transcription factors (ERN1 to ERN3) that are able to bind specifically to the NF box. ERN1 is identical to an ERF-like factor identified recently. Expression analysis has revealed that ERN1 and ERN2 genes are upregulated in root hairs following NF treatment and that this activation requires a functional NFP gene. Transient expression assays in Nicotiana benthamiana have further shown that nucleus-targeted ERN1 and ERN2 factors activate NF box–containing reporters, whereas ERN3 represses ERN1/ERN2-dependent transcription activation. A model is proposed for the fine-tuning of NF-elicited gene transcription in root hairs involving the interplay between repressor and activator ERN factors.
INTRODUCTION
Legumes have the unique capacity to establish symbiotic associations with nitrogen-fixing soil bacteria known as rhizobia, thereby facilitating growth in nitrogen-limiting soils. This endosymbiotic interaction leads to the formation of specialized root organs known as nodules, within which rhizobia reduce atmospheric nitrogen for the benefit of the host plant. Rhizobial infection in temperate legumes such as Medicago and Lotus is usually initiated in root hairs and is concomitant with nodule organogenesis in the root cortex. Rhizobial attachment to the tip of a growing root hair induces curling around the entrapped bacteria, which subsequently enter the plant cell via a plasma membrane invagination and the formation of an intracellular tubular structure called the infection thread. Bacteria divide within the infection thread, which progresses through the outer root cortex toward the newly formed nodule primordium, within which bacteria are released into host cells and subsequently differentiate into nitrogen-fixing bacteroids (Timmers et al., 1999; Gage, 2004).
The success of this symbiotic interaction requires specific and reciprocal signaling between the two partners. In response to flavonoids secreted by host roots, rhizobia produce specific lipochitooligosaccharide signal molecules called Nod factors (NFs) that play a pivotal role in recognition and controlled host root infection (D'Haeze and Holsters, 2002). Purified NFs are sufficient to induce many of the early plant symbiotic responses normally observed during preinfection stages of the interaction. These include rapid ion fluxes, membrane depolarization, specific calcium oscillations, root hair morphogenetic changes, and specific activation of early nodulin genes (Geurts et al., 2005; Oldroyd et al., 2005; Stacey et al., 2006).
The specificity and subnanomolar activity of NFs argue that receptors mediate NF signal transduction pathways in host roots. Based on rhizobial genetic studies, it has been proposed that an initial signaling receptor pathway is required for early preinfection responses, followed by a second entry receptor pathway that is necessary for bacterial root infection and involving a different specificity for NF recognition (Ardourel et al., 1994). Genetic studies conducted with the model legumes Lotus japonicus and Medicago truncatula have led to the identification and cloning of host genes essential for early NF perception/transduction (Geurts et al., 2005; Oldroyd et al., 2005; Stacey et al., 2006). These studies have revealed that NFs are likely to be perceived via LysM domain–containing receptor kinases such as L. japonicus NFR1/NFR5 and M. truncatula NFP (Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006). Following perception, NF signal transduction requires a number of essential genes encoding putative ion channels (DMI1/CASTOR/POLUX) (Ané et al., 2004; Imaizumi-Anraku et al., 2005), LRR receptor–like kinases (DMI2/SYMRK/NORK) (Endre et al., 2002; Stracke et al., 2002), and nucleoporins (NUP85 and NUP133) (Kanamori et al., 2006; Saito et al., 2007), all acting upstream of the NF-induced calcium spiking response in root hairs (Miwa et al., 2006). This specific calcium signal is potentially decoded by the calcium- and calmodulin-dependent kinase DMI3 (Lévy et al., 2004; Mitra et al., 2004a), which is required for subsequent signaling steps that lead to the transcriptional activation of early nodulin genes (Charron et al., 2004; Mitra et al., 2004b).
Although significant progress has been made in deciphering the primary signal perception/transduction components of NF signaling, little is known about the mechanisms leading to transcriptional gene activation in response to NFs. Genetic studies have identified two GRAS-type transcription regulators, NSP1 and NSP2, lying downstream of DMI3 and required for subsequent NF-induced gene expression in both Medicago and Lotus (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2007). Although a direct transcriptional regulatory function has not yet been demonstrated for these factors, it is proposed that they could be coactivators of other DNA binding transcription factors (Udvardi and Scheible, 2005). Most recently, a genetic approach has identified an ERF transcription factor named ERN (for ERF Required for Nodulation) that is necessary for NF-elicited gene activation and could potentially be involved in the direct transcriptional activation of early nodulin genes (Middleton et al., 2007).
In addition to genetic approaches, the direct identification of NF-responsive cis-regulatory elements and trans-acting factors should also contribute to elucidating the molecular mechanisms regulating NF-elicited gene expression. To this end, we have made use of the well-characterized M. truncatula Mt ENOD11 gene as a model to determine the molecular mechanisms of NF-dependent gene regulation (Charron et al., 2004). Mt ENOD11, which encodes an atypical repetitive Pro-rich protein, is a widely used marker gene for both the early preinfection and subsequent infection stages of the symbiotic association (Journet et al., 2001). During preinfection stages, Mt ENOD11, along with Mt ENOD12 and rip1, are the earliest M. truncatula genes characterized to date that are specifically activated in root hairs in response to NFs (Journet et al., 1994; Cook et al., 1995; Charron et al., 2004). NF-elicited Mt ENOD11 transcriptional activation in the root epidermis is both rapid and strong and is dependent on the genetically defined NFP/DMI/NSP NF signaling pathway (Catoira et al., 2000; Ben-Amor et al., 2003; Oldroyd and Long, 2003; Charron et al., 2004; Mitra et al., 2004b; Sauviac et al., 2005).
In a previous article, we showed that the 411-bp Mt ENOD11 promoter sequence directly upstream of the start codon is sufficient to confer both preinfection (NF-mediated) and infection-related expression (Boisson-Dernier et al., 2005). By contrast, the −257-bp truncated promoter is only capable of driving infection-related expression. This finding suggested that the Mt ENOD11 regulatory sequences required for early NF-elicited activation lie within the −411 to −257 promoter region and can be dissociated from the infection-responsive regulatory region. In this study, we have performed a detailed analysis of the −411 to −257 Mt ENOD11 promoter region using deletion, point mutation, and gain-of-function approaches. This has led to the definition of a novel NF-responsive regulatory unit called the NF box and associated regulatory motifs that are necessary and sufficient to confer NF-induced activation in root hairs. Yeast one-hybrid screening of root hair–specific cDNA libraries has identified three NF box binding AP2/ERF transcription factors, including the recently described ERN factor essential for nodulation (Middleton et al., 2007). These transcription factors, named ERN1, ERN2, and ERN3, are highly expressed in root hairs and upregulated by NFs only in the presence of a functional NFP-dependent signaling pathway. Importantly, ERN1 and ERN2 act as transcriptional activators, while ERN3 acts as a putative repressor of NF box–containing target reporters in transiently expressing Nicotiana benthamiana cells. These results led us to propose that ERN factors are key players in the transcriptional regulation of NF-activated early nodulin genes operating via a complex regulatory mechanism involving both repressor and activator factors.
RESULTS
The −411 to −358 Mt ENOD11 Promoter Region Mediates NF-Elicited Root Hair Expression
In order to characterize cis-regulatory sequences required for NF-elicited expression in root hairs, we performed a detailed functional analysis of the −411 to −257 Mt ENOD11 promoter region fused to the β-glucuronidase (GUS) reporter gene in transgenic M. truncatula roots. Because NF-elicited gene activation is restricted to a specific zone of the root epidermis (Journet et al., 2001), a quantitative measurement of GUS activity in entire root extracts frequently underestimates tissue-specific GUS expression in root hairs. For this reason, each construct was analyzed by both fluorimetric and histochemical GUS assays using a large number of independent transgenic roots as described (see Methods) (Boisson-Dernier et al., 2005). In transgenic roots carrying a 2.3-kb Mt ENOD11 promoter fused to the GUS reporter gene (PMt ENOD11-GUS), histochemical GUS activity is totally absent in root hairs of control water-treated roots, whereas a basal activity level is observed in lateral root primordia (Journet et al., 2001; Charron et al., 2004; Boisson-Dernier et al., 2005; Sauviac et al., 2005). This nonsymbiotic expression, detected by fluorimetric assays, has proved useful as an internal control in this study.
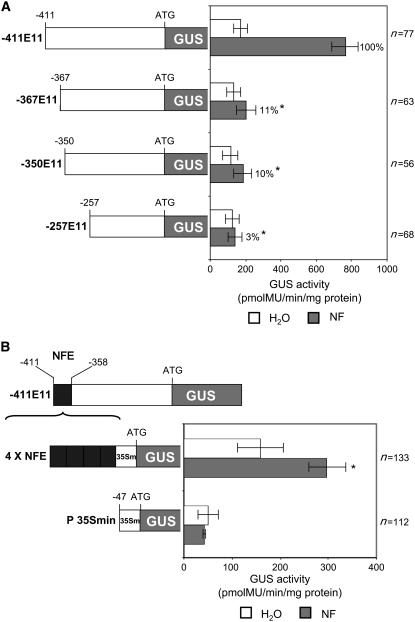
Figure 1A illustrates the results of fluorimetric assays performed on a series of 5′ promoter deletions between positions −411 and −257 of the Mt ENOD11 promoter. While GUS activity corresponding to basal expression in nonepidermal root tissues was similar for all deletions studied, the capacity to drive NF-elicited expression was dramatically reduced in the −367E11 deletion. This indicates that sequences between −411 and −367 are important for NF-elicited gene activation in root hairs. Furthermore, the fact that the −411E11 promoter deletion lacking sequences between −357 and −258 was still capable of driving NF-elicited gene activation (data not shown) suggests that this region is not essential for the NF-elicited response.
Figure 1.
Identification of a 54-bp Nod Factor–Responsive Element (NFE) within the Mt ENOD11 Promoter Sufficient to Drive Root Hair–Specific Expression.
GUS activity driven by Mt ENOD11 5′ promoter deletions and gain-of-function constructs fused to the GUS reporter gene were analyzed in transgenic M. truncatula roots.
(A) Schematic representation of Mt ENOD11 5′ promoter deletion fusions. Numbers above the promoters indicate the positions of 5′ deletions relative to the translation initiation codon ATG. Fluorimetric GUS activities were quantified in root extracts following either water (white bars) or NF (10−9 M) (gray bars) treatment. The percentage values refer to the proportion of NF-induced GUS activity (i.e., after subtraction of the water control value) of a given construct in relation to the −411E11 reference construct. Asterisks indicate statistically significant differences (P < 0.05, Student's t test) compared with the reference −411E11 construct (see Supplemental Table 1 online).
(B) Fluorimetric analyses of reporter GUS activities driven by the gain-of-function 4xNFE construct and the control P35S min construct following NF treatments (10−9 M). The asterisk indicates a statistically significant difference (P < 0.05, Student's t test) compared with the reference P35S min construct.
Error bars represent ±sd of mean activity values derived from three independent experiments performed with n number of individual roots.
To determine whether the −411 to −357 Mt ENOD11 promoter region can, by itself, direct NF-elicited epidermal expression in a heterologous context, a gain-of-function construct comprising a tetramer of this sequence named NFE (for Nod Factor–Responsive Element) fused to the −47-bp cauliflower mosaic virus (CaMV) 35S minimal promoter (here named P35S min) (Pontier et al., 2001) was analyzed in transgenic M. truncatula roots. A tetramer of NFE was used in this gain-of-function approach, since multimers have been shown to confer expression to minimal promoters more efficiently than monomers, probably by compensating for the absence of general positive regulatory elements (Wu et al., 1998; Pontier et al., 2001; Rushton et al., 2002). In water-treated samples, transgenic roots carrying the 4xNFE construct exhibited a higher basal GUS activity than roots carrying the P35S min construct, suggesting that sequences within 4xNFE can act as general positive transcriptional enhancers. Following NF application, control roots with the P35S min construct showed no induction of GUS activity in the root epidermis, whereas roots carrying 4xNFE exhibited clear NF-elicited GUS activity in root hairs (Figure 1B), although at lower levels than the reference −411E11 construct (Figure 1A). This gain-of-function promoter analysis demonstrates that the −411 to −357 NFE sequence is sufficient to confer NF-elicited root hair–specific expression.
NF-Mediated Gene Activation Requires a Conserved GCC-Like Motif
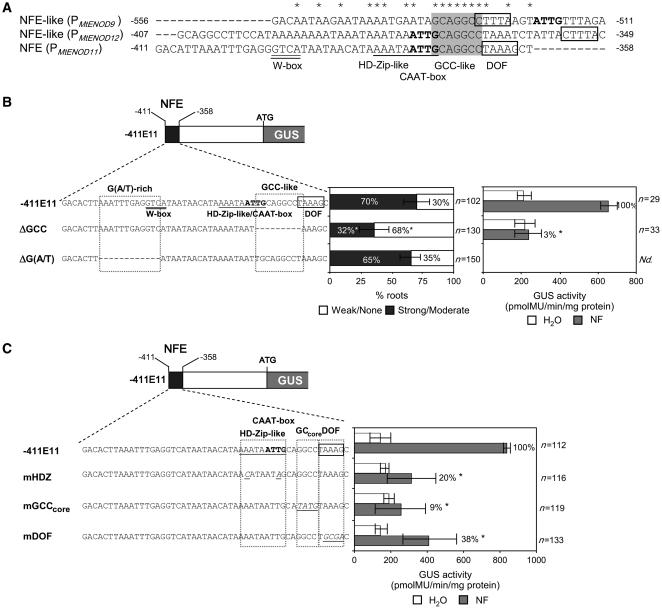
In order to identify putative cis elements within the NFE region that could be involved in NF-regulated gene expression, we searched for conserved sequences within NFE that are common to other NF-responsive gene promoters. Comparison between NFE and 5′ upstream sequences of two other epidermal NF-activated genes, Mt ENOD12 (Journet et al., 1994) and Mt ENOD9 (D. Barker, unpublished data), allowed us to identify NFE-homologous regions within both promoters (Figure 2A). Promoter deletion analysis further revealed that truncated −534-bp Mt ENOD12 and −552-bp Mt ENOD9 promoters including the NFE-homologous regions were able to direct NF-responsive reporter expression, whereas deletions lacking the NFE-homologous region (−323 bp for Mt ENOD12 and −525 bp for Mt ENOD9) were no longer responsive to NFs (data not shown). As shown in Figure 2A, a striking feature of NFE and NFE-homologous regions is the presence of a perfectly conserved GCAGGCC motif, which is reminiscent of, but not identical to, GCC box motifs (AGCCGCC) (Ohme-Takagi and Shinshi, 1995). This conserved GCC-like motif is located within a particularly AT-rich region, and in the case of NFE, it is flanked by consensus binding sites for CAAT binding and DOF transcription factors (Kusnetsov et al., 1999; Yanagisawa and Schmidt, 1999). Interestingly, DOF and CAAT box sites are also found in the vicinity of the GCC-like motif in the NFE-homologous sequences of both the Mt ENOD12 and Mt ENOD9 promoters. In addition, an HD-ZIP–like binding site overlapping the CAAT box motif and a conserved W box binding site for WRKY transcription factors can be identified within the Mt ENOD11 NFE (Figure 2A).
Figure 2.
Functional Analyses of Conserved cis Motifs Involved in NF-Elicited Expression.
(A) Partial sequence alignment of NFE and NFE-like sequences within the Mt ENOD11, -9, and -12 promoters. Asterisks indicate conserved nucleotides at the given positions. The fully conserved 8-bp GCC-like motif is shaded. Putative W box (GTCA), HD-ZIP–like (AAATAATTG), CAAT box (CAAT or ATTG), and DOF (TAAAG or CTTTA) sequences are indicated by double underlining, underlining, boldface lettering, and boxing, respectively.
(B) GUS activity of the −411E11 reference and derived constructs block-deleted within the conserved GCC-like and/or G(A/T)-rich motifs of NFE. The relative strength of root hair–specific GUS activity was visually screened in individual NF-treated roots (see Methods) and is represented as the percentage of roots exhibiting clear-cut GUS activity in the root hairs. GUS activity levels were also measured by fluorimetric assays in protein extracts from either water-treated (white bars) or NF-treated (gray bars) roots. The percentage values refer to NF-elicited GUS activities (i.e., after subtraction of the water control value) relative to that obtained with the −411E11 reference construct.
(C) Quantitative analysis of GUS activity driven by the −411E11 construct and its mutated derivatives. Substituted nucleotides are shown in italics and underlined. The percentage values refer to NF-induced GUS activities relative to the reference construct.
In (B) and (C), error bars represent ±sd and asterisks indicate significant differences compared with the reference construct (P < 0.05, Student's t test) (see Supplemental Table 1 online). n represents the number of individual transgenic roots analyzed.
To investigate the importance of the conserved GCC-like motif for NF-elicited gene activation, we deleted the entire motif from within the −411E11 promoter region to create the ΔGCC construct (Figure 2B). As a control, a block deletion of an upstream G(A/T)-rich sequence, overlapping the W box motif, was also generated (ΔG[A/T]). As shown in Figure 2B, block deletion of the GCC-like motif alone resulted in a significant reduction in the proportion of transgenic roots exhibiting epidermal NF-elicited histochemical GUS activity, whereas deletion of the upstream G(A/T)-rich sequence did not alter NF-induced GUS activity. Quantitative fluorimetric assays confirmed the drastic effect of deleting the GCC-like motif in relation to NF-elicited GUS activation, whereas nonsymbiotic expression in control roots remained unaltered (Figure 2B; see Supplemental Table 1 online). These results indicate that within NFE, the GCC-like motif and/or its flanking sequences, including the potential CAAT box and HD-ZIP–like motif, are important for NF-elicited gene activation.
In order to address the importance of individual cis elements within the NFE region, base substitutions were introduced at sites overlapping or in the proximity of the GCC-like motif. For this purpose, we generated −411E11 derivative constructs mutated in the central GGCC core (GCcore) as well as in the HD-ZIP–like/CAAT box and DOF sites (see Methods) (Figure 2C). These experiments confirmed that, as for the block deletion, the specific mutation of the GCcore (mGCcore) results in a major reduction (>10 fold) in NF-elicited GUS activity, similar to the levels for control roots (Figure 2C; see Supplemental Table 1 online). Although less striking than with the GCcore mutation, a significant threefold to fivefold reduction of the NF-mediated response was also observed in roots transformed with the mDOF and mHDZ constructs, suggesting that these flanking sequences also partially influence the efficiency of the NF-dependent response. Importantly, none of these point mutations modified the nonsymbiotic GUS activity levels in roots (Figure 2C; see Supplemental Table 1 online).
Taken together, these results underline the importance of the GCC-like GCAGGCC motif with its GCcore sequence for NF-elicited expression in root hairs. This central GC-rich sequence may function in cooperation with neighboring cis elements, which together form a fully functional regulatory unit. This particular combination of cis motifs, therefore, could constitute a promoter signature for NF-elicited gene expression in root hairs.
The NF Box Defines a Novel Regulatory Unit Sufficient for NF-Elicited Gene Activation
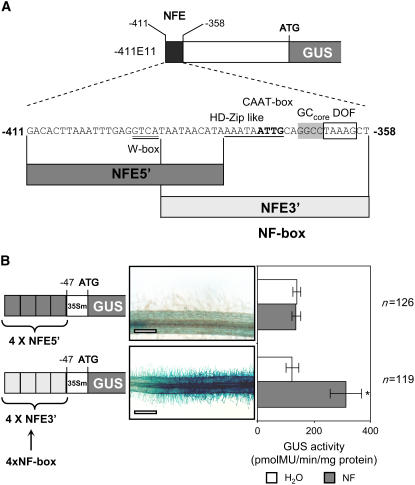
To further evaluate to what extent these motifs are sufficient to direct NF-elicited expression in root hairs, additional gain-of-function experiments were performed. For this, tetramers of two promoter fragments of ∼30 bp in length derived from either the 5′ or the 3′ end of NFE were fused to the P35Smin-GUS reporter (Figure 3A). In agreement with the deletion and point mutation experiments, NF-elicited GUS activity was only observed in roots carrying the 4xNFE3′ construct, which includes the GCC-like, HD-ZIP–like/CAAT, and DOF motifs (Figure 3B). Interestingly, a similar level of GUS reporter activity was observed with either the 4xNFE or the 4xNFE3′ construct, indicating that NFE3′ contains all of the regulatory elements required for the root hair–specific NF response (cf. Figures 1B and 3B). Furthermore, following Sinorhizobium meliloti inoculation, the 4xNFE3′ construct driving the P35Smin-GUS reporter was activated during preinfection (NF-dependent) symbiotic stages (data not shown). However, as expected, this construct was unable to drive infection-related expression in either root tissues or the infection zone of the nodule (Boisson-Dernier et al., 2005) (see Supplemental Figure 1 online). Nonsymbiotic expression in lateral root primordia was also absent in roots transformed with this construct (data not shown). Thus, we conclude that the 30-bp NFE3′ sequence, renamed NF box, constitutes a novel regulatory unit sufficient to confer NF-dependent root hair–specific expression.
Figure 3.
The NF Box Confers an NF-Specific Response in the Root Epidermis.
(A) Schematic representation of the NFE sequence and associated cis motifs. The sequences NFE5′ and NFE3′ (NF box) were used to generate the new gain-of-function constructs tested in (B).
(B) Histochemical and quantitative analyses of GUS activity directed by gain-of-function 4xNFE5′ and 4xNFE3′ (4xNF box) constructs. Data are represented as mean values obtained by the analysis of n individual roots from three independent experiments. Error bars represent ±sd. The asterisk indicates a significant difference compared with the reference construct (P < 0.05, Student's t test) (see Supplemental Table 1 online). Bars = 200 μm.
A Yeast One-Hybrid Screen Identifies NF Box Binding Proteins
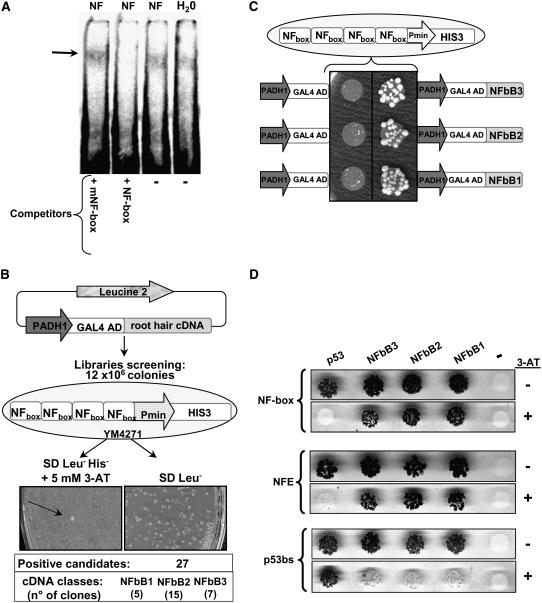
Initial experiments to determine whether nuclear proteins can bind specifically to the NF box regulatory unit were performed using the gel shift technique. Since experiments with extracts from whole roots had not been conclusive, we increased the sensitivity of the gel shift assay using protein extracts from isolated root hairs. As shown in Figure 4A, a weak but reproducible NF box–specific retardation signal was observed in these assays, with a slightly enhanced level in NF-treated root hair samples. More importantly, competition studies showed that this retardation signal was dependent on the presence of the GCcore motif (Figure 4A), thus confirming the importance of this motif within the NF box for the specific binding of potential trans-acting factors present in M. truncatula root hairs.
Figure 4.
Isolation of NF Box Binding Proteins.
(A) Band shift experiments using an NF box–containing DNA probe with root hair protein extracts from 6-h water- or NF-treated roots. In competitor lane reactions, 50 to 100 molar excesses of cold NF box or mNF box probes were included.
(B) Schematic representation of the one-hybrid strategy used to screen cDNA libraries from NF-treated root hairs using a HIS3 reporter gene under the control of a minimal HIS3 promoter fused to a tetramer of the NF box and expressed in the yeast YM4271 reporter strain. The number of positive clones corresponding to each of the three cDNA classes (NFbB1, NFbB2, and NFbB3) isolated after two rounds of screening and able to grow on 5 mM 3-AT is indicated.
(C) Confirmation of the interaction of NFbB1, NfbB2, and NFbB3 with the NF box by retransformation of yeast YM4271 containing the tetramer NF box-HIS3 reporter with the isolated plasmids expressing GAL4-NFbB fusions and control GAL4 plasmids. Yeast cells were grown on synthetic dropout (SD) Leu−His− + 5 mM 3-AT medium.
(D) Binding specificity of NFbB proteins. YM4721 reporter strains carrying tetramer NF box, tetramer NFE, and trimer p53 cis (p53bs) sequences were transformed with plasmids expressing the mouse GAL4-p53 factor that interacts with the p53 binding site, the NFbBs GAL4-NFbBs proteins, and the water control (−). Yeast growth was examined under either nonselective SD Leu− conditions (−3-AT) or selective SD Leu−His− conditions on medium supplemented with 5 mM 3-AT (+3-AT).
In order to identify the root hair transcription factors mediating NF-elicited gene activation, a yeast one-hybrid screen was performed using the NF box as bait. A YM4271 yeast reporter strain containing a HISTIDINE3 (HIS3) gene under the control of a chimeric promoter comprising a tetramer of the NF box sequence fused to the HIS3 minimal promoter was used to screen GAL4-fused cDNA libraries derived from NF-elicited root hairs (see Methods) (Figure 4B). This NF box reporter yeast strain is unable to grow on selective medium (without His and supplemented with 5 mM 3-aminotriazole [3-AT]), thus allowing efficient screening. After initially screening 12 × 106 yeast transformants, 67 candidates were selected for a subsequent screening on more stringent selective medium, followed by retransformation of yeast with isolated plasmid DNAs. This resulted in 27 confirmed positive clones that could be classified into three closely related groups designated NFbB1 (5 clones), NFbB2 (15 clones), and NFbB3 (7 clones) for NF box binding proteins (Figure 4B).
To examine the specificity of the interactions of NFbB1, NFbB2, and NFbB3 with the NF box bait sequence in yeast, the corresponding plasmids were used to transform individual yeast strains harboring HIS3 reporters fused to either the tetramer NF box or the NFE tetramer. As a negative control, we made use of a trimer of the unrelated p53 human binding site (Matchmaker; Clontech) (Figures 4C and 4D). Significant yeast growth was only detected when the three NFbB clones were expressed in the presence of the NF box or NFE HIS3 reporters, but not with the p53 HIS3 reporter (Figure 4D). This shows that reporter yeast growth is dependent on the direct interaction of NFbB1, NFbB2, and NFbB3 with NF box–containing target sequences.
NF Box–Interacting Factors Belong to the ERF Transcription Factor Family
Sequence analysis revealed that all positive NFbB clones corresponded to in-frame GAL4AD fusions. NFbB2 and NFbB3 cDNAs are 1098 and 885 bp in length and contain open reading frames of 313 and 238 amino acids, respectively. Positive NFbB1 cDNA clones (857 bp) are all 5′ truncated at approximately the 60-bp position and encode a protein of 248 amino acids missing the 20 N-terminal residues. This indicates that the 5′ truncated NFbB1 is perfectly able to bind to NF box–containing target sequences in yeast.
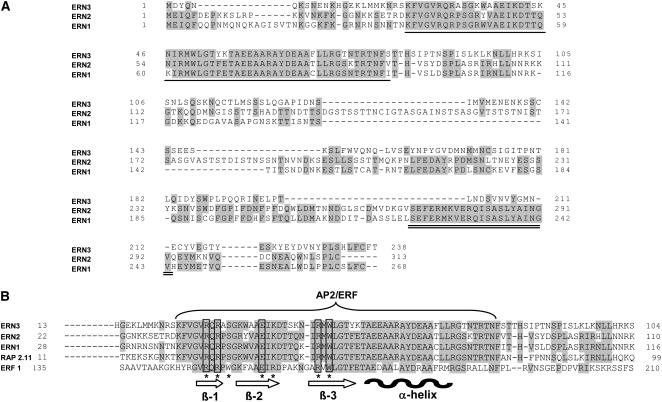
Subsequent BLAST analysis revealed that NFbB1, -2, and -3 all belong to the large family of AP2/ERF transcription factors (Riechmann and Meyerowitz, 1998). Furthermore, it turns out that NFbB1 is identical to the symbiosis-associated ERF-like transcription factor called ERN that was recently characterized by Middleton et al. (2007). For simplicity, therefore, we have renamed our three AP2/ERF transcription factors ERN1 (NFbB1), ERN2 (NFbB2), and ERN3 (NFbB3). ERN1, -2, and -3 all contain the highly conserved AP2/ERF domain putatively involved in DNA binding (Wessler, 2005) (Figure 5). ERN1 and ERN2 are the most closely related and contain, in addition to the conserved AP2/ERF domain, an identical 22–amino acid stretch at the C terminus that is absent in the ERN3 sequence (Figure 5A). Phylogenetic analysis of the conserved ERN AP2/ERF domain in relation to all known Arabidopsis thaliana family members (http://datf.cbi.pku.edu.cn) shows that the three ERNs, together with Arabidopsis RAP2.11, form a distinct group within the ERF family, as reported for ERN1 (data not shown) (Middleton et al., 2007). A very high level of sequence conservation (∼80 to 90%) is observed between the AP2/ERF DNA binding domains of all three ERNs and RAP2.11, while there is only ∼50% conservation between ERN AP2/ERF DNA binding domains and other ERF proteins such as At ERF1 (Figure 5B).
Figure 5.
NF Box Binding Proteins Named ERN1, ERN2, and ERN3 Belong to the ERF Transcription Factor Family.
Comparison of the deduced amino acid sequences of the three entire ERN1, ERN2, and ERN3 proteins (A) and the corresponding AP2/ERF DNA binding domains aligned by ClustalW (Thompson et al., 1997) with those of the related Arabidopsis RAP2.11 proteins (B). Identical amino acid residues are shaded gray. The conserved AP2/ERF DNA binding domain in (A) is underlined, and the 20–amino acid stretch conserved in ERN1 and ERN2 is double underlined. In (B), the conserved AP2/ERF DNA binding domain is indicated. The black wavy bar and arrows represent predicted α-helix and β-sheet regions, respectively, within the AP2/ERF domain of At ERF1 (Allen et al., 1998). Asterisks correspond to conserved amino acid residues required for DNA binding to GCC box motifs.
Importantly, the critical amino acid residues within the AP2/ERF DNA binding domain of the Arabidopsis At ERF1 factor required for binding to its target DNA GCC box are only partially conserved in ERN and RAP2.11 AP2/ERF DNA binding domains (Figure 5B) (Allen et al., 1998). Therefore, we predict that these differences are related to the specific binding of ERNs to the novel NF box GCcore motif.
NF-Elicited Upregulation of ERN Genes in M. truncatula Root Hairs
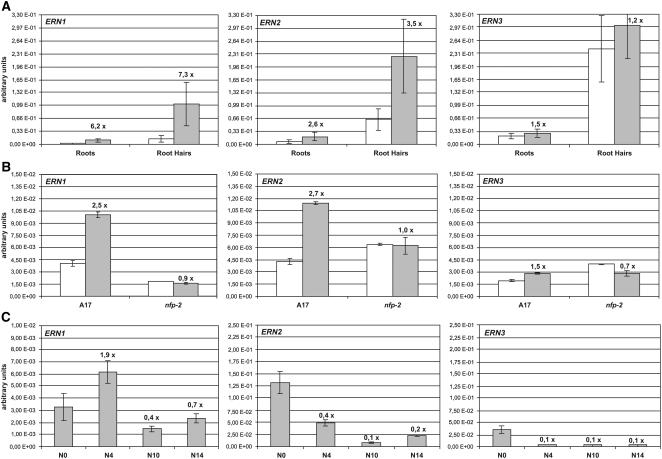
To study ERN gene expression in M. truncatula root hairs, quantitative RT-PCR analysis was performed on RNA isolated from purified root hairs (Sauviac et al., 2005). As shown in Figure 6A, all three genes are constitutively expressed in untreated roots and root hairs. In all cases, transcript levels are ∼10-fold higher in root hairs compared with whole root samples. Following NF treatment, both ERN1 and ERN2 genes are upregulated in root hairs and in entire roots, while the level of ERN3 transcripts is not significantly modified. On the other hand, no NF induction of these genes was detected for the nodulation-defective nfp-2 mutant (Figure 6B). This shows that, as for their target gene Mt ENOD11, NF-elicited activation of the ERN genes is dependent on the NFP locus, an essential component of NF perception and signaling in root hairs (Ben Amor et al., 2003; Arrighi et al., 2006).
Figure 6.
The ERN1, ERN2, and ERN3 Genes Are Upregulated in NF-Treated M. truncatula Root Tissues.
(A) Quantitative RT-PCR for ERN1, ERN2, and ERN3 transcripts in total RNA samples extracted from 3-d in vitro grown wild-type intact roots or isolated root hairs treated with either water (white bars) or 10−8 M NFs (gray bars) for 6 h.
(B) Quantitative RT-PCR for ERN transcripts in total RNA samples extracted from 3-d aeroponically grown wild-type (A17) or nfp-2 mutant roots treated with either water (white bars) or 10−9 M NFs (gray bars) for 6 h.
(C) Quantitative RT-PCR for ERN transcripts in total RNA samples extracted from nitrogen-starved 10-d aeroponically grown wild-type roots (N0) or isolated nodules harvested at 4 d (N4), 10 d (N10), or 14 d (N14) after inoculation.
Values represent averages of either two or three independent biological experiments after normalization against EF1α transcript levels (see Methods). Multiplication factors above the shaded bars refer to NF:water expression ratios. Error bars represent ±sd.
To determine whether ERN genes are expressed at later stages of the nodulation process, quantitative RT-PCR analysis was performed on RNA extracted from roots and isolated nodules of aeroponically grown plants. Similar ERN1 transcript levels were observed in noninoculated root samples from plants grown for 3 d (Figure 6B) and 10 d (Figure 6C) in aeroponic conditions, while ERN2 and ERN3 showed significantly enhanced transcript levels in root samples of 10-d aeroponically grown plants. This upregulation of ERN2 and ERN3 may be associated with the development of lateral roots, which are not yet visible in 3-d aeroponically grown plants. During nodulation, the ERN1 transcript level is higher in young 4-d-old nodules compared with roots, whereas ERN2 and ERN3 transcript levels are lower in nodules compared with roots. However, for the moment, we cannot rule out that a localized low-level induction in nodules may be masked by the high basal level of ERN2 and ERN3 expression in roots. Finally, ERN genes display a similar expression profile in different M. truncatula organs, for which transcript levels were mainly detected in root and leaf tissues (see Supplemental Figure 2 online).
Nuclear Localization of ERNs in N. benthamiana Cells
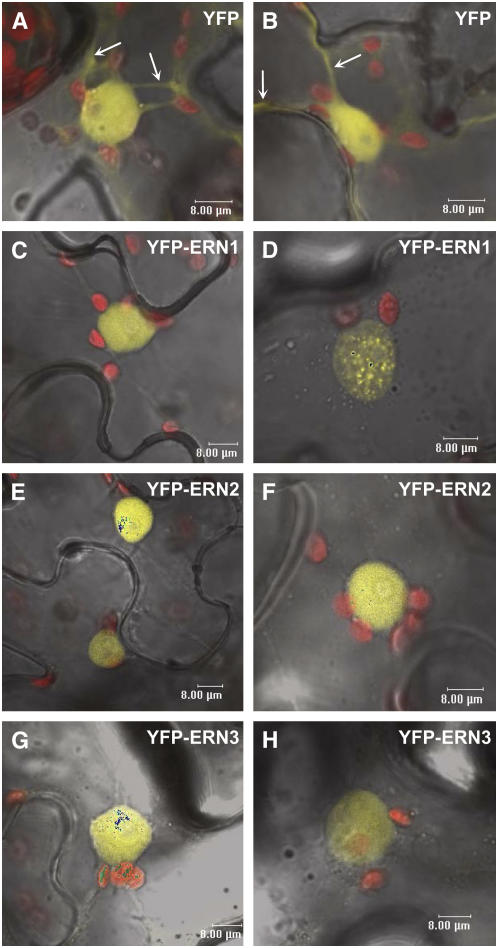
To examine the subcellular localization of the ERN transcription factors, yellow fluorescent protein (YFP) N-terminally fused ERN proteins under the control of the 35S promoter were analyzed in N. benthamiana leaves. As expected, controls expressing a 35S-YFP construct showed fluorescence labeling in both cytoplasmic strands and nucleoplasm (Figures 7A and 7B). Although ERNs do not have a typical nuclear localization signal, our experiments clearly show that YFP-ERN fusion proteins are targeted to the nucleus of transformed cells (Figures 7C to 7H). This is also the case for other AP2/ERF proteins (Lee et al., 2004; Krizek and Sulli, 2006). In addition to the homogeneous nucleoplasm localization observed for all three fusions, the YFP-ERN1 fusion protein was also frequently associated with subnuclear particles (Figure 7D). These subnuclear particles were very dynamic, with an appearance that often varied with time and with a variable frequency between experiments.
Figure 7.
Nuclear Localization of ERN1-, ERN2-, and ERN3-YFP Fusion Proteins.
Confocal images of epidermal N. benthamiana cells expressing control YFP ([A] and [B]), YFP-ERN1 ([C] and [D], YFP-ERN2 ([E] and [F]), and YFP-ERN3 ([G] and [H]) fusion proteins under the control of the 35S promoter. Note the exclusive nuclear localization for the ERN proteins in the nucleoplasm ([C] and [E] to [H]) or in discrete nuclear bodies (D) compared with the cytoplasm (arrows) and the nucleus-localized YFP control ([A] and [B]). In all panels, chloroplast fluorescence appears red. Similar results were obtained with green fluorescent protein (GFP) fusion proteins (data not shown).
ERN3 Negatively Modulates ERN1/ERN2 trans-Activation of a Target NF Box Reporter in N. benthamiana Cells
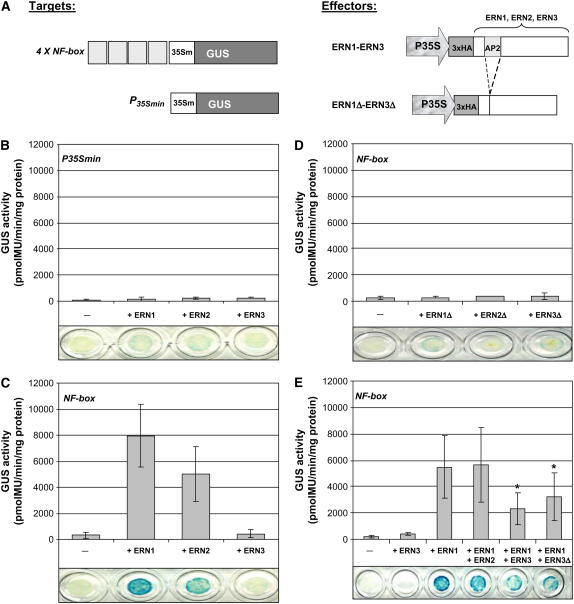
To determine whether ERN1, -2, and -3 proteins are capable of trans-activating Mt ENOD11 in plant cells, transient expression assays in N. benthamiana were performed. Leaves were coinfiltrated with either target 4xNF box or control P35Smin-GUS reporters in combination with effectors comprising 3HA-tagged ERN proteins under the control of the 35S promoter (Figure 8A). As shown in Figures 8B and 8C, ERN1 and ERN2 effectors specifically trans-activated the target 4xNF box reporter but not the control P35Smin-GUS reporter. Importantly, ERN1 and ERN2 effectors deleted for their respective AP2/ERF DNA binding domains (ERN1Δ and ERN2Δ, respectively) were no longer able to induce the target 4xNF box GUS reporter, indicating that specific DNA binding is required for trans-activation (Figure 8D). By striking contrast, the ERN3 effector was unable to activate the target 4xNF box reporter, despite the accumulation of significant quantities of the tagged ERN3 protein (Figure 8C; see Supplemental Figure 3 online).
Figure 8.
Transactivation and Repressor Properties of ERN Factors in N. benthamiana Cells.
(A) Schematic representation of the 3xHA-tagged ERN effector proteins and the target GUS reporters used for trans-activation studies in A. tumefaciens–infiltrated N. benthamiana cells. The P35Smin-GUS reporter construct alone or fused to the tetramer of the NF box (4xNF box) was used as target GUS reporter. The effector constructs were under the control of the 35S promoter (P35S), and all contained a 3xHA 5′ tag upstream of the ERN sequences, deleted (ERN1Δ, ERN2Δ, and ERN3Δ) or not (ERN1, ERN2, and ERN3), for their respective AP2/ERF (AP2) DNA binding domains.
(B) to (E) Histochemical and fluorimetric GUS activities of N. benthamiana leaf discs at 24 h following infiltration with the reporters alone (−) or in combination with the different effectors. ERN1 to -3 or ERN1Δ to -3Δ effectors were coinfiltrated with P35Smin (B) or 4xNF box ([C] and [D]) target GUS reporters. Transcriptional activation of the 4xNF box reporter in (E) was measured following expression of individual ERN1 or ERN3 effectors or combined expression of ERN1/ERN2, ERN1/ERN3, or ERN1/ERN3Δ effectors. GUS activity levels were measured using 1 μg of total protein extracts. Error bars represent ±sd of mean GUS activity values derived from either three or four ([B] to [D]) or seven to nine (E) independent experiments. In (E), data are compiled from measurements of 15 to 18 individual samples that were used for Student's t test statistical analysis. Asterisks indicate statistically significant differences (P < 0.05, Student's t test) compared with the reference ERN1 effector construct (see Supplemental Table 2 online).
To address the question of whether ERN3 can modulate the trans-activation properties of the ERN1/ERN2 transcriptional activators, a combination of effector proteins was expressed in N. benthamiana using the target 4xNF box reporter. Coexpression of both ERN1 and ERN2 resulted in similar GUS activity levels to those observed when expressing ERN1 alone (Figure 8E). However, coexpression of ERN3 with either ERN1 or ERN2 led to a significant reduction in GUS activity levels compared with those obtained with ERN1 or ERN2 (Figure 8E; see Supplemental Figure 4 online). Strikingly, the ERN3 protein lacking its AP2/ERF DNA binding domain (ERN3Δ) was equally capable of attenuating the transcriptional activation of the target 4xNF box reporter by ERN1 (Figure 8E; see Supplemental Figure 4 online). This suggests that ERN3-mediated repression is an active mechanism that does not absolutely require the DNA binding domain.
DISCUSSION
NF-Elicited Gene Activation in Root Hairs Requires a Regulatory Unit with a Novel GCC-Like Motif
Mt ENOD11 is a well-characterized molecular marker that has been widely used to study NF signaling in the root epidermis (Charron et al., 2004; Miwa et al., 2006; Sun et al., 2006; Marsh et al., 2007). In this study, we performed a detailed analysis of the Mt ENOD11 promoter in order to elucidate the molecular mechanisms of NF-regulated gene expression. A gain-of-function approach has shown that a 33-bp promoter sequence that we call the NF box is sufficient to direct specific NF-mediated expression, which is elicited during the early preinfection stage of the S. meliloti association. On the other hand, the NF box alone is not sufficient to confer Mt ENOD11 expression during the later stage of rhizobial infection, which we have previously shown to depend upon a proximal 257-bp promoter region lying downstream of the NF box (Boisson-Dernier et al., 2005). We propose that NF box–mediated gene expression is activated through initial NF perception (via the NFP/DMI/NSP signaling pathway) and that the infection-related expression requires a second distinct signaling pathway that is presumably activated at a later stage before bacterial entry. The NF box sequence is also found in the promoters of other NF-responsive M. truncatula genes such as Mt ENOD12 and Mt ENOD9, which also encode putative Pro-rich cell wall–associated proteins presumably involved in root hair cell wall modification during preinfection stages (Pichon et al., 1992; Journet et al., 2001). It should be noted that the NF box is not present in the 200-bp promoter region of the pea (Pisum sativum) Ps ENOD12B gene that Vijn et al. (1995) have reported to confer NF-elicited expression in dividing cortical cells of the nodule primordia. We suggest that the NF box may be a regulatory unit specifically associated with early NF signaling in the root epidermis. The NF box is also distinct from the recently identified conserved Root Hair Element comprising the strictly conserved CACG motif, which is essential for root hair–specific activation of genes encoding angiosperm cell wall–related proteins (Kim et al., 2006). We thus propose that the NF box acts through an alternative regulatory pathway and therefore may be of interest for engineering novel inducible synthetic promoters for legume-centric research and technology development (Rushton et al., 2002).
Block deletion and point mutation analysis has revealed that a GCC-like (GCAGGCC) motif within the NF box is essential for NF-dependent expression in root hairs and for binding of root hair–specific nuclear protein factors. This motif is reminiscent of, although not identical to, the well-characterized GCC box (GCCGCC) recognized by ERF transcription factors. Despite the lack of the GCC box consensus sequence, a number of essential nucleotides for specific protein binding, indicated in boldface in the GCC box core (GCCGCC) (Hao et al., 1998), can be located to identical relative positions within the NF box (GCAGGCC). Additional studies will now be required to determine the DNA binding specificities of NF box–interacting ERN factors (discussed in more detail below) for this novel variant of the classical GCC box.
Mutations within the CAAT box and DOF-like motifs flanking or partially overlapping the GCC-like motif significantly reduce NF-responsive gene activation, but not as dramatically as mutations within the GCC-like motif. These flanking or overlapping nucleotides can contribute to the binding affinity of transcription factors to the central GC-rich region, as reported previously for other factors (Tournier et al., 2003; Xue and Loveridge, 2004). Alternatively, the GCC-like motif may act in cooperation with neighboring cis motifs to specify expression, as has been reported for a number of plant genes (Buttner and Singh, 1997; Riechmann and Meyerowitz, 1998; Diaz et al., 2002; Brown et al., 2003; Yamamoto et al., 2006). The future characterization of the core cis-regulatory elements specifically recognized by ERN factors should make it possible to scan the M. truncatula genome to discover NF-dependent regulons.
Novel ERF Factors Interacting with the NF Box
Yeast one-hybrid screening using the NF box sequence as bait resulted in the identification of three closely related AP2/ERF transcription factors specifically interacting with this NF-responsive sequence. Interestingly, one of these NF box–interacting ERF factors is identical to the ERN transcription factor very recently identified by a genetic approach (Middleton et al., 2007). The ERN deletion mutant bit1-1 is defective in root infection and nodulation as well as in NF-elicited expression of Mt ENOD11 in root hairs. The three NF box–interacting factors, now named ERN1 (previously named ERN), ERN2, and ERN3, all contain the typical AP2/ERF DNA binding domain and belong to the same phylogenetically related group V of the ERF subfamily that includes Arabidopsis RAP2.11 (Okamuro et al., 1997; Nakano et al., 2006; Middleton et al., 2007). We have shown here that ERN factors bind specifically to the NF box sequence, which contains the essential GCC-like sequence motif (GCAGGCC). Although >107 yeast clones were screened for NF box binding, it is significant that we were only able to identify the three ERN factors, despite the fact that other AP2/ERF clones are represented in the root hair–specific cDNA libraries used for the one-hybrid screen (F. de Carvalho-Niebel and A. Niebel, unpublished data). Taken together, these results argue that ERN-type transcription factors specifically recognize the atypical GCC-like motif within the NF box.
Detailed structural information concerning the interaction between the transcription factor and its target DNA is only available for a few members of the AP2/EREBP family. NMR analysis of the solution structure of Arabidopsis ERF1 bound to the GCC box has revealed that an antiparallel three-stranded β-sheet binds within the DNA major groove (Allen et al., 1998). This interaction involves direct base contacts with a total of seven amino acids present in the β-sheet of the conserved AP2 domain of ERF1 (Arg-150, Arg-152, Trp-154, Glu-160, Arg-162, Arg-170, and Trp-172). The AP2 domain of the three ERN factors shares a high level of amino acid identity with that of RAP2.11 but significantly less with ERF1 (Figure 5B). A comparison between the AP2 domains of ERF1 and ERN factors reveals amino acid substitutions at two key positions involved in DNA binding (Trp-154→Ser and Arg-162→Lys). We propose that these amino acid substitutions modify the binding specificity of the ERN factors and presumably correlate with the atypical NF box GCC-like motif. For example, Fukazawa et al. (2000) showed that the substitution of a highly conserved Arg residue by Lys changes the optimal binding site of the yeast bZIP protein GCN4 from the palindromic site (GACGTC) to the pseudopalindromic site (TGACTCA).
We have demonstrated that the ERN genes are preferentially expressed in root hairs (10-fold higher compared with the whole root) and that in ERN1 and ERN2 are significantly upregulated (7.3- and 3.5-fold, respectively) following the application of purified NFs. Importantly, NF-elicited ERN gene induction is dependent on the NFP genetic locus, an essential component of the NF signaling pathway in the root epidermis, which likely encodes a component of the NF receptor (Ben Amor et al., 2003). Thus, in agreement with similar findings by Middleton et al. (2007), our data support the proposition that ERN factors constitute novel downstream components of the epidermal NF signaling pathway. Interestingly, we note that ERN factors and the Arabidopsis ortholog RAP2.11 appear to be preferentially expressed in root hairs and the differentiation zone of the root epidermis (this study and Genevestigator Gene Atlas; Zimmermann et al., 2004). Furthermore, WIN1/SHN1, another member of the group V Arabidopsis ERF factors, is involved in epidermal surface cuticle biosynthesis and cell wall structural modification (Aharoni et al., 2004; Broun et al., 2004). It is conceivable, therefore, that ERNs may have been recruited from ERF regulatory proteins that function in processes associated with root epidermal cell differentiation.
ERN Factor Interplay May Coordinate NF-Elicited ENOD Gene Activation
As a complement to the yeast one-hybrid–based in vivo system, we studied the transactivation properties of individual ERN factors using a heterologous plant transient expression assay. We have shown that ERN-GFP/YFP fusion proteins transiently expressed in N. benthamiana cells localize to the nucleus, despite the fact that typical nuclear localization signal domains are not present in the predicted protein sequences. It is possible that basic amino acid clusters are responsible for nuclear targeting of ERN factors, as reported for other AP2/ERF proteins (Chen et al., 2003; Krizek and Sulli, 2006; Saleh et al., 2006). We observed that the ERN1 fusion protein is also associated with discrete nuclear bodies (Figure 7). Previous studies have described a similar subnuclear compartmentalization for certain transcription factors and have associated this with functional regulation, including phosphorylation and targeted degradation via the proteasome pathway (Yang et al., 2005; Al-Sady et al., 2006; Saleh et al., 2006). Additional experiments in Medicago roots using native promoter and nuclear domain–associated markers will now be required to further study distinct ERN1 subnuclear localization.
We have found that the nucleus-targeted ERN1 and ERN2 act as positive regulators of the NF box in N. benthamiana cells and that this activity is dependent on the presence of the AP2/ERF DNA binding domain. It should also be noted that ERN1 and ERN2 possess a well-conserved C-terminal sequence that is similar to the C-terminal CMV-4 domain of Arabidopsis and rice (Oryza sativa) RAP2.11 clade members (Nakano et al., 2006), whereas ERN3 lacks this C-terminal sequence and is unable to activate reporter gene expression. Furthermore, when ERN3 is expressed together with either ERN1 or ERN2, there is a clear reduction in transcriptional activation of the 4xNF box reporter, suggesting that ERN3 may function as a repressor-type ERF factor. Coordinated induction of both activator- and repressor-type AP2/ERFs has been proposed to fine-tune the regulation of gene expression and to modulate antagonistic signaling pathways (Fujimoto et al., 2000; McGrath et al., 2005; Nakano et al., 2006).
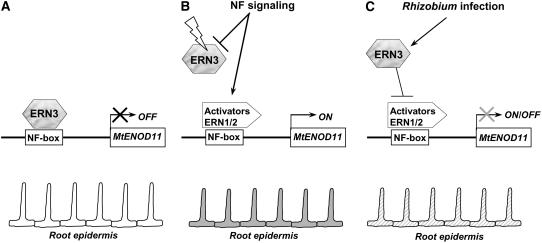
Our results suggest that activator and repressor ERN proteins function to fine-tune NF-mediated root hair expression of plant genes regulated via NF boxes. In the model presented in Figure 9, we propose that, in the absence of rhizobia, the highly expressed ERN3 repressor limits the expression of target host genes by binding to the NF box (Figure 9A). Following Rhizobium inoculation, NF signal perception and transduction via the NFP-dependent pathway results in enhanced transcription and protein accumulation of the ERN1 and ERN2 transcriptional activators (Figure 9B). Middleton et al. (2007) has reported that while the ERN1 deletion mutant bit1-1 is defective in nodulation, infection, and Mt ENOD11 gene activation, it is still capable of partial transduction of the NF signal and of limited infection thread initiation. This could be due to partial redundancy with respect to ERN2, which may also be required to regulate NF-elicited ENOD gene activation. The NF-dependent accumulation of ERN1/ERN2 activators may also be accompanied by a change in the level or activity of the ERN3 repressor protein (Figure 9B), although any model to explain this must be consistent with our demonstration that ERN3 transcript levels remain constant following NF treatment. In this respect, it is interesting that ubiquitin-conjugated downregulation of a tobacco (Nicotiana tabacum) repressor-type ERF3 factor has been proposed by Koyama et al. (2003). Additional regulatory steps must be postulated to account for the expression of Mt ENOD11 and other genes in the subsequent steps of infection. Once rhizobia are enclosed within the curled root hair, activation of the entry receptor pathway, essential for bacteria root infection (Ardourel et al., 1994), requires transcriptional factor(s) that mediate Mt ENOD11 expression via the 257-bp proximal promoter region (Boisson-Dernier et al., 2005). We suggest that de novo accumulation of active ERN3 factors may displace ERN1/ERN2 activators from the NF box by an active repressor mechanism (Fujimoto et al., 2000; Ohta et al., 2001; Song et al., 2005), as reported for other class II ERF factors, thus turning off gene expression in adjacent noninfected epidermal cells (Figure 9C). Our findings argue that ERN family members contribute to both positive and negative regulatory fine-tuning of host gene expression during preinfection stages of the legume/Rhizobium symbiotic association.
Figure 9.
Model for ERN Factor–Mediated Regulation of Early Nodulin Gene Expression in M. truncatula Root Hairs.
In the absence of the rhizobial partner, we propose that constitutive binding of ERN3 to the NF box sequence represses Mt ENOD11 gene expression in the root epidermis (A). During the early preinfection stages of the symbiotic association (B), NFs secreted by S. meliloti are perceived and transduced by the NFP-dependent NF signaling pathway. Transcription activation and accumulation of the ERN1/2 activators, probably accompanied by modifications of ERN3 protein levels/activities, lead to transactivation of the Mt ENOD11 gene in root hairs via the NF box regulatory unit (B). Upon bacterial root infection, de novo accumulation of active ERN3 presumably leads to the progressive repression of ERN1/ERN2 transcription activities.
METHODS
Plant Material, Growth Conditions, and NF Treatments
Medicago truncatula cv Jemalong A17 and the NF signaling mutant nfp-2 (Arrighi et al., 2006) were used in this study. Seeds were surface-sterilized and germinated according to Boisson-Dernier et al. (2001) and either used for Agrobacterium rhizogenes transformation or transferred to nitrogen-starved medium for 3 d before subsequent NF treatment. Aeroponically grown wild-type and mutant nfp-2 seedlings were transferred to Falcon tubes (Becton Dickinson Labware) containing either water (control) or a 10−9 M aqueous solution of purified Sinorhizobium meliloti NFs (Roche et al., 1991) and incubated for 6 h before tissue harvest for subsequent RNA extraction. NF treatment of seedling roots grown on pouch paper support (Mega International) was performed as described by Sauviac et al. (2005) by immersing roots for 16 to 18 h in either water (control) or an aqueous solution of purified NFs (10−8 M). Seedling roots were then used for RNA extraction from intact roots or from purified root hair fractions as described by Sauviac et al. (2005).
Treatments of M. truncatula Roots Generated by A. rhizogenes Transformation
Germinated seedlings were inoculated with A. rhizogenes as described by Boisson-Dernier et al. (2001). At ∼3 weeks after inoculation, composite kanamycin-resistant plants were transferred to pouch paper/Farhaeus agar plates (nitrogen- and antibiotic-free) as described by Boisson-Dernier et al. (2005). Transgenic roots growing on the paper support were treated with either water (control) or an aqueous solution of purified NFs (10−9 M) or inoculated with a liquid culture of S. meliloti strain RCR2011 (GMI6526) (Ardourel et al., 1994) harboring the hemA-lacZ transgene (5 × 10−5 bacteria/mL), as described by Boisson-Dernier et al. (2005).
Chimeric Promoter–GUS Constructs
Promoter–GUS Fusion and Deletion Constructs
Mt ENOD12 promoter deletions −531E12 and −327E12 were generated using ScaI and SphI internal restriction sites, respectively. Mt ENOD9 promoter deletions −558E9 and −532E9 were generated by PCR using the 5′ primers −549E9-for and −523E9-for (Table 1) in combination with the T7 universal primer as 3′ primer. Mt ENOD11 promoter deletions −411E11, −367E11, and −257E11 were described previously (Boisson-Dernier et al., 2005). The −350E11 deletion construct was generated by PCR using 350E11-for and 0E11-rev primers (Table 1). The −411E11 construct lacking the −357 to −258 promoter region was obtained by cloning the −411 to −358 promoter region (generated by PCR with primer pairs −411E11EcoRI-for and −358E11HindIII-rev) (Table 1) between the EcoRI and HindIII sites upstream from the −257E11 deletion construct.
Table 1.
Primers Used in This Study
| Name | Sequence 5′ to 3′ |
|---|---|
| −549E9-for | 5′-GGAAGTCAGACAATAAGAATTCAATGAAATAGCAGG-3′ |
| −523E9-for | 5′-GGCCTTTAAGAATTCTTTAGACCAAGCTTATGG-3′ |
| −411E11EcoRI-for | 5′-AATAAAGAATTCGACATTAAATTTGAGGG-3′ |
| 411E11XbaI-for | 5′-TAAAAATCTAGACACTTAAATTTGAGG-3′ |
| −411E11ΔGC-for | 5′-GAGGGTCATAATAACATAAAATAAT*AAAGCTATTATTTTACCAGCC-3′ |
| −411E11ΔGC-rev | 5′-GGCTGGTAAAATAATAGCTTT*ATTATTTTATGTTATTATGACCCTC-3′ |
| −411E11ΔGAT-for | 5′-GGCCAGTGAATTCGACACTTA*ATAATAACATAAAATAATTGCAG-3′ |
| −411E11ΔGAT-rev | 5′-CTGCAATTATTTTATGTTATTAT*TAAGTGTCGAATTCACTGGCC-3′ |
| −411E11dledel-for | 5′-GATTAATAAAAGAATTCGACACTTA*ATAATAACATAAAATAATAAAGC-3′ |
| −411E11dbledel-rev | 5′-GCTTTATTATTTTATGTTATTAT*TAAGTGTCGAATTCTTTATTAATC-3′ |
| −392E11EcoRI-for | 5′-TGGAATTCATAATAACATAAAATAATTG-3′ |
| −392E11XbaI-for | 5′-TGTCTAGAATAATAACATAAAATAATTG-3′ |
| −380E11NheI-rev | 5′-CTGCAAGCTAGCTATGTTATTATGACC-3′ |
| −380E11HindIII-rev | 5′-CTGCAAAAGCTTTATGTTATTATGACC-3′ |
| −367E11mutDOF-for | 5′-AGGCCTGCGACTATTATTTTACC-3′ |
| −367E11mutGC-for | 5′-CATATGTAAAGCTATTATTTTACC-3′ |
| −367E11mutGC-rev | 5′-CATATGCAATTATTTTATGTTATTATGAC-3′ |
| −367E11mutHDZ-rev | 5′-AGGCCTGCACTTAGTTTATGTTATTATGAC-3′ |
| −358E11HindIII-rev | 5′-GGCTGGTAAAATAAAAGCTTTAGGCC-3′ |
| −358E11NheI-rev | 5′-GGCTGGTAAAATGCTAGCTTTAGG-3′ |
| −350E11-for | 5′-GGTACCAGCCTTTTAATTTGACC-3′ |
| −257E11-rev | 5′-GGAATCTAGATTCTTGGC-3′ |
| −257E11-rev | 5′-GGAATCTAGATTCTTGGC-3′ |
| −0E11-rev | 5′-TGCCCACAGGCCGTCGAG-3′ |
| ERN1-for | 5′-GGAAGATGGTGCTGTTGCTT-3′ |
| ERN1-rev | 5′-TGTTGGATTGTGAACCTGACTC-3′ |
| ERN2-for | 5′-ATGATTCCCCTCTCGCTTCT-3′ |
| ERN2-rev | 5′-CACTGGCTGTGCCAATACAG-3′ |
| ERN3-for | 5′-TGGCCATTACCTCAACAAAGG-3′ |
| ERN3-rev | 5′-CCTTCCACATAGCACTCATTCA-3′ |
| ERN1delAP2-for | 5′-CACTCGAACCAGAATTCTCACTCATG-3′ |
| ERN1delAP2-rev | 5′-GCCTTTGCCGAATTCCAACAAAC-3′ |
| ERN2delAP2-for | 5′-CTCGTACCAGAATTCTCACACATG-3′ |
| ERN2delAP2-rev | 5′-CTTTGTCTCAGAATTCCAAACTTGTCTC-3′ |
| ERN3delAP2-for | 5′-TACTCGTAGAATTCTCTCTACTACTC-3′ |
| ERN3delAP2-rev | 5′-GCTCTTTGTCGAATTCCAACAAAC-3′ |
Nucleotides in boldface correspond to substitutions in order to generate new restriction sites (underlined).
Internal and Block Deletion Constructs
Block deletions of the two E11 motifs, named GCC-like (TGCAGGCCT) or G(A/T)-rich (AAATTTGAGGGTC), were generated using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Single block deletions were generated using a −411E11-containing plasmid as a template and the corresponding complementary primer pairs −411E11ΔGC-for/−411E11ΔGC-rev (GC-rich motif deletion) and −411E11ΔGAT-for/−411E11ΔGAT-rev (G[A/T]-rich motif deletion) (Table 1).
Site-Directed Mutagenesis
Motif substitutions within the NF-box sequence were introduced into the −411E11 deletion construct. Point mutations in the HD-ZIP–like motif (AAATAATTG to ACATAAGTG) were obtained by PCR using −411E11EcoRI-for and −367mutHDZ-rev primers (Table 1). The resulting −411 to −367 mutated PCR fragment was cloned into the EcoRI and StuI sites in front of a −367E11 deletion to reconstitute the −411E11mHDZ construct. Substitution in the GC-rich motif (TGCAGGCCT to TGCATATGT) was generated by PCR using primer pairs −411E11EcoRI-for/−367mutGC-rev and −367mutGC-for/−257E11-rev (Table 1). PCR with these primers generated two DNA fragments (−411 to −367 and −367 to −257, respectively) that after restriction digestion with EcoRI/NdeI and NdeI/XbaI enzymes, respectively, were ligated together with the EcoRI/XbaI sites in front of the −257E11 deletion to reconstitute the −411E11mGCcore construct. Substitution in the DOF motif (TAAAG to TGCAG) was introduced by PCR using −367mutDOF-for and −257E11-rev primers (Table 1). The mutated −367 to −257 PCR fragment was subsequently inserted between StuI/XbaI sites of a −411E11 deletion construct to generate the −411E11mDOF construct.
Gain-of-Function Constructs
The tetramers of the NFE region (−411 to −358), of NFE5′ (−411 to −380), and of NFE3′ (NF box) (−392 to −358) were inserted into EcoRI/HindIII sites upstream from the minimal CaMV 35S promoter (−47) (Pontier et al., 2001). Four tandem copies of each promoter region were obtained as described by Rushton et al. (2002). Briefly, each promoter region (NFE region, NFE5′, or NF box) was amplified by PCR using the primer pairs listed in Table 1 to generate for each sequence three PCR fragments named A, B, and C bordered by EcoRI/NheI, XbaI/NheI, and XbaI/HindIII sites. After cloning into the pGEM-T vector (Promega), the resulting plasmids were digested with ScaI (which cuts outside the Mt ENOD11 promoter sequences) together with either NheI (A and B) or XbaI (B and C). Ligation of the digested plasmids 5′-ScaI/NheI-3′ and 5′-XbaI/ScaI-3′ recreated an intact plasmid with dimers AB and BC of the respective promoter regions. Plasmids AB and BC, which have lost the internal NheI/XbaI sites after ligation, were digested with ScaI/NheI and ScaI/XbaI as before. Ligation of two such fragments then creates the tetrameric ABBC constructs, which were then inserted into the EcoRI and HindIII sites upstream from the minimal −47 CaMV 35S promoter.
All deletion, mutation, and gain-of-function promoter constructs were fused to GUS in the binary vector pLP100 (Szabados et al., 1995) and verified by sequencing using the GUS−1 primer (5′-TGCCCACAGGCCGTCGAG-3′). pLP100 binary vectors were introduced into A. rhizogenes strain ARqua1 by the freeze-thaw method as described by Höfgen and Willmitzer (1998).
Histochemical and Fluorimetric GUS Assays
Histochemical GUS staining was performed as described previously (Journet et al., 2001) by incubation of transgenic root samples in X-glcA buffer solution (5-bromo-4-chloro-3-indolyl glucuronide, cyclohexylammonium salt; Biosynth). Stained samples were examined with a stereomicroscope (Leica Microsystems) and a Zeiss Axiophot microscope (Carl Zeiss). Double staining for both GUS and β-galactosidase activities after inoculation with a S. meliloti strain carrying a constitutive hemA-lacZ fusion was performed as described by Vernoud et al. (1999). For enzymatic GUS assays, pools of 7 to 10 independent transgenic roots were ground in liquid nitrogen and homogenized in 1× GUS extraction buffer (50 mM sodium phosphate, pH 7.5, 10 mM 2-mercaptoethanol, 10 mM Na2EDTA, 0.1% Triton X-100, and 0.1% sodium lauryl-sarcosine) for total protein extraction. GUS activities were measured fluorimetrically using 1 μg of total protein extract as described previously (Boisson-Dernier et al., 2005).
A visual-based quantification method adapted from Sidorenko et al. (2000) was used to evaluate GUS staining intensities for different promoter deletion constructs. Briefly, roots were analyzed at 6 h after staining and classified according to overall visual GUS staining intensities into two distinct categories: moderate/strong staining (+) or undetectable/weak staining (−). For each construct, ∼40 to 150 independent roots from three separate experiments were analyzed by histochemical and fluorimetric methods. For all constructs tested, there is a linear correlation between the results obtained from the two quantification methods (0.95 < r < 0.98). Statistical data analyses were performed using Student's t test, in which a significant difference was defined for P < 0.05.
Electrophoretic Mobility Shift Assay
A pGEM-T vector containing the NF box sequence or its mutated derivative was digested by EcoRI/SstI restriction enzymes, and the respective DNA fragments were purified using the JETSORB gel extraction kit (Genomed). The NF box probe was radiolabeled with [α-33P]dCTP (3000 Ci mmol−1) by filling in with the Klenow fragment of DNA polymerase I (Promega) and purified on MicroSpin columns (Amersham Biosciences), according to the manufacturer's instructions. Unlabeled competitor DNA was filled in with nonradioactive nucleotides. Total protein extract from isolated root hair was prepared according to Busk and Pages (1997). One microgram of protein extract was incubated with the radioactive probe (50,000 cpm) in 12 μL of 1× binding buffer (25 mM HEPES, pH 7.8, 75 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.2 mM DTT, and 10% glycerol) and 0.1 μg of poly(dA-dT) for 30 min at 4°C before loading on a 0.5× Tris-borate-EDTA, 6% polyacrylamide gel. Electrophoresis was run at 90 V for 1 h at 4°C. In competition assays, the protein was incubated first with unlabeled oligonucleotide for 10 min prior to adding the radioactive probe and continuing with incubation for a further 30 min.
Quantitative RT-PCR Analysis
For NF-treated samples, total RNA was extracted from intact roots or isolated root hairs of M. truncatula using the Macherey-Nagel total RNA isolation kit according to the manufacturer's instructions and the technique described by Sauviac et al. (2005). M. truncatula organs and isolated nodule samples used for the quantitative RT-PCR analysis were described previously (Godiard et al., 2007). Genomic DNA was removed via on-column DNase treatment following basic kit instructions, and the absence of DNA was confirmed by PCR amplification of the rip1 gene (Cook et al., 1995) using primer pairs spanning an intronic region (Sauviac et al., 2005). The DNA-free RNA samples were quantified, and RNA integrity was checked on a 1% (w/v) agarose gel. First-strand cDNA synthesis was performed using 0.7 to 1 μg of total RNA with an anchored oligo(dT) (17T+V) and SuperScript II reverse transcriptase (Invitrogen) following the manufacturer's protocol. Thirty nanograms of an in vitro transcribed nebulin RNA was included in each first-strand cDNA reaction in order to control the efficiency of the reverse transcriptase (data not shown). Quantitative RT-PCR was performed with the Light Cycler Fast Start Reaction Mix MasterPLUS SYBR Green I (Roche) on a Roche light cycler real-time PCR machine according to the manufacturer's instructions. Each reaction was performed with 2 μL of a 1:20 (v/v) dilution of the first cDNA strand, with 0.5 μM of each primer in a total reaction volume of 10 μL. The cycling conditions were as follows: 95°C for 9 min followed by 45 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 10 s, and extension at 72°C for 10 to 15 s. PCR amplification specificity was verified by analysis of a dissociation curve at the end of the PCR cycles by heating samples from 55 to 60°C to 95°C as well as by agarose gel resolution and sequencing of the corresponding PCR products. Primer pairs to amplify the elongation factor EF1-α were described previously (Sauviac et al., 2005), and the primers used to amplify ERN1, ERN2, and ERN3 are listed in Table 1 (ERN1-for, ERN1-rev, ERN2-for ERN2-rev, ERN3-for, and ERN3-rev primers). Transcript levels for each of the target genes (ERN1, ERN2, and ERN3) were normalized to the endogenous elongation factor EF1-α transcript level. The data shown represent means of values obtained from either two or three independent biological experiments and quantitative RT-PCR.
Yeast One-Hybrid Screening
DNA fragments containing four tandem copies of NFE (−411 to −358) or the NF box (−392 to −358) were inserted into the EcoRI/MluI sites of the pHISi vector (Clontech) to create the tetramer NFE-HIS3 and tetramer NF-box-HIS3 constructs. These and the control p53-HIS3 (Matchmaker one-hybrid system; Clontech) plasmids were linearized by XhoI digestion and used to transform the YM4271 yeast strain. Genome integration of the corresponding constructs was verified by both the complementation of the HIS− auxotrophic phenotype and the ability to PCR-amplify insert-specific DNA fragments. Yeast colony PCR was performed using 2 μL of yeast suspension obtained after 30 min of incubation at 37°C within a Lyticase/TE solution (catalog No. L4025; Sigma-Aldrich). A positive yeast recombinant showing basal low HIS level, for which growth was blocked at 2 to 5 mM 3-AT, was chosen for screening of the M. truncatula root hair cDNA libraries. cDNA libraries were prepared using the Creator Smart cDNA library construction kit (Clontech) from pooled samples of 10−8 M NF-treated root hairs (2, 6, and 18 h) by directional cloning in SfiI sites of a pGADT7-modified vector (Sauviac et al., 2005; F. de Carvalho-Niebel, L. Sauviac, and L. Deslandes, unpublished data). Two types of libraries were generated, using either the Powerscript reverse transcriptase (RNaseH−), producing preferentially full-length cDNAs, or the wild-type Moloney murine leukemia virus reverse transcriptase, with its RNaseH activity, giving rise to shorter average size cDNAs. Primary libraries were Seaprep-amplified following the manufacturer's instructions (Cambrex Bio Science). Powerscript and Moloney murine leukemia virus libraries had 106 independent clones with an average insert size of 1.1 kb and 2.8 × 106 clones with an average insert size of 0.7 kb, respectively. cDNA libraries were screened with selection set at 5 mM 3-AT, using the YM4271 (tetramer NF box-HIS3) yeast strain. HIS+ candidates able to grow at 5 and 10 mM 3-AT were selected, amplified by PCR, and sequenced. Plasmids extracted from HIS+ colonies were amplified in Escherichia coli before retransformation into YM4271 (tetramer NF box-HIS3), YM4271 (tetramer NFE-HIS3), and YM4271 (p53-HIS3) strains.
ERN Plasmid Constructs and Transient Expression in N. benthamiana Leaves
In vitro BP and LR recombination reactions were performed according to the manufacturer's instructions (Invitrogen) between ERN1, ERN2, and ERN3 pGADT7 clones and the respective donor plasmid pDONR 207 (Invitrogen Life Sciences) and destination vectors PAM-PAT35S-GFP/YFP-GTW and PAM-PAT35S-3xHA-GTW (a kind gift of L. Deslandes, Laboratory of Plant Microbe Interactions) to generate the respective 35S-GFP-ERN, 35S-YFP-ERN, and 35S-3xHA-ERN fusion constructs. A PCR strategy was used to create ERNs deleted for the respective AP2/ERF DNA binding domains. For each ERN, PCR was performed using ERN-PDONR 207 plasmids as templates in two independent PCRs with PDONR 207 forward and reverse primers (Invitrogen Life Sciences) combined, respectively, with the ERNdelAP2-rev or ERNdelAP2-for primers listed in Table 1. These reactions generated two DNA fragments lacking the region containing the AP2/ERF domain of each ERN, which after restriction digestion with SfiI and EcoRI, respectively, were ligated together within SfiI sites of PDONR 207 plasmids to reconstitute the ERN1Δ, ERN2Δ, and ERN3Δ PDONR 207 plasmids. These ERN-AP2–deleted donor plasmids were recombined with PAM-PAT35S-YFP-GTW and PAM-PAT35S-3xHA-GTW destination vectors to generate 35S-YFP-ERNΔ and 35S-3xHA-ERNΔ fusion constructs. These binary vectors were introduced into Agrobacterium tumefaciens strains GV3101 or GV3103 by electroporation following the Invitrogen protocol. A. tumefaciens strains harboring the various constructs with or without p19 (Voinnet et al., 2003) were grown in Luria-Bertani medium under antibiotic selection at 28°C before harvesting and resuspension in 10 mM MgCl2, 10 mM MES, pH 5.6, with 100 μM acetosyringone (catalog No. D13440-6; Sigma-Aldrich). After incubation at room temperature for 2 h, A. tumefaciens cultures were infiltrated into leaves of 3-week-old Nicotiana benthamiana plants using a 1-mL needleless syringe. For coagroinfiltration, equal volumes of each A. tumefaciens culture adjusted to an OD600 of 0.5 were mixed prior to infiltration. The subcellular distribution patterns of YFP- or YFP/GFP-fused proteins were analyzed by fluorescence optical and confocal microscopy from 30 to 60 h after infiltration. Each construct was analyzed in three distinct infiltrated leaves of six to eight plants in at least three independent experiments. For transactivation studies, leaf discs were collected at 24 h after inoculation and either used directly for histochemical GUS assays or frozen in liquid nitrogen and stored at −80°C until processing. For the protein gel blot analysis, one-tenth volume of proteins extracted from two leaf discs per sample was loaded per lane. For enzymatic GUS assays, total proteins were extracted and quantified as described above, and 1 μg of total protein was used in replicates to measure enzymatic activities of individual samples.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU038802 (ERN1), EU038803 (ERN2), and EU038804 (ERN3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Histochemical Analysis of GUS Activity Driven by the Gain-of-Function NF Box Construct at 4 d after Inoculation with S. meliloti in Transgenic Roots and in Young Nodules.
Supplemental Figure 2. Expression Profiles of ERN1, ERN2, and ERN3 Genes in Different Plant Organs.
Supplemental Figure 3. Expression of ERN Proteins in A. tumefaciens–Infiltrated N. benthamiana Cells.
Supplemental Figure 4. Histochemical and Fluorimetric GUS Activities of N. benthamiana Leaf Discs following Infiltration with the 4xNF Box Reporter Alone or in Combination with ERN2, ERN3, ERN2/ERN3, or ERN2/ERN3Δ Effectors.
Supplemental Table 1. Statistical Analysis of GUS Activity Directed by Different Promoter Constructs.
Supplemental Table 2. Statistical Analysis of GUS Activity Measured in Transactivation Studies Described in Figure 8E and Supplemental Figure 4.
Supplementary Material
Acknowledgments
We thank Fabienne Maillet and Jean Denarié for providing S. meliloti Nod factors and Etienne Journet for providing seeds of the M. truncatula nfp-2 mutant line. We thank Laurent Deslandes for helpful advice concerning yeast and N. benthamiana transformation and for providing binary PAM-PAT destination vectors, and members of Pascal Gamas's group at the Laboratory of Plant Microbe Interactions for providing M. truncatula organs and nodule samples for quantitative RT-PCR analyses. We also thank Giles Oldroyd for providing the ERN paper (Middleton et al., 2007) before publication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Fernanda de Carvalho-Niebel (fniebel@toulouse.inra.fr).
Online version contains Web-only data.
References
- Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23 439–446. [DOI] [PubMed] [Google Scholar]
- Ané, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Ardourel, M., Demont, N., Debellé, F., Maillet, F., de Billy, F., Promé, J.C., Dénarié, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi, J.F., et al. (2006). The Medicago truncatula LysM motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor, B., Shaw, S.L., Oldroyd, G.E., Maillet, F., Penmetsa, R.V., Cook, D., Long, S.R., Dénarié, J., and Gough, C. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34 495–506. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier, A., Andriankaja, A., Chabaud, M., Niebel, A., Journet, E.P., Barker, D.G., and de Carvalho-Niebel, F. (2005). MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol. Plant Microbe Interact. 18 1269–1276. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier, A., Chabaud, M., Garcia, F., Bécard, G., Rosenberg, C., and Barker, D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700. [DOI] [PubMed] [Google Scholar]
- Broun, P., Poindexter, P., Osborne, E., Jiang, C.Z., and Riechmann, J.L. (2004). A transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 30 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.L., Kazan, K., McGrath, K.C., Maclean, D.J., and Manners, J.M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk, P.K., and Pages, M. (1997). Protein binding to the abscisic acid-responsive element is independent of VIVIPAROUS1 in vivo. Plant Cell 9 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 27 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira, R., Galera, C., de Billy, F., Penmetsa, R.V., Journet, E.P., Maillet, F., Rosenberg, C., Cook, D., Gough, C., and Dénarié, J. (2000). Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12 1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron, D., Pingret, J.L., Chabaud, M., Journet, E.P., and Barker, D.G. (2004). Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 136 3582–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.Q., Dong, Y., Wang, Y.J., Liu, Q., Zhang, J.S., and Chen, S.Y. (2003). An AP2/EREBP-type transcription-factor gene from rice is cold-inducible and encodes a nuclear-localized protein. Theor. Appl. Genet. 107 972–979. [DOI] [PubMed] [Google Scholar]
- Cook, D.R., Dreyer, D., Bonnet, D., Howell, M., Nony, E., and VandenBosch, K. (1995). Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haeze, W., and Holsters, M. (2002). Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12 79R–105R. [DOI] [PubMed] [Google Scholar]
- Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Martinez, M., Isabel-La Moneda, I., and Carbonero, P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29 453–464. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kaló, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa, J., Sakai, T., Ishida, S., Yamaguchi, I., Kamiya, Y., and Takahashi, Y. (2000). Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, D.J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts, R., Fedorova, E., and Bisseling, T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8 346–352. [DOI] [PubMed] [Google Scholar]
- Godiard, L., Niebel, A., Micheli, F., Gouzy, J., Ott, T., and Gamas, P. (2007). Identification of new potential regulators of the Medicago truncatula-Sinorhizobium meliloti symbiosis using a large-scale suppression subtractive hybridization approach. Mol. Plant Microbe Interact. 20 321–332. [DOI] [PubMed] [Google Scholar]
- Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273 26857–26861. [DOI] [PubMed] [Google Scholar]
- Heckmann, A.B., Lombardo, F., Miwa, H., Perry, J.A., Bunnewell, S., Parniske, M., Wang, T.L., and Downie, J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku, H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531. [DOI] [PubMed] [Google Scholar]
- Journet, E.P., El-Gachtouli, N., Vernoud, V., de Billy, F., Pichon, M., Dedieu, A., Arnould, C., Morandi, D., Barker, D.G., and Gianinazzi-Pearson, V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14 737–748. [DOI] [PubMed] [Google Scholar]
- Journet, E.P., Pichon, M., Dedieu, A., de Billy, F., Truchet, G., and Barker, D.G. (1994). Rhizobium meliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic alfalfa. Plant J. 6 241–249. [DOI] [PubMed] [Google Scholar]
- Kaló, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kanamori, N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.W., Lee, S.H., Choi, S.B., Won, S.K., Heo, Y.K., Cho, M., Park, Y.I., and Cho, H.T. (2006). Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T., Okada, T., Kitajima, S., Ohme-Takagi, M., Shinshi, H., and Sato, F. (2003). Isolation of tobacco ubiquitin-conjugating enzyme cDNA in a yeast two-hybrid system with tobacco ERF3 as bait and its characterization of specific interaction. J. Exp. Bot. 54 1175–1181. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A., and Sulli, C. (2006). Mapping sequences required for nuclear localization and the transcriptional activation function of the Arabidopsis protein AINTEGUMENTA. Planta 224 612–621. [DOI] [PubMed] [Google Scholar]
- Kusnetsov, V., Landsberger, M., Meurer, J., and Oelmuller, R. (1999). The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J. Biol. Chem. 274 36009–36014. [DOI] [PubMed] [Google Scholar]
- Lee, J.H., Hong, J.P., Oh, S.K., Lee, S., Choi, D., and Kim, W.T. (2004). The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: Possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Mol. Biol. 55 61–81. [DOI] [PubMed] [Google Scholar]
- Lévy, J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- Marsh, J.F., Rakocevic, A., Mitra, R.M., Brocard, L., Sun, J., Eschstruth, A., Long, S.R., Schultze, M., Ratet, P., and Oldroyd, G.E. (2007). Medicago truncatula NIN is essential for Rhizobium-independent nodule organogenesis induced by autoactive CCaMK. Plant Physiol. 144 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, K.C., Dombrecht, B., Manners, J.M., Schenk, P.M., Edgar, C.I., Maclean, D.J., Scheible, W.R., Udvardi, M.K., and Kazan, K. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton, P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R.M., Gleason, C.A., Edwards, A., Hadfield, J., Downie, J.A., Oldroyd, G.E., and Long, S.R. (2004. a). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R.M., Shaw, S.L., and Long, S.R. (2004. b). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101 10217–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, H., Sun, J., Oldroyd, G.E., and Downie, J.A. (2006). Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48 883–894. [DOI] [PubMed] [Google Scholar]
- Murakami, Y., Miwa, H., Imaizumi-Anraku, H., Kouchi, H., Downie, J.A., Kawaguchi, M., and Kawasaki, S. (2007). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 31 255–265. [DOI] [PubMed] [Google Scholar]
- Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E., Harrison, M.J., and Udvardi, M.K. (2005). Peace talks and trade deals. Keys to long-term harmony in legume-microbe symbioses. Plant Physiol. 137 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Long, S.R. (2003). Identification and characterization of NODULATION-SIGNALING PATHWAY 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon, M., Journet, E.P., Dedieu, A., de Billy, F., Truchet, G., and Barker, D.G. (1992). Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier, D., Balagué, C., Bezombes-Marion, I., Tronchet, M., Deslandes, L., and Roby, D. (2001). Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J. 26 495–507. [DOI] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 633–646. [DOI] [PubMed] [Google Scholar]
- Roche, P., Lerouge, P., Ponthus, C., and Promé, J.C. (1991). Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J. Biol. Chem. 266 10933–10940. [PubMed] [Google Scholar]
- Rushton, P.J., Reinstadler, A., Lipka, V., Lippok, B., and Somssich, I.E. (2002). Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, A., Lumbreras, V., Lopez, C., Dominguez-Puigjaner, E., Kizis, D., and Pages, M. (2006). Maize DBF1-interactor protein 1 containing an R3H domain is a potential regulator of DBF1 activity in stress responses. Plant J. 46 747–757. [DOI] [PubMed] [Google Scholar]
- Sauviac, L., Niebel, A., Boisson-Dernier, A., Barker, D.G., and de Carvalho-Niebel, F. (2005). Transcript enrichment of Nod factor-elicited early nodulin genes in purified root hair fractions of the model legume Medicago truncatula. J. Exp. Bot. 56 2507–2513. [DOI] [PubMed] [Google Scholar]
- Sidorenko, L.V., Li, X., Cocciolone, S.M., Chopra, S., Tagliani, L., Bowen, B., Daniels, M., and Peterson, T. (2000). Complex structure of a maize MYB gene promoter: Functional analysis in transgenic plants. Plant J. 22 471–482. [DOI] [PubMed] [Google Scholar]
- Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., and Geurts, R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791. [DOI] [PubMed] [Google Scholar]
- Song, C.P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., and Zhu, J.K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, G., Libault, M., Brechenmacher, L., Wan, J., and May, G.D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9 110–121. [DOI] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Sun, J., Cardoza, V., Mitchell, D.M., Bright, L., Oldroyd, G.E., and Harris, J.M. (2006). Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 46 961–970. [DOI] [PubMed] [Google Scholar]
- Szabados, L., Charrier, B., Kondorosi, A., de Bruijn, F.J., and Ratet, P. (1995). New plant promoter and enhancer testing vectors. Mol. Breed. 1 419–423. [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 15 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers, A.C., Auriac, M.C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628. [DOI] [PubMed] [Google Scholar]
- Tournier, B., Sanchez-Ballesta, M.T., Jones, B., Pesquet, E., Regad, F., Latche, A., Pech, J.C., and Bouzayen, M. (2003). New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 28 149–154. [DOI] [PubMed] [Google Scholar]
- Udvardi, M.K., and Scheible, W.R. (2005). Plant science. GRAS genes and the symbiotic green revolution. Science 17 1749–1750. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Journet, E.P., and Barker, D.G. (1999). MtENOD20, a Nod factor-inducible molecular marker for root cortical cell activation. Mol. Plant Microbe Interact. 12 604–614. [Google Scholar]
- Vijn, I., Christiansen, H., Lauridsen, P., Kardailsky, I., Quandt, H.J., Broer, I., Drenth, J., Ostergaard Jensen, E., van Kammen, A., and Bisseling, T. (1995). A 200 bp region of the pea ENOD12 promoter is sufficient for nodule-specific and Nod factor induced expression. Plant Mol. Biol. 28 1103–1110. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Wessler, S.R. (2005). Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 10 54–56. [DOI] [PubMed] [Google Scholar]
- Wu, C.Y., Suzuki, A., Washida, H., and Takaiwa, F. (1998). The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J. 14 673–683. [DOI] [PubMed] [Google Scholar]
- Xue, G.P., and Loveridge, C.W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37 326–339. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M.P., Onodera, Y., Touno, S.M., and Takaiwa, F. (2006). Synergism between RPBF DOF and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 141 1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa, S., and Schmidt, R.J. (1999). Diversity and similarity among recognition sequences of DOF transcription factors. Plant J. 17 209–214. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Tian, L., Latoszek-Green, M., Brown, D., and Wu, K. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58 585–596. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.