Abstract
Efforts to understand nuclear organization in plant cells have received little assistance from the better-studied animal nuclei, because plant proteomes do not contain recognizable counterparts to the key animal proteins involved in nuclear organization, such as lamin nuclear intermediate filament proteins. Previous studies identified a plant-specific insoluble nuclear protein in carrot (Daucus carota), called Nuclear Matrix Constituent Protein1 (NMCP1), which contains extensive coiled-coil domains and localizes to the nuclear periphery. Here, we describe a genetic characterization of two NMCP1-related nuclear proteins in Arabidopsis thaliana, LITTLE NUCLEI1 (LINC1) and LINC2. Disruption of either gene caused a reduction in nuclear size and altered nuclear morphology. Moreover, combining linc1 and linc2 mutations had an additive effect on nuclear size and morphology but a synergistic effect on chromocenter number (reduction) and whole-plant morphology (dwarfing). The reduction in nuclear size in the linc1 linc2 double mutant was not accompanied by a corresponding change in endopolyploidy. Rather, the density of DNA packaging at all endopolyploid levels in the linc1 linc2 mutants was increased significantly. Our results indicate that the LINC coiled-coil proteins are important determinants of plant nuclear structure.
INTRODUCTION
The nucleus is the most visible organelle of the eukaryotic cell and, in terms of function, arguably the most critical. In addition to harboring the chromosomes and supporting the coordinated expression of a host of genes, the nucleus also participates in the transport of a variety of macromolecules to and from the cytoplasm. The nucleus is surrounded by the nuclear envelope, which is composed of at least two key components: the inner and outer nuclear membranes and nuclear pore complexes (Hetzer et al., 2005). In addition, a mesh-like structure internal to the inner membrane, called the nuclear lamina, is present at the nuclear periphery in metazoans (Gruenbaum et al., 2005), although a distinct nuclear lamina has not been unequivocally identified in yeast or plants.
Nuclei are remarkably dynamic and differentiate into an array of shapes and sizes within a single organism. In the flowering plant Arabidopsis thaliana, differentiated nuclei are common and thus provide a good system to study this highly dynamic process. To get a sense of nuclear morphology variation, Galbraith and colleagues (Chytilova et al., 1999) used a nucleus-targeted green fluorescent protein to investigate the shapes and sizes of living nuclei from several Arabidopsis tissues and cell types via confocal microscopy. They noted that spherical nuclei—the most abundant type overall and the predominant nuclear shape in the meristems—were just one of many nuclear shapes found within the epidermal and pavement cells. Furthermore, spindle-shaped nuclei were found in differentiated root epidermal and cortex tissue, oriented along the long axis of the cell, and rod-like nuclei were located within vascular tissues. The largest nuclei were located in leaf hairs (trichomes), and some of the smallest were observed in guard cells. This survey and related studies in other plants indicate that nuclear morphology is diverse and is largely associated with particular tissue and cell types (Sheen et al., 1995; Chytilova et al., 2000). Very little is known about the proteins or pathways that lead to the observed diversity in nuclear morphology or about the mechanisms that link the differentiated state of a cell to changes in nuclear morphology.
DNA content is one factor influencing nuclear size and varies widely within a single plant due to endoreduplication, the process whereby DNA is replicated without an intervening mitosis (Galbraith et al., 1991). For example, Arabidopsis behaves genetically as a diploid, but vegetative adult tissues are composed of a mixture of cells with nuclei ranging in ploidy levels from 2C (where C = haploid genome complement) to 64C. The absence of nuclear division leads to larger endoreduplicated nuclei with expanded nuclear envelopes. Studies have documented a linear relationship between ploidy level (DNA content) and nuclear volume (Jovtchev et al., 2006), suggesting that there is a fixed amount of DNA that can be packaged per unit volume in the nucleus.
Another important consideration is the interplay among nuclear morphology, chromatin dynamics, and gene expression (Lanctot et al., 2007). The genomic era has provided an abundance of sequence information that is now being superimposed with profiles of epigenetic information, such as DNA methylation and histone modification, to yield rich epigenome maps that contextualize the primary sequence information (Zhang et al., 2006; Esteller, 2007; Vaughn et al., 2007; Zilberman et al., 2007). Other tiers of information, such as spatial and temporal information about chromosome position within the nucleus, must be integrated onto epigenomic maps. Some broad spatial patterns have emerged, including the packaging of heterochromatic sequences at the nuclear periphery and sequestration of the majority of euchromatin within the nuclear interior (Fransz et al., 2002; Fang and Spector, 2005). Relatively little is understood about the molecular determinants and pathways that organize nuclei.
To advance our understanding of the processes and components regulating nuclear morphology, we initiated a reverse genetic study in Arabidopsis on a protein family sharing similarity to a nuclear protein, Nuclear Matrix Constituent Protein1 (NMCP1), first identified in carrot (Daucus carota) (Masuda et al., 1997). This 134-kD carrot protein purifies with an insoluble nuclear fraction and localizes exclusively to the nuclear periphery (Masuda et al., 1997). Carrot NMCP1 contains a predicted coiled-coil domain, thought to enable dimerization as a first step toward higher order filament formation. Here, we describe a genetic characterization of related coiled-coil proteins in Arabidopsis. We found that disruption of two of the genes in this family, LITTLE NUCLEI1 (LINC1) and LINC2, causes a reduction in nuclear size. Moreover, linc1 linc2 double mutants show dramatic changes in nuclear morphology (more circular shape and reduced chromocenter number) and an abnormal whole-plant developmental phenotype. The reduction in nuclear size in linc1 linc2 mutants was accompanied by an altered spectrum of polyploid levels, but higher ploidy levels typical of wild-type plants (32C and 64C) were still observed. A correlation of nuclear size and fluorescence indicated that the DNA packaging density in the linc1 linc2 mutants is much higher than that observed in wild-type cells. We discuss the potential roles of the LINC proteins in plant nuclear morphology.
RESULTS
An Arabidopsis Gene Family Encoding Coiled-Coil Proteins Related to the Carrot Peripheral Nuclear Protein NMCP1
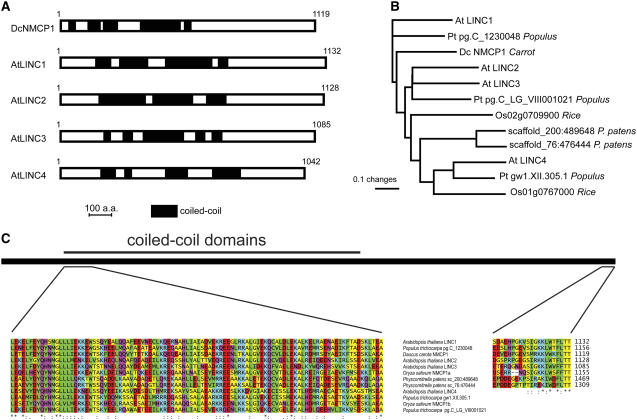
We undertook a reverse genetics study in Arabidopsis to study the functions of genes encoding proteins related to the carrot NMCP1 (Dc NMCP1) protein as part of a larger effort to dissect determinants of plant nuclear architecture. Amino acid similarity searches using Dc NMCP1 as a query against the Arabidopsis proteome revealed four highly related proteins. As shown in Figure 1A, all four Arabidopsis proteins, which we designated LINC1 to LINC4 (based on genetic phenotypes; see below), contain a large central coiled-coil region (indicated in black) and smaller N- and C-terminal domains (Meier, 2007). The C- and N-terminal domains have, on average, a lower level of amino acid similarity; however, there are motifs within the flanking domain that are highly conserved between all Dc NMCP1 homologs (e.g., the extreme C terminus; Figure 1C). A phylogenetic analysis including Dc NMCP1 homologs from Arabidopsis, Oryza sativa (rice), Populus trichocarpa (poplar), and Physcomitrella patens (moss) is shown in Figure 1B.
Figure 1.
The LINC Protein Family.
(A) The protein organization of Dc NMCP1 is compared with that of Arabidopsis LINC1 to LINC4. Putative coiled-coiled domains are shown in black, and the lengths of the proteins are shown (number of amino acids [a.a.]).
(B) An unrooted phylogenetic distance tree showing the probable relationships among 12 homologs of Dc NMCP1.
(C) Selected regions of the multiple sequence alignment used to construct the phylogenetic tree shown in (B) are detailed.
We consulted public genomics and proteomics resources to investigate LINC proteins and draw some initial functional inferences. First, we searched public microarray expression data sets using the tool sets found at the Botany Array Resource (http://bbc.botany.utoronto.ca/) for genes that are coexpressed (r > 0.75) with each of the four LINC genes based on tissue and developmental program specificity. A subset of genes (n = 91) was correlated with the expression of all four of the LINC genes. The majority of these proteins (79 of the 91) were assigned to a functional class, revealing a 2.5-fold overrepresentation of proteins in the cell cycle and DNA processing and transcription functional categories (see Supplemental Figure 1 online). Second, we looked for available information on the subcellular localization of LINC proteins. LINC4 has been identified as a plastid protein in a proteomic study of Arabidopsis chloroplasts (Kleffmann et al., 2006).The remaining LINC proteins are predicted by computation methods to localize to the nucleus, consistent with their similarity to Dc NMCP1, which is localized to the nucleus. More direct evidence for a nuclear location was obtained by the proteomic study of Pendle et al. (2005), which identified LINC3 as a nucleolar protein.
LINC1 Is Localized to the Nuclear Periphery and LINC2 Is Localized to the Nucleoplasm
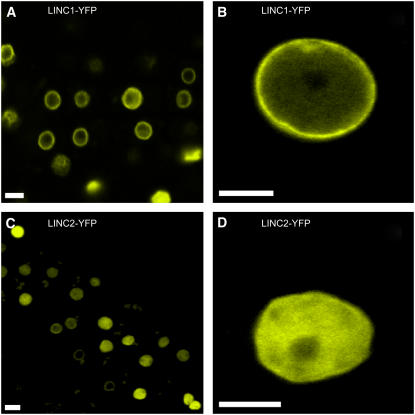
We began our characterization by determining the subcellular localization of LINC1 and LINC2 using a yellow fluorescent protein (YFP) reporter in transgenic plants. We fused a genomic fragment encompassing the entire predicted coding region of either LINC1 or LINC2 upstream of YFP within a plant transformation vector behind the constitutive cauliflower mosaic virus promoter, 35S. Each construct was introduced into both wild-type and linc1-1 linc2-1 mutant plants via Agrobacterium tumefaciens–mediated transformation. Selected T1 transformants (hemizygous) were self-pollinated, and T2 plants segregating the transgene were examined. Both the P35S:LINC1g-YFP and P35S:LINC2g-YFP transgenes complemented, to different degrees, the morphological phenotypes displayed by linc1-1 linc2-1 plants (see below), demonstrating that the transgenes are functional (see Supplemental Figure 2 online). Seven-day-old roots from seedlings of the T2 transgenic plants in either a wild-type or a linc1-1 linc2-1 background were examined by epifluorescence microscopy to determine the subcellular localization of LINC1-YFP and LINC2-YFP. All roots examined displayed a strong fluorescence signal found exclusively within the nucleus (confirmed using differential interference contrast optics or counterstaining with propidium iodide), but the pattern differed between LINC1 and LINC2. To obtain a more accurate determination of the localization pattern(s) of LINC1-YFP and LINC2-YFP, we performed laser scanning confocal microscopy on undifferentiated meristem and differentiated epidermal root nuclei. The results show that in all types of examined nuclei, LINC1-YFP is predominantly found concentrated at the nuclear periphery (Figure 2) and also, to a lesser extent, in the nucleoplasm excluding the nucleolus. The LINC2-YFP signal was primarily diffuse throughout the nucleoplasm (except the nucleolus) in all examined nuclei, although in a small percentage of nuclei LINC2-YFP was concentrated at the nuclear periphery (Figure 2).
Figure 2.
Nuclear Localization of LINC1 and LINC2.
Localization of LINC1-YFP and LINC2-YFP fusion proteins in transgenic linc1-1 linc2-1 Arabidopsis root cells expressing either a LINC1-YFP or LINC2-YFP transgene under the control of the constitutive 35S viral promoter.
(A) and (B) YFP signals from transgenic LINC1-YFP roots at the root meristem (A) and from a differentiated cell (B).
(C) and (D) YFP signals from transgenic LINC2-YFP roots from the meristem (C) and from a differentiated cell (D).
LINC1-YFP is localized predominantly at the nuclear periphery, and LINC2-YFP is localized diffusely throughout the nucleoplasm and is partially excluded from the nucleolus. Bars = 5 μm.
Genetic Analysis of linc Mutations
To study the function of LINC genes and their gene products, we undertook a reverse genetic approach. For this initial study, we chose to focus on LINC1 and LINC2, as these genes were most similar to each other and shared the most similarity with the prototypic Dc NMCP1. We anticipated that there might be functional redundancy between these LINC proteins; therefore, we set out to examine linc1 and linc2 single mutants as well as linc1 linc2 double mutant combinations. We focused on predicted loss-of-function or hypomorphic alleles containing large insertions of an engineered Agrobacterium T-DNA into the coding regions of the targeted gene. The positions of insertions and RT-PCR data demonstrating that the alleles caused a severe reduction in transcript accumulation are shown in Supplemental Figure 3 online. We intercrossed linc1-1 and linc2-1 homozygotes (in strain Columbia), creating an F1 linc1-1/+ linc2-1/+ plant that was self-pollinated to generate an F2 family, which was examined for developmental phenotypes and mutation segregation ratios. The observed progeny ratios did not deviate significantly from the expected segregation ratios (wild type:linc1-1:linc2-1:linc1-1 linc2-1, 14:12:9:8). None of the F2 plants displayed a developmental phenotype, with the notable exception of the eight linc1-1 linc2-1 double mutants, all of which exhibited a moderate dwarf and leaf-curling phenotype (Figure 3). The dwarfing phenotype was also observed in an independent linc1 linc2 double mutant line constructed using a different linc1 T-DNA insertional allele (linc1-2).
Figure 3.
Morphological Phenotypes of linc1 and linc2 Mutants.
Representative plants from an F2 family segregating linc1-1 and linc2-1. Genotypes are indicated above the plants. Only linc1-1 linc2-1 plants have an obvious whole-plant phenotype.
linc1 linc2 Mutants Have Reduced Cell Size but Endoreduplication Occurs
The switch from normal mitotic cell cycles to endocycles (endoreduplication) is a tightly regulated process coinciding with a decrease in the activity of cyclin-dependent kinases at M-phase (Sugimoto-Shirasu and Roberts, 2003). Mutations causing more or fewer endocycles lead to dramatic effects on whole-plant morphology, including syndromes similar to that displayed by linc1 linc2 double mutants. A variety of cell types, including epidermal and leaf hair (trichomes), depend on endoreduplication for normal cell size and morphology (Sugimoto-Shirasu et al., 2005). At maturity, trichomes are single cells that normally develop three to four branches and are sensitive to endoreduplication, such that extra or fewer endocycles are associated with overbranching or underbranching, respectively (Perazza et al., 1999). In addition, reduced endoreduplication is associated with leaves that have small epidermal cells. Thus, small epidermal cells or malformed trichome cells (underbranched or overbranched) is a straightforward proxy for identifying endoreduplication defects.
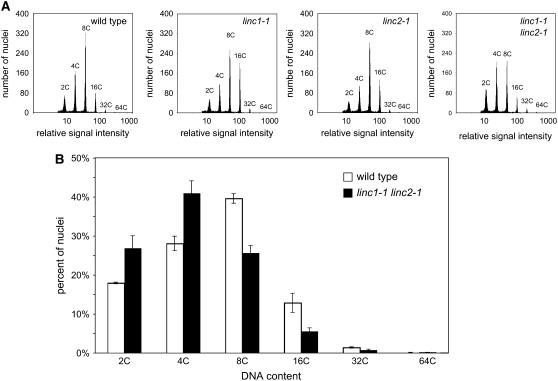
To determine whether such cell morphological changes were occurring in the linc mutants, we first investigated epidermal leaves of 4-week-old wild-type and linc1-1 linc2-1 plants by measuring epidermal cell size from agarose leaf impressions. We found that the epidermal cells of linc1-1 linc2-1 are approximately one-fourth the size of wild-type epidermal cells (see Supplemental Figure 4 online). Next, we examined the trichomes of linc1-1 linc2-1 plants and found that they developed normally (see Supplemental Figure 5 online), each typically having three to four branches, which indicates that the nuclei of these cells have normal ploidy levels. We then measured ploidy levels of the linc1 and linc2 mutants by flow cytometry to directly analyze nuclei isolated from 4-week-old leaf tissue. The data indicate (Figure 4A) that all of the linc mutants are capable of reaching the upper tiers of polyploidy (16C, 32C, and 64C) observed in wild-type Arabidopsis cells. No significant differences in endoreduplication were observed in either of the linc single mutants (Figure 4B), but the linc1-1 linc2-1 mutants showed a shift in the numbers of cells with 2C and 4C ploidy levels, away from the higher ploidy levels (8C, 16C, and 32C). Nonetheless, these shifts in ploidy, from an average of 7.3C to 5.5C, cannot account easily for the dramatic reduction in cell size seen in linc1-1 linc2-1 double mutants.
Figure 4.
Endopolyploidy Measurements in linc1 and linc2 Mutants.
Nuclear flow cytometry analysis of wild-type and linc mutant nuclei.
(A) Representative flow cytometry histogram plots of nuclei isolated from 4-week-old leaf tissue. From left are data for the wild type, linc1-1, linc2-1, and linc1-1 linc2-1.
(B) Aggregate data from individual histograms of independent wild-type (white columns; n = 4) and linc1-1 linc2-1 mutant (black columns; n = 12) samples.
Altered Nuclear Morphology Caused by linc Mutations
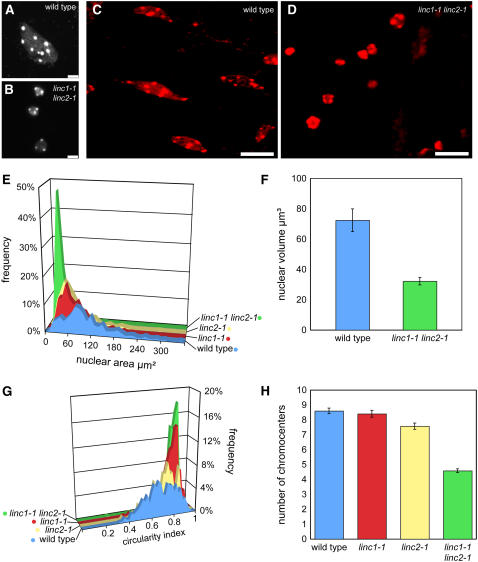
We extended our phenotypic characterization of the linc mutants by inspecting nuclear morphology. Figures 5A to 5D show representative nuclei from 2-week-old root epidermal cells and anther filament cells. Nuclei in linc1-1 linc2-1 mutants were noticeably smaller and appeared more homogenous with respect to size and shape than nuclei from wild-type and single mutant plants. These effects were not specific to the above-mentioned tissues but were also observed in nuclei from leaf tissues. We quantified the nuclear effects of the linc mutations by measuring two-dimensional images of 4′,6-diamidino-2-phenylindole (DAPI)–stained, fixed nuclei. Most strikingly, linc1-1 linc2-1 nuclei were ∼20% of the size of wild-type nuclei (wild type = 100 ± 2.6 μm2, linc1-1 linc2-1 = 17.7 ± 0.5 μm2). Furthermore, the nuclei of linc1-1 and linc2-1 single mutants, which do not have a whole-plant phenotype, are significantly smaller than wild-type nuclei (wild type = 100 ± 2.6 μm2, linc1-1 = 62.8 ± 1.5 μm2, linc2-1 = 56.2 ± 1.73 μm2). The frequency histogram shown in Figure 5E illustrates that wild-type nuclei displayed a wide range of nuclear sizes, while linc1-1, linc2-1, and particularly linc1-1 linc2-1 genotypes displayed a reduction in nuclear size as well as size heterogeneity.
Figure 5.
Nuclear Morphology Changes in linc1 and linc2 Mutants.
(A) and (B) Epifluorescence images of representative DAPI-stained nuclei isolated from 4-week-old leaf tissue from wild-type (A) or linc1-1 linc2-1 (B) plants. Bars = 5 μm.
(C) and (D) Confocal projection images of propidium iodide–stained nuclei in whole tissue anther filament cells from wild-type (C) or linc1-1 linc2-1 (D) plants. Bars = 10 μm.
(E) Histogram of nuclear areas (μm2) as measured from DAPI-stained nuclei isolated from 4-week-old leaves. Blue, wild type (n = 518); red, linc1-1 (n = 526); yellow, linc2-1 (n = 479); green, linc1-1 linc2-1 (n = 657).
(F) Average nuclear volume (μm3) ± se of propidium iodide–stained nuclei within anther filament tissue. Blue, wild type (n = 14); green, linc1-1 linc2-1 (n = 30).
(G) Histogram of nuclear circularity indices (4πA/P2) as measured from DAPI-stained nuclei isolated from 4-week-old leaves. Blue, wild type (n = 518); red, linc1-1 (n = 526); yellow, linc2-1 (n = 479); green, linc1-1 linc2-1 (n = 657).
(H) Bar graph displaying the average number of chromocenters ± se observed in propidium iodide–stained nuclei isolated from 2-week-old root tissue. Blue, wild type (n = 150); red, linc1-1 (n = 116); yellow, linc2-1 (n = 117); green, linc1-1 linc2-1 (n = 220).
To avoid confounding issues inherent in two-dimensional measurements and in nuclei isolation, we also measured the volumes of nuclei in whole tissue. We chose anther filaments because of the homogeneity of the tissue and the presence of large, differentiated nuclei. First, we observed fixed nuclei in whole tissue and confirmed that nuclei in linc1-1 linc2-1 mutants were morphologically distinct, smaller, and more spherical than nuclei in matched wild-type tissue, consistent with our observations in isolated leaf nuclei (Figures 5C and 5D). Second, we used laser scanning confocal microscopy to measure the three-dimensional volume of anther filament nuclei stained with propidium iodide. Figure 5F shows that the average volume of nuclei from wild-type anther filament cells is 72.4 ± 7.5 μm3, while that of linc1-1 linc2-1 is 32.2 ± 2.5 μm3. Our observations of three-dimensional nuclei from linc1 linc2 mutants are consistent with our two-dimensional measurements, suggesting that unusually small and spherical nuclear morphology is a bona fide phenotype caused by the loss of LINC1 and LINC2 function, not an artifact of nuclear isolation.
The reduction in nuclear size in linc1 linc2 mutants was accompanied by a near absence of spindle-shaped nuclei. This alteration in nuclear morphology was apparent in nuclei from many tissues, including root (epidermal, cortex, and root hair cells), leaves (epidermal and pavement cells), and flowers (anther filament cells) (Figures 5A to 5D). We quantified these observations by measuring the circularity index of nuclei obtained from 4-week-old whole leaves. The circularity index was calculated using the equation 4πA/P2 (where A = area and P = perimeter) and indicates how closely each nucleus corresponds to a spherical shape (a perfect sphere has a circularity index of 1). Any deviation from a circular shape (e.g., an elongated, lobulated, or spindle shape) causes the index to decrease. Figure 5G shows a distribution of circularity indices (mean = 0.68 ± 0.006) that reflects the extensive variety of nuclear shapes typically found in wild-type leaf cells. The circularity indices of linc2-1 nuclei were only slightly different from those of the wild type (mean = 0.72 ± 0.005), while linc1-1 nuclei had a greater effect (mean = 0.80 ± 0.004). Finally, the linc1-1 linc2-1 double mutant combination resulted in an increased circularity index (mean = 0.83 ± 0.003), a value close to the combined differences of linc1-1 and linc2-1 from the wild type, suggesting that the effects of these two mutations are additive. There was no clear association between nuclear size and shape (see Supplemental Figure 6 online). For instance, it is possible to find both large and small circular nuclei as well as large and small spindle-shaped nuclei. We conclude that separate processes determine the two nuclear parameters—size and shape—and that linc1-1 and linc2-1 differentially affect both nuclear morphology parameters.
One prominent feature of Arabidopsis nuclei is the presence of chromocenters, which are densely staining heterochromatic regions that correspond to the centromeres and the nuclear organizing regions (Fransz et al., 2002). Chromocenters are enriched in epigenetic marks of silent chromatin, such as dimethylated histone H3-Lys 9 and cytosine methylation (Soppe et al., 2002). We compared the number of chromocenters in wild-type and linc mutant root nuclei to determine whether the alteration in overall nuclear morphology extended to changes in internal nuclear architecture. We found that the average number of chromocenters per wild-type nucleus is 8.6 ± 0.2, while the average chromocenter number was reduced to 4.6 ± 0.1 in linc1-1 linc2-1 double mutants (Figure 5H). The single mutants, linc1-1 and linc2-1, had slightly fewer chromocenters, but this difference was only significant for linc2-1 (7.6 ± 0.2). These findings indicate that many complex changes in nuclear morphology occur in the absence of LINC1 and LINC2.
Increased Nuclear Density in linc1 linc2 Mutant Nuclei
The results from the flow cytometry experiments (Figure 4) and the nuclear size measurements (Figure 5D) indicate that linc1-1 linc2-1 mutants have a dramatic reduction in nuclear size that cannot be accounted for by a commensurate reduction in endoreduplication levels. These observations suggest that linc1-1 linc2-1 mutants have increased nuclear DNA packaging densities relative to wild-type plants. To investigate this hypothesis further, we compared the size of nuclei in the wild-type and linc1-1 linc2-1 mutants from diploid guard cells, which do not undergo endoreduplication (Melaragno et al., 1993). The area of the spherical linc1-1 linc2-1 guard cell nuclei (3.8 ± 0.1 μm2; n = 44) was approximately three times smaller than the area of nuclei in wild-type guard cells (11.6 ± 0.3 μm2; n = 40).
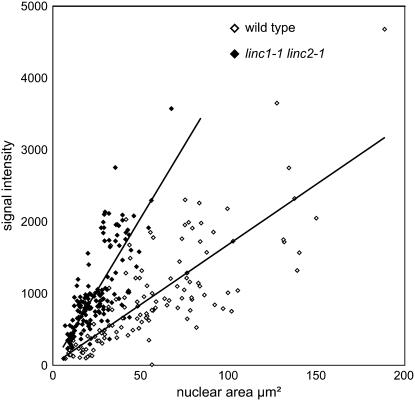
We also measured directly the relationship between the fluorescence signal and nuclear area of individual DAPI-stained nuclei from a population of wild-type and linc1-1 linc2-1 leaf cells of varying ploidy levels. Figure 6 shows that there is a positive correlation between the quantity of DNA staining, reflecting endopolyploid levels, and nuclear area in wild-type nuclei, as has been shown by previous investigators (Jovtchev et al., 2006). DNA staining and nuclear area are also positively correlated in linc1-1 linc2-1 nuclei; however, the ratio of DNA content per unit area is higher than in wild-type nuclei. We note that the range and distribution of DNA staining are comparable between the linc1-1 linc2-1 and wild-type samples. Collectively, these data demonstrate that the loss of LINC1 and LINC2 function results in an increase in nuclear DNA packaging density.
Figure 6.
Increased DNA Density in linc1-1 linc2-1 Mutants.
Scatterplot of nuclear area (μm2; x axis) versus total DAPI signal per nucleus (y axis). Nuclei were isolated from 4-week-old leaf tissue. Open diamonds, wild type; closed diamonds, linc1-1 linc2-1.
DISCUSSION
We present a functional characterization of members of a family of Arabidopsis genes encoding long coiled-coil proteins with structural and amino acid similarity to the carrot nuclear peripheral protein Dc NMCP1. Loss-of-function mutations in either LINC1 or LINC2 caused a significant decrease in nuclear size (area measurements; Figure 5) without causing abnormal morphological phenotypes at the whole-plant level (Figure 3). Combining linc1-1 and linc2-1 mutations had a synergistic effect on whole-plant morphology (dwarfing) and an additive effect on nuclear morphology. Nuclei in linc1-1 linc2-1 double mutants are smaller than those found in wild-type plants or either linc single mutant. The reduction in nuclear volume in the linc1-1 linc2-1 double mutants was estimated to be approximately twofold to fivefold. Our direct volume measurements of anther filament nuclei in intact tissues yielded the lower twofold figure (Figure 5F). The higher estimate is based in part upon the approximately fivefold difference in area measured in leaf cell nuclei (Figure 5E), assuming that these nuclei are significantly flattened during isolation and imaging (Berr and Schubert, 2007) (the estimate would be higher if we assume that the nuclei are spherical). The fivefold figure for nuclear volume reduction is corroborated by our area measurements of the spherical, diploid guard cell nuclei in linc1-1 linc2-1 compared with wild-type leaf tissue (a threefold smaller area corresponds to a fivefold reduction in volume). Nuclei in linc1-1 and linc1-1 linc2-1 mutants were altered in shape as well as size, being more circular and lacking the distinctive elongated spindle shape commonly seen in wild-type cells. Although the linc2-1 mutation, by itself, did not alter nuclear shape significantly, loss of LINC2 function did enhance the shape phenotype of the linc1-1 mutation. These data indicate that LINC1 and LINC2 have overlapping functions in the determination of nuclear size and shape in Arabidopsis.
Nuclear morphology changes caused by LINC1 and LINC2 loss-of-function mutations also affect internal nuclear organization. The number of chromocenters in interphase nuclei was decreased significantly in linc1-1 linc2-1 mutants but not in either linc single mutant (Figure 5H). Chromocenters are aggregates of constitutive heterochromatin, and in Arabidopsis they are composed of a variety of repetitive DNA sequences, including the rRNA gene clusters and the centromeric repeat arrays containing 180-bp tandem repeats and various transposable elements. In wild-type nuclei, the number of chromocenters approaches 10, corresponding to the diploid chromosome number, but the actual number varies and trends to a lower number due to the coalescence of previously distinct chromocenters. Chromocenters are dynamic structures, as has been reported in a recent cytogenetic analysis of Arabidopsis protoplasts and floral tissue, in which certain sequences became dispersed during dedifferentiation and flowering (Tessadori et al., 2007a, 2007b). Moreover, chromocenter number and composition change over time and in response to epigenetic modifier mutations (e.g., ddm1 and met1) (Soppe et al., 2002). One simple explanation for reduced chromocenter number is that the space constraints imposed by the smaller nuclear dimensions in linc1-1 linc2-1 mutants lead to more frequent encounters between individual chromocenters, which result in their coalescence. An alternative explanation is that the underlying nuclear architecture is disrupted in the linc1-1 linc2-1 mutants, such that barriers against chromocenter association are lowered. Recent work by Berr and Schubert (2007) suggests that the arrangement of interphase chromosomes is constrained predominantly by morphological features, such as nuclear shape, volume, and endopolyploid level.
A shift in the spectrum of endopolyploidy is another component of the pleiotropic linc1 linc2 phenotype. As shown in Figure 4, the double mutant contains the normal range of endopolyploidy, consistent with the observation that leaf trichome morphology, which is dependent on the highest levels of endoreduplication, is normal (see Supplemental Figure 5 online). Nevertheless, there is an overall reduction in endopolyploidy levels of leaf cell populations (Figure 4). It is tempting to ascribe the reduction in nuclear size, and perhaps the alteration in nuclear shape, to this shift in endopolyploidy, but three considerations argue against this interpretation. First, the ∼25% reduction in overall endopolyploidy levels seen in Figure 4B (wild type = 7.3C, linc1-1 linc2-1 = 5.5C) cannot explain the dramatic reduction in leaf cell nuclear size (approximately fivefold), assuming the operation of the established relationship between endopolyploidy and nuclear size (Jovtchev et al., 2006). Second, when we restricted our measurements to diploid guard cells, the projected area of linc1-1 linc2-1 nuclei was threefold smaller than that observed for wild-type nuclei. Third, the linc1-1 and linc2-1 single mutants also showed a reduction in nuclear size (Figure 5E) in the absence of an alteration of endopolyploidy (Figure 4A). Therefore, it is probable that the ploidy shift seen in linc1 linc2 mutants is a secondary effect of the reduction in nuclear size.
Regardless of whether nuclear size changes are driving endopolyploidy changes or these ploidy shifts constitute an independent synthetic phenotype of the linc1-1 linc2-1 double mutants, it is clear that there is a new relationship between DNA content and nuclear size in the double mutants, reflected by the increased slope of the regression line in Figure 6. Our results demonstrate that the linc1-1 linc2-1 mutation combination increases the chromatin packaging ratio: DNA content per unit of nuclear volume. This phenotype is particularly intriguing in light of studies demonstrating that there is a fixed relationship between DNA content and nuclear volume that applies to a range of plant species with genome sizes spanning >2 orders of magnitude (Fujimoto et al., 2005; Jovtchev et al., 2006). Despite violating this fixed chromatin packaging ratio, the linc1 and linc2 single mutants do not exhibit abnormal developmental phenotypes. Even the dwarf linc1 linc2 double mutants proceed through their life cycle with relative few developmental abnormalities beyond those that can be attributed to a reduction in cell size secondary to a decrease in nuclear size. These considerations suggest that the alterations in nuclear morphology and interphase chromatin packaging caused by the linc mutations do not disrupt fundamental gene regulatory mechanisms.
The separation of nuclear assembly into two phases provides an explanation for the apparent discrepancy between nuclear form and function in the linc mutants. Studies using cell-free extracts, mostly in nonplant systems, have demonstrated that nuclear assembly is organized into two basic stages: (1) enclosure of chromatin within a functional nuclear envelope, and (2) growth and expansion of the nuclear envelope coincident with the incorporation of nuclear pore complexes. Chromatin enclosure involves the positioning of partially assembled nuclear pore complexes on the condensed chromosome surface and the independent recruitment and fusion of membrane vesicles to form the nuclear envelope (Wozniak and Clarke, 2003). In the second phase, nuclear volume increases with the insertion of nuclear pore complexes into an expanding nuclear envelope. We propose that it is in this second phase of nuclear assembly that LINC proteins might play an important role. In particular, it appears that LINC proteins affect a differentiation phase of nuclear development that follows nuclei formation. This hypothesis is supported by the lack of obvious ultrastructural defects visible in transmission electron micrographs of linc1-1 linc2-1 nuclei in the meristematic cells in the shoot apex (data not shown). In meristem tissues, nuclei are predominantly 2C or 4C (G1 or G2) and are uniformly small and circular in appearance. By contrast, nuclei found within most adult tissues have differentiated into an array of noncircular shapes, and many of the cells are polyploid due to endoreduplication. The preponderance of small circular nuclei in all examined tissues of linc1-1 linc2-1 mutants suggests that differentiation from the baseline nuclear form is blocked. This hypothesis does not exclude the possibility that the underlying nuclear architecture of meristematic 2C linc1-1 linc2-1 nuclei differs from that seen in wild-type nuclei. It is possible that the LINC proteins are incorporated early on as a requisite foundation for later nuclear differentiation. This notion is in agreement with the transcript profile of the LINC genes, which are most highly expressed in the meristem tissues.
Some insight into the mechanisms by which LINC proteins might participate in nuclear expansion and differentiation can be gained from other experimental systems. Several proteins have been identified in metazoans that are involved in nuclear expansion during nuclear assembly, including the nuclear intermediate filament protein lamin A and two recently described novel proteins, MEL-28 (in Caenorhabditis elegans) and KUGELKERN (in Drosophila melanogaster) (Brandt et al., 2006; Galy et al., 2006). A common structural feature between MEL-28, KUGELKERN, and the lamins is the presence of a coiled-coil domain. The possible parallels between mammalian lamins and LINC proteins are intriguing. LINC proteins, like lamins, are segmented into a tripartite structure, consisting of a central coiled-coil domain flanked by putative globular N- and C-terminal domains. Although LINC proteins lack other motifs found in lamins (e.g., C-terminal CAAX box) and do not share primary protein sequence similarity with lamins, Meier and colleagues (Rose et al., 2005) suggest that the Arabidopsis LINC proteins could be analogs of lamins, based on the nuclear peripheral localization of Dc NMCP1 as well as overall protein organization. This hypothesis is supported by our localization results of LINC1, which is concentrated at the nuclear periphery. A functional parallel between plant LINC proteins and animal lamins is also suggested by the nuclear morphological changes caused by defects or alterations in lamin genes (Capell and Collins, 2006). For example, interphase fibroblasts cultured from patients carrying lamin A mutations display nuclear abnormalities, including altered nuclear shape, nuclear blebbing, chromatin disorganization, and mislocalization of nuclear envelope proteins (Vigouroux et al., 2001; Cao et al., 2007). Similarly, an engineered disruption of lamin B1 in mice leads to nuclear malformation in addition to an array of severe developmental phenotypes (Vergnes et al., 2004). Furthermore, depleting lamin A from nuclear assembly extracts does not disturb nuclear envelope formation but is necessary for additional nuclear envelope growth and proper polymerization of lamins at the periphery (Moir et al., 2000). Thus, while individual lamins are necessary to maintain nuclear architecture, they are dispensable for assembling a minimal functional nucleus. Further work is required to determine whether LINC proteins serve structural and signal integration roles analogous to lamins or whether these plant proteins influence nuclear architecture through a unique mechanism.
METHODS
Plant Materials and Growth Conditions
All SALK T-DNA insertion alleles (Alonso et al., 2003) used in this study are in the Arabidopsis thaliana Columbia ecotype background and were obtained through the ABRC stock center. Plants were grown in long-day lighting conditions (16 h of light/8 h of dark) on soil in growth chambers. Genotyping of individual T-DNA alleles was performed by standard PCR using KlenTaq LA (Sigma-Aldrich) and the allele-specific primers SALK_025347 (linc1-1), LP, 5′-TGCCTTCTCCTCGCTTTTCAA-3′, and RP, 5′-TGCGTGAATGGGAAAGAAAGTTG-3′; SALK_076653 (linc2-1), LP, 5′-GAAGCTCATTGCTAGAGAAGGGG-3′, and RP, 5′-AACGCTGATCGTTCATGTTCCA-3′; in combination with the T-DNA–specific primer LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′.
Cloning and Plant Transformation
Genomic fragments corresponding to At1g67230 (LINC1) and At1g13220 (LINC2) were amplified from genomic DNA using PfuUltra II enzyme (Clontech) and primers 5′-CACCATGTCCACGCCGTTGAAGGTGTGGC-3′ and 5′-CGTCGTCAAGAAAGTCCAAAGC-3′ (for LINC1) or 5′-CACCATGACGCCGAGAAGCGAGACGC-3′ and 5′-TGTAGTGAGAAAAGTCCAAAGC-3′ (for LINC2) and cloned into pENTR/D-TOPO (Invitrogen). Each construct was sequenced and the genomic insert was then transferred into the plant expression vector pEarleyGate101 (Earley et al., 2006) using LR Clonase (Invitrogen). The resulting vector was then introduced into plants via Agrobacterium tumefaciens–mediated transformation, as described previously (Clough and Bent, 1998).
Imaging
All fluorescence images were acquired using a Nikon Eclipse epifluorescence microscope, with appropriate filter sets, coupled to Phyllum software. Nuclear area measurements were performed using NIH ImageJ software. Confocal images were collected using a Leica TCS SP2 microscope, and nuclear volume was determined using Imaris software.
RNA Isolation and RT-PCR
RNA was prepared from 2-week-old whole seedlings grown on 0.5× Murashige and Skoog medium (Sigma-Aldrich) using TRIzol reagent (Invitrogen), as described previously (Rangwala et al., 2006). Subsequently, RNA was treated with DNaseI (Promega) and reverse-transcribed with SuperScript II reverse transcriptase (Invitrogen) using oligo(d)T primers according to the manufacturer's specifications. First-stand cDNA was used as template in conventional PCR using KlenTaq LA (Sigma-Aldrich) DNA polymerase and the following primer pairs: Actin2F (5′-TGATATTCAACCAATCGTGTGTGAC-3′) and Actin2R (5′-AAGCAAGAATGGAACCACCGATCC-3′); At1g67230F (5′-GGCCGAATCTGAAACTGGCACAAA-3′) and At1g67230R (5′-CATGCTGCACCACATTCGTGCTTA-3′); and At1g13220F (5′-AAGAGAGTGCTTTGAGGGTGCAGA-3′) and At1g13220R (5′-GCTTGCCAGTGTCAATCCCATGTT-3′).
Nuclei Isolation and Imaging
Nuclei were isolated from 4-week-old plants by removing ∼1-cm distal leaf tips and fixing tissue in 3:1 ethanol:acetic acid overnight at −20°C. Tissues were rehydrated in 100 mM sodium citrate, pH 4.8, buffer for 15 min and incubated in digestion buffer (0.03% cytohelicase [Sigma-Aldrich], 0.03% pectolyase [Sigma-Aldrich], and 0.03% cellulase Onozuka RS [Serva] in 100 mM sodium citrate, pH 4.8, buffer) for 45 min at 37°C. Digested tissue was briefly homogenized by pipetting with a wide-bore pipette, centrifuged at 1000g, washed with 1 mL of 100 mM sodium citrate, pH 4.8, buffer three times, and resuspended in 3:1 ethanol:acetic acid. Nuclei were dropped onto Superfrost microscope slides, dried for ∼30 min, and stained with 10 μg/mL of DAPI.
Flow Cytometry
The ploidy levels of leaf cell nuclei from 4-week-old plants were determined by flow cytometry using a PA-11 ploidy analyzer (Partec) with UV light excitation at 366 nm from a mercury arc lamp. Leaves were chopped with a razor blade in Cystain extraction buffer (Partec), filtered through a 30-μm CellTrics filter (Partec) into a sample tube, and stained with Cystain fluorescent buffer (Partec). Data were collected for ∼20,000 nuclei per run and were presented on a logarithmic scale. The number of data points collected per genotype was as follows: wild type = 4 (2 technical replicates × 2 biological samples); linc1-1 = 2 (2 technical replicates × 1 biological sample); linc2-1 = 2 (2 technical replicates × 1 biological sample); and linc1-1 linc2-1 = 12 (6 technical replicates × 2 biological samples).
Scanning Electron Microscopy
Arabidopsis leaflets were immersed in drops of fixative (2.5% glutaraldehyde in 0.1 M phosphate buffer), cut into small pieces, and transferred to vials of fresh fixative. Samples were allowed to fix for 1.5 h, washed several times with buffer, and postfixed with 1% aqueous osmium tetraoxide for 4 h. Samples were further processed using the osmium-thiocarbohydrazide-osmium method (Kelley et al., 1973), dehydrated in ethyl alcohol, and critical point dried with liquid CO2. Mounted samples were sputter-coated with 50 nm of gold and examined in a Hitachi S-450 scanning electron microscope operated at 20-kV accelerating voltage.
Computational Analysis
Analysis of the LINC protein sequences was performed using the MultiCoil program (http://multicoil.lcs.mit.edu/cgi-bin/multicoil) (Wolf et al., 1997). Identification and classification of genes coexpressed with LINC1 to LINC4 was performed using the tools found at http://bbc.botany.utoronto.ca/ (Toufighi et al., 2005). Microarray data from the AtGenExpress tissue set were cross-referenced using the Expression Angler tool, and identified genes were classified using the Classification Superviewer tool based on the Munich Information Center of Protein Sequences annotations (Schoof et al., 2002; Schmid et al., 2005).
Phylogenetic Analysis
A pairwise amino acid alignment was produced using ClustalX (Thompson et al., 1997) with the BLOSUM cost matrix, followed by manual adjustment. For the phylogenetic analysis, the entire alignment was used to create an unrooted neighbor-joining tree using PAUP* 4.0 (Swofford, 2003), with 1000 bootstrap replicates.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative and GenBank/DDBJ/EMBL databases under the following accession numbers: LINC1 (At1g67230), LINC2 (At1g13220), LINC3 (At1g68790), and LINC4 (At5g65770).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genes Coexpressed with LINC1 to LINC4.
Supplemental Figure 2. Complementation of linc1-1 linc2-1 Mutants with the P35S:LINC1g-YFP or P35S:LINC2g-YFP Transgene Construct.
Supplemental Figure 3. Genomic Organization and T-DNA Insertional Alleles of LINC1 and LINC2.
Supplemental Figure 4. Reduced Leaf Epidermal Cell Size in linc1-1 linc2-1 Mutants.
Supplemental Figure 5. Leaf Trichomes Develop Normally in linc1-1 linc2-1 Mutants.
Supplemental Figure 6. The linc1-1 and linc2-1 Mutations Differentially Affect Nuclear Area and Circularity.
Supplementary Material
Acknowledgments
We are grateful for the microscopy work of Howard Berg at the Donald Danforth Plant Science Center (transmission electron microscopy) and Michael Veith (scanning electron microscopy) in the Washington University Biology Department. We thank Sanjida Rangwala, Hankuil Yi, Ashley Galant, and Hye-Ryun Woo for helpful comments on the manuscript. The different Arabidopsis strains used in the study were obtained from the ABRC at Ohio State University. This work was supported by grants from the National Science Foundation to E.J.R. (Grants MCB-0321990 and MCB-0548597) and by Biotechnology and Biological Science Research Council David Phillips Fellowships to K.S.-S. Additional support was provided by the Danforth Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eric J. Richards (richards@wustl.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Berr, A., and Schubert, I. (2007). Interphase chromosome arrangement in Arabidopsis thaliana is similar between differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics 176 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, A., et al. (2006). Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr. Biol. 16 543–552. [DOI] [PubMed] [Google Scholar]
- Cao, K., Capell, B.C., Erdos, M.R., Djabali, K., and Collins, F.S. (2007). A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc. Natl. Acad. Sci. USA 104 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell, B.C., and Collins, F.S. (2006). Human laminopathies: Nuclei gone genetically awry. Nat. Rev. Genet. 7 940–952. [DOI] [PubMed] [Google Scholar]
- Chytilova, E., Macas, J., Sliwinska, E., Rafelski, S.M., Lambert, G.M., and Galbraith, D.W. (2000). Nuclear dynamics in Arabidopsis thaliana. Mol. Biol. Cell 11 2733–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytilova, E.V.A., Macas, J., and Galbraith, D.W. (1999). Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann. Bot. (Lond.) 83 645–654. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Esteller, M. (2007). Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8 286–298. [DOI] [PubMed] [Google Scholar]
- Fang, Y., and Spector, D.L. (2005). Centromere positioning and dynamics in living Arabidopsis plants. Mol. Biol. Cell 16 5710–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz, P., De Jong, J.H., Lysak, M., Castiglione, M.R., and Schubert, I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, S., Ito, M., Matsunaga, S., and Fukui, K. (2005). An upper limit of the ratio of DNA volume to nuclear volume exists in plants. Genes Genet. Syst. 80 345–350. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Harkins, K.R., and Knapp, S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, V., Askjaer, P., Franz, C., Lopez-Iglesias, C., and Mattaj, I.W. (2006). MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr. Biol. 16 1748–1756. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Margalit, A., Goldman, R.D., Shumaker, D.K., and Wilson, K.L. (2005). The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6 21–31. [DOI] [PubMed] [Google Scholar]
- Hetzer, M.W., Walther, T.C., and Mattaj, I.W. (2005). Pushing the envelope: Structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21 347–380. [DOI] [PubMed] [Google Scholar]
- Jovtchev, G., Schubert, V., Meister, A., Barow, M., and Schubert, I. (2006). Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114 77–82. [DOI] [PubMed] [Google Scholar]
- Kelley, R.O., Dekker, R.A., and Bluemink, J.G. (1973). Ligand-mediated osmium binding: Its application in coating biological specimens for scanning electron microscopy. J. Ultrastruct. Res. 45 254–258. [DOI] [PubMed] [Google Scholar]
- Kleffmann, T., Hirsch-Hoffmann, M., Gruissem, W., and Baginsky, S. (2006). plprot: A comprehensive proteome database for different plastid types. Plant Cell Physiol. 47 432–436. [DOI] [PubMed] [Google Scholar]
- Lanctot, C., Cheutin, T., Cremer, M., Cavalli, G., and Cremer, T. (2007). Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat. Rev. Genet. 8 104–115. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Xu, Z.J., Takahashi, S., Ito, A., Ono, M., Nomura, K., and Inoue, M. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp. Cell Res. 232 173–181. [DOI] [PubMed] [Google Scholar]
- Meier, I. (2007). Composition of the plant nuclear envelope: Theme and variations. J. Exp. Bot. 58 27–34. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir, R.D., Yoon, M., Khuon, S., and Goldman, R.D. (2000). Nuclear lamins A and B1: Different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle, A.F., Clark, G.P., Boon, R., Lewandowska, D., Lam, Y.W., Andersen, J., Mann, M., Lamond, A.I., Brown, J.W., and Shaw, P.J. (2005). Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza, D., Herzog, M., Hulskamp, M., Brown, S., Dorne, A.M., and Bonneville, J.M. (1999). Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152 461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala, S.H., Elumalai, R., Vanier, C., Ozkan, H., Galbraith, D.W., and Richards, E.J. (2006). Meiotically stable natural epialleles of Sadhu, a novel Arabidopsis retroposon. PLoS Genet. 2 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A., Schraegle, S.J., Stahlberg, E.A., and Meier, I. (2005). Coiled-coil protein composition of 22 proteomes—Differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 5 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Zaccaria, P., Gundlach, H., Lemcke, K., Rudd, S., Kolesov, G., Arnold, R., Mewes, H.W., and Mayer, K.F. (2002). MIPS Arabidopsis thaliana Database (MAtDB): An integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 30 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J., Hwang, S., Niwa, Y., Kobayashi, H., and Galbraith, D.W. (1995). Green-fluorescent protein as a new vital marker in plant cells. Plant J. 8 777–784. [DOI] [PubMed] [Google Scholar]
- Soppe, W.J., Jasencakova, Z., Houben, A., Kakutani, T., Meister, A., Huang, M.S., Jacobsen, S.E., Schubert, I., and Fransz, P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., Roberts, G.R., Stacey, N.J., McCann, M.C., Maxwell, A., and Roberts, K. (2005). RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc. Natl. Acad. Sci. USA 102 18736–18741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6 544–553. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). (Sunderland, MA: Sinauer Associates).
- Tessadori, F., Chupeau, M.C., Chupeau, Y., Knip, M., Germann, S., van Driel, R., Fransz, P., and Gaudin, V. (2007. a). Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell Sci. 120 1200–1208. [DOI] [PubMed] [Google Scholar]
- Tessadori, F., Schulkes, R.K., Driel, R.V., and Fransz, P. (2007. b). Light-regulated large-scale reorganization of chromatin during the floral transition in Arabidopsis. Plant J. 50 848–857 [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi, K., Brady, S.M., Austin, R., Ly, E., and Provart, N.J. (2005). The Botany Array Resource: E-northerns, expression angling, and promoter analyses. Plant J. 43 153–163. [DOI] [PubMed] [Google Scholar]
- Vaughn, M.W., et al. (2007). Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5 e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes, L., Peterfy, M., Bergo, M.O., Young, S.G., and Reue, K. (2004). Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA 101 10428–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, C., Auclair, M., Dubosclard, E., Pouchelet, M., Capeau, J., Courvalin, J.C., and Buendia, B. (2001). Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell Sci. 114 4459–4468. [DOI] [PubMed] [Google Scholar]
- Wolf, E., Kim, P.S., and Berger, B. (1997). MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci. 6 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, R., and Clarke, P.R. (2003). Nuclear pores: Sowing the seeds of assembly on the chromatin landscape. Curr. Biol. 13 R970–R972. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E., and Ecker, J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T., and Henikoff, S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.