Abstract
Cotranslational and posttranslational modifications are increasingly recognized as important in the regulation of numerous essential cellular functions. N-myristoylation is a lipid modification ensuring the proper function and intracellular trafficking of proteins involved in many signaling pathways. Arabidopsis thaliana, like human, has two tightly regulated N-myristoyltransferase (NMT) genes, NMT1 and NMT2. Characterization of knockout mutants showed that NMT1 was strictly required for plant viability, whereas NMT2 accelerated flowering. NMT1 impairment induced extremely severe defects in the shoot apical meristem during embryonic development, causing growth arrest after germination. A transgenic plant line with an inducible NMT1 gene demonstrated that NMT1 expression had further effects at later stages. NMT2 did not compensate for NMT1 in the nmt1-1 mutant, but NMT2 overexpression resulted in shoot and root meristem abnormalities. Various data from complementation experiments in the nmt1-1 background, using either yeast or human NMTs, demonstrated a functional link between the developmental arrest of nmt1-1 mutants and the myristoylation state of an extremely small set of protein targets. We show here that protein N-myristoylation is systematically associated with shoot meristem development and that SnRK1 (for SNF1-related kinase) is one of its essential primary targets.
INTRODUCTION
N-terminal protein maturation involves a series of cotranslational modifications of the N termini of proteins (Meinnel et al., 1993; Bradshaw et al., 1998). N-myristoylation (MYR), catalyzed by N-myristoyltransferase (NMT), is one such modification. It involves the addition of the saturated C:14 fatty acid myristate to the α N terminus of a subset of proteins (reviewed in Bhatnagar et al., 2001). MYR is known to affect the membrane binding properties of several families of proteins, such as calcium-dependent protein kinases and small GTPases of the ADP-ribosylation factor family, many of which are involved in transduction pathways (Boutin, 1997). Disruption of the associated NMT gene strongly impairs cell growth in unicellular organisms (Duronio et al., 1989; Price et al., 2003). However, it remains unclear whether cell survival requires the MYR of all or only a subset of the myristoylated proteins comprising the so-called N-myristoylome. Characterization of the N-myristoylome is the key starting point for addressing this issue, which remains a challenge due to the extremely complex nature of the protein sequence recognized by NMT. Recognition is based on the chemical properties of the first eight residues of the peptide sequence, with only the nature of the first residue (Gly) strictly defined and fixed. Moreover, several studies have indicated that orthologous NMTs have overlapping but different substrate specificities (Towler et al., 1988; Maurer-Stroh et al., 2002a, 2004). For instance, a higher eukaryotic NMT has been shown to acylate several substrates in vitro that the Saccharomyces cerevisiae NMT is unable to modify (Boisson et al., 2003). Structural analyses of the interaction between NMT and peptide substrates confirmed these findings. Potent peptide substrate-based inhibitors of pathogenic NMTs have been designed and shown to have only weak activity against animal NMTs (Devadas et al., 1998). However, several NMTs from human (Homo sapiens) and plants have been shown to complement a yeast nmt-deficient mutant (Duronio et al., 1992; Boisson et al., 2003). Attempts to define the corresponding subproteome in the complete Arabidopsis thaliana proteome have shown that a large number of protein targets (>1.7% of the proteome; i.e., 422 proteins) undergo this modification. Complete predicted proteome annotation is now available online at the genomic bioinformatics resource facility for Arabidopsis (The Arabidopsis Information Resource; http://www.arabidopsis.org/servlets/TairObject?type=keywordandid=6894.
Mammals and higher plants have two NMT homologs, NMT1 and NMT2, but it remains unclear whether the corresponding enzymes have redundant, similar, or different functions (Giang and Cravatt, 1998; Qi et al., 2000; Bhatnagar et al., 2001; Boisson et al., 2003). Difficulties in predicting the substrate specificity of a given NMT—even for fungal NMTs, the most thoroughly studied NMTs—make it impossible to determine whether the genes encoding different NMTs are redundant or play different roles within the organism at different developmental stages or within different organs, tissues, or cell types. The mouse NMT1 gene has recently been shown to be essential for early embryonic development (Yang et al., 2005). These data indicate that the NMT genes of mammals are nonredundant, as also suggested by substrate specificity and small interfering RNA injection analyses (Giang and Cravatt, 1998; Ducker et al., 2005). The deleterious effects of NMT1 knockout early in embryogenesis have prevented investigation of the possible role of NMT1 at later stages of development.
We used a functional genomics approach in various knockout and inducible transgenic plant lines to investigate the roles of the two Arabidopsis NMT genes. NMT1 was found to be essential throughout organ development, at both early and later stages. NMT2 was found to be essential only for the transition to flowering. We tried to identify the primary protein targets of At NMT1 by in vivo complementation experiments with the nmt1-1 line and orthologous (yeast and human) NMT genes, together with in vitro myristoylation experiments with the same purified NMTs. Our data indicate that the developmental phenotype of the nmt1-1 line results from a lack of myristoylation of at most four of the 422 potential target proteins of the Arabidopsis N-myristoylome. The target proteins identified included two β-subunits (yeast SIP2p ortholog) of the SnRK1 (SnRK1) kinase. Based on characterization of the impact of MYR on the SnRK1 pathway, through expression, activity, and subcellular localization experiments, we conclude that improper signaling due to SnRK1 kinase dysfunction is involved in the developmental arrest observed in the nmt1-1 line.
RESULTS
Two Expressed NMT Genes in Arabidopsis and an Intriguing Small Open Reading Frame Flanking NMT2
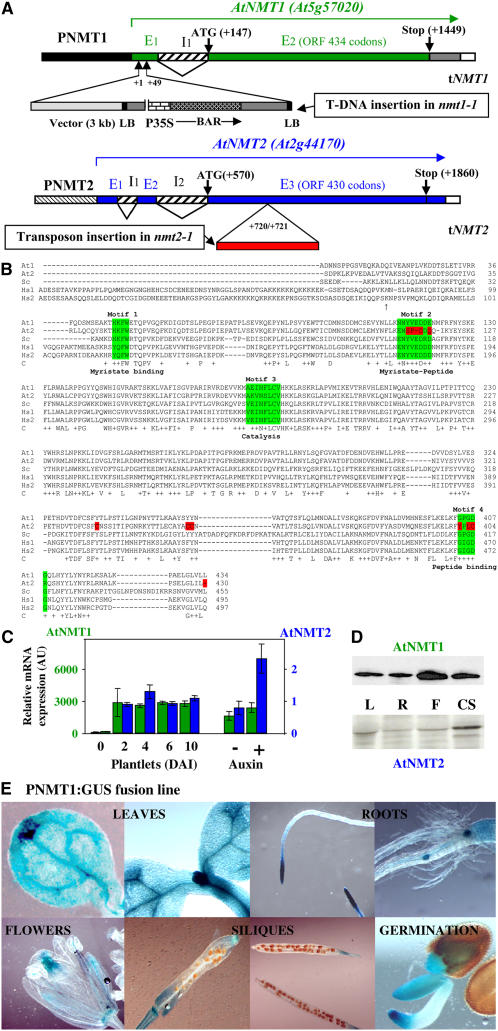
Screening of the complete genome of Arabidopsis with various NMTs originating from yeast or animals led to the identification of two loci, At5g57020 (NMT1) and At2g44170 (NMT2) (Figure 1A). Full-length cDNAs corresponding to the two loci were amplified and sequenced (GenBank accession numbers AF250956 and AF250957). Sequence analysis of the open reading frame (ORF) showed very strong sequence identity between these sequences and NMT sequences from other plants, including both monocots and dicots (73 to 76%). Sequence alignment with other well-characterized NMT genes, those from S. cerevisiae (Sc NMT) and both human NMTs (Hs NMT1 and Hs NMT2), showed levels of sequence identity of 50, 55, and 56%, respectively (Figure 1B). These findings strongly suggest that both At NMT genes encode enzymes with NMT activity. In previous studies, At NMT1 activity has been shown to be similar to that of known NMTs, with strong dependence on myristoyl-CoA and a peptide starting with an N-terminal Gly (Qi et al., 2000; Boisson and Meinnel, 2003). The complete proteome of At NMT1 substrates (422 substrates in the December 2006 update; see http://www.isv.cnrs-gif.fr/tm/maturation/myristoylome2007am.htm) was determined by a proteomic approach (Boisson et al., 2003). Unlike At NMT1, At NMT2 was unable to complement the heat sensitivity of the yeast nmt-181 strain (Boisson et al., 2003). Moreover, the low solubility of At NMT2 made it impossible to purify this protein at high yield from bacteria or yeast in vivo. We conclude that At NMT2 is probably an NMT, but further studies are required to identify its peptide substrates.
Figure 1.
ORFs, Genes, and Expression Patterns of NMT1 and NMT2 in Various Organs and during Development.
(A) Schematic representation of the wild-type At NMT gene structures and their disruption in Arabidopsis lines nmt1-1 and nmt2-1. The exon (En)–intron (In) structure of the gene is shown. Translation initiation (ATG) and termination codons (stop) are indicated. At NMT1: The tandem T-DNA insertion in the nmt1-1 line is shown, with the border sequences of the insert (left border [LB] and cloning vector) labeled to indicate the orientation of the insertion. BAR encodes the BASTA resistance gene. P35S is the 35S promoter. The exact location of the T-DNA insertion (+1/+49 from the origin of the transcript, as indicated in GenBank under accession number AF250956) was checked by DNA sequencing, PCR amplification, and restriction fragment length analysis. At NMT2: The location of the inserted transposon in line nmt2-1 is indicated from the origin of the transcript (as indicated in GenBank under accession number AF250957). The location of the inserted transposon end was checked by DNA sequencing, PCR amplification, and restriction fragment length analysis.
(B) The full-length ORFs of each NMT used in this study were aligned using ClustalX (Jeanmougin et al., 1998). The numbering of each of the five amino acid sequences is indicated below the sequences for each block of 100 residues. Amino acids shown with an arrow at the N terminus of Hs NMT1 indicate the alternative translation start sites of each isoform. Line C shows strictly conserved residues within the catalytic core are shown below the amino acid sequence. Conservative changes are indicated with a plus sign. In the last line, the region highlighted in green corresponds to the binding sites of each substrate (Motifs 1 to 4). Residues shown in red are not conserved in At NMT2. At, Arabidopsis NMTs (At1 and At2); Sc, S. cerevisiae NMT; Hs, H. sapiens NMTs (Hs1 and Hs2).
(C) Levels of transcripts for cytoplasmic NMTs expressed relative to actin transcript levels. Measurements were made by real-time PCR. Left: NMT1 or NMT2 levels in wild-type seedlings DAI 6 were taken as 1. The synthetic auxin variant used was 2,4-D (see Results). AU, arbitrary units.
(D) Relative levels of At NMT1 and At NMT2 proteins. Left: Immunoblot analysis performed in leaves (L), roots (R), flowers (F), and cell suspensions (CS) with specific antibodies against each of these NMTs.
(E) Expression of PNMT1:GUS in seedlings. Various NMT1 promoter-GUS (PNMT1:GUS) lines were analyzed. A representative image is shown in each case.
A more thorough analysis revealed that the ORF preceding At NMT2, At2g44175, encodes a 113–amino acid protein. At2g44175 displays a high level of identity to both At NMT1 (53%) and At NMT2 (48%) over an 82–amino acid residue overlap corresponding to the sequence covering the extreme N terminus of both At NMTs (covering the sequence between motifs 1 and 2, see Figure 1B). No cDNA corresponding to this ORF has yet been found, and it is unclear whether this ORF is translated. RT-PCR analysis with several pairs of specific primers failed to detect the corresponding transcript. Moreover, the product of this 340-nucleotide ORF, if expressed, would be unlikely to have NMT activity, as it lacks the peptide binding domain. With its 82% identity to At NMT1 over an overlap of 128 nucleotides and 84% identity to At NMT2 over an overlap of 66 nucleotides, it seemed possible that At2g44175 might encode small interfering RNAs targeting both NMTs. An analysis with mfold RNA secondary structure prediction tools (Zuker, 2003) showed that At2g44175 was unlikely to encode typical small interfering RNA structures targeting either or both NMTs. Finally, fine sequence analysis of At NMT2 showed meristem-specific regulation regions, including an LBS/WBS3 motif in intron I2 (13 of 14 nucleotides conserved), an LBS/WBS1 motif in the 3′ untranslated region (10 of 12 nucleotides conserved), and a CArG box1 in the proximal region of the promoter, suggesting that this gene is subject to transcriptional regulation in meristems. LBS/WBS motifs are binding sites for transcription factors specific to floral meristems, such as the protein products of WUSCHEL and LEAFY (Hong et al., 2003).
Temporal and Spatial Expression Patterns of the Two NMT Genes in Arabidopsis
We used real-time RT-PCR to determine patterns of mRNA production for both At NMT genes. On day 6 after imbibition (DAI 6), NMT2 was expressed significantly less strongly than NMT1 (Figure 1C). The NMT1 transcript was more abundant than other transcripts of the cytosolic N-terminal maturation pathway, such as those of MAP1A, MAP2A, and MAP2B (Ross et al., 2005), whereas the NMT2 transcript was significantly less abundant. We next investigated the expression of the two NMT genes in various organs. NMT2 was strongly induced in flowers, consistent with the presence of LBS/WBS floral meristem motifs, whereas both NMT genes were expressed only weakly in fruits. The levels of both mRNAs increased strongly at the time of germination (DAI 2) and showed no further increase during development (Figure 1C, left). Finally, if cell division was stimulated with a synthetic auxin (2,4-D), or in cell suspensions, NMT2 displayed significant induction, whereas NMT1 did not (Figure 1C). Antibodies were raised against purified At NMT1 or specific peptides of At NMT2. These antibodies detected the corresponding proteins, at the expected size, in various Arabidopsis extracts (Figure 1D). An analysis of cross-reactivity with NMT1 antibodies also indicated that NMT2 was produced in much smaller amounts than NMT1 (one to two orders of magnitude less NMT2), consistent with the results of mRNA analysis and transcriptional or posttranscriptional regulation. Green fluorescent protein (GFP) fusion analysis indicated that both NMTs were present in the cytosol.
We investigated the spatial expression patterns of the two NMT genes by fusing the corresponding promoters to the β-glucuronidase (GUS) gene. The length of each promoter was defined such that it did not encompass the proximal gene: At2g44175 in the case of NMT2 and At5g57015 in the case of NMT1. The promoters of NMT1 and NMT2 were each 2 kb long. Complementation experiments confirmed these findings (see below). No GUS staining was observed with the NMT2 promoter, consistent with the low levels of mRNA detected. By contrast, strong staining was observed for fusions of GUS with the NMT1 promoter (PNMT1:GUS, Figure 1E). Strong NMT1 expression appeared to be associated with the metabolically active areas of vegetative organs, such as developing leaves, hydathodes in mature leaves, and root apical and lateral meristems. In flowers, the NMT1 promoter was found to be highly active in pollen grains, within stamens, or in female areas. The NMT1 promoter was also highly active in growing areas of young siliques but not in the embryo. During seed germination, the NMT1 promoter was found to be highly active in the tips of shoots and root tips, which displayed the highest levels of cell division.
Thus, both NMT1 and NMT2 appear to display tightly regulated expression in plants. NMT1 is expressed significantly more strongly than NMT2, but both genes are induced at the time of germination and in zones requiring cell division.
NMT1 Knockout Leads to Late-Embryo Abortion Due to Early Developmental Defects Affecting Shoot Apical Meristem Differentiation
We investigated the functions of the two NMTs in Arabidopsis by searching for knockout mutants of these two genes. We screened the Versailles T-DNA insertion collection and identified one line with an insertion in the first intron of NMT1. This line remains the only putative candidate for a knockout in NMT1. The T-DNA encodes the BAR gene, conferring resistance to the herbicide BASTA, making it possible to select transgenic lines. Fine analysis of the insertion region around NMT1 indicated that it actually corresponded to a tandem T-DNA insertion and part of the original shuttle vector (Figure 1A). This large insertion resulted in a small (50 bases) deletion within the first exon of NMT1. We grew the heterozygous line on soil in the greenhouse but were unable to recover homozygous lines with the insertion in NMT1. A segregation ratio of 1 wild type:2 nmt1/NMT1 was observed, suggesting an essential role of NMT1. The progeny of the heterozygous line was cultured in vitro in sucrose-supplemented control medium. In these conditions, T-DNA segregation on DAI 4 perfectly followed Mendelian (1:2:1) rules. All wild-type lines were sensitive to BASTA, consistent with a single insertion. The homozygous line with this insertion had a late (after germination) developmental arrest phenotype. This phenotype could only be detected after DAI 4, when the cotyledons of the wild type were growing and becoming green (Figure 2A). After DAI 3, the homozygous lines ceased to grow and did not green, whereas heterozygous and wild-type lines originating from the same seed lot grew normally (Figure 2A). The mutant line displayed the same phenotype whether grown in the dark or in the light. In the absence of sucrose as a reduced carbon source, the nmt1-1 seedlings greened but grew no further. Finally, the nmt1-1 line was much more sensitive to high glucose concentration than the wild type. Thus, the nmt1-1 line responded to light stimuli but displayed impaired sugar sensing.
Figure 2.
Phenotypes Associated with NMT1 Knockout in the nmt1-1 Line and Rescue with At NMT1.
(A) Global phenotype of the nmt1-1 line. Left: Phenotypes of various independent nmt1-1 lines at various time points and their comparison with the wild type. A close-up of the nmt1-1 line is shown. The seedling was dissected to uncover the cotyledons, which are normally covered by the seed coat (see left part).
(B) RT-PCR analysis of the NMT1 transcript in the nmt1-1 line. The EF transcript was used as the positive control probe. The reasons for transcript overproduction are given in Supplemental Figure 1 online.
(C) Immunoblot analysis of seedlings 10 DAI, showing the absence of At NMT1 in the nmt1-1 line. We analyzed 80-μg aliquots of total protein. The band migrated at the expected Mr of ∼50 kD.
(D) Complementation of the nmt1-1 line with the N1At1 construct leads to reversion to the wild type. Left: Comparison of the phenotypes of wild-type and nmt-1 plants stably expressing the N1At1 transgene (nmt-1-N1At1). Growth 35 DAI is shown. Right: Immunoblot analysis of the amount of At NMT1 in wild-type, nmt-1, or nmt-1-N1At1 lines. Proteins were extracted 7 DAI. PEPC, phosphoenolpyruvate carboxylase. Antibodies were provided by J. Vidal.
(E) Analysis of the cytological phenotype of the nmt1-1 line and comparison with the wild type as observed 4 DAI. Left panel: Confocal imaging of the root meristem region stained with FM464. Center panel: Thin cross section of the SAM region stained with toluidine blue. The circle indicates the SAM. Right panel: Whole-mount cleared seeds observed with differential interference contrast microscopy at the heart embryo stage. The position of the SAM is indicated with an arrow.
The observed phenotype of the homozygous line was entirely consistent with the above analysis of NMT1 expression, with a strong increase in mRNA levels after germination (Figures 1C to 1E). We then investigated the consequences of this insertion. RT-PCR analysis unexpectedly suggested that NMT1 was overexpressed in this line (Figure 2B; see Supplemental Figure 1A online). Further RNA gel blot and RT-PCR experiments revealed that the excess NMT1 transcripts were produced from a BAR:NMT1 gene fusion and that expression was driven by the 35S promoter (P35S) directing BAR gene expression in the T-DNA (see Supplemental Figures 1B and 1C online). As NMT was the second gene in the fusion, with many stop codons (14) between the BAR and NMT1 genes, and as internal ribosome entry sites are exceptional in eukaryotes, it was considered highly unlikely that this transcript would be translated to produce NMT1 protein. If such translation had occurred, the protein generated would have been identical to the wild type. The NMT1 protein was undetectable on immunoblots of the homozygous line (Figure 2C), confirming the strong specificity of the antibody. We checked that the observed phenotype was associated with the absence of NMT1 by conducting complementation experiments with At NMT1 as a transgene. As expression of this gene appeared to be tightly regulated (see above), we generated a construct including the complete At NMT1 gene (the full-length gene, from its own promoter, PNMT1, to its transcription terminator). The corresponding DNA fragment spanned a 5-kb region (PNMT1:At NMT1 transgene in Supplemental Figure 2A online). Stable insertion of this transgene fully complemented the developmental defect of the NMT1 insertion mutant (Figure 2D, left). Moreover, the NMT1 protein was detected on immunoblots of the transgenic line (Figure 2D, right). We therefore concluded that the homozygous line with the T-DNA insertion was a true NMT1 knockout mutant, which we named nmt1-1.
We investigated the defects associated with NMT1 knockout in the nmt1-1 line, at both morphological and tissue levels. The seed coat remained intact in this line. The seed coat was removed and dissected, unmasking apparently normal white cotyledons (Figure 2A, right). These findings indicated unusually late growth arrest. We analyzed the tissues of the root tip. The root meristem was perfectly developed, with no tissue differences visible. As shown by FM464 staining and confocal microscopy, all developmental zones (root cap, meristematic zone, elongation zone, and maturation zone) were perfectly defined in both the nmt1-1 line and the wild type (Figure 3E, left). Permeability to propidium iodide was similar in the nmt1-1 line and the wild type, indicating that the root cells were fully viable. By contrast, an analysis of thin longitudinal sections revealed complete disorganization of the rib zone of the shoot apical meristem (SAM). The tunica with anticlinal cell division and the corpus were unrecognizable in the nmt1-1 line, consistent with the observed developmental arrest (Figure 3E, center). Finally, an analysis of heart-shaped embryos in the siliques of the heterozygous nmt1-1/NMT1 line showed that some embryos were asymmetric, possibly indicating defects at early embryonic stages (Figure 3E, right).
Figure 3.
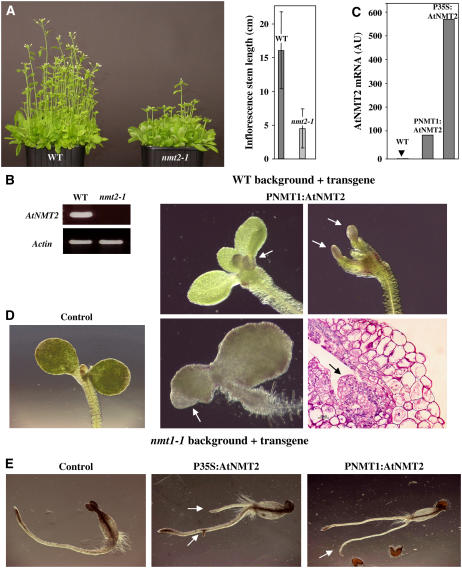
Variations in NMT2 Protein Levels in Various Backgrounds Induce Developmental Abnormalities in the Root and SAM.
(A) Effect of At NMT2 knockout in the nmt2-2 line. Left: A set of 50 seedlings grown for 14 d on soil. Right: Measurements of the length of the inflorescence stems.
(B) RT-PCR analysis showing the absence of NMT2 transcripts in line nmt2-1. The standard deviation on each of the measurements was 3.0, 2.6, and 12% for the wild type, PNMT1:At NMT2, and P35S:At NMT2, respectively.
(C) Real-time PCR analysis of NMT2 transcript levels in the wild type as a function of the promoter used. An arbitrary value of 1 was assigned to At NMT2 levels in the wild type.
(D) Phenotypes induced by NMT2 overexpression 4 DAI in the wild-type background. Arrows indicate unusual extra buds appearing at the SAM. Bottom right: Thin cross section of the SAM region stained as in Figure 2E, center panel. For the wild-type control, see Figure 3D.
(E) Phenotypes observed in the nmt1-1 background. The arrows indicate the unusual additional roots observed in this background.
Thus, NMT1 inactivation resulted in the production of a viable embryo, late developmental arrest being associated with a defect due to malformation of the SAM.
NMT2 Knockout Affects Flowering Time, and a Double NMT Knockout Does Not Strengthen the nmt1-1 Phenotype
We characterized a transposon insertion line (see Figure 1A) corresponding to a true NMT2 knockout (Figure 3B). This line was named nmt2-1. In contrast with what was observed with the nmt1-1 line, the only effect of this insertion was a reproducible delay in flowering (Figure 3A). This phenotype is consistent with the presence of regulatory floral meristem motifs in the NMT2 promoter and the significant induction of expression in flowers. It suggests that NMT2 is involved in pathways controlling the transition between the vegetative and floral stages and possibly in floral meristem differentiation. No data concerning the activity of NMT2 are available, and this enzyme accumulates in only small amounts. We investigated the effect of NMT2 overexpression in the wild-type background. Two transgenes were generated: NMT2 under control of the 35S promoter (transgene P35S:At NMT2 in Supplemental Figure 2B online) and NMT2 under control of the NMT1 promoter (PNMT1:AtNMT2 transgene in Supplemental Figure 2A online). The PNMT1:AtNMT2 transgene was similar to that used for complementation by NMT1 in the nmt1-1 line (see above). The PNMT1-driven NMT2 construct was expressed at similar levels to NMT1, whereas the P35S:AtNMT2 transgene resulted in levels of expression 10 times higher (Figure 3C). Following stable expression of the PNMT1:AtNMT2 construct in the wild type, developmental abnormalities were observed in the SAM and the tip of the cotyledons. No such abnormalities were observed with the P35S:AtNMT2 construct, consistent with ectopic expression of this promoter (see details below). This pattern of abnormalities is consistent with the expression pattern associated with the NMT1 promoter. These results also indicate that NMT2 is functional and involved in meristem activity or the determination of stem cell identity.
We investigated whether NMT2 could compensate for the absence of NMT1 in the nmt1-1 line. The nmt1-1 and nmt2-1 lines were crossed. The phenotype of the double homozygous line (nmt1-1-nmt2-1) was found to be identical to that of the nmt1-1 line. Thus, the late developmental arrest observed in the nmt1-1 line is not associated with NMT2 and must have been caused by specific target proteins for NMT1 activity. As the PNMT1:At NMT2 transgene caused an abnormal meristem phenotype, we investigated the capacity of various transgene constructs overexpressing NMT2, to levels similar to or higher than those for NMT1, to rescue the developmental defect of the nmt1-1 line. We inserted the P35S:At NMT2 and PNMT1:At NMT2 transgenes into the nmt1-1 background (Figure 3E). Unlike the control transgenic line without NMT2 transgene-containing constructs, both lines overexpressing NMT2 had lateral roots or root tips initiated at the crown. Some of the lines also displayed secondary roots (Figure 3E).
Thus, NMT2 is a functional gene: its knockout leads to late flowering and its overexpression to developmental defects in meristematic zones. However, the activities of NMT2 and NMT1 do not overlap in terms of developmental arrest in the nmt1-1 line.
Reduced NMT1 Expression under Control of the Ethanol-Inducible AlcA Promoter in the nmt1-1 Background Leads to Abnormal Morphogenesis
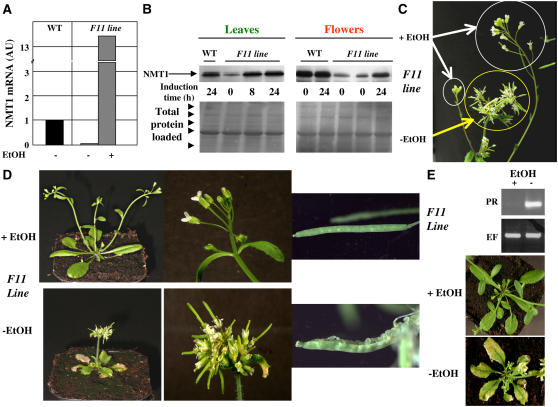
NMT1 inactivation is associated with strong SAM developmental defects and seedling growth arrest. This phenotype has made it impossible to assess the impact of MYR at later developmental stages in animals (Ntwasa et al., 2001; Yang et al., 2005). We investigated the role of MYR at later stages by constructing an inducible transgene in which NMT1 was placed under the control of the Aspergillus nidulans AlcA promoter and the AlcR gene was coexpressed under the control of the 35S promoter (Roslan et al., 2001). This construct (see Supplemental Figure 2C online) therefore results in the expression of NMT1 only in the presence of ethanol. Two transgenic lines were constructed, one in the wild-type background and the other in the nmt1-1 background. The lack of viability of the nmt1-1 line made it necessary to begin by establishing the transgene in a heterozygous NMT1/nmt1 line grown in the presence of ethanol. We obtained only a few lines able to complement the homozygous nmt1-1 line on soil in the presence of ethanol (experimental conditions for plant growth are shown in Supplemental Figure 3 online). This was probably due to the pattern of AlcR expression under control of the 35S promoter, which is only weakly active early in development (Sunilkumar et al., 2002). Full characterization of one line (F11) showed ethanol-dependent NMT1 gene induction (Figure 4A). In the absence of ethanol, this line had much lower than normal levels of NMT1 mRNA, 5% those in the wild type. Eight hours of treatment with ethanol was sufficient to induce overexpression. The NMT1 protein was induced similarly in flowers and leaves, with NMT1 levels in the absence of ethanol one-tenth those in its presence, consistent with quantitative PCR data. Ethanol induced the production of NMT1 protein to levels at least equivalent to those in the wild type (Figure 4B). The F11 line proved to be a useful tool for studying decreases in NMT1 levels during development. Ethanol could be used at any time to induce NMT1 expression or reversion.
Figure 4.
Further Developmental Defects Are Revealed with an NMT1 Ethanol-Inducible Line.
(A) Real-time PCR analysis of the level of NMT1 transcripts in wild-type or F11 plants grown in the presence or absence of ethanol (EtOH). This analysis was based on leaf mRNA. The standard deviation on each of the measurements was 4, 4, and 7% for the wild type, F11-ethanol, and F11+ethanol, respectively.
(B) Top: Immunoblot analysis of the amount of NMT1 in wild-type or F11 plants grown in the presence or absence of ethanol for the indicated time. The analysis was based on proteins from leaves and flowers. Bottom: Ponceau red staining of the corresponding gel sample confirming equal protein loading. The arrowheads indicate the position of the Mr marker (175, 94, 67, 46, and 30 kD). The major band corresponds to the large chain of ribulose-1,5-bisphosphate carboxylase/oxygenase.
(C) Reversal of the phenotype of a given seedling induced by ethanol. F11 seedlings were grown in the absence of ethanol for 2 weeks (yellow circle). NMT1 was induced in the presence of ethanol vapor for 1 week. The new flower shoots appearing as a result of this induction are circled in white.
(D) New phenotypes associated with the lack of NMT1 as discovered in F11 plants grown in the presence or absence of ethanol. Wild-type plants grown in the presence or absence of ethanol had phenotypes similar to that of F11 grown in the presence of ethanol.
(E) PR-1 transcripts are induced in the absence of ethanol in line F11. Top: RT-PCR analysis of PR-1 (PR) and EF-1 (EF) transcripts. A mixture of 10 distinct plants was used and only technical replicates performed in this case. Bottom: Phenotype of the corresponding flowers in plants with necrotic leaves.
When grown in the absence of ethanol (i.e., low NMT1 levels), the F11 line had a dwarf, bushy phenotype, with highly abnormal flower buds (Figure 4C). Fruit development was also impaired, as siliques opened too early and immature seeds were unable to develop further. Plants with low levels of NMT1 were virtually sterile. Interestingly, if initially grown in the absence of ethanol, with ethanol later added to the medium for a couple of weeks, new inflorescence stems arising from new floral meristems grew and looked normal (Figure 4D). This internal control on the same seedling provides strong evidence for an association between the observed phenotype and NMT1 expression. Finally, the leaves of the plant were yellow and necrotic when NMT1 levels were low (Figure 4E). PR-1 is a marker gene for stress response and aging in Arabidopsis (reviewed in van Loon et al., 2006). We found that PR-1 transcript levels were much higher in the F11 line than in the wild type (Figure 4E), indicative of a plant stress response.
We conclude that NMT1 is essential, not only for SAM development, but also at later developmental stages, for flower differentiation, fruit maturation, and stress response. These data are consistent with the dwarf phenotype induced by antisense RNA directed against At NMT1 (Qi et al., 2000).
Expression of Either of the Hs NMTs, Unlike That of Sc NMT, Compensates in Vivo for the Absence of At NMT1 in nmt1-1 Mutants
An updated search for NMT sequences in various data libraries and associated phylogenetic trees identified three main classes of NMT: those from protists (including yeast), plants, and animals (see Supplemental Figure 4 online). Higher eukaryotes often have two NMTs. There are therefore five classes of NMTs (one in protists, two in plants, and two in animals), reflecting variations in substrate specificity (see Maurer-Stroh et al., 2002b, 2002a). We hypothesized that complementation of the nmt1-1 knockout with NMTs from each of the five classes could be used to identify the protein subsets involved in each of the defects displayed by this mutant in early development and fruit and flower development.
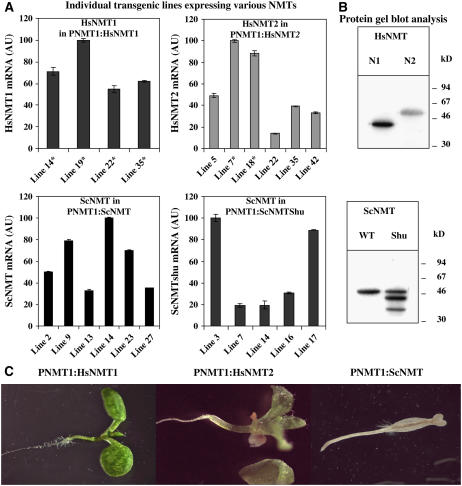
We constructed transgenes from the S. cerevisiae NMT (Sc NMT), H. sapiens NMT1 (Hs NMT1) and NMT2 (Hs NMT2), similar to those already generated for At NMT1 and At NMT2, to ensure that the data obtained were comparable with those for plant NMT genes. The cloning of the various cDNAs is described in Methods. The At NMT1 ORF was replaced with the orthologous NMT ORF of the PNMT1 family (i.e., with an NMT expressed under control of the promoter of At NMT1; see Supplemental Figure 2A online). The corresponding transgenes (PNMT1:Sc NMT, PNMT1:Hs NMT1, and PNMT1:Hs NMT2) were transferred to plants, and various transgenic plant lines were established in the heterozygous nmt1/NMT1 background. We assessed expression of the NMT transgene in each transgenic line. Variations (20 to 50 lines screened) between lines were observed, and mRNA levels were 10 to 500% those of the normal At NMT1 mRNA, consistent with normal levels of expression, as expected (Figure 5A). Expression to 5% the normal level for At NMT1 was sufficient to rescue the developmental phenotype of the SAM in line F11 (Figure 3). We then checked that each of the four orthologous NMT ORF mRNAs was efficiently translated in plant extracts (Figure 5B).
Figure 5.
Complementation with Orthologous NMT1s in the nmt1-1 Background.
(A) Real-time PCR analysis of the level of each NMT transcript in the wild type under control of the At NMT1 promoter. An asterisk indicates that the corresponding transgene complemented the nmt1-1 line. Values for individual lines generated in the nmt1/NMT1 background are reported. An arbitrary value (AU) of 100 was assigned to the highest value obtained with each NMT construct. The vertical bar indicates the sd.
(B) Immunoblot analysis showing the translation of each NMT in plant extracts. Proteins with an N-terminal fusion to the poly-His tag were produced in wheat germ extract, separated by electrophoresis, blotted, and detected with anti-His antibodies.
(C) Phenotypes observed in the nmt1-1 background (10 DAI).
Homozygous nmt1-1 plants expressing an orthologous NMT transgene were selected. The Hs NMT1 transgene systematically rescued the developmental defect of the nmt1-1 line (Figure 5C, left panel). All rescued lines appeared normal at later stages and were fertile. Rescue was more difficult to achieve with Hs NMT2 but was nonetheless observed (Figure 5C, center panel). Only transgenic lines with higher levels of Hs NMT2 mRNA, such as lines 7 and 18 (Figure 5A), showed sufficient complementation for the plant to be viable and fertile. We concluded that the two Hs NMTs displayed substrate specificity, facilitating the MYR of essential substrates of At NMT1, particularly those involved in SAM differentiation. By contrast, nmt1-1 rescue was not observed with the Sc NMT transgene (PNMT1:Sc NMT), regardless of the level of the corresponding mRNA (Figure 5A). We investigated whether optimization of the translational efficiency of this transgene could be used to make complementation possible. We constructed another transgene, with codon preference adjusted to match that of Arabidopsis more closely (Kliman and Henry, 2005). This transgene encoded the same protein, but the nucleotide sequence of the ORF was modified so as to improve gene codon usage. This Sc NMT transgene was identical for 1051 of the 1362 nucleotides of the ORF (77% identity; see Supplemental Methods online). Nevertheless, the corresponding synthetic transgene (PNMT1:Sc NMT-Shu), like the original sequence, was unable to rescue the nmt1-1 line. Indeed, translation efficiency was slightly lower with this construct (Figure 5B), indicating that translation with the wild-type sequence was not a limiting factor.
Thus, unlike the two Hs NMT genes, Sc NMT cannot complement the SAM defect in the nmt1-1 line. By contrast, we and others have previously reported that both At NMT1 and Hs NMT1 complement the heat-sensitive yeast Sc NMT nmt1-181 mutant (Duronio et al., 1992; Boisson et al., 2003).
In Vitro Assays of the Myristoylation of Protein Candidates Reveal That a Small Subset of Myristoylome Proteins Is Involved in the Early Embryo Defect of the nmt1-1 Line
We investigated the cause of the lack of complementation by Sc NMT by purifying all four NMTs used for in vivo complementation experiments to homogeneity. All four NMTs proved to be active against a set of myristoylatable peptides originating from Arabidopsis (Table 1; Table 4 in Boisson et al., 2003; Boisson and Meinnel, 2003). With the reference substrates (derived from the SOS3 calcium sensor or the GPA1 G-protein; Table 1), the NMT with the highest specific activity was Sc NMT, as shown by its kcat/Km value. For this crucial parameter for in vivo MYR (see discussion in Boisson and Meinnel, 2003), the NMTs were ranked in the following order Sc NMT > At NMT1 > Hs NMT1s > Hs NMT2 (Table 1). Thus, taking into account the similar levels of expression of all the corresponding genes (as shown in Figure 5B), the absence of complementation by Sc NMT was not due to low catalytic efficiency. By contrast, the need for higher levels of Hs NMT2 for complementation is entirely consistent with the significantly lower catalytic efficiency of Hs NMT2 than of At NMT1 (Table 1).
Table 1.
In Vitro Myristoylation of Various Peptides by Several NMTs
| Predicted Protein Functionb | At NMT1
|
Hs NMT1
|
Hs NMT2
|
Sc NMT1
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TAIR Entrya | N-Terminal Sequence | Km (mM) | kcat (s−1) | Relative kcat/Kmc | Km (mM) | kcat (s−1) | Relative kcat/Kmc | Km (mM) | kcat (s−1) | Relative kcat/Kmc | Km (mM) | kcat (s−1) | Relative kcat/Kmc | |
| At5g2470 | GCSVSKKK | SOS3 calcium sensor | 0.043 ± 0.01 | 0.51 ± 0.03 | 100 | 0.06 ± 0.02 | 0.37 ± 0.04 | 56 | 0.027 ± 0.006 | 0.06 ± 0.01 | 18 | 0.015 ± 0.007 | 0.65 ± 0.03 | 367 |
| G2A variant | ACSVSKKK | SOS3 (G2A variant) | nm | nm | <0.1 | nm | nm | <0.1 | nm | nm | <0.1 | nm | nm | <0.1 |
| At2g26300 | GLLCSRSRR | GPA1 α- subunit | 0.5 ± 0.1 | 1.7 ± 0.2 | 29 | 0.18 ± 0.01 | 0.86 ± 0.01 | 40 | 0.16 ± 0.01 | 0.10 ± 0.01 | 6 | 0.04 ± 0.01 | 0.99 ± 0.05 | 213 |
| At4g33400 | GASHSHED | DEM1 | 0.5 ± 0.1 | 0.7 ± 0.1 | 12 | >0.3 | >0.13 | 4 | >0.3 | >0.014 | 0.3 | nm | nm | <0.1 |
| At3g19240 | GTSQSRED | DEM2 | 0.18 ± 0.03 | 0.38 ± 0.03 | 18 | 0.15 ± 0.02 | 0.20 ± 0.01 | 11 | 0.39 ± 0.03 | 0.06 ± 0.01 | 1.2 | nm | nm | <0.1 |
| At2g07180 | GICFSAED | Protein kinase APK1-related | 0.09 ± 0.02 | 0.28 ± 0.02 | 26 | 0.14 ± 0.01 | 0.16 ± 0.01 | 9 | 0.038 ± 0.016 | 0.010 ± 0.001 | 3 | nm | nm | <0.1 |
| At5g01020 | GNCGTRDE | Protein kinase APK1-related | 0.39 ± 0.03 | 0.05 ± 0.01 | 1 | nm | nm | <0.1 | nm | nm | <0.1 | nm | nm | <0.1 |
| At5g21170 | GNANGKDED | AKINβ1 subunit of SnRK1 | 0.99 ± 0.18 | 0.7 ± 0.1 | 6 | 0.28 ± 0.04 | 0.15 ± 0.01 | 4 | >0.3 | >0.02 | 0.9 | nm | nm | <0.1 |
| At4g16360 | GNVNAREE | AKINβ2 subunit of SnRK1 | 0.64 ± 0.17 | 0.70 ± 0.03 | 9 | >0.3 | >0.11 | 4 | >0.3 | >0.022 | 0.7 | nm | nm | <0.1 |
| At3g01650 | GGGNSKEE | Copine-related | >0.3 | >0.20 | 7 | >0.3 | >0.142 | 4 | >0.3 | >0.018 | 0.7 | nm | nm | <0.1 |
| At4g13540 | GGSTSKDE | Unknown | >0.1 | >0.09 | 4 | >0.1 | >0.002 | 2 | >0.1 | >0.002 | 0.1 | nm | nm | <0.1 |
| At5g39590 | GASSSTDD | Unknown | 0.10 ± 0.02 | 0.010 ± 0.001 | 1 | nm | nm | <0.1 | nm | nm | <0.1 | nm | nm | <0.1 |
| At5g56460 | GNCWCRFE | Protein kinase APK1-related | 0.011 ± 0.002 | 0.34 ± 0.01 | 263 | 0.018 ± 0.003 | 0.21 ± 0.01 | 95 | 0.012 ± 0.006 | 0.060 ± 0.001 | 43 | >0.6 | >0.09 | 2 |
| At1g67800 | GGSSSKES | Copine-related | 0.005 ± 0.001 | 0.10 ± 0.01 | 164 | 0.024 ± 0.008 | 0.11 ± 0.02 | 39 | 0.032 ± 0.003 | 0.06 ± 0.01 | 16 | >0.3 | >0.06 | 2 |
The N-terminal octapeptide sequence was derived from several target proteins of interest (see text and http://www.isv.cnrs-gif.fr/tm/maturation/myristoylome2007am.htm).
The functional annotation associated with each entry was retrieved from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/index.jsp).
A value of 100 was assigned to the catalytic efficiency (kcat/Km) of myristoylation of the reference peptide SOS3 by At NMT1 (see sequence and original data in Boisson and Meinnel, 2003). Standard deviations were systematically <10% of the determined value. The G2A change in any peptide results in no reaction with NMTs and was used as a control. G2 is considered to be residue 2 because it must be unmasked by NME-dependent Met1 removal. A value below 0.1 (<0.1) was considered negative for MYR (see Boisson et al., 2003). Values between 0.1 and 0.3 are in the so-called twilight zone (Boisson et al., 2003). nm, nonmeasurable, indicating a complete absence of signal even at the highest NMT concentration used.
The substrate specificity of Sc NMT has been described elsewhere (Boisson et al., 2003; Boisson and Meinnel, 2003), building on the detailed, pioneering work of J.I. Gordon and colleagues (reviewed in Bhatnagar et al., 2001). Our data are entirely consistent with published results (Maurer-Stroh et al., 2002a, 2002b). The main difference between Sc NMT and At NMT1 is the more restricted substrate specificity of Sc NMT due to the inability of this enzyme to acylate protein substrates with acidic residues at both positions 8 and 9. We built on our previous studies of the proteome of proteins undergoing MYR in Arabidopsis (422 substrates in the December 2006 version; see http://www.isv.cnrs-gif.fr/tm/maturation/myristoylome2007am.htm). We selected all proteins with acidic residues (i.e., Asp or Glu) at both positions 8 and 9. Nine proteins were identified. Overall, the peptides derived from the N termini of these candidates showed no significant sequence bias with respect to the complete set of 422 sequences, other than the two acidic residues at positions 8 and 9 (Table 1). We assessed the acylation (i.e., MYR) of the corresponding peptides by purified At NMT1, Sc NMT, Hs NMT1, and Hs NMT2 (Table 1). At NMT1 acylated all nine peptides. Hs NMT1 modified only seven peptides of the subset, consistent with the partial overlap of substrate specificity between NMTs from plants and animals. Hs NMT2 acylated the same set, but At4g13540 showed only borderline modification, suggesting that it is probably not acylated in vivo. Finally, none of the nine peptides was acylated by Sc NMT, as expected.
These data indicate that the MYR of only six of the initial nine protein candidates is required in vivo to rescue the developmental phenotype of the nmt1-1 line. These proteins include two noncatalytic β-subunits of the SNF1-related (SnRK1) heterotrimeric kinase (At5g21170, AKINβ1; At4g16360, AKINβ2). SnRK1 is known to regulate various stress responses and glucose metabolism in plants. This finding is consistent with (1) the hypersensitivity of the nmt1-1 line to glucose and (2) the crucial involvement of MYR in the yeast and mammalian systems homologous to SnRK1, SNF1, and AMPK.
SnRK1 Relocalization Depends on Both the MYR State of the Two AKINβ Subunits and the Origin of the Expressed NMT
The two AKINβ subunits, β1 and β2, correspond to two of the three β-subunits of the heterotrimeric Ser/Thr kinase SNF1-related protein kinase (SnRK1) in plants (Polge and Thomas, 2007). The β3-subunit is not myristoylated (Gissot et al., 2004). SnRK1 is involved in regulating many global cellular responses, including glucose starvation, stress, sugar signaling, cell cycle control, and aging. In Arabidopsis, SnRK1 β-subunit levels increase between DAI 2 and DAI 8 (Bouly et al., 1999), and SnRK1 has been identified as an early marker of the SAM (Pien et al., 2001). The β-subunits are known to regulate the subcellular distribution of the SNF1 kinase in yeast and mammals (Vincent et al., 2001; Warden et al., 2001). SnRK1 is therefore a candidate for the MYR-sensitive protein involved in SAM development.
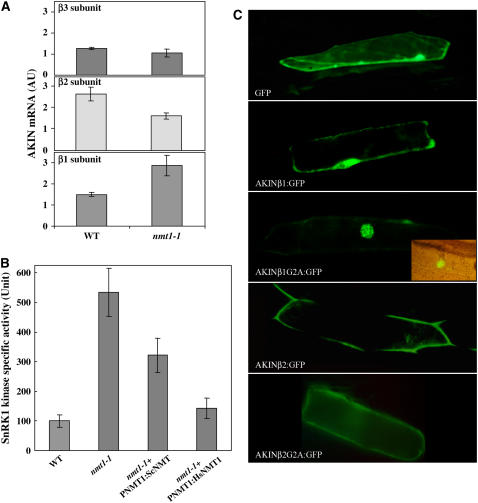
We investigated whether SnRK1 was a critical target by abolishing MYR. We assessed the expression of the three β-subunits of SnRK1 in both the nmt1-1 line and the wild type on DAI 3 (Figure 6A). AKINβ1 expression levels in the mutant nmt1-1 line were twice those in the wild type, whereas the β2- and β3-subunits were expressed slightly less strongly. We next measured the kinase activity associated with SnRK1 (Figure 6B). SnRK1-associated kinase activity was five times stronger in the nmt1-1 line than in the wild type on DAI 3. We finally investigated the impact of MYR on the subcellular distribution of the Arabidopsis AKINβ1 and -β2 subunits. The ORFs of the two cDNAs were fused N-terminally to the GFP sequence. The corresponding Gly2Ala variants were produced to inhibit MYR. The distribution of GFP obtained with each construct was observed in plant cells transfected with the constructs (Figure 6C). The fluorescence associated with the GFP of either the wild-type GFP and AKINβ:GFP fusions was mostly associated with the plasma membrane. Following MYR inhibition due to the presence of the Gly2Ala substitution, significant relocalization of the GFP fusions was observed, away from the plasma membrane toward (1) the nucleus for the β1-subunit and (2) the cytosol for the β2-subunit. This effect of the Gly2Ala substitution on the AKINβ1:GFP and AKINβ2:GFP fusions mimicked the impact of At NMT1 knockout on AKINβ1 and -β2 in the nmt1-1 line. MYR inhibition therefore leads to relocalization of the SnRK1 complex, driven by either or both of the β1- and β2-subunits from the plasma membrane to a soluble fraction, such as the nucleus or cytosol.
Figure 6.
Increased Kinase Activity and Relocalization of AKINβ1 upon MYR Inhibition.
(A) Real-time PCR analysis of the level of each AKINβ1, -β2, and -β3 transcript at DAI 3 in the wild type and nmt1-1. An arbitrary value of 1 corresponds to similar mRNA content in all three cases. The vertical bar indicates the sd.
(B) SnRK1 activity assay in various genetic backgrounds including the wild type, nmt1-1+PNMT1:Hs NMT1, nmt1-1+PNMT1:Sc NMT, and nmt1-1. Each measurement was started with soluble protein extracts prepared from seedlings at a developmental stage corresponding to DAI 3. Protein concentration was measured in each case, and the same amount of protein was added to each assay. Three biological replicates were performed, and the sd is shown in each case. The data correspond to those obtained with the AMARA peptide as the substrate. We obtained similar results with the SAMS peptide. Activity unit is expressed as nanomoles of radioactive phosphate incorporated per 45 min and per microgram of protein at 30°C. A value of 100 was assigned to the wild type.
(C) Fluorescence microscopy analysis of the expression of various GFP fusions in plant cells. From top to bottom, as indicated in each image: GFP (control), AKINβ1:GFP, AKINβ1[G2A]:GFP, AKINβ2:GFP, and AKINβ2[G2A]:GFP fusions. The inset in the AKINβ1[G2A]:GFP figure corresponds to the Nomarski image of the same cell, showing the nucleus.
The associated kinase activity, reflecting both the distribution and MYR state of SnRK1, was measured in the nmt1-1+PNMT1:Hs NMT1 and nmt1-1+PNMT1:Sc NMT backgrounds (Figure 6B). Hs NMT1 production in the nmt1-1 line restored kinase activity to wild-type levels. By contrast, Sc NMT production did not significantly decrease the high levels of SnRK1 kinase activity found in the nmt1-1 line. These data provide strong in vivo support for the results obtained in vitro (Table 1), defining the small number of critical targets of MYR, including both AKINβ subunits.
DISCUSSION
MYR is an important type of acylation, highly specific for proteins with an N-terminal Gly residue (Bhatnagar et al., 2001). Several hundreds of proteins are N-myristoylated in higher eukaryotes. MYR is required for embryonic development in animals (Ntwasa et al., 2001; Yang et al., 2005). This absolute requirement for MYR has made it impossible to determine the importance of this lipid modification for further development of the organism. We performed a complete functional characterization of the two NMT genes of Arabidopsis and provide a description of the impact of MYR not only during embryonic development but also at later stages of development. Using logic derived from trans-kingdom complementation experiments in the nmt1-1 background, and the in vitro substrate specificity of equivalent NMTs, we conclude that growth arrest is caused by a very limited subset of the 422 protein substrates of At NMT1 (http://www.isv.cnrs-gif.fr/tm/maturation/myristoylome2007am.htm) and that SnRK1 is a highly sensitive target.
NMT1 and NMT2 Both Are Functional Genes in Arabidopsis, and NMT1 Plays a Crucial Role during Seedling Development
We show here that there are two functional NMT genes in Arabidopsis. Such duplication was expected in an organism known to have undergone whole-genome duplication (Arabidopsis Genome Initiative, 2000). Nevertheless, several other dicotyledonous plants have at least two different NMT genes in their genome (see Supplemental Figure 4 online), showing that Arabidopsis is not unique among plants in having two NMT genes. Both NMT genes and the proteins they encode have particular expression characteristics or functions, as deduced from gene knockout experiments. Thus, although both NMTs play a role in plant development, NMT2 cannot compensate for NMT1, even when driven by the NMT1 promoter. However, this protein does induce several developmental phenotypes (Figure 4). This indicates that the two genes are functional but involved at different developmental stages and that they probably have different protein substrates. Closer examination of the amino acid sequence showed that At NMT2 was very similar to other NMTs from animals, yeast, and plants (see Supplemental Figure 4 online), suggesting that it probably acts as an NMT. Nevertheless, At NMT2 has several substitutions in motifs 2 to 4, which are involved in peptide substrate binding and acylation (shown in red in Figure 1C). This would probably result in NMT2 having a substrate specificity different from that of NMT1, potentially accounting for the data obtained. At this stage, we cannot exclude the possibility that NMT2 is a poorly active form that sequesters NMT1 substrates by binding to them, without modifying them. Unfortunately, our failure to obtain a highly purified form of this protein and our inability to model the type of substrate that could fit the peptide binding pocket of NMT2 made it impossible to test this hypothesis. This system, involving two NMT isozymes with unique functional roles, is reminiscent of that in mammals (Ducker et al., 2005; Yang et al., 2005). The Arabidopsis nmt1-1-nmt2-1 double mutant had a phenotype similar to that of the nmt1-1 line, indicating that NMT1 is the most important NMT gene during seedling development.
The NMT1 Null Mutant Displays Severe SAM Defects
We focused on the late embryonic developmental arrest of the nmt1-1 line, as this is the strongest, primary phenotype associated with a lack of MYR. In the absence of NMT1, the embryo develops normally to very late stages. The seed germinates and shows normal morphogenesis, with the exception of the SAM, but fails to develop beyond cotyledon development and greening, consistent with the expression pattern of NMT1 (Figures 1C to 1E). Before DAI 4, the nmt1-1 and wild-type lines could not be distinguished. Unlike eukaryotic protists and worms, in which NMT has an essential function (Duronio et al., 1989; Kamath et al., 2003; Price et al., 2003), higher eukaryotes have developed an NMT that is essential for embryonic development but not strictly required for cell viability. Similar phenotypes have been reported in animals, including (1) fruitflies depleted of their only NMT gene (Ntwasa et al., 2001), and (2) mice, in which the NMT1 gene was shown to be dispensable for cell viability but essential for embryonic development (Yang et al., 2005). The Arabidopsis nmt1-1 mutant line responds to synchronization and germination signals (humidity, cold, or heat) and further develops and grows with the setting of a normal root meristem. Thus, the embryo may develop until a particular stage, at which one or several key myristoylated proteins play an essential role in establishing the SAM. Alternatively, cells may divide but display subtle abnormalities due to the general lack of MYR of many proteins of the myristoylome, eventually leading to late embryonic development arrest. Our complementation experiments with Sc NMT indicate that this hypothesis can be excluded and that essential targets for establishment of the SAM correspond to only a very small fraction of the myristoylome.
The major defect in the line lacking NMT1 concerns SAM formation, with root meristem formation being normal (Figures 2D and 2E). Shoot and root development is arrested on DAI 4. Root meristem growth and lateral root development probably depend on SAM development and vice versa, accounting for the eventual cessation of root development in the nmt1-1 line. A number of SAM mutants have been described, but none has been reported to have a phenotype identical to that of the nmt1-1 line. These mutants usually correspond to genes such as WUSCHEL or SHOOT MERISTEMLESS, which are involved in transcriptional or posttranscriptional regulation. The SAM is under both hormonal and sugar/nutrient sensing control (reviewed in Carraro et al., 2006; Francis and Halford, 2006), consistent with the observed greater sensitivity to sugar of the nmt1-1 line than of the wild type.
NMT1 Is Required for SnRK1 Regulation
NMT1 plays a crucial role in plant development at different stages: establishment of the SAM, flower buds, fruit maturation, fertility, and cell defenses. The diversity of phenotypes observed during plant development suggests that different NMT1 target proteins block the activity of one or a family of essential proteins specifically expressed or essential at each of these essential development steps in the absence of MYR. Moreover, a number of myristoylome proteins are thought to be dispensable, because they belong to duplicated genes or gene families. In addition, several myristoylated proteins are known to be fully active even in the absence of MYR (Zhu et al., 1995; Tao et al., 2006), which does not seem to be essential for the functions of these proteins. Most of the genes encoding proteins of the N-myristoylome are induced by biotic or abiotic stresses (for example, see Ishitani et al., 2000), suggesting that these genes probably correspond to nonessential functions. In S. cerevisiae, in which MYR depends on genes essential for cell division, systematic gene inactivation experiments have shown most (73%) of the genes encoding myristoylated proteins (64) to be nonessential (Ashrafi et al., 1998). These data suggest that the genes involved in Arabidopsis SAM development probably correspond to only a small subset of the 422 proteins of the N-myristoylome.
Our approach, using in vivo complementation combined with in vitro acylation experiments with Sc NMT, Hs NMT1, and Hs NMT2 and in vivo validation (Figures 5 and 6) demonstrated that a small set of proteins triggers the phenotype of the nmt1-1 line. Two critical targets for the MYR-sensitive protein involved in SAM development were AKINβ subunits β1 and β2. AKIN (for AMP-dependent protein kinase) is the plant ortholog of the yeast SNF1 and the mammalian AMPK heterotrimeric complexes (for a review, see Polge and Thomas, 2007). AKIN is known as SnRK1 (for SNF1-related protein kinase 1) in plants. SnRK1 is a Ser/Thr kinase regulating global cellular responses to glucose starvation. SNF1 has several β-subunits, known as SIP1, SIP2, and GAL83 in yeast. AKINβ1 and -β2 are functional homologs of SIP1 and SIP2 in plants, as suggested by (1) their level of sequence identity and (2) the fact that both SIP1 and SIP2 also undergo MYR (Ashrafi et al., 1998; Hedbacker et al., 2004). Sip2 knockout increases SNF1 activity in yeast and specifically regulates aging (Ashrafi et al., 2000). Sip2 is one of only 12 genes encoding essential myristoylated proteins in yeast (Ashrafi et al., 1998) and was the only candidate protein showing an overlap with the small number of functions identified here. It has also been shown that MYR-dependent trafficking from the plasma membrane to the nucleus of SNF1, driven by SIP2, induces histone kinase activation and chromatin structure remodeling (Lin et al., 2003). AKINβ1 appears to be homologous to SIP2, as it is found in the nucleus when its MYR is inhibited. SnRK1 signal transduction mechanisms are extremely complex, respond to many signals, and remain poorly understood in plants (for reviews, see Rolland et al., 2002, 2006; Francis and Halford, 2006; Polge and Thomas, 2007). Our data suggest that a MYR defect of the AKINβ subunit of SnRK1 would lead to incorrect sugar signaling, preventing SAM differentiation, and contributing to the developmental arrest observed in the nmt1-1 line. Thus, whatever the downstream pathway regulated, the myristoylation status of SnRK1 probably plays a major role in SAM development in plants. Our characterization of the impact of β-subunit MYR on the SnRK1 pathway suggests that MYR of the β1-subunit negatively regulates nuclear SnRK1 activity by sequestering the complex at the plasma membrane. The loss of MYR results in relocalization of the complex to the nucleus, as reported for SIP2 in yeast (Lin et al., 2003). Even though the nature of the developmental phenotype associated with the nmt1-1 line clearly deserves further investigation, SnRK1 relocalization in the nucleus alone could well account for this phenomenon.
Concluding Remarks
Covalent fatty acylation has received considerable attention in recent years because of its involvement in essential biological processes, many of which are now considered excellent targets for the treatment of diseases. Proteomics and bioinformatic tools have recently advanced the study of target proteins. We recently showed that 422 proteins of the Arabidopsis proteome are myristoylated. We show here that MYR, like prenylation, is essential for various plant development processes and that a very small number of proteins of the N-myristoylome directly link MYR with one of these processes: SnRK1 kinase dysfunction and SAM development. In the filamentous fungus A. nidulans, a point mutation in an NMT gene reducing the affinity of myristoyl-CoA for NMT induces aberrant cell polarity (Shaw et al., 2002). Recently, a target of this mechanism during cell morphogenesis was proposed to correspond to the 26S proteosome (Lee and Shaw, 2007). This approach to MYR function also identified a unique target in the myristoylome as potentially involved in a specific process involving lipid modification. In this context, it is interesting to note that SnRK1 probably associates with the 26S proteasome in Arabidopsis (Rolland et al., 2006).
These data suggest that, for each pathway involving MYR, only a small proportion of the myristoylated proteins are likely to be directly involved. We intend to test this hypothesis by identifying primary targets in processes involving MYR.
METHODS
Materials
Seeds of Arabidopsis thaliana ecotypes Wassilewskija (Ws-4), Landsberg erecta (Ler), and Columbia were surface-sterilized and sown on 0.5× Murashige and Skoog (Sigma-Aldrich) medium supplemented with 0.8% agarose and 1% sucrose in Petri dishes. Seeds were imbibed for 2 d in the dark at 4°C for vernalization. Petri dishes were transferred to a growth chamber (21°C, 16 h of daylight; light intensity 100 μE·m−2·s−1) for up to 5 weeks. Plant growth kinetics were assessed as a function of days after imbibition, corresponding to the number of days of incubation in the growth chamber at 21°C. Arabidopsis lines were propagated under greenhouse conditions, as previously described (Giglione et al., 2000). We followed the recommendations of Meinke and Koornneef (1997) for naming knockout plant lines.
All chemicals were purchased from Sigma-Aldrich. Stock Myr-CoA solutions (200 μM) were prepared in 1% Triton X-100 in 10 mM sodium acetate buffer, pH 5.6. Peptides were custom-synthesized by Genscript with the exception of AMARA (Millipore). All peptides were >95% pure and were dissolved in water at a final concentration of 4 mM. Restriction enzymes and T4 DNA ligase were purchased from New England BioLabs. The buffers used were those recommended by the manufacturer. Oligonucleotides were synthesized by MWG-Biotech. Nucleotide sequences were determined by the Big-Dye Terminator V3 method, with a 16-capillary ABI PRISM 3100 genetic analyzer (PE-Applied Biosystems), using DNA templates purified with the Nucleospin plasmid (Macherey-Nagel) or the QIAquick gel extraction kit (Qiagen). PCR was performed with Eurobiotaq II (Eurobio) on an Omn-E thermal cycler (Hybaid), unless otherwise stated. All PCR-derived fragments for cloning were amplified with Pfu DNA polymerase (Stratagene) and sequenced.
Cloning
The cloning of the complete cDNAs for At NMT1 and At NMT2 has been described elsewhere (Boisson et al., 2003), and the sequences of these clones are available from GenBank under accession numbers AF250956 and AF250957, respectively. We obtained pBB131, which encodes Saccharomyces cerevisiae NMT (Sc NMT), from Jeffrey I. Gordon. The cDNAs for both NMTs of Homo sapiens (Hs NMT1 and Hs NMT2) were cloned by rapid amplification of cDNA ends from a human fetus RACE library (Clontech). We cloned the longest cDNAs corresponding to GenBank entries AF020500 (Hs NMT1) and AF043325 (Hs NMT2) (Glover et al., 1997; Giang and Cravatt, 1998). Among the three described length variants of Hs NMT1 (Duronio et al., 1992; Glover et al., 1997; McIlhinney et al., 1998), we were able to distinguish the shortest form (Hs NMT1s) as the main form, with smaller amounts of the longest form (Hs NMT1l), whereas the medium-length form (Hs NMT1m) was undetectable, as previously reported (McIlhinney et al., 1998). We deduced that the medium-length form probably originated from a truncated rather than corresponding to a true form. We chose to work only with the major form, Hs NMT1s, which has been described and fully characterized (Duronio et al., 1992), unlike Hs NMT1l. The Hs NMT1s form has also been demonstrated to complement the heat-sensitive nmt yeast derivative (nmt-181). Hs NMT1s translation starts 80 amino acids downstream from that of Hs NMT1l (see arrow in Figure 1B). The Sc NMT nucleotide sequence, adapted to respect the codon usage of Arabidopsis, was custom-synthesized by Epoch-Biolabs using the most likely codon for each amino acid. The Sc NMT protein sequence (456 amino acids) was reverse-translated to give a 1365-nucleotide sequence with the most probable codon usage for Arabidopsis (see http://www.kazusa.or.jp/codon/). In total, 311 of the 1365 bases were changed, improving the mean codon usage value significantly from 0.397 to 0.464. The nucleotide sequence of the reshuffled Sc NMT sequence is available in the Supplemental Methods online.
GFP fusions were constructed in vectors pSmGFP and pSmRSGFP expressing GFP under the control of the 35S promoter, as previously described (Giglione et al., 2000). AKINβ1 and -β2 cDNAs (kindly provided by Martine Thomas) were cloned in frame with the gfp gene between the single XbaI and BamHI sites to generate N-terminal protein fusions with GFP. Gly-to-Ala (G2A) codon substitutions at position 2 were created to inhibit MYR. Oligonucleotide sequences are available in the Supplemental Methods online.
T-DNA Mutant Screening
The collection of T-DNA mutants screened for an insertion in At NMT1 (encoding At5g57020) was generated in the Ws-4 ecotype at the Institut National de la Recherche Agronomique (Bouchez and Hofte, 1998). In total, 36,864 genomic DNA extracts from T-DNA–tagged lines, grouped into 48 hyperpools, were tested by PCR, as described elsewhere (Geelen et al., 2000), with a combination of four At NMT1 primers (sequences available from the authors on request) and four T-DNA–specific primers to cover the At NMT1 locus. Insertion in At NMT2 (encoding At2g44170) was identified after in silico screening at http://signal.salk.edu/cgi-bin/tdnaexpress. The nmt2-1 (GT_5_74071) line was obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk). This line originated from a project (http://www.jic.bbsrc.ac.uk/science/cdb/exotic/index.htm) in which gene trap transposon tagging was used to generate insertions in the genome of the Arabidopsis Ler ecotype (Sundaresan et al., 1995). Detailed information on the constructs, primers, and screening methods used is available (http://www.jic.bbsrc.ac.uk/science/cdb/exotic/Exotic_Handbook.pdf). Screenings for At NMT2 insertion mutants were performed at the Arabidopsis Knockout Facility as previously described (Sussman et al., 2000) using the screening protocol available from http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/. DNA pools from libraries of Arabidopsis T-DNA insertion mutants were obtained from the ABRC.
Segregation Analysis
For segregation analysis and the selection of plants homozygous or heterozygous for the T-DNA or transposon insertion, the germination medium was supplemented with 5 μg/mL of 5-phosphinotricine (Sigma-Aldrich), or seedlings on compost were sprayed with 1 mL/L of BASTA herbicide (AgrEvo France), or with 50 μg/mL of hygromycin (Sigma-Aldrich) or 50 μg/mL of kanamycin. Most constructs were transferred in binary vectors derived from the pCAMBIA series: pCAMBIA1390 and pCAMBIA1305.1 (Hellens et al., 2000). Details concerning the cloning strategies used are provided in the Supplemental Methods online. GUS fusions were made in pPR97 (kindly provided by Szabados et al., 1995) for studies of the At NMT1 promoter and pCAMBIA1305.1 for the At NMT2 promoter. For ethanol-inducible At NMT1, the cloning vectors were pSRN4 and pGPTV (Roslan et al., 2001). Constructs were used for the electrotransformation of Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993), which was then used to transform Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Transgenic plants (T1) were generated by germinating seeds and were subjected to selection in vitro in the presence of the herbicide or an antibiotic. Resistant plants were transferred to soil at the two-leaf stage and grown in the greenhouse.
Quantitative Real-Time PCR
Total RNA was extracted from Arabidopsis seedlings, organs, or cell suspensions as previously described (Kay et al., 1987). Poly(A)+ RNA was isolated on oligo(dT) 25 Dynabeads (Dynal). We subjected 200 ng of mRNA to reverse transcription. The cDNA (∼2 ng) was then amplified by PCR, using specific oligonucleotides. Real-time PCR was optimized with the LightCycler FastStart DNA Master SYBR Green kit (Roche) for each primer pair (amplification efficiency was always >90%), using a standard cDNA. Each cDNA sample was quantified at least twice (including two biological and two technical replicates), with respect to a standard cDNA quantified in the same conditions. All experiments were performed with a LightCycler, and the data were processed with LightCycler version 1.0 quantitative software (Roche). Oligonucleotide sequences are available in the Supplemental Methods online. The standard deviation [d(x/y)] corresponding to each x/y (e.g., NMT/actin) ratio was calculated for each value, using the following formula, where d(x) and d(y) correspond to the individual deviations of each x and y measurement, respectively: 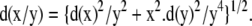
Microscopy
GUS staining was performed as previously described (Malamy and Benfey, 1997). Plant material was stained for 1 to 3 d and observed under the stereomicroscope (Leica MZ FLIII). For the analysis of apical meristems, seedlings were fixed 5 DAI, dehydrated as previously described (Beeckman and Viane, 1999), and embedded in Historesin, according to the manufacturer's instructions (Historesin embedding kit; Leica). We cut 1-μm serial sections on an ultramicrotome (LKB Ultrotome III). These sections were stained with 0.5% toluidine blue and observed under a REICHERT Polyvar microscope. FM464 staining was analyzed by confocal microscopy with a Leica-spz upright microscope (laser beam He/Ne 543 nm). For GFP studies, we bombarded onion epidermal cells with DNA constructs using the PDS-1000/He instrument (Bio-Rad) as previously described (Giglione et al., 2000). Transient GFP production was examined with a REICHERT B Polyvar fluorescence microscope equipped with a binocular CCD camera (Wild).
Biochemistry
All NMT ORFs were inserted into pET16b (Novagen) as N-terminal fusions with a 6xHis tag as previously described (Boisson et al., 2003). NMT was produced in Escherichia coli by transforming BL21-pRares (Rosetta; Novagen) cells with an appropriate plasmid. Cells were grown at 22°C for 6 h in 2YT medium supplemented with 50 μg/mL of ampicillin and 34 μg/mL of chloramphenicol to an OD600 of 0.9. They were then induced with 0.4 mM isopropylthio-β-galactoside and incubated for another 12 h (Boisson et al., 2003). In all cases, cells were harvested by centrifugation and resuspended in 10 to 20 mL of buffer A (20 mM sodium phosphate buffer, pH 7.3, and 500 mM NaCl plus 10 mM 2-mercaptoethanol). Samples were subjected to sonication, and cell debris was removed by centrifugation. The supernatant (5 to 15 mL) was applied to a Hi-Trap chelating HP nickel affinity column (0.7 × 2.5 cm; GE Healthcare) equilibrated in buffer A. Elution was performed at a flow rate of 0.5 mL/min in two steps: buffer B (buffer A plus 0.5 M imidazole) followed by a linear 0.35 mM/min imidazole gradient. The pool of purified protein (5 mL) was first dialyzed against buffer A for 12 h and then against buffer A plus 55% glycerol for 24 h before storage at −20°C. In vitro–coupled transcription-translation was performed for each NMT gene inserted in pIVEX 1.4 WG using RTS-100 CECF wheat germ lysate kits (Roche). Protein concentration was determined with the Bio-Rad kit. BSA was used as the protein standard. NMT activity was assayed at 30°C by continuously monitoring the absorbance at 340 nm of NADH by coupling the reaction to pyruvate dehydrogenase activity (Boisson et al., 2003). The standard assay was performed in a final volume of 200 μL in quartz cuvettes with a 1-cm optical path. Changes in absorbance over time were followed using an Ultrospec-4000 spectrophotometer (AP Biotech) equipped with a temperature control unit and a six-position Peltier-heated cell changer. The reaction mixture contained 50 mM Tris, pH 8.0, 1 mM MgCl2, 0.193 mM EGTA, 0.32 mM DTT, 0.2 mM thiamine pyrophosphate, 2 mM pyruvate, 0.1 mg/mL of BSA, 0.1% Triton X-100, 5 to 1000 μM peptide, 2.5 mM NAD+, and 0.125 units/mL of porcine heart PDH (0.33 units/mg). The kinetic parameters kcat and Km were obtained with Enzyme Kinetics module 1.2 of Sigma Plot (version 9.0) by nonlinear Michaelis-Menten equation fitting. Confidence limits for the data set are given. The kinetic parameter kcat/Km was obtained by iterative nonlinear least square fits of the Michaelis-Menten equation using the experimental data (Dardel, 1994). Confidence limits for the fitted kcat/Km values were determined by 100 Monte Carlo iterations, using the experimental standard deviations on individual measurements.
The SnRK1 complex was recovered after 40% saturation ammonium sulfate precipitation of the soluble proteins of a plant extract (Radchuk et al., 2006). SnRK1 kinase activity was assayed in the presence of [γ-32P]ATP as previously described (Radchuk et al., 2006) using 200 μM of two selective peptides: AMARA (AMARAASAAALARRR) or SAMS (HMRSAMSGLHLVKRR) (Sugden et al., 1999). Activity is expressed as nanomoles of radioactive phosphate incorporated into peptide for 45 min and per microgram of proteins at 30°C. Protein concentration was determined with the Bio-Rad kit.
Rabbit antisera against purified At NMT1 were produced at Eurogentec and purified before use by differential ammonium sulfate precipitation and diethylaminoethyl chromatography. Antibodies specific for At NMT2 were produced in chickens using a specific peptide derived from the N terminus of the amino acid sequence (3-17, DPKLPVEDALVTVAC; see alignment with At NMT1 in Figure 1C) at US-Biological. Rabbit antibodies against the Hs NMTs were generated with two peptides (RAMELLSACQGPAR and TSHGAIEPDKDNVR) at Production d'Anticorps et Services. Antibodies against Sc NMT were obtained from J.I. Gordon. Polyacrylamide gel electrophoresis in SDS-containing denaturing gels (0.75-mm-thick 10% polyacrylamide gels) was performed using the Mini-PROTEAN III system (Bio-Rad). Gels were stained with Bio-Safe Coomassie Brilliant Blue stain (Bio-Rad) and blotted onto membranes, which were probed with anti-His antibodies (GE Healthcare) as previously described (Giglione et al., 2000). Immunoblots were probed with anti-NMT antibodies (1/1000 to 3000), mouse anti-His-tag antibodies (dilution 1/2000), and peroxidase-conjugated anti-mouse antibodies from sheep or donkey (dilution 1/5000) and developed with ECL detection reagents (GE Healthcare). Membranes were placed against x-ray films for signal detection (Kodak).
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes analyzed in this study are At5g57020 (NMT1) and At2g44170 (NMT2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. T-DNA Insertion in Line nmt1-1 Causes BAR:AtNMT1 Transcriptional Fusions and At NMT1 Overexpression.
Supplemental Figure 2. Transgenes Used in This Study.
Supplemental Figure 3. Sensitization of Arabidopsis Seedlings to Ethanol Vapor.
Supplemental Figure 4. The Phylogenetic Tree of NMT Sequences Suggests That There Are Three Main NMT Families.
Supplemental Methods. Transgene Construct Cloning Strategies, the Shuffled Sc NMT Sequence, and a List of Oligonucleotide Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank J.I. Gordon (Washington University School of Medicine, St. Louis, MO), M. Thomas (Univ Paris-Sud, Orsay, France), and P. Ratet (Centre National de la Recherche Scientifique) for generously providing living and/or molecular material. We thank Marie-Noelle Soler (Institut Fédératif de Recherche 87, Gif/Yvette, France) for expert microscopy support. We also thank F. Granier (Institut National de la Recherche Agronomique) for providing access to the Versailles T-DNA collection and A. Martinez (Unité Propre de Recherche 2355) for the updated version of the Arabidopsis N-myristoylome. This work was supported by Centre National de la Recherche Scientifique Grant PGP04-11, by Grants BCMS-275 and IMPB-022 (Fonds National de la Science, France), and by Grant 3603 from the Association pour la Recherche sur le Cancer (Villejuif, France). J.A.T. was supported by a postdoctoral fellowship from the Centre National de la Recherche Scientifique. B.B. held a studentship from Fondation pour la Recherche Médicale (France). The Imaging and Cell Biology facility of the Institut Fédératif de Recherche 87 (FR-W2251) “La plante et son environnement” is supported by Action de Soutien à la Technologie et la Recherche en Essonne, Conseil de l'Essonne (France).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Thierry Meinnel (thierry.meinnel@isv.cnrs-gif.fr).
Online version contains Web-only data.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Ashrafi, K., Farazi, T.A., and Gordon, J.I. (1998). A role for Saccharomyces cerevisiae fatty acid activation protein E in regulating N-myristoylation during entry into stationary phase. J. Biol. Chem. 273 25864–25874. [DOI] [PubMed] [Google Scholar]
- Ashrafi, K., Lin, S.S., Manchester, J.K., and Gordon, J.I. (2000). Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14 1872–1885. [PMC free article] [PubMed] [Google Scholar]
- Beeckman, T., and Viane, R. (1999). Embedding thin plant specimens for oriented sectioning. Biotech. Histochem. 75 23–26. [DOI] [PubMed] [Google Scholar]
- Bhatnagar, R.S., Ashrafi, K., Futterer, K., Waksman, G., and Gordon, J.I. (2001). Biology and enzymology of protein N-myristoylation. In The Enzymes, F. Tamanoi and D.S. Sigman, eds (San Diego, CA: Academic Press), pp. 241–286.
- Boisson, B., Giglione, C., and Meinnel, T. (2003). Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278 43418–43429. [DOI] [PubMed] [Google Scholar]
- Boisson, B., and Meinnel, T. (2003). A continuous assay of myristoyl-CoA:protein N-myristoyltransferase for proteomic analysis. Anal. Biochem. 322 116–123. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., and Hofte, H. (1998). Functional genomics in plants. Plant Physiol. 118 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly, J.P., Gissot, L., Lessard, P., Kreis, M., and Thomas, M. (1999). Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINalpha1, an SNF1-like protein kinase. Plant J. 18 541–550. [DOI] [PubMed] [Google Scholar]
- Boutin, J.A. (1997). Myristoylation. Cell. Signal. 9 15–35. [DOI] [PubMed] [Google Scholar]
- Bradshaw, R.A., Brickey, W.W., and Walker, K.W. (1998). N-terminal processing: The methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci. 23 263–267. [DOI] [PubMed] [Google Scholar]
- Carraro, N., Peaucelle, A., Laufs, P., and Traas, J. (2006). Cell differentiation and organ initiation at the shoot apical meristem. Plant Mol. Biol. 60 811–826. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dardel, F. (1994). MC-Fit: Using Monte-Carlo methods to get accurate confidence limits on enzyme parameters. Comput. Appl. Biosci. 10 273–275. [DOI] [PubMed] [Google Scholar]
- Devadas, B., Freeman, S.K., McWherter, C.A., Kishore, N.S., Lodge, J.K., Jackson-Machelski, E., Gordon, J.I., and Sikorski, J.A. (1998). Novel biologically active nonpeptidic inhibitors of myristoylCoA:protein N-myristoyltransferase. J. Med. Chem. 41 996–1000. [DOI] [PubMed] [Google Scholar]
- Ducker, C.E., Upson, J.J., French, K.J., and Smith, C.D. (2005). Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol. Cancer Res. 3 463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio, R.J., Reed, S.I., and Gordon, J.I. (1992). Mutations of human myristoyl-CoA:protein N-myristoyltransferase cause temperature-sensitive myristic acid auxotrophy in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89 4129–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio, R.J., Towler, D.A., Heuckeroth, R.O., and Gordon, J.I. (1989). Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science 243 796–800. [DOI] [PubMed] [Google Scholar]
- Francis, D., and Halford, N.G. (2006). Nutrient sensing in plant meristems. Plant Mol. Biol. 60 981–993. [DOI] [PubMed] [Google Scholar]
- Geelen, D., Lurin, C., Bouchez, D., Frachisse, J.M., Lelievre, F., Courtial, B., Barbier-Brygoo, H., and Maurel, C. (2000). Disruption of putative anion channel gene AtCLC-a in Arabidopsis suggests a role in the regulation of nitrate content. Plant J. 21 259–267. [DOI] [PubMed] [Google Scholar]
- Giang, D.K., and Cravatt, B.F. (1998). A second mammalian N-myristoyltransferase. J. Biol. Chem. 273 6595–6598. [DOI] [PubMed] [Google Scholar]
- Giglione, C., Serero, A., Pierre, M., Boisson, B., and Meinnel, T. (2000). Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 19 5916–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissot, L., Polge, C., Bouly, J.P., Lemaitre, T., Kreis, M., and Thomas, M. (2004). AKINbeta3, a plant specific SnRK1 protein, is lacking domains present in yeast and mammals non-catalytic beta-subunits. Plant Mol. Biol. 56 747–759. [DOI] [PubMed] [Google Scholar]
- Glover, C.J., Hartman, K.D., and Felsted, R.L. (1997). Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J. Biol. Chem. 272 28680–28689. Erratum. J. Biol. Chem. 273: 5988. [DOI] [PubMed] [Google Scholar]
- Hedbacker, K., Townley, R., and Carlson, M. (2004). Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24 1836–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R., Mullineaux, P., and Klee, H. (2000). Technical Focus: A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 5 446–451. [DOI] [PubMed] [Google Scholar]
- Hong, R.L., Hamaguchi, L., Busch, M.A., and Weigel, D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, E.E., Gelvin, S.B., Melchers, L.S., and Hoekema, A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2 208–218. [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.S., Shi, W., and Zhu, J.K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G., and Gibson, T.J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23 403–405. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237. [DOI] [PubMed] [Google Scholar]
- Kay, R., Chan, A., Daly, M., and McPherson, J. (1987). Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236 1299–1302. [DOI] [PubMed] [Google Scholar]
- Kliman, R.M., and Henry, A.N. (2005). Inference of codon preferences of Arabidopsis thaliana. Int. J. Plant Sci. 166 3–11. [Google Scholar]
- Lee, S.C., and Shaw, B.D. (2007). A novel interaction between N-myristoylation and the 26S proteasome during cell morphogenesis. Mol. Microbiol. 63 1039–1053. [DOI] [PubMed] [Google Scholar]
- Lin, S.S., Manchester, J.K., and Gordon, J.I. (2003). Sip2, an N-Myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J. Biol. Chem. 278 13390–13397. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh, S., Eisenhaber, B., and Eisenhaber, F. (2002. a). N-terminal N-myristoylation of proteins: Refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 317 523–540. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh, S., Eisenhaber, B., and Eisenhaber, F. (2002. b). N-terminal N-myristoylation of proteins: Prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 317 541–557. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh, S., Gouda, M., Novatchkova, M., Schleiffer, A., Schneider, G., Sirota, F.L., Wildpaner, M., Hayashi, N., and Eisenhaber, F. (2004). MYRbase: Analysis of genome-wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins. Genome Biol. 5: R21. [DOI] [PMC free article] [PubMed]
- McIlhinney, R.A., Young, K., Egerton, M., Camble, R., White, A., and Soloviev, M. (1998). Characterization of human and rat brain myristoyl-CoA:protein N-myristoyltransferase: Evidence for an alternative splice variant of the enzyme. Biochem. J. 333 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke, D., and Koornneef, M. (1997). Community standards for Arabidopsis genetics. Plant J. 12 247–253. [Google Scholar]
- Meinnel, T., Mechulam, Y., and Blanquet, S. (1993). Methionine as translation start signal: A review of the enzymes of the pathway in Escherichia coli. Biochimie 75 1061–1075. [DOI] [PubMed] [Google Scholar]
- Ntwasa, M., Aapies, S., Schiffmann, D.A., and Gay, N.J. (2001). Drosophila embryos lacking N-myristoyltransferase have multiple developmental defects. Exp. Cell Res. 262 134–144. [DOI] [PubMed] [Google Scholar]
- Pien, S., Wyrzykowska, J., and Fleming, A.J. (2001). Novel marker genes for early leaf development indicate spatial regulation of carbohydrate metabolism within the apical meristem. Plant J. 25 663–674. [DOI] [PubMed] [Google Scholar]
- Polge, C., and Thomas, M. (2007). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12 20–28. [DOI] [PubMed] [Google Scholar]
- Price, H.P., Menon, M.R., Panethymitaki, C., Goulding, D., McKean, P.G., and Smith, D.F. (2003). Myristoyl-CoA:protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J. Biol. Chem. 278 7206–7214. [DOI] [PubMed] [Google Scholar]
- Qi, Q., Rajala, R.V., Anderson, W., Jiang, C., Rozwadowski, K., Selvaraj, G., Sharma, R., and Datla, R. (2000). Molecular cloning, genomic organization, and biochemical characterization of myristoyl-CoA:protein N-myristoyltransferase from Arabidopsis thaliana. J. Biol. Chem. 275 9673–9683. [DOI] [PubMed] [Google Scholar]
- Radchuk, R., Radchuk, V., Weschke, W., Borisjuk, L., and Weber, H. (2006). Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 140 263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57 675–709. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14(Suppl): S185–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslan, H.A., Salter, M.G., Wood, C.D., White, M.R., Croft, K.P., Robson, F., Coupland, G., Doonan, J., Laufs, P., Tomsett, A.B., and Caddick, M.X. (2001). Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28 225–235. [DOI] [PubMed] [Google Scholar]
- Ross, S., Giglione, C., Pierre, M., Espagne, C., and Meinnel, T. (2005). Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 137 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, B.D., Momany, C., and Momany, M. (2002). Aspergillus nidulans swoF encodes an N-myristoyl transferase. Eukaryot. Cell 1 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden, C., Donaghy, P.G., Halford, N.G., and Hardie, D.G. (1999). Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 120 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Sunilkumar, G., Mohr, L., Lopata-Finch, E., Emani, C., and Rathore, K.S. (2002). Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol. Biol. 50 463–474. [DOI] [PubMed] [Google Scholar]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados, L., Charrier, B., Kondorosi, A., de Bruijn, F.J., and Ratet, P. (1995). New plant promoter and enhancer testing vectors. Mol. Breed. 1 419–423. [Google Scholar]
- Tao, J., Shumay, E., McLaughlin, S., Wang, H.Y., and Malbon, C.C. (2006). Regulation of AKAP-membrane interactions by calcium. J. Biol. Chem. 281 23932–23944. [DOI] [PubMed] [Google Scholar]
- Towler, D.A., Adams, S.P., Eubanks, S.R., Towery, D.S., Jackson-Machelski, E., Glaser, L., and Gordon, J.I. (1988). Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263 1784–1790. [PubMed] [Google Scholar]
- van Loon, L.C., Rep, M., and Pieterse, C.M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162. [DOI] [PubMed] [Google Scholar]
- Vincent, O., Townley, R., Kuchin, S., and Carlson, M. (2001). Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden, S.M., Richardson, C., O'Donnell, J., Jr., Stapleton, D., Kemp, B.E., and Witters, L.A. (2001). Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem. J. 354 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.H., Shrivastav, A., Kosinski, C., Sharma, R.K., Chen, M.H., Berthiaume, L.G., Peters, L.L., Chuang, P.T., Young, S.G., and Bergo, M.O. (2005). N-myristoyltransferase 1 is essential in early mouse development. J. Biol. Chem. 280 18990–18995. [DOI] [PubMed] [Google Scholar]
- Zhu, D., Cardenas, M.E., and Heitman, J. (1995). Myristoylation of calcineurin B is not required for function or interaction with immunophilin-immunosuppressant complexes in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270 24831–24838. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.