Abstract
The selective replacement of photodamaged D1 protein within the multisubunit photosystem II (PSII) complex is an important photoprotective mechanism in chloroplasts and cyanobacteria. FtsH proteases are involved at an early stage of D1 degradation, but it remains unclear how the damaged D1 subunit is recognized, degraded, and replaced. To test the role of the N-terminal region of D1 in PSII biogenesis and repair, we have constructed mutants of the cyanobacterium Synechocystis sp PCC 6803 that are truncated at the exposed N terminus. Removal of 5 or 10 residues blocked D1 synthesis, as assessed in radiolabeling experiments, whereas removal of 20 residues restored the ability to assemble oxygen-evolving dimeric PSII complexes but inhibited PSII repair at the level of D1 degradation. Overall, our results identify an important physiological role for the exposed N-terminal tail of D1 at an early step in selective D1 degradation. This finding has important implications for the recognition of damaged D1 and its synchronized replacement by a newly synthesized subunit.
INTRODUCTION
Visible light–induced damage to the photosynthetic apparatus in the thylakoid membrane is an important factor in limiting biomass production in both terrestrial and aquatic oxygenic photosynthesis (Long and Humphries, 1994). A major target for irreversible photodamage is photosystem II (PSII), which is the pigment protein reaction center complex of the thylakoid membrane involved in catalyzing the light-induced splitting of water to molecular oxygen and the injection of electrons into the photosynthetic electron transport chain (Prášil et al., 1992; Adir et al., 2003). An inevitable consequence of PSII activity and its associated electron transfer reactions is the generation of highly oxidizing species such as Tyr radicals, chlorophyll cations, and singlet oxygen, which can all cause irreversible oxidative damage to the PSII enzyme, leading to loss of activity (reviewed in Barber and Andersson, 1992).
Of the >20 subunits found within PSII, the D1 reaction center subunit is the most prone to damage and is selectively replaced following partial disassembly of the complex in a process called the PSII repair cycle (for recent reviews, see Nishiyama et al., 2006; Yokthongwattana and Melis, 2006), with the remaining undamaged subunits being recycled (Komenda et al., 2004). PSII repair occurs at all light intensities (Jansen et al., 1999), but only when repair is unable to match the rate of damage to PSII is overall photosynthetic activity reduced (Prášil et al., 1992).
Despite its physiological importance, the molecular details of PSII repair and especially D1 degradation are still poorly understood. Homologs of the bacterial ATP-dependent FtsH protease, a member of the large AAA+ (for ATPases associated with diverse cellular activities) protein family, were recently shown to participate at an early stage in the repair-related degradation of D1 in both chloroplasts (Bailey et al., 2002) and cyanobacteria (Silva et al., 2003; Komenda et al., 2006). However, it remains unclear how FtsH might recognize damaged D1 and where in D1 degradation might be initiated.
A variety of early studies have suggested that damaged D1 is cleaved first within the stroma-exposed D-E loop connecting the fourth and fifth transmembrane helices of the protein in both plants (Greenberg et al., 1987; Haussühl et al., 2001) and cyanobacteria (Kanervo et al., 2003), and indeed, Escherichia coli FtsH can act as an endoproteinase (Okuno et al., 2006). However, E. coli FtsH is also able to degrade membrane proteins processively from either the N- or C-terminal end of target molecules (Chiba et al., 2002). For N-terminal proteolysis, there is a structural requirement that the tail be longer than 20 amino acid residues (Chiba et al., 2000). In the case of D1, the N-terminal (but not C-terminal) tail is on the same side of the membrane as the protease domain of FtsH (Lindahl et al., 1996); significantly, it protrudes from the cyanobacterial PSII complex in recent crystal structures (Figure 1A) (Ferreira et al., 2004; Loll et al., 2005) and is of sufficient length in both cyanobacteria and higher plants to engage with FtsH (Figure 1B). Consequently, the processive degradation of D1 by FtsH starting from the N terminus (Nixon et al., 2005) seems to be a plausible alternative to earlier models that emphasized cleavage in the D-E loop (Greenberg et al., 1987; Spetea et al., 1999; Lindahl et al., 2000; Haussühl et al., 2001; Huesgen et al., 2005). In the case of chloroplasts, it has also been suggested that D1 degradation by FtsH might be facilitated by cleavage of D1 by Deg proteases on the opposite lumenal side of the membrane (Kapri-Pardes et al., 2007; Sun et al., 2007).
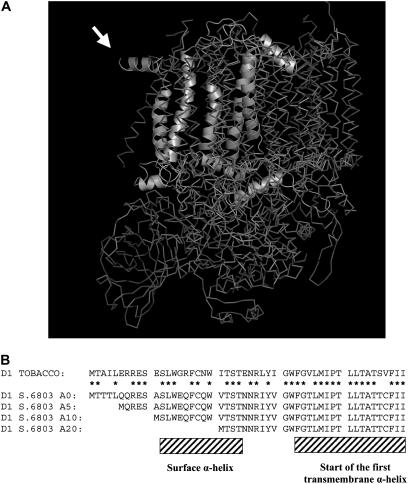
Figure 1.
Cyanobacterial PSII Structure and the N Terminus of D1 in the Constructed N-Terminal Truncation Mutants of Synechocystis sp PCC 6803.
(A) Crystal structure of PSII from Thermosynechococcus elongatus showing the N-terminal region of the D1 subunit exposed at the periphery of the complex. The D1 polypeptide chain is shown only in one of the PSII monomers in the dimer. This image is modeled from the coordinates determined by Ferreira et al. (2004). Note that the first nine residues of D1 cannot be resolved in the structure. Amino acid residues 11 to 20, forming the parallel helix protruding from the structure, are indicated by the arrow.
(B) Comparison of the decoded N-terminal sequences (residues 1 to 50) of D1 from Synechocystis sp PCC 6803 and tobacco (Nicotiana tabacum). Shown are the positions of the surface α-helix and the beginning of the first transmembrane α-helix plus the amino acid residues deleted in the A5, A10, and A20 mutants. Stars indicate residues conserved in chloroplast and cyanobacterial sequences.
Here, we have used mutagenesis in combination with functional assays to test the involvement of the exposed N-terminal tail of D1 in PSII function and, in particular, the selective degradation of damaged D1 during PSII repair in the cyanobacterium Synechocystis sp PCC 6803. Our results, together with the recent analysis of cyanobacterial and Arabidopsis thaliana FtsH mutants, provide clear support for the N-terminal FtsH-mediated degradation of damaged D1. Our work has a number of important implications for the mechanism and regulation of PSII repair.
RESULTS
Synechocystis Mutants Lacking 5 or 10 Amino Acid Residues in the Exposed N-Terminal Region of the D1 Protein Are Blocked in the Synthesis of D1 Protein
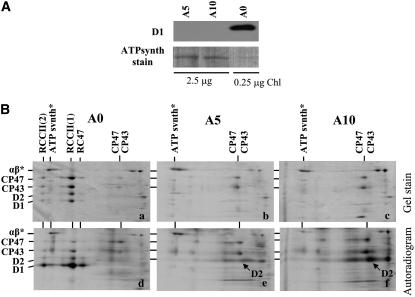
To test the role of the N-terminal region of D1 in the assembly and repair of PSII, we constructed a series of Synechocystis sp PCC 6803 mutants containing a D1 protein with the N-terminal tail shortened by 5 to 20 amino acid residues. Removal of 5 and 10 residues in mutants A5 (ΔT2-Q6) and A10 (ΔT2-A11) (Figure 1B), respectively, generated nonphotoautotrophic strains unable to accumulate D1 in the membrane (Figure 2A). Two-dimensional analysis of radioactively labeled thylakoid proteins indicated the lack of D1 synthesis in both strains (Figure 2B). As observed previously for a psbA triple deletion mutant unable to synthesize D1, the other major PSII subunits, D2, CP43, and CP47, were still synthesized in A5 and A10 but could not assemble into larger PSII complexes (Komenda et al., 2004, 2006). As expected, the control strain A0 was able to assemble monomeric [RCCII(1)] and dimeric [RCCII(2)] PSII core complexes and smaller amounts of a non-oxygen-evolving PSII subcomplex lacking CP43 (termed RC47) (Figure 2B, panels a and d) (Komenda et al., 2004, 2006). Using quantitative RT-PCR, the level of the psbA2 transcript in the A0, A5, and A10 strains was determined to be 47, 50, and 57% of that found in the wild type, respectively. Thus, the lack of D1 accumulation in the A5 and A10 mutants appears to be due to effects at the posttranscriptional level.
Figure 2.
Immunodetection of the D1 Protein and Two-Dimensional Analysis of Thylakoid Protein Complexes in the Control Strain of Synechocystis sp PCC 6803 A0 and the D1-Truncated Nonautotrophic Mutants A5 and A10.
(A) Thylakoid proteins from cells of the strains A0, A5, and A10 were separated by denaturing SDS-PAGE on a 12 to 20% polyacrylamide gel containing 7 M urea, electroblotted onto a polyvinylidene difluoride (PVDF) membrane, and immunodecorated using an antibody raised against amino acid residues 59 to 76 of the D1 protein from Synechocystis. A total of 0.25 μg of chlorophyll for the A5 and A10 strains and 2.5 μg of chlorophyll for the A0 strain were loaded onto the gel. Loading is documented by Ponceau staining of the α and β subunits of ATP synthase (ATPsynth stain).
(B) Thylakoid protein complexes from cells of the Synechocystis strains A0 (a and d), A5 (b and e), and A10 (c and f) were analyzed by two-dimensional blue-native SDS-PAGE. The gel was stained by Coomassie blue (a to c; gel stain), dried, and exposed to a phosphorimager plate to detect the radioactive labeling of the proteins (d to f; autoradiogram). A total of 6 μg of chlorophyll per sample was loaded onto the gel, and α and β subunits of ATP synthase (αβ*) were used as internal standards to check correct loading of the gels.
A Mutant Lacking 20 Amino Acid Residues at the N Terminus of the D1 Protein Grows Photoautotrophically and Contains Functional PSII Complexes
Somewhat surprisingly, removal of 20 residues in mutant A20 (ΔT2-V21) (Figure 1B) restored photoautotrophic growth in assays conducted at a cultivation irradiance of 25 μmol·m−2·s−1, although at a slower rate than the control strain, A0 (Table 1). However, when the strain was cultivated at a five times higher irradiance, its growth was abolished, while the control strain A0 was still able to grow (Table 1).
Table 1.
Characteristics of the Synechocystis sp PCC 6803 Control Strain A0 and N-Terminal Truncation Mutant A20 Grown at Irradiance of 25 (LL) or 125 (ML) μmol·m−2·s−1
| Strain | Autotrophic Doubling Time (h)a | Oxygen Evolution (μmol O2·mg−1 chlorophyll·h−1)b | TL B Band (°C)c | TL Q Band (°C)d | RCCII(2) (%)e | RCCII(1) (%)e | RC47 (%)e |
|---|---|---|---|---|---|---|---|
| A0 LL | 9.8 ± 0.1 | 640 ± 20 | 28 ± 1 | 10 ± 2 | 48 | 48 | 4 |
| A0 ML | 10.4 ± 0.2 | 820 ± 20 | 28 ± 1 | 9 ± 2 | 48 | 45 | 7 |
| A20 LL | 13.9 ± 0.2 | 560 ± 30 | 28 ± 2 | 9 ± 1 | 36 | 36 | 28 |
| A20 ML | No growth | 430 ± 40 | 28 ± 1 | 10 ± 2 | 18 | 46 | 36 |
Measured on microtitration plates. Values shown are means of 11 measurements ± sd. Initial OD750 nm of the cultures was 0.005.
Light-saturated rate of oxygen evolution measured in the presence of 1 mM p-benzoquinone and 5 mM potassium ferricyanide. Values shown are means of three measurements ± sd. The A20 ML culture for measurement was obtained as described in Methods.
Temperature corresponding to the peak of the thermoluminescence glow curve measured in the range −10 to 70°C after a single-turnover saturating flash given at 5°C in the absence of diuron. Values shown are means of three measurements ± sd.
Temperature corresponding to the peak of the thermoluminescence glow curve measured in the range −10 to 60°C after a single-turnover saturating flash given at −10°C in the presence of 10−5 M diuron. Values shown are means of three measurements ± sd.
Amount obtained by gel densitometry of the CP47 bands from dimeric [RCCII(2)] and monomeric [RCCII(1)] PSII core complexes and core lacking CP43 (RC47) separated by two-dimensional blue-native SDS-PAGE in Figure 5, as quantified by ImageQuant software. Values shown are means of three measurements; sd did not exceed 8%.
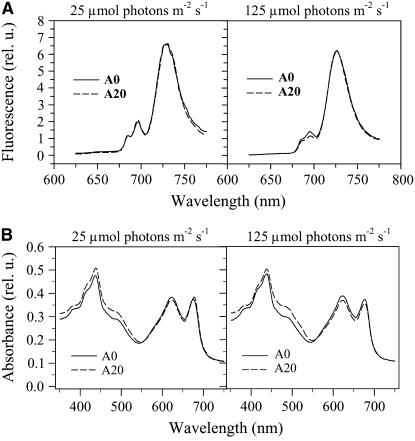
The low-temperature fluorescence emission spectra following pigment excitation at 435 nm showed the same ratio of PSII to PSI in cells of A0 and A20, as judged by the ratio of fluorescence intensities at 695 nm (from PSII) and 720 nm (from PSI) (Mann et al., 2000), when cultivated under low-light conditions (25 μmol·m−2·s−1) (Figure 3A). The cellular content of phycobilisomes and chlorophyll was also similar in both strains, although the A20 mutant contained a slightly higher amount of carotenoids, as deduced from the increased absorption in the 450- to 490-nm region of the room temperature visible absorption spectrum (Figure 3B).
Figure 3.
77K Fluorescence Emission Spectra and Room Temperature Absorption Spectra of Cells of the Control Strain A0 and the Truncation Mutant A20 Cultivated at 25 and 125 μmol·m−2·s−1 White Light.
(A) 77K fluorescence spectra of A0 (solid lines) and A20 (dotted lines) cells. The spectra were obtained using an Aminco Bowman series 2 luminescence spectrometer (Spectronic Unicam). The cells were excited at 435 nm, and the spectra were corrected for the sensitivity of the photomultiplier and normalized to the maximum PSI emission at 725 nm.
(B) Absorption spectra of A0 (solid lines) and A20 (dotted lines) cells. The spectra were obtained using the UV-3000 spectrophotometer (Shimadzu) with cell suspensions of equal OD753nm.
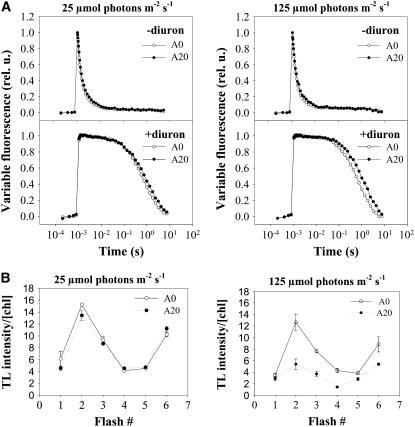
To test for potential effects of the N-terminal deletion on PSII function in A20, we conducted a number of measurements of PSII activity in vivo. These included the following: (1) the rate and extent of reoxidation of the reduced primary quinone electron acceptor (QA−) by forward electron transfer to the secondary quinone electron acceptor (QB) in single-turnover experiments (Figure 4A); (2) the rate of charge recombination between QA− and the S2 state of the water-oxidizing complex when QB reduction was blocked by the addition of the herbicide diuron (Figure 4A); (3) the peak positions of the thermoluminescence (TL) B and Q bands, which give information on the energy stored in the S2QB− and S2QA− redox states, respectively (Table 1); and (4) the TL signal intensity oscillations of the B band, which give information on S state cycling of the water-oxidizing complex (Vass and Govindjee, 1996) (Figure 4B). In all cases, there were no significant differences between A0 and A20. Therefore, these data argue against perturbations in A20 to the structure of the D-E loop of D1, which is involved in binding QB and regulating its redox properties (Ohad and Hirschberg, 1992). However, the lower rate of PSII oxygen evolution (Table 1) and the lower amplitude of TL B band oscillations on a chlorophyll basis (Figure 4B) indicate fewer photochemically active PSII complexes in the A20 mutant, despite similar levels of immunodetectable D1 subunit (Figure 6A).
Figure 4.
Relaxation of the Variable Fluorescence and Oscillations of the Flash-Induced Thermoluminescence B Band in Cells of Strains A0 and A20 Cultivated at 25 and 125 μmol·m−2·s−1 White Light.
(A) Cells of A0 (open circles) and A20 (closed circles) at 2.5 μg chlorophyll/mL were dark-adapted for 5 min, and the decay in variable fluorescence was followed before and after a saturating flash using the PSI double modulated fluorimeter in the absence (left panel) or presence (right panel) of the PSII inhibitor diuron (10 μM final concentration).
(B) Cells of A0 (open circles) and A20 (closed circles) at 25 μg chlorophyll/mL were filtered onto the nitrocellulose membrane and dark-adapted for 5 min, one to six flashes were applied at 3°C, and thermoluminescence in the range 3 to 70°C was measured using the PSI thermoluminometer. The area below the TL glow curve was plotted against the number of flashes. Values are means of three measurements ± sd.
Figure 6.
Detection of PSII and Other Thylakoid Proteins in Strains A0 and A20 Cultivated at 25 μmol·m−2·s−1 White Light.
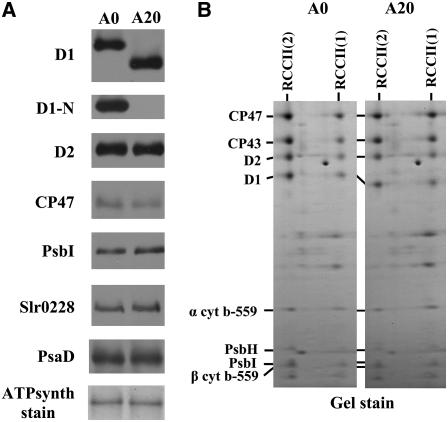
(A) Thylakoid proteins from cells of the A0 and A20 strains were separated by denaturing SDS-PAGE, electroblotted onto a PVDF membrane, and immunodecorated using antibodies raised against the following: amino acid residues 59 to 76 (D1); amino acid residues 2 to 15 (D1-N) of the D1 protein from Synechocystis; the last 12 amino acid residues of D2 from Synechocystis; amino acid residues 380 to 394 of CP47 protein from barley; the last 14 amino acid residues of PsbI from Synechocystis; the whole protein PsaD from Synechocystis; and amino acid residues 98 to 115 of FtsH (slr0228) from Synechocystis. Equal protein loading was checked by Ponceau staining and documented by the staining intensity of the α and β subunits of ATP synthase (ATPsynth stain). A total of 0.5 μg of chlorophyll was loaded onto the gel.
(B) Protein composition of dimeric and monomeric PSII core complexes from thylakoids of A0 and A20 strains resolved by two-dimensional blue-native SDS-PAGE. The designated proteins were identified by immunodetection and mass spectrometry.
Mutant A20 Is Able to Assemble Monomeric and Dimeric PSII Complexes but Has Increased Levels of the RC47 Complex
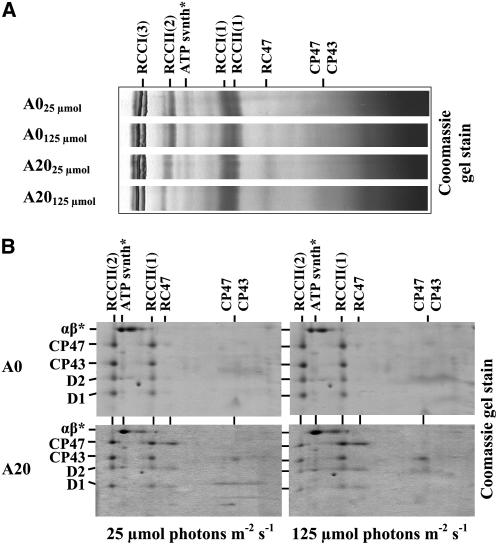
To test whether the assembly status of PSII was perturbed in A20, thylakoid complexes were analyzed by two-dimensional gel electrophoresis (Figure 5). A20 showed the presence of the same protein complexes as A0, including RCCII(2) and RCCII(1) (Figure 5B). A striking difference between A0 and A20 was an increase in the amount of RC47 (Figure 5B), which is an intermediate in both the assembly and repair of PSII (Table 1). This increased level of RC47 in A20 is in agreement with the observed reduction in PSII activity in the A20 mutant. Comparison of the levels of several PSII subunits, the PSI subunit PsaD, and the FtsH homolog slr0228 (sometimes called FtsH2) showed no difference between A0 and A20 cells when cultivated under low light conditions (25 μmol·m−2·s−1) (Figure 6A). Immunoblots using D1-specific antibodies confirmed that D1 had a faster electrophoretic mobility in A20 than in A0 and was truncated at the N terminus (Figure 6A). No differences between the strains were found in the content of small subunits PsbE, PsbF, PsbH, and PsbI in RCCII(2) and RCCII(1) separated by blue-native electrophoresis (Figure 6B), indicating that the N-terminal truncation had not prevented the binding of these low-molecular-mass subunits to PSII. This is particularly important for PsbI, which is the closest subunit to the N-terminal region of D1 (Ferreira et al., 2004; Loll et al., 2005).
Figure 5.
Protein Complexes Resolved by Blue-Native PAGE and Separation of Large PSII Proteins of PSII Complexes Revealed by Two-Dimensional Electrophoresis from Thylakoids of Strains A0 and A20 Cultivated at 25 and 125 μmol·m−2·s−1 White Light.
(A) Thylakoid protein complexes from cells of A0 and A20 were analyzed by blue-native PAGE. Designation of complexes is as follows: RCCI(3) and RCCI(1), trimeric and monomeric PSI complexes, respectively; RCCII(2) and RCCII(1), dimeric and monomeric PSII core complexes, respectively; RC47, the monomeric PSII core complex lacking CP43; CP47 and CP43, unassembled PSII antennae; ATPsynth, ATP synthase. A total of 6 μg of chlorophyll per sample was loaded onto the gel.
(B) SDS-PAGE analysis of large subunits D1, D2, CP47, and CP43 in the PSII complexes resolved by blue-native PAGE in (A). The two-dimensional gel was stained with Coomassie blue. A total of 6 μg of chlorophyll per sample was loaded onto the gel, and α and β subunits of ATP synthase (αβ*) were used as internal standards to check correct loading of the gels. The identity of proteins was verified by immunodetection.
PSII Activity in A20 Is Irradiance Dependent
The growth data in Table 1 raised the possibility that the level of active PSII complexes in A20 might be light dependent. To obtain support for this hypothesis, we compared PSII activity in low-light-grown cells of A0 and A20 with that in cells that were transferred to a five times greater irradiance (125 μmol·m−2·s−1). It should be noted that while the A20 strain was unable to grow at this irradiance in growth assays starting with a low initial inoculum of cells (OD750nm ∼ 0.005) (Table 1), it could be grown when inoculated at a higher cell density (OD750nm ∼ 0.75) (see Supplemental Figure 1 online). The higher growth irradiance did not affect PSII activity in the A0 control strain, as reflected by the rate of oxygen evolution and by the relative contents of RCCII(2), RCCII(1), and RC47 (Table 1, Figures 3 to 5). By contrast, the A20 cells grown at 125 μmol·m−2·s−1 showed a marked decrease in PSII activities, as demonstrated by a lower rate of oxygen evolution (Table 1), a lower 77K fluorescence peak at 695 nm (Figure 3A), and reduced TL intensities (Figure 4B). Importantly, A20 displayed a redistribution in the levels of the different types of PSII complex at the higher irradiance: there was a clear increase in the amounts of the RCCII(1) complex and the RC47 complex at the expense of RCCII(2) (Figure 5, Table 1). The A20 cells also contained increased levels of carotenoid at the higher irradiance, as judged by the higher absorption in the 450- to 490-nm region (Figure 3B), possibly as a consequence of increased light stress.
Growth of cells at the increased irradiance caused only a minor slowing in the kinetics of electron transfer in the total population of active PSII complexes either in the absence (Figure 4A, −diuron) or the presence (Figure 4A, +diuron) of diuron. In addition, the positions of the TL maxima were not changed by exposure to higher irradiance (Table 1). Although the amplitude of the TL B band oscillations decreased at the higher irradiance (Figure 4B), the oscillations maintained the typical wild-type pattern, with maxima after the second and sixth flashes. Overall, these data indicated a further reduction in PSII activity in the A20 mutant at the higher irradiance but excluded the formation of a significant population of partially damaged oxygen-evolving complexes with drastically modified acceptor and donor side properties.
PSII Repair and Selective D1 Degradation Are Inhibited in the A20 Truncation Mutant
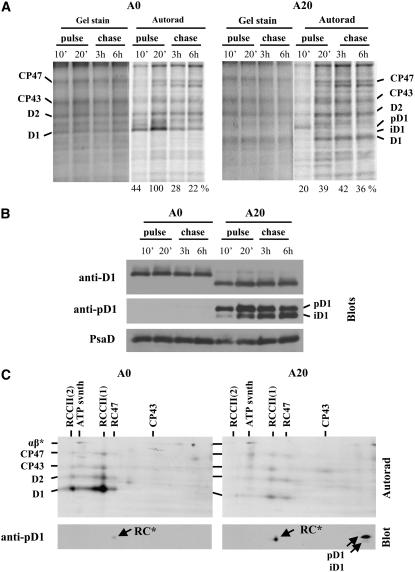
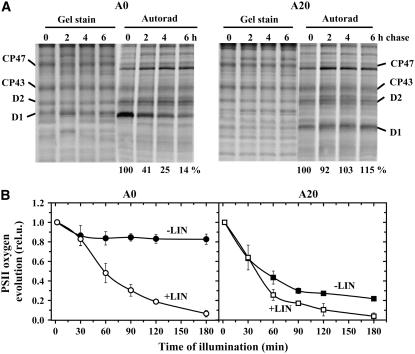
To investigate the dynamics of PSII assembly and disassembly in the A20 mutant, we assessed the synthesis and degradation of PSII proteins in a radioactive pulse-chase experiment. At an irradiance of 125 μmol·m−2·s−1, the D1 protein in A0 was preferentially synthesized and degraded in response to light damage, as demonstrated in Figure 7A. The rate of D1 synthesis in A20 was about half of that in A0, and after an initial 10 min of pulse, the preferentially labeled protein was mainly in the form of the precursor form of D1, pD1. An immunoblot using an antibody raised against the cleavable C-terminal extension of the D1 precursor revealed the presence of unprocessed and partially processed forms of D1, pD1 and iD1, in thylakoids of A20 but not A0 (Figure 7B). Interestingly, a two-dimensional gel analysis showed that pD1 in A20 was not incorporated into any PSII complex but was detected in the low-molecular-weight region of the gel, consistent with it being unassembled (Figure 7C). After 20 min of labeling, the labeled D1 in A20 was found mostly in its fully mature form in PSII complexes and was now labeled to a similar extent as other large PSII proteins, D2, CP47, and CP43 (Figure 7C). During the chase, also conducted at 125 μmol·m−2·s−1, all of the radiolabeled PSII proteins, including D1, were stable in A20, while the labeling of D1 in A0 was selectively decreased after 3 h of the chase (Figure 7A). A similar stability of labeled D1 was also observed in A20 at a higher irradiance of 500 μmol·m−2·s−1 (Figure 8A), while the radiolabeled D1 in A0 was degraded with a half-life of ∼90 min. Overall, these results indicated that the N terminus of D1 played a key role in the removal of damaged D1 during the PSII repair cycle and that replacement of D1 was not limited by the lack of D1 synthesis.
Figure 7.
Synthesis and Degradation of PSII Proteins in A0 and A20 at 125 μmol·m−2·s−1 White Light, Corresponding Accumulation of D1 (iD1 and pD1) Forms, and Their Distribution in Various PSII Complexes.
(A) Cells of the A0 and A20 strains (chlorophyll concentration, 75 μg/mL) were pulse-labeled at 125 μmol·m−2·s−1 white light in the presence of radioactive Met/Cys for 10 and 20 min (pulse, 10' and 20'), then the cells were washed and incubated at the same irradiance for 3 and 6 h (chase, 3 h and 6 h) in the presence of unlabeled Met/Cys. Thylakoid proteins from labeled cells were separated by denaturing SDS-PAGE and stained with Coomassie blue (Gel stain), and radioactive labeling of the proteins was visualized using a phosphorimager (Autorad). Quantification of radioactivity was performed by ImageQuant software with all samples equally loaded (see Gel stain) on a single gel. The radioactivity incorporated into D1 of A0 after the 20 min pulse was taken as 100%. Values at bottom show means of three measurements; sd did not exceed 7%.
(B) Thylakoid proteins from cells labeled as in (A) were separated by denaturing SDS-PAGE, electroblotted onto a PVDF membrane, and immunodecorated using antibodies raised against the following: amino acid residues 59 to 76 (anti-D1); the last 16 amino acid residues of the D1 precursor from Synechocystis (anti-pD1); and the whole protein PsaD from Synechocystis, which was also used as a loading control. A total of 0.5 μg of chlorophyll was loaded onto the gel.
(C) Thylakoid proteins from cells pulse-labeled for 20 min (A) were separated by two-dimensional blue-native SDS-PAGE and electroblotted onto a PVDF membrane. The membrane was exposed to a phosphorimager plate (Autorad) or immunodecorated using antibodies raised against the last 16 amino acid residues of the D1 precursor from Synechocystis (Blot, anti-pD1). RC indicates the complex containing iD1 together with D2 and cytochrome b559 but lacking CP47 and CP43 (Komenda et al., 2004).
Figure 8.
Effect of High Irradiance on the Turnover of PSII Proteins in Cells of A0 and A20, and PSII Activity in Cells of A0 and A20 Untreated or Treated with Lincomycin.
(A) Pulse-chase radioactive labeling of A0 and A20 cells. Cells (chlorophyll concentration, 5 μg/mL) were subjected to 250 μmol·m−2·s−1 white light for 20 min in the presence of a radioactively labeled mixture of Met and Cys (0 h). The cells were washed, unlabeled Met and Cys were added, and the cells were subjected to 500 μmol·m−2·s−1 white light for 2, 4, and 6 h. Thylakoids were isolated, their proteins were separated by SDS-PAGE, gels were stained with Coomassie blue (Gel stain), and the radioactive labeling of the proteins was visualized using a phosphorimager (Autorad). Quantification of radioactivity was performed as described in the legend to Fig. 7 and normalized to the amount of signal measured after the pulse period (0 h), which was taken as 100%. Values at bottom show means of three measurements; sd did not exceed 7%.
(B) Time course of PSII photoinactivation in cells of A0 and A20 subjected to high irradiance. Cells of A0 (left panel, circles) and A20 (right panel, squares) (chlorophyll concentration, 75 μg/mL) were illuminated with 500 μmol·m−2·s−1 white light for 180 min either in the absence (closed symbols) or presence (open symbols) of 100 μg/mL lincomycin (LIN), and PSII oxygen-evolving activity was assayed in whole cells. Values in the plot represent means of four measurements ± sd. Initial values were in the range shown in Table 1.
To further demonstrate the inhibitory effect of truncating D1 on PSII repair, PSII-catalyzed oxygen evolution was assessed in A0 and A20 cells subjected to a high irradiance of white light (500 μmol·m−2·s−1) either in the presence or the absence of the protein synthesis inhibitor lincomycin (Figure 8B). Under these conditions, both A0 and A20 cultures (OD750nm ∼ 1.2) were still capable of photoautotrophic growth in the absence of lincomycin (see Supplemental Figure 1 online). The measurements showed that the truncation mutant was indeed more sensitive to photoinhibition than the control strain containing full-length D1. In A0, the irradiance caused a small decrease in the activity not exceeding 20%, while in A20 cells, this decrease reached almost 80% (Figure 8B, −LIN). The rate of PSII photodamage, estimated from the high light–induced decrease in PSII activity in the presence of lincomycin, which blocks PSII repair, was consistently slightly slower in A0 due to the presence of a previously observed initial slow phase or lag, which might reflect a larger pool of assembled PSII in A0 awaiting photoactivation (Kuviková et al., 2005; Komenda et al., 2007). More importantly, the small difference between the rate of photoinactivation in the presence and absence of lincomycin in the A20 strain indicated that PSII repair was strongly inhibited at the increased irradiance. The inhibition of repair was further confirmed when the cells of both A0 and A20 were subjected to 2000 μmol·m−2·s−1 for 30 min to inhibit PSII activity and then transferred to low light (50 μmol·m−2·s−1). The recovery of PSII activity in A0 was again much faster than in A20 (see Supplemental Figure 2 online).
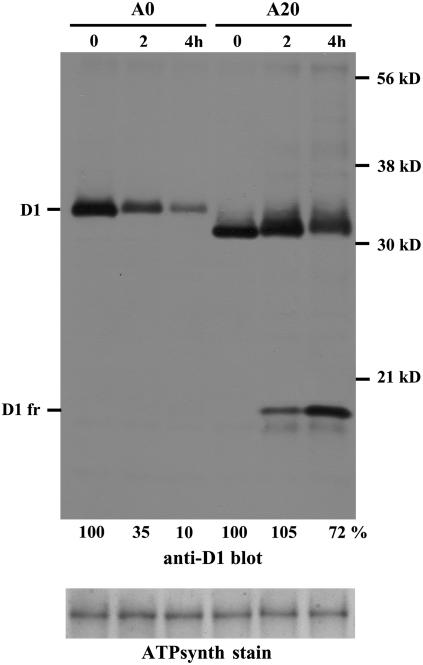
The inhibitory effect of the truncation on the rate of D1 degradation could also be demonstrated in cells of A20 treated with lincomycin and exposed to 500 μmol·m−2·s−1 (Figure 9). Immunoblotting using the anti-D1 antibody showed a much lower rate of D1 degradation in A20 compared with cells of A0 treated under identical conditions. Moreover, in A20, the D1 band became more diffuse during the light treatment and exhibited a marked reduction in its mobility, indicative of oxidative modifications to the protein (Komenda et al., 2002). Interestingly, we detected the formation of an N-terminal 17-kD D1 fragment in light-treated cells of A20 but not in A0 (Figure 9, D1 fr). This finding suggested that other enzymic or nonenzymic degradation pathways might come into action when the N terminus is too short and when protein synthesis is blocked.
Figure 9.
Degradation of the D1 Protein in Cells of A0 and A20 during High-Light Treatment in the Presence of Lincomycin.
Thylakoid proteins from cells of A0 and A20 exposed to 500 μmol·m−2·s−1 white light for 4 h in the presence of the protein synthesis inhibitor lincomycin were separated by SDS-PAGE, electroblotted onto a PVDF membrane, and immunodecorated using antibodies raised against amino acid residues 59 to 76 (anti-D1) of the D1 protein from Synechocystis. D1 fr designates the D1 fragment induced by light in cells of A20. Equal protein loading was checked by Ponceau staining and documented by the staining intensity of the α and β subunits of ATP synthase (ATPsynth stain). A total of 0.5 μg of chlorophyll was loaded onto the gel. Quantification of the D1 bands was performed by ImageQuant software, and signal at time 0 of A0 and A20 was taken as 100%. Values at bottom show means of three measurements; sd did not exceed 5%.
Degradation of the D1 Protein in the Truncation Mutant A20 Is Blocked after Formation of the Repair Cycle Intermediate RC47
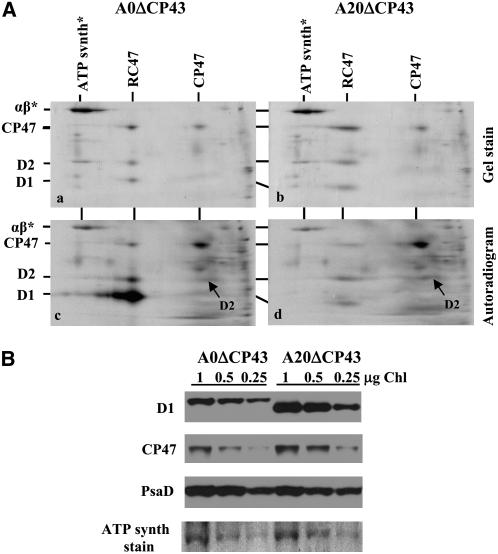
Two-dimensional gel electrophoresis showed a marked increase in the level of the RC47 complex relative to the amount of RCCII(1) and RCCII(2) in A20 cultivated at 125 μmol·m−2·s−1 (Figure 5). RC47 is both a PSII assembly intermediate and the likely substrate for the selective replacement of D1 during repair (Komenda et al., 2004). To substantiate that removal of D1 from RC47 was impaired in A20, D1 turnover was examined in mutants of A0 and A20 in which the gene encoding CP43 was additionally inactivated, so that PSII assembly was unable to progress beyond the formation of RC47. In the case of A0ΔCP43, autoradiograms revealed that the D1 subunit was turned over in RC47 much more rapidly than D2 and CP47 and hence was preferentially radiolabeled in a pulse-labeling experiment, as shown previously (Figure 10A, panel c) (Komenda et al., 2004, 2006). By contrast, in the A20ΔCP43 strain, selective turnover of D1 in the RC47 complex was inhibited, and instead there was a low and equal incorporation of [35S]Met/Cys into D1, D2, and CP47 (Figure 10A, panel d). Impaired degradation of D1 in the A20ΔCP43 mutant was also reflected by a twofold to threefold increase in the accumulation of RC47 on two-dimensional gels (Figure 10A, panel b) and by increased levels of both D1 and CP47 detected by immunoblotting (Figure 10B).
Figure 10.
Turnover of PSII Proteins in the RC47 Complex and Their Accumulation in A0 and Truncation Mutant A20, Both Lacking the psbC Gene Encoding the Inner PSII Antenna CP43.
(A) Thylakoid proteins and their radioactive labeling in cells of the CP43-less strains A0ΔCP43 (a and c) and A20ΔCP43 (b and d). Proteins were separated by two-dimensional blue-native SDS-PAGE and stained with Coomassie blue (a and b), and radioactive proteins were visualized by autoradiography (c and d). A total of 6 μg of chlorophyll per sample was loaded onto the gel, and α and β subunits of ATP synthase (αβ*) were used as internal standards to check correct loading of the gels.
(B) Quantification of the D1 protein in cells of A0ΔCP43 and A20ΔCP43. Thylakoid proteins were separated by denaturing SDS-PAGE on a 12 to 20% polyacrylamide gel containing 7 M urea, electroblotted onto a PVDF membrane, and immunodecorated using the antibodies raised against amino acid residues 59 to 76 of the D1 protein from Synechocystis, CP47, and PsaD. Correct protein loading was checked by Ponceau staining and documented by the staining intensity of the α and β subunits of ATP synthase (ATPsynth stain). A total of 1, 0.5, or 0.25 μg of chlorophyll was loaded onto the gel.
DISCUSSION
A long-standing question in plant and cyanobacterial biology has been the mechanism of rapid D1 turnover in vivo (Adir et al., 2003). Here, we provide experimental evidence to support the hypothesis that the N-terminal region of D1 is needed for selective degradation of damaged D1. The N-terminal tail of D1 is located at the periphery of PSII and includes a flexible stretch of 9 N-terminal amino acid residues, which was unable to be resolved in the recent crystal structures of cyanobacterial PSII, followed by a region of ∼10 amino acids, which forms a parallel helix to the membrane (Figure 1A) (Ferreira et al., 2004; Loll et al., 2005). Both of these regions are missing in mutant A20 (Figure 1B). Interestingly, the initial 10 amino acids seem to be also important for the synthesis of D1, as removal of these residues results in a phenotype similar to that of psbA deletion mutants (Komenda et al., 2004). The surprising restoration of D1 synthesis in A20 might be related to the removal of the parallel helix, resulting in the reappearance of a sufficiently long flexible stretch of amino acids at the N terminus that might be crucial for the elongation of D1 during synthesis, or to effects on the secondary structure of the psbA mRNA to allow efficient translation.
A20 Assembles Functional PSII Complexes
To investigate whether shortening of the N-terminal tail in A20 had a major impact on the structure of the PSII complex, we used a variety of noninvasive biophysical techniques to assess PSII function in both A0 and A20. These included measurement of the rate of electron transfer between the bound plastoquinones, QA and QB, on the acceptor side of the complex and the rate of charge recombination between the reduced acceptor side and oxidized water-oxidizing complex in chlorophyll flash fluorescence assays (Figure 4A); measurement of the thermoluminescence Q and B bands (Table 1); and cycling of the S states in the water-oxidizing complex using TL measurements (Figure 4B). These assays are very sensitive to subtle changes in the redox properties of the electron-transfer components on both the donor and acceptor sides of PSII (Allahverdiyeva et al., 2004). In all cases, the properties of the photochemically active PSII complexes found in A20 were very similar to those in A0. Therefore, we conclude that the N-terminal tail of D1 missing in A20 does not play an important role in modulating electron transfer in PSII, which is entirely consistent with its location on the periphery of PSII.
Our data also confirmed that A20 was able to assemble and accumulate dimeric PSII complexes (Figures 5 and 6), which again is consistent with the recent crystal structures that place the N-terminal tail outside the contact region between the two monomers (Ferreira et al., 2004; Loll et al., 2005). Importantly, we were able to confirm the presence of a number of low-molecular-mass PSII subunits in dimeric PSII complexes in A20 by mass spectrometry and immunoblotting, including PsbI (Figure 6B). This subunit interacts with the second transmembrane helix of D1 and participates in the ligation of an accessory chlorophyll (ChlZD1) in PSII and is the closest interacting subunit to the N-terminal region of D1 (Ferreira et al., 2004; Loll et al., 2005). The presence of PsbI in A20 PSII argues strongly against a major disruption to this region of D1.
Degradation of D1 Is Impaired in A20
The major effect observed in the A20 mutant was a reduced ability to repair photodamaged PSII complexes in vivo (Figure 8). In principle, impaired PSII repair might be due to effects on one or more of the following steps: the partial disassembly of the damaged PSII complex to form the RC47 complex, the removal of the damaged D1 subunit from RC47, and the synthesis and integration of the newly synthesized D1 subunit into the complex. Assessing which of these steps is affected is also complicated by the possibility that the synthesis and degradation of D1 are coordinated so that impaired synthesis might impair degradation and vice versa (Komenda et al., 2000). However, assays involving the use of lincomycin enable degradation to be studied independently of protein synthesis.
Although pulse-labeling experiments indicated that there was some effect on D1 synthesis in A20, possibly because of effects on D1 degradation, the major conclusion from our experiments is that it is the removal of damaged D1 from the RC47 that is most impaired in A20. Pulse-chase and immunoblotting experiments clearly demonstrated that D1 degradation is much slower in A20 even at low light intensities, despite the fact that there is precursor D1 available in the membrane for integration into the complex. The increased levels of RC47 in cells of A20 at higher irradiances, plus the inhibition of D1 degradation in the RC47 complex in the A20/ΔCP43 strain, indicated that RC47 can be formed in the mutant but that it is the subsequent step of D1 degradation that is impaired. This was shown most clearly when D1 levels were assayed by immunoblotting in the presence of lincomycin to prevent any possible influence of reduced D1 synthesis on degradation (Figure 9).
The Phenotype of A20 Resembles That of ftsH (slr0228) Null Mutants
Strikingly, the phenotype of A20 is very similar to that of ftsH (slr0228) null mutants impaired in D1 degradation (Silva et al., 2003; Komenda et al., 2006). Both types of mutant show light-sensitive photoautotrophic growth, are impaired in PSII repair, are unable to degrade D1 promptly following photodamage, contain elevated levels of the RC47 complex, and contain elevated levels of precursor D1 in the membrane. However, in the A20 truncation mutant, the level of FtsH (slr0228) (sometimes called FtsH2) is the same or even higher than in the control strain; therefore, depletion of the protease cannot explain the observed phenotype (Figure 6A). Additional deletion of ftsH (slr0228) in A20 resulted in an even more light-sensitive phenotype, as the strain was not able to grow even at 25 μmol·m−2·s−1. This enhanced sensitivity to visible light stress is likely to be due to an additional role of FtsH (slr0228) in alternative, less effective pathways for D1 degradation and/or by its involvement in biochemical processes outside PSII repair that are needed for the survival of cells in the light (Mann et al., 2000; Komenda et al., 2006). Nevertheless, the strain was able to assemble PSII complexes at 5 μmol·m−2·s−1, with most PSII proteins being in the form of RC47 complex (see Supplemental Figure 3 online).
The FtsH-Mediated N-Terminal–Dependent Model for D1 Degradation
In theory, a number of factors might contribute to impaired D1 degradation in the A20 mutant, including effects on trafficking of damaged PSII complexes in the membrane and recognition by the proteolytic apparatus. Given what is known about the mode of action of FtsH complexes in the literature (for review, see Ito and Akiyama, 2005), the simplest model to explain our results is one in which FtsH complexes recognize the N terminus of damaged D1 and degrade damaged D1 in a highly processive reaction initiated from the N terminus. FtsH complexes are encoded by a multigene family in both cyanobacteria and plants and are likely to be a mixture of homooligomers and heterooligomers (Sakamoto et al., 2003; Adam et al., 2006; Aluru et al., 2006). In agreement with the mechanism of E. coli FtsH (Chiba et al., 2000), impaired D1 degradation in A20 is likely to be due to the N-terminal tail being too short to initiate ATP-dependent translocation of damaged D1 into the proteolytic chamber, where processive degradation of D1 would occur. An attractive feature of this N-terminal model for D1 degradation is that it provides a mechanism for synchronizing the degradation of D1 with the insertion, possibly cotranslationally, of a newly synthesized D1 subunit into the RC47 complex. This repair process probably occurs in the granal margins and stromal lamellae in chloroplasts (Danielsson et al., 2006) and close to perforations in the thylakoid membrane in cyanobacteria (Nevo et al., 2007).
Implications for the Recognition of Damaged D1
The question arises of how FtsH complexes might be able to differentiate between damaged and undamaged D1 subunits. It is likely that oxidative damage to amino acid side chains and cofactors within PSII triggers the partial disassembly of PSII and destabilization of the D1 subunit in the membrane (Sharma et al., 1997). The detachment of CP43 and lumenally exposed extrinsic subunits from damaged PSII would allow space for docking of FtsH to the RC47 complex and/or access to the N-terminal tail of D1 (Figure 11). In addition, the N-terminal region of D1 might undergo a conformational change following PSII damage to allow engagement with FtsH. As FtsH complexes are considered to be poor at unfolding proteins (Herman et al., 2003), it is unlikely that they will be able to pull fully folded undamaged D1 subunits out of the membrane even if they should engage with the N-terminal degradation tag. ATP-driven removal of damaged D1 from the membrane by FtsH probably relies on prior destabilization or partial unfolding of damaged D1 subunit, achieved for instance by partial disassembly of the damaged PSII complex, photooxidative damage to amino acid side chains, and loss of pigments bound to D1 (Nixon et al., 2005). The presence of a lipid belt around D1 might also facilitate D1 exchange (Loll et al., 2007).
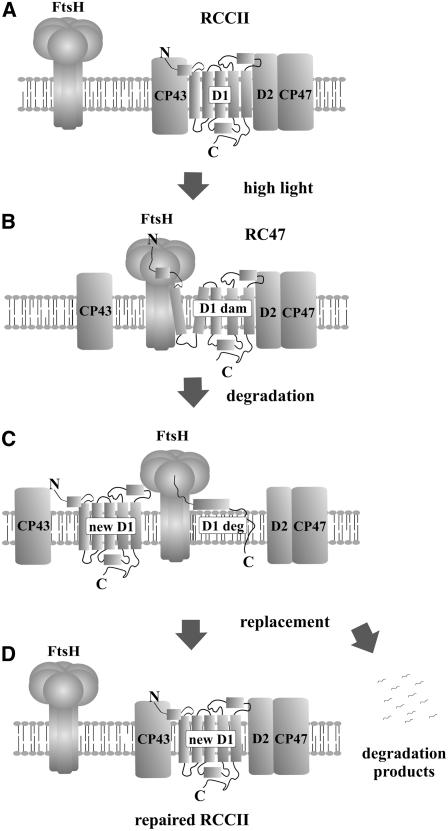
Figure 11.
Proposed Model for Selective D1 Replacement during PSII Repair in Synechocystis.
For clarity, just one of the monomers in the PSII dimer is shown, and the extrinsic and small transmembrane subunits of PSII are omitted.
(A) Intact PSII core complex with the functional and correctly folded D1 protein.
(B) High light–induced inactivation of PSII is followed by the release of CP43 and extrinsic proteins. In the resulting core complex lacking CP43 (RC47), the structure of damaged D1 protein (D1 dam) is destabilized, the protein is recognized by FtsH, and its released N terminus is caught by the protease.
(C) The damaged D1 subunit is degraded (D1 deg) by FtsH processively from the N to the C terminus, releasing short oligopeptides but no distinct larger fragments.
(D) Insertion of the new D1 molecule and reassembly of the active dimeric PSII core complex (RCCII).
An important consequence of this model is that FtsH will be able to remove damaged D1 no matter the site of damage, as long as the N-terminal degradation tag is accessible and the D1 subunit is sufficiently destabilized (Nixon et al., 2005). Consistent with this, recent work has shown that Synechocystis FtsH (slr0228) is also important for removing D1 and D2 following UV-B irradiation, which damages PSII through a quite different mechanism from the acceptor-side mechanism of visible light damage (Cheregi et al., 2007).
Comparison with Previous Models of D1 Degradation
Until now, D1 degradation following so-called acceptor-side photoinhibition has been considered to occur in a two-step process with the first cleavage event occurring in the D-E stromal loop (reviewed in Adir et al., 2003; Yokthongwattana and Melis, 2006). The main evidence in support of cleavage in this region has come largely from analysis of low-abundance D1 breakdown fragments, such as in Spirodela treated with cycloheximide (Greenberg et al., 1987), in plants whose repair capacity was unable to cope with the rate of photodamage (Canovas and Barber, 1993; Kettunen et al., 1996), and in isolated thylakoid membranes and detergent-solubilized PSII complexes (reviewed in Barber and Andersson, 1992). In all cases, the possibility that the observed cleavage was unrelated to physiological D1 turnover could not be excluded (Lupínková and Komenda, 2004); indeed, mutants with extensive deletions in the D-E loop still showed selective D1 turnover (Nixon et al., 1995; Mulo et al., 1997). Analysis of electron transfer and QB function in the A20 mutant argue strongly against the possibility that truncation of the N terminus impairs D1 degradation through effects on the conformation of the D-E loop, a scenario that is especially unlikely given the determined structure for PSII.
There have also been various reports based on in vitro experiments to suggest that D1 might be cleaved on the lumenal side of the membrane under conditions of so-called donor-side photoinhibition, when electron donation to PSII is impaired (reviewed in Huesgen et al., 2005). The possibility of such cleavages in vivo has been highlighted by recent studies on mutants of Arabidopsis either lacking one or both of the lumenal Deg5 and Deg8 proteases (Sun et al., 2007) or containing reduced levels of the lumenal Deg1 protease (Kapri-Pardes et al., 2007). In the case of a deg5 deg8 double mutant, the D1 subunit still showed selective synthesis and degradation at low light intensities, and the inhibition of D1 degradation at high light was only partial and much less extensive than in the ftsH2 (var2) Arabidopsis mutant (Bailey et al., 2002), which would suggest only an auxiliary and nonessential function of the Deg5 and Deg8 proteases in D1 degradation. In the case of Deg1, which appears to be essential for cell viability, rates of D1 synthesis and degradation have not yet been assessed in knockdown mutants in planta, so it remains unclear what role DegP1 plays (Kapri-Pardes et al., 2007).
Partially Redundant Pathways of D1 Degradation Might Exist
While we propose that the main pathway for selective D1 degradation starts at the N terminus, we do not exclude the presence of additional, less rapid, backup pathways, possibly including FtsH-mediated cleavage of the D-E loop, which operate in response to extensive light damage or alternative forms of abiotic stress such as heat denaturation (Yoshioka et al., 2006). The presence of these minor pathways would explain why A20 is still able to maintain a high level of functional PSII complexes under low-light growth conditions but not at higher light intensities, when the PSII repair cycle must be more active. In agreement with this possibility, the appearance of a D1 fragment was detected in the high light–treated cells of the truncation mutant but not the control strain (Figure 9). However, this degradation fragment was not detected in the mutant in the absence of lincomycin, so the physiological relevance is unclear. In situations in which there is cleavage of damaged D1 in the stromally exposed loops, FtsH might also be involved in the subsequent removal of breakdown fragments, possibly from the N terminus of D1 and also from the newly exposed N and C termini.
N-Terminally Mediated D1 Degradation in Chloroplasts
We also believe that the N-terminal pathway is the main route for the fast, repair-related degradation of D1 in chloroplasts. This hypothesis obviously needs direct experimental confirmation in vivo; nevertheless, it is supported by several facts. First, FtsH proteases are involved at an early stage in D1 degradation in both chloroplasts (Bailey et al., 2002; Sakamoto et al., 2003; Aluru et al., 2006) and cyanobacteria (Silva et al., 2003; Komenda et al., 2006). By contrast, the Deg2 protease, which was previously considered a likely candidate to cleave D1 in the D-E loop (Haussühl et al., 2001), was recently found not to be required for D1 turnover in planta (Huesgen et al., 2006). Likewise, none of the three Deg proteases present in Synechocystis is essential for D1 turnover (Barker et al., 2006). Second, inactivation of PSII and partial PSII disassembly precede D1 degradation in both cyanobacteria and plants (Nixon et al., 2005; Nishiyama et al., 2006; Yokthongwattana and Melis, 2006). Third, N-terminal phosphorylation of D1 at the initial Thr residue of mature D1 inhibits D1 degradation in chloroplasts (Koivuniemi et al., 1995). Although not essential for PSII repair (Bonardi et al., 2005), D1 phosphorylation seems to be a fine-tuning mechanism to prevent the premature degradation of D1 in membrane regions where the newly synthesized D1 molecule is not available (Rintamäki et al., 1996). If selective degradation of chloroplast D1 starts at the N terminus, then its phosphorylation would be an excellent tool to regulate the process. By contrast, a more sophisticated regulatory mechanism would have to be invoked to explain the inhibition of D1 cleavage in the D-E loop, which is distant from the N terminus.
METHODS
Strains and Culture Conditions
The glucose-tolerant strain of Synechocystis sp PCC 6803 (Williams, 1988) was grown in BG-11 medium containing 5 mM glucose, and the newly constructed autotrophic strains were grown without glucose. Solid media contained in addition 10 mM TES/NaOH, pH 8.2, 1.5% agar, and 0.3% sodium thiosulfate (Pakrasi et al., 1988). Fifty- to 100-mL liquid cultures were shaken in 250-mL conical flasks at 29°C with a surface irradiance of 25 μmol·m−2·s−1 white light until the cultures reached the late exponential phase (a chlorophyll concentration ∼4 to 5 μg/mL). For characterization of the strains grown at 25 and 125 μmol·m−2·s−1, the culture was diluted to 2 μg chlorophyll/mL and transferred to plan-parallel cuvettes in which the cultures were bubbled with 2% CO2. The same cuvettes were used for exposure of cultures (5 μg chlorophyll/mL) to photoinhibitory light (500 μmol·m−2·s−1) in the presence and absence of lincomycin (100 μg/mL). For this purpose, cultures were grown in a double-wall, thermoregulated cultivation cylinder (internal diameter, 35 mm) at a chlorophyll concentration of 6 to 8 μg/mL and maintained by diluting regularly with ∼10 mL of medium every 150 min. The culture was bubbled with air containing 2% CO2 and illuminated with white light of 50 μmol·m−2·s−1 at 29°C. Measurement of growth curves was performed on a 96-well microtitration plate illuminated at 25 or 125 μmol·m−2·s−1 and shaken intensely at 29°C. The cultures were initially diluted to OD750 nm = 0.005 (chlorophyll concentration, 0.015 μg/mL), and OD750 nm was measured every 6 h using a microplate reader (Tecan Sunrise). Values plotted against time were used for calculation of the doubling time.
Mutant Construction by Megaprimer Mutagenesis
Site-directed mutagenesis of the psbA2 gene was based on the PCR megaprimer method (Ke and Madison, 1997) using two rounds of PCR that employ two flanking primers with different melting temperatures and one internal mutagenic primer. For the autotrophic transformant A20, the resulting 1.8-kb PCR product containing the mutated psbA2 gene with upstream and downstream regions was used directly to transform a psbA triple deletion strain, psbAII-KS, with the psbA1 and psbA3 genes inactivated by chloramphenicol and spectinomycin resistance cassettes, respectively, and the psbA2 gene replaced by a kanamycin resistance/sacB cartridge (Lagarde et al., 2000). The sacB gene encodes levansucrase and confers sucrose sensitivity to this strain. The transformants expressing truncated psbA genes were selected either for autotrophy on BG-11 plates without glucose or for growth on BG-11 plates containing 5% sucrose and 5 mM glucose. Sucrose-resistant colonies were additionally checked for kanamycin sensitivity. The control strain A0 expressing the wild-type form of D1 was obtained by a transformation of the psbAII-KS strain using the same 1.8-kb PCR product without mutation. Sequencing of the entire psbA2 region confirmed that in all strains the particular gene was correctly inserted under the control of the psbA2 promoter.
Luminescence and Polarographic Methods
Photosynthetic thermoluminescence was measured with cells filtered onto the nitrocellulose membrane (6 μg of chlorophyll) using a commercial apparatus (Photon Systems Instruments). Cells were covered with 25 mM HEPES/NaOH, pH 7.5, containing 0.5 M sorbitol and dark-adapted for 5 min at 25°C. Then, the temperature was decreased to −10°C with the flash given either at 3°C in the absence of the PSII inhibitor diuron for measurement of the B band or at −10°C in the presence of diuron (10−5M), and thermoluminescence was measured during heating from −10 to 70°C. For the measurement of flash-induced oscillations of the B band, cells were illuminated by one to six single-turnover saturating flashes at 3°C, TL intensity was measured from 3 to 70°C, and the area below the glow curve was plotted against the number of flashes. For the measurement of chlorophyll concentrations, cells were sedimented by centrifugation and extracted with 100% methanol. The concentration of chlorophyll was calculated from the absorbance values of the extract at 666 and 720 nm (Wellburn and Lichtenthaler, 1984) using a Shimadzu UV3000 spectrophotometer. Absorption spectra of cells in vivo were measured using the same spectrophotometer in a compartment close to the photomultiplier with a slit width of 2 nm. Fluorescence emission spectra at 77K were assessed by an Aminco spectrofluorimeter. Cells containing 1 μg of chlorophyll in 50 μL of BG-11 were frozen in liquid nitrogen. Fluorescence was excited at 435 nm, and fluorescence emission was recorded in the range 600 to 800 nm with a slit width of 4 nm. Spectra were corrected for the wavelength sensitivity of the photomultiplier. The light-saturated steady state rate of oxygen evolution in cell suspensions was measured polarographically in the thermostatted chamber at 29°C (Bartoš et al., 1975) in the presence of the artificial electron acceptors p-benzoquinone (0.5 mM final concentration) and potassium ferricyanide (1 mM final concentration).
Radioactive Labeling of the Cells
Cells were radiolabeled using a mixture of l-[35S]Met and l-[35S]Cys (>1000 Ci/mmol, 400 μCi/mL final activity; Trans-label; MP Biochemicals) (Tichý et al., 2003) at the irradiances and temperatures indicated in the figure legends.
Thylakoid Preparation and Protein Analyses
Thylakoid membranes were prepared by breakage of the cells with glass beads at 4°C (Komenda et al., 2004) with the following modification: cells were washed, broken, and finally resuspended in 25 mM MES/NaOH, pH 6.5, containing 10 mM CaCl2, 10 mM MgCl2, and 25% glycerol. The protein composition of thylakoids was assessed by electrophoresis on a denaturing 12 to 20% linear gradient polyacrylamide gel containing 7 M urea (Komenda et al., 2004). The thylakoids were solubilized with SDS and DTT and loaded with equal amounts of chlorophyll. The gel was run overnight at 18°C, and proteins on the gel were electroblotted onto a PVDF membrane (Amersham Biosciences). The membrane was incubated with specific antibodies and then with a secondary antibody–peroxidase conjugate. Proteins were visualized with an enhanced chemiluminiscence kit (Roche). The specific antibodies were raised against the following: (1) residues 58 to 86 of Synechocystis D1; (2) residues 2 to 17 of Synechocystis D1; (3) the last 16 amino acid residues of the D1 precursor from Synechocystis; (4) the last 12 residues of plant D2 (Komenda et al., 2004); (5) residues 380 to 394 of barley (Hordeum vulgare) CP47; (6) the last 14 residues of the Synechocystis PsbI protein; (7) the whole isolated PsaD from Synechocystis; and (8) residues 98 to 115 of FtsH (slr0228) from Synechocystis. For autoradiography, the membrane was exposed to the sensitive film for 48 to 96 h. Assembly of the PSII complexes was assessed by two-dimensional electrophoresis consisting of blue-native PAGE in one direction (6 to 12% linear gradient polyacrylamide gel) (Schägger and Von Jagow, 1991) and SDS-PAGE (12 to 20% linear gradient polyacrylamide gel containing 7 M urea) (Komenda et al., 2004) in the second direction. Samples of thylakoid membranes solubilized with dodecyl maltoside and containing 6 μg of chlorophyll were used for each analysis. The two-dimensional gels were stained with Coomassie Brilliant Blue R, destained, dried, and exposed to a phosphorimager plate.
RNA Isolation, Reverse Transcription, and Quantitative PCR
Total RNA was isolated from frozen cells following the hot Trizol protocol described by Caldo et al. (2004), purified with the RNeasy MinElute Cleanup Kit (Qiagen), and treated with TURBO DNase (Ambion). Twenty nanograms of purified RNA was used for cDNA synthesis using random primers and SuperScript II reverse transcriptase (Invitrogen). Real-time quantitative PCR was performed on the Rotor-Gene 3000 using the iQ SYBR Green Supermix (Bio-Rad). Each quantitative PCR experiment was performed in two replicates for two independent RNA isolations from the same culture. rps1 (encoding 30S ribosomal protein S1) was used as a reference gene. Its level was found to be proportional to total RNA (measured spectrophotometrically) in all strains. The ΔΔCt method was used to calculate psbA gene expression normalized against rps1. The ΔCt values were reproducible to within 0.5 cycle.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Growth of Strains A0 and A20 during Incubation of Cells at 125 and 500 μmol·m−2·s−1.
Supplemental Figure 2. Recovery of the PSII Activity in Strains A0 and A20 after the Photoinhibitory Treatment.
Supplemental Figure 3. Protein Complexes Resolved by Blue-Native PAGE and Separation of Large PSII Proteins of PSII Complexes Revealed by Two-Dimensional Electrophoresis from Thylakoids of Strains A0Δslr0228 and A20Δslr0228 Cultivated at 5 μmol·m−2·s−1.
Supplementary Material
Acknowledgments
We are grateful to E.-M. Aro and L.A. Eichacker for the donation of specific antisera, to James Murray for help preparing Figure 1A, to M. Dobáková for help with measurements of growth curves and two-dimensional protein analyses, and to Jana Hofhanzlová for TL measurements. This work was supported by the Grant Agency of the Czech Republic (Projects 203/04/0800 and 206/06/0322), by the Ministry of Education, Youth, and Sports of the Czech Republic (Projects LN00A141 and MSM6007665808), by the Czech Academy of Sciences (Institutional Research Concept AV0Z50200510), and by the Biotechnology and Biological Science Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Josef Komenda (komenda@alga.cz).
Online version contains Web-only data.
References
- Adam, Z., Rudella, A., and van Wijk, K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9 234–240. [DOI] [PubMed] [Google Scholar]
- Adir, N., Zer, H., Shochat, S., and Ohad, I. (2003). Photoinhibition—A historical perspective. Photosynth. Res. 76 343–370. [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva, Y., Deák, Z., Szilárd, A., Diner, B.A., Nixon, P.J., and Vass, I. (2004). The function of D1-H332 in photosystem II electron transport studied by thermoluminescence and chlorophyll fluorescence in site-directed mutants of Synechocystis 6803. Eur. J. Biochem. 271 3523–3532. [DOI] [PubMed] [Google Scholar]
- Aluru, M.R., Yu, F., Fu, A., and Rodermel, S. (2006). Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J. Exp. Bot. 57 1871–1881. [DOI] [PubMed] [Google Scholar]
- Bailey, S., Thompson, E., Nixon, P.J., Horton, P., Mullineaux, C.W., Robinson, C., and Mann, N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277 2006–2011. [DOI] [PubMed] [Google Scholar]
- Barber, J., and Andersson, B. (1992). Too much of a good thing—Light can be bad for photosynthesis. Trends Biochem. Sci. 17 62–66. [DOI] [PubMed] [Google Scholar]
- Barker, M., de Vries, R., Nield, J., Komenda, J., and Nixon, P.J. (2006). The Deg proteases protect Synechocystis PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem II repair cycle. J. Biol. Chem. 281 30347–30355. [DOI] [PubMed] [Google Scholar]
- Bartoš, J., Berková, E., and Šetlík, I. (1975). A versatile chamber for gas exchange measurements in suspensions of algae and chloroplasts. Photosynthetica 9 395–406. [Google Scholar]
- Bonardi, V., Pesaresi, P., Becker, T., Schleiff, E., Wagner, R., Pfannschmidt, T., Jahns, P., and Leister, D. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437 1179–1182. [DOI] [PubMed] [Google Scholar]
- Caldo, R.A., Nettleton, D., and Wise, R.P. (2004). Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell 16 2514–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas, P.M., and Barber, J. (1993). Detection of a 10 kDa breakdown product containing the C-terminus of the D1-protein in photoinhibited wheat leaves suggests an acceptor side mechanism. FEBS Lett. 324 341–344. [DOI] [PubMed] [Google Scholar]
- Cheregi, O., Sicora, C., Kos, P.B., Barker, M., Nixon, P.J., and Vass, I. (2007). The role of the FtsH and Deg proteases in the repair of UV-B radiation-damaged photosystem II in the cyanobacterium Synechocystis PCC 6803. Biochim. Biophys. Acta 1767 820–828. [DOI] [PubMed] [Google Scholar]
- Chiba, S., Akiyama, Y., and Ito, K. (2002). Membrane protein degradation by FtsH can be initiated from either end. J. Bacteriol. 184 4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, S., Akiyama, Y., Mori, H., Matsuo, E., and Ito, K. (2000). Length recognition at the N-terminal tail for the initiation of FtsH-mediated proteolysis. EMBO Rep. 1 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson, R., Suorsa, M., Paakkarinen, V., Albertsson, P.-A., Styring, S., Aro, E.-M., and Mamedov, F. (2006). Dimeric and monomeric organization of photosystem II. Distribution of five distinct complexes in the different domains of the thylakoid membrane. J. Biol. Chem. 20 14241–14249. [DOI] [PubMed] [Google Scholar]
- Ferreira, K.N., Iverson, T.N., Maglaoui, K., Barber, J., and Iwata, S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303 1831–1838. [DOI] [PubMed] [Google Scholar]
- Greenberg, B.M., Gaba, V., Mattoo, A.K., and Edelman, M. (1987). Identification of a primary in vivo degradation product of the rapidly-turning-over 32 kD protein of photosystem II. EMBO J. 6 2865–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussühl, K., Andersson, B., and Adamska, I. (2001). A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 20 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, C., Prakash, S., Lu, C.Z., Matouschek, A., and Gross, C.A. (2003). Lack of robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol. Cell 11 659–669. [DOI] [PubMed] [Google Scholar]
- Huesgen, P.F., Schuhmann, H., and Adamska, I. (2005). The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol. Plant. 123 413–420. [Google Scholar]
- Huesgen, P.F., Schuhmann, H., and Adamska, I. (2006). Photodamaged D1 protein is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Lett. 580 6929–6932. [DOI] [PubMed] [Google Scholar]
- Ito, K., and Akiyama, Y. (2005). Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59 211–231. [DOI] [PubMed] [Google Scholar]
- Jansen, M.A.K., Mattoo, A.K., and Edelman, M. (1999). D1-D2 protein degradation in the chloroplast. Complex light saturation kinetics. Eur. J. Biochem. 260 527–532. [DOI] [PubMed] [Google Scholar]
- Kanervo, E., Spetea, C., Nishiyama, Y., Murata, N., Andersson, B., and Aro, E.-M. (2003). Dissecting a cyanobacterial proteolytic system: Efficiency in inducing degradation of the D1 protein of photosystem II in cyanobacteria and plants. Biochim. Biophys. Acta 1607 131–140. [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes, E., Naveh, L., and Adam, Z. (2007). The thylakoid lumen protease Deg 1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19 1039–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, S.H., and Madison, E.L. (1997). Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 25 3371–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen, R., Tyystjärvi, E., and Aro, E.-M. (1996). Degradation pattern of photosystem II reaction center protein D1 in intact leaves. Plant Physiol. 111 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi, A., Aro, E.-M., and Andersson, B. (1995). Degradation of the D1 and D2 proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry 34 16022–16029. [DOI] [PubMed] [Google Scholar]
- Komenda, J., Barker, M., Kuviková, S., DeVries, R., Mullineaux, C.W., Tichý, M., and Nixon, P.J. (2006). The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis PCC 6803. J. Biol. Chem. 281 1145–1151. [DOI] [PubMed] [Google Scholar]
- Komenda, J., Hassan, H.A.G., Diner, B.A., Debus, R.J., Barber, J., and Nixon, P.J. (2000). Degradation of the photosystem II D1 and D2 proteins in different strains of the cyanobacterium Synechocystis PCC 6803 varying with respect to the type and level of psbA transcript. Plant Mol. Biol. 42 635–645. [DOI] [PubMed] [Google Scholar]
- Komenda, J., Kuviková, S., Granvogl, B., Eichacker, L.A., Diner, B.A., and Nixon, P.J. (2007). Cleavage after residue Ala352 in the C-terminal extension is an early step in the maturation of the D1 subunit of photosystem II in Synechocystis PCC 6803. Biochim. Biophys. Acta 1767 829–837. [DOI] [PubMed] [Google Scholar]
- Komenda, J., Lupínková, L., and Kopecký, J. (2002). Absence of the psbH gene product destabilizes photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur. J. Biochem. 269 610–619. [DOI] [PubMed] [Google Scholar]
- Komenda, J., Reisinger, V., Müller, B.C., Dobáková, M., Granvogl, B., and Eichacker, L.A. (2004). Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J. Biol. Chem. 279 48620–48629. [DOI] [PubMed] [Google Scholar]
- Kuviková, S., Tichý, M., and Komenda, J. (2005). A role of the C-terminal extension of the photosystem II D1 protein in sensitivity of the cyanobacterium Synechocystis PCC 6803 to photoinhibition. Photochem. Photobiol. Sci. 4 1044–1048. [DOI] [PubMed] [Google Scholar]
- Lagarde, D., Beuf, L., and Vermaas, W. (2000). Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, M., Spetea, C., Hundal, T., Oppenhaim, A.B., Adam, Z., and Andersson, B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the PSII D1 protein. Plant Cell 12 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, M., Tabak, S., Cseke, L., Pichersky, E., Andersson, B., and Adam, Z. (1996). Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 271 29329–29334. [DOI] [PubMed] [Google Scholar]
- Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. (2005). Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438 1040–1044. [DOI] [PubMed] [Google Scholar]
- Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. (2007). Lipids in photosystem II: Interactions with proteins and cofactors. Biochim. Biophys. Acta 1767 509–519. [DOI] [PubMed] [Google Scholar]
- Long, S., and Humphries, S. (1994). Photoinhibition of photosynthesis in nature. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45 633–662. [Google Scholar]
- Lupínková, L., and Komenda, J. (2004). Oxidative modifications of the photosystem II D1 protein by reactive oxygen species: From isolated protein to cyanobacterial cells. Photochem. Photobiol. 79 152–162. [DOI] [PubMed] [Google Scholar]
- Mann, N.H., Novac, N., Mullineaux, C.W., Newman, J., Bailey, S., and Robinson, C. (2000). Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479 72–77. [DOI] [PubMed] [Google Scholar]
- Mulo, P., Tyystjärvi, T., Tyystjärvi, E., Govindjee, Mäenpää, P., and Aro, E.-M. (1997). Mutagenesis of the D-E loop of photosystem II reaction center protein D1. Function and assembly of photosystem II. Plant Mol. Biol. 33 1059–1071. [DOI] [PubMed] [Google Scholar]
- Nevo, R., Charuvi, D., Shimoni, E., Schwartz, R., Kaplan, A., Ohad, I., and Reich, Z. (2007). Thylakoid membrane perforations and connectivity enable intracellular traffic in cyanobacteria. EMBO J. 26 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, Y., Allakhverdiev, S.I., and Murata, N. (2006). Regulation by environmental conditions of the repair of photosystem II in cyanobacteria. In Advances in Photosynthesis and Respiration, Photoprotection, Photoinhibition, Gene Regulation, and Environment, B. Demmig-Adams, W.W. Adams, and A. Matoo, eds (Dordrecht, The Netherlands: Springer), pp. 193–203.
- Nixon, P.J., Barker, M., Boehm, M., de Vries, R., and Komenda, J. (2005). FtsH-mediated repair of the photosystem two complex in response to light stress in the cyanobacterium Synechocystis sp. PCC 6803. J. Exp. Bot. 56 347–356. [DOI] [PubMed] [Google Scholar]
- Nixon, P.J., Komenda, J., Barber, J., Deak, Z., Vass, I., and Diner, B.A. (1995). Deletion of the PEST-like region of photosystem II modifies the QB-binding pocket but does not prevent rapid turnover of D1. J. Biol. Chem. 270 14919–14927. [DOI] [PubMed] [Google Scholar]
- Ohad, N., and Hirschberg, J. (1992). Mutations in the D1 subunit of the photosystem II distinguish between quinone and herbicide binding sites. Plant Cell 4 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno, T., Yamanaka, K., and Ogura, T. (2006). An AAA protease FtsH can initiate proteolysis from internal sites of a model substrate, apo-flavodoxin. Genes Cells 11 261–268. [DOI] [PubMed] [Google Scholar]
- Pakrasi, H.B., Williams, J.G.K., and Arntzen, C.J. (1988). Targeted mutagenesis of the psbE and psbF genes blocks photosynthetic electron transport: Evidence for a functional role of cytochrome b-559 in photosystem II. EMBO J. 7 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prášil, O., Adir, N., and Ohad, I. (1992). Dynamics of photosystem II: Mechanism of photoinhibition and recovery process. In The Photosystems: Structure, Function and Molecular Biology, J. Barber, ed (Amsterdam: Elsevier), pp. 295–348.
- Rintamäki, E., Kettunen, R., and Aro, E.-M. (1996). Differential D1 dephosphorylation in functional and photodamaged photosystem II centers. Dephosphorylation is a prerequisite for degradation of damaged D1. J. Biol. Chem. 271 14870–14875. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., and Von Jagow, G. (1991). Blue native electrophoresis for isolation of membrane-protein complexes in enzymatically active form. Anal. Biochem. 199 223–231. [DOI] [PubMed] [Google Scholar]
- Sharma, J., Panico, M., Shipton, C.A., Nilsson, F., Morris, H.R., and Barber, J. (1997). Primary structure characterization of the photosystem II D1 and D2 subunits. J. Biol. Chem. 272 33158–33166. [DOI] [PubMed] [Google Scholar]
- Silva, P., Thompson, E., Bailey, S., Kruse, O., Mullineaux, C.W., Robinson, C., Mann, N.H., and Nixon, P.J. (2003). FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. Plant Cell 15 2152–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetea, C., Hundal, T., Lohmann, F., and Andersson, B. (1999). GTP bound to chloroplast thylakoid membranes is required for light-induced, multienzyme degradation of the photosystem II D1 protein. Proc. Natl. Acad. Sci. USA 96 6547–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Peng, L., Guo, J., Chi, W., Ma, J., Lu, C., and Zhang, L. (2007). Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19 1388–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichý, M., Lupínková, L., Sicora, C., Vass, I., Kuviková, S., Prášil, O., and Komenda, J. (2003). Synechocystis 6803 mutants expressing distinct forms of the photosystem II D1 protein from Synechococcus 7942: Relationship between the psbA coding region and sensitivity to visible and UV-B radiation. Biochim. Biophys. Acta 1605 55–66. [DOI] [PubMed] [Google Scholar]
- Vass, I., and Govindjee. (1996). Thermoluminescence from the photosynthetic apparatus. Photosynth. Res. 48 117–126. [DOI] [PubMed] [Google Scholar]
- Wellburn, A.R., and Lichtenthaler, K. (1984). Formulae and programme to determine total carotenoids and chlorophyll a and b of leaf extracts in different solvents. In Advances In Photosynthesis Research, C. Sybesma, ed (Dordrecht, The Netherlands: Martinus Nijjhoff), pp. 10–12.
- Williams, J.G.K. (1988). Construction of specific mutations in PSII photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167 766–778. [Google Scholar]
- Yokthongwattana, K., and Melis, A. (2006). Photoinhibition and recovery in oxygenic photosynthesis: Mechanism of a photosystem II damage and repair. In Advances in Photosynthesis and Respiration, Photoprotection, Photoinhibition, Gene Regulation, and Environment, B. Demmig-Adams, W.W. Adams, and A. Matoo, eds (Dordrecht, The Netherlands: Springer), pp. 175–191.
- Yoshioka, M., Uchida, S., Mori, H., Komayama, K., Ohira, S., Merita, N., Nakanishi, T., and Yamamoto, Y. (2006). Quality control of photosystem II. Cleavage of reaction center D1 protein in spinach thylakoids by FtsH protease under moderate heat stress. J. Biol. Chem. 281 21660–21669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.