Abstract
We demonstrate that the Arabidopsis thaliana MYB46 transcription factor is a direct target of SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1), which is a key transcriptional activator regulating the developmental program of secondary wall biosynthesis. The MYB46 gene is expressed predominantly in fibers and vessels in stems, and its encoded protein is targeted to the nucleus and can activate transcription. MYB46 gene expression was shown to be regulated by SND1, and transactivation analysis demonstrated that SND1 as well as its close homologs were able to activate the MYB46 promoter. Electrophoretic mobility shift assays and chromatin immunoprecipitation experiments revealed that SND1 binds to the MYB46 promoter. Dominant repression of MYB46 caused a drastic reduction in the secondary wall thickening of fibers and vessels. Overexpression of MYB46 resulted in an activation of the biosynthetic pathways of cellulose, xylan, and lignin and concomitantly led to ectopic deposition of secondary walls in cells that are normally nonsclerenchymatous. In addition, the expression of two secondary wall–associated transcription factors, MYB85 and KNAT7, was highly upregulated by MYB46 overexpression. These results demonstrate that MYB46 is a direct target of SND1 and is another key player in the transcriptional network involved in the regulation of secondary wall biosynthesis in Arabidopsis.
INTRODUCTION
The ability of vascular plants to produce the reinforcing secondary walls was acquired when they first appeared on land during the Silurian period. It enabled vascular plants to build strong mechanical tissues for water transport and support, which provides the basis for their dominance on dry land (Raven et al., 1999). Because secondary walls are mainly composed of cellulose, hemicellulose, and lignin, it is apparent that vascular plants evolved to be able to coordinately activate the biosynthetic pathways of these secondary wall components (Ye et al., 2006). Unraveling the molecular mechanisms underlying the activation of secondary wall biosynthesis will not only help our understanding of the evolution of vascular plants but also have significant implications in genetic modifications of the quality and quantity of wood, an important raw material for pulping and papermaking, construction, and potentially for biofuel production.
Several transcription factors belonging to NAC (for NAM, ATAF1/2, and CUC2) and MYB families have recently been shown to be key players in regulating secondary wall biosynthesis. The endothecium layer in anthers deposits secondary walls, which are essential for the rupture of stomium to release pollens during anther dehiscence. In Arabidopsis thaliana, the deposition of secondary walls in anther endothecium was found to be regulated by two NAC genes, NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1; At2g46770) and NST2 (At3g61910) (Mitsuda et al., 2005). Simultaneous mutations of NST1 and NST2 cause a loss of secondary wall thickening in the endothecium, and overexpression of NST1 or NST2 results in ectopic deposition of secondary walls in cells that are normally parenchymatous. In addition to NACs, a MYB transcription factor, MYB26 (At3g13890), is also essential for secondary wall thickening in anther endothecium. Mutation of MYB26/MALE STERILE35 leads to a loss of secondary wall deposition (Steiner-Lange et al., 2003), and its overexpression causes ectopic secondary wall thickening in parenchyma cells (Yang et al., 2007). It was suggested that MYB26 may act upstream of NST1 and NST2 based on the observations that overexpression or mutation of MYB26 upregulates or downregulates, respectively, the expression levels of NST1 and NST2 (Yang et al., 2007).
The most abundant cell types that deposit secondary walls are fibers and tracheary elements. Recent studies have revealed important roles of NAC domain proteins in the regulation of secondary wall deposition in these cell types. Two NAC domain proteins, VASCULAR-RELATED NAC DOMAIN6 (VND6; At5g62380) and VND7 (At1g71930), have been shown to be required for vessel development in primary roots of Arabidopsis (Kubo et al., 2005; Demura and Fukuda, 2007). The VND6 and VND7 genes are specifically expressed in developing metaxylem and protoxylem cells, respectively, and dominant repression of VND6 or VND7 functions results in a loss of metaxylem or protoxylem development. Overexpression of VND6 or VND7 leads to ectopic formation of metaxylem- or protoxylem-like vessels. It was suggested that VND6 and VND7 are involved in regulating the differentiation of metaxylem and protoxylem, respectively (Kubo et al., 2005).
NAC domain proteins have also been found to regulate secondary wall biosynthesis in fibers. The Arabidopsis SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1; At1g32770) gene is specifically expressed in interfascicular fibers and xylary fibers, and dominant repression of SND1 functions causes a drastic reduction in secondary wall thickening in fibers (Zhong et al., 2006). Overexpression of SND1 leads to activation of secondary wall biosynthetic pathways and concomitantly causes ectopic deposition of secondary walls in cells that are normally parenchymatous. Recently, it was found that SND1 (also named NST3) together with NST1 act redundantly in regulation of secondary wall biosynthesis in fibers (Mitsuda et al., 2007; Zhong et al., 2007). Simultaneous knockout of SND1/NST3 and NST1 results in a severe reduction in the expression of secondary wall biosynthetic genes and a concomitant loss of all three major secondary wall components, including cellulose, xylan, and lignin. Theses studies demonstrate that SND1 and NST1 are key transcriptional activators for the developmental program of secondary wall biosynthesis.
Overexpression of SND1 and simultaneous inhibition of SND1 and NST1 have been shown to cause upregulation or downregulation, respectively, of several secondary wall–associated transcription factors (Zhong et al., 2006, 2007). This suggests that SND1 regulates the expression of these transcription factors, which in turn activate the secondary wall biosynthetic pathways. To further understand the transcriptional network involved in the regulation of secondary wall biosynthesis, it is essential to find the target genes regulated by SND1. In this study, we report that the MYB46 transcription factor is a direct target of SND1. We show that the MYB46 gene is predominantly expressed in fibers and vessels and that its encoded protein is a transcriptional activator. We demonstrate that SND1 directly binds to the promoter of the MYB46 gene and activates its expression. We further show that dominant repression of MYB46 causes a dramatic reduction in secondary wall thickening, and its overexpression leads to ectopic secondary wall deposition and upregulation of secondary wall biosynthetic genes and two secondary wall–associated transcription factors. Our study reveals an important player in the SND1-mediated transcriptional regulation of secondary wall biosynthesis.
RESULTS
The MYB46 Gene Is Predominantly Expressed in Fibers and Vessels in Arabidopsis Inflorescence Stems
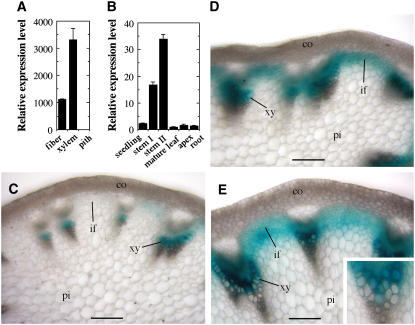
The Arabidopsis inflorescence stems develop interfascicular fibers with secondary walls, which have been used as a model to study the molecular mechanisms underlying secondary wall biosynthesis (Zhong et al., 2001). To investigate the transcriptional regulation of secondary wall biosynthesis, we isolated developing fiber cells using laser microdissection (Zhong et al., 2006) and analyzed transcription factors that were upregulated during secondary wall thickening in fiber. In this report, we studied a MYB transcription factor, MYB46 (At5g12870), which was found to be expressed in fibers and xylem cells undergoing secondary wall thickening but not in parenchymatous pith cells (Figure 1A). At the organ level, the MYB46 gene was predominantly expressed in inflorescence stems in which the secondary wall–containing interfascicular fibers and xylem cells are produced (Figure 1B).
Figure 1.
Expression Pattern of the MYB46 Gene.
The expression pattern of the MYB46 gene was studied using quantitative PCR and GUS reporter gene analysis in transgenic Arabidopsis plants expressing MYB46:GUS. co, cortex; if, interfascicular fiber; pi, pith; xy, xylem. Bars = 80 μm.
(A) Quantitative PCR analysis of MYB46 expression in laser-dissected cells showing its high level expression in interfascicular fibers and xylem cells. The expression level of MYB46 in pith cells is set to 1. Error bars represent se of three biological replicates.
(B) Quantitative PCR analysis showing the predominant expression of MYB46 in stems. The expression level of MYB46 in mature leaf is set to 1. Error bars represent se of three biological replicates.
(C) Cross section of a rapidly elongating internode of MYB46:GUS plants showing GUS staining in developing vessels in protoxylem. Note that the GUS staining was nearly absent in interfascicular fibers, which were undergoing rapid elongation but no secondary wall thickening.
(D) Cross section of an internode near the cessation of elongation showing GUS staining in both interfascicular fibers and metaxylem. At this stage, the interfascicular fibers initiated secondary wall thickening.
(E) Cross section of a nonelongating internode showing GUS staining in interfascicular fibers and metaxylem, both undergoing secondary wall thickening. Inset is a high-magnification view of a vascular bundle showing GUS signals in both developing vessels and xylary fibers.
To investigate its developmental expression pattern, the MYB46 gene was fused with the β-glucuronidase (GUS) reporter gene and transformed into wild-type Arabidopsis plants. To ensure that the reporter gene expression analysis represents the endogenous MYB46 expression pattern, a 3-kb 5′ upstream sequence, the entire exon and intron region, and a 2-kb 3′ downstream sequence of the MYB46 gene were used. Examination of the stems of transgenic plants revealed that in rapidly elongating internodes in which protoxylem is the only secondary wall–containing cells being produced, the GUS staining was seen in developing vessels in the protoxylem (Figure 1C). At this stage, interfascicular fiber cells are undergoing rapid elongation and no secondary wall thickening is evident (Ye et al., 2002). Concomitantly, little GUS staining was present in the interfascicular fiber cells (Figure 1C). In internodes that are near the cessation of elongation and in nonelongating internodes, intensive GUS staining was observed in both interfascicular fibers and developing metaxylem (Figures 1D and 1E). Close examination of metaxylem revealed that the GUS staining was present in both developing vessels and xylary fibers (inset in Figure 1E). These results demonstrate that the expression of the MYB46 gene is associated with secondary wall thickening in interfascicular fibers, xylary fibers, and vessels.
MYB46 is a member of the MYB transcription factor family. In the Arabidopsis genome, a large number of MYB genes have been annotated, and some of them have been characterized for their biological functions (Stracke et al., 2001). MYB46 is most closely related to Eg MYB2, a Eucalyptus MYB gene that has been proposed to regulate the coordinated expression of monolignol-specific pathway genes (Goicoechea et al., 2005). MYB46 belongs to the R2R3-type MYBs that contain two MYB repeats, each of which forms a helix-turn-helix structure (Stracke et al., 2001). The MYB46 gene is 2001 bp long from the start codon to the stop codon and consists of two exons and one intron. The longest open reading frame in the MYB46 cDNA is 843 bp, and it encodes a protein of 280 amino acid residues with a predicted molecular mass of 31.5 kD and a predicted pI of 5.78.
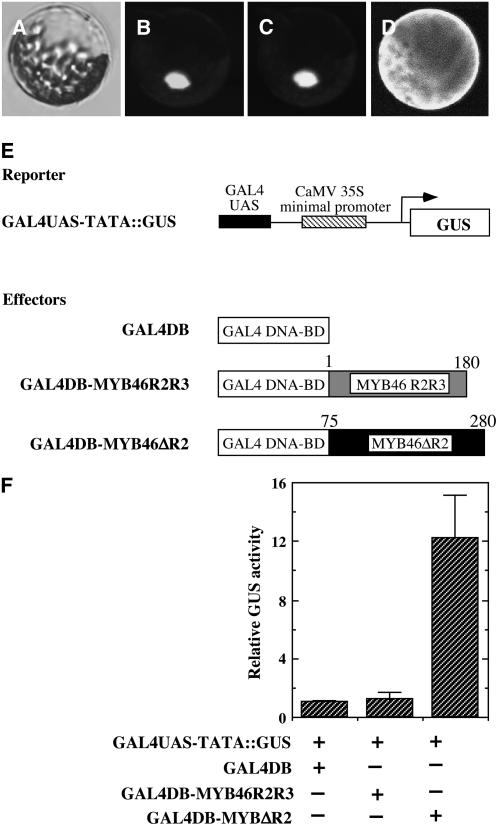
Consistent with its predicted role as a transcription factor, green fluorescent protein (GFP)–tagged MYB46 was shown to be targeted to the nucleus when expressed in Arabidopsis leaf protoplasts (Figures 2A to 2D). Transcriptional analysis revealed that MYB46 fused with the GAL4-DNA binding domain was able to activate transcription of the GUS reporter gene driven by the GAL4 upstream binding sequence (Figures 2E and 2F), indicating that MYB46 is a transcriptional activator.
Figure 2.
MYB46 Is Localized in the Nucleus and Is a Transcriptional Activator.
The subcellular location of MYB46 was examined in Arabidopsis leaf protoplasts expressing yellow fluorescent protein (YFP)–tagged MYB46 protein. The transcription activation activity of MYB46 was studied by cotransfecting Arabidopsis leaf protoplasts with the GUS reporter construct and various effector constructs containing different parts of MYB46 fused with the GAL4 DNA binding domain.
(A) to (C) Differential interference contrast (DIC) image (A), the MYB46-YFP signal (B), and nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI) (C) of an Arabidopsis protoplast expressing MYB46-YFP. Note that MYB46-YFP is targeted to the nucleus.
(D) An Arabidopsis protoplast expressing YFP alone showing the ubiquitous presence of fluorescence throughout the cytoplasm.
(E) Diagrams of the reporter and effector constructs used for transcriptional activation analysis. The reporter construct contains the GUS reporter gene driven by the GAL4 upstream activation sequence (UAS) linked with the CaMV 35 minimal promoter sequence (GAL4UAS-TATA∷GUS). The GAL4 DNA binding domain (GAL4DB) alone and GAL4DB fused with the MYB46 R2R3 DNA binding domain (GAL4DB-MYB46R2R3) or the MYB46 protein missing the R2 motif (GAL4DB-MYB46ΔR2) were constructed under the CaMV 35 promoter to create effector constructs for testing their ability to activate expression of the GUS reporter gene.
(F) MYB46 is able to activate expression of the GUS reporter gene in Arabidopsis protoplasts. The effector and reporter constructs together with a construct expressing the luciferase gene were cotransfected into Arabidopsis leaf protoplasts. The GUS activity in individual samples was normalized against the luciferase activity. Error bars represent se of three biological replicates.
Expression of the MYB46 Gene Is Regulated by SND1
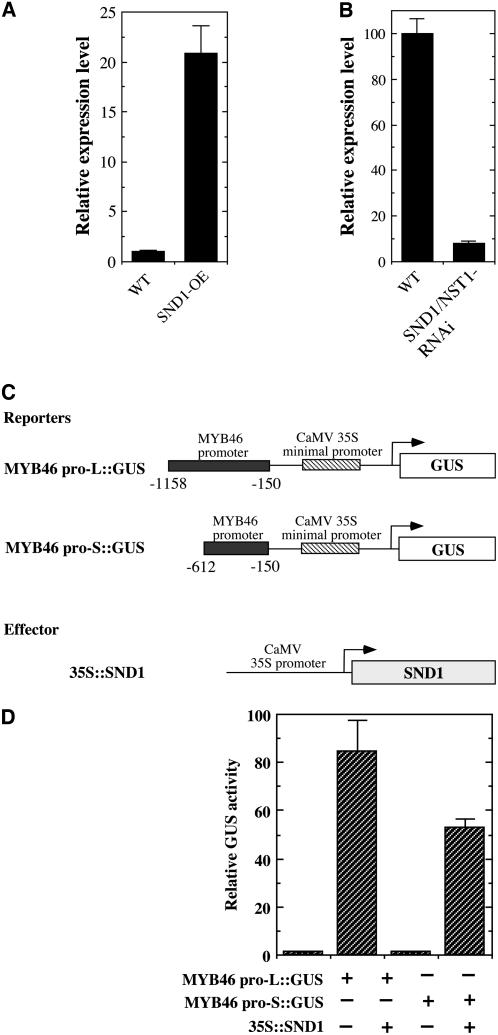
Because the MYB46 gene is associated with secondary wall thickening, in a similar way to SND1, we wished to study whether its expression is regulated by SND1. Examination of the expression level of MYB46 in SND1 overexpressors revealed that MYB46 was induced ∼21-fold compared with the wild type (Figure 3A). In addition, the MYB46 expression was found to be downregulated to 7.8% of that of the wild type by simultaneous inhibition of SND1 and NST1 (Figure 3B). These results suggest that MYB46 may be a target of SND1.
Figure 3.
SND1 Regulates the Expression of the MYB46 Gene.
(A) Real-time quantitative PCR analysis showing an induction of MYB46 expression by SND1 overexpression (SND1-OE). The expression level of MYB46 in the wild type is set to 1. Error bars represent se of three biological replicates.
(B) Real-time quantitative PCR analysis showing a marked reduction in MYB46 expression by RNA interference (RNAi) inhibition of SND1 and NST1 (SND1/NST1-RNAi). The expression level of MYB46 in the wild type is set to 100. Error bars represent se of three biological replicates.
(C) Diagrams of the reporter and effector constructs used for transactivation analysis. The effector construct contains the SND1 cDNA driven by the CaMV 35S promoter (35S∷SND1). The reporter constructs consist of the GUS reporter gene driven by the MYB46 promoter linked with the CaMV 35S minimal promoter sequence. The MYB46 promoter sequence used is between −150 and −1158 (MYB46 pro-L∷GUS) or −150 and −612 (MYB pro-S∷GUS) relative to the start codon of the MYB46 gene.
(D) Transactivation analysis showing that SND1 activates MYB46 promoter-driven expression of GUS reporter gene. The reporter and effector constructs together with a construct expressing the luciferase gene were cotransfected into Arabidopsis leaf protoplasts. The GUS activity in individual samples was normalized against the luciferase activity. Error bars represent se of three biological replicates.
SND1 Binds to and Activates the MYB46 Promoter
We next investigated whether SND1 could activate the transcription of MYB46 promoter-driven GUS reporter gene. Cotransfection of Arabidopsis leaf protoplasts with the cauliflower mosaic virus (CaMV) 35S promoter–driven SND1 expression construct and the MYB46 promoter-linked GUS reporter gene demonstrated that SND1 was able to transactivate the expression of MYB46 promoter-driven GUS reporter gene (Figures 3C and 3D).
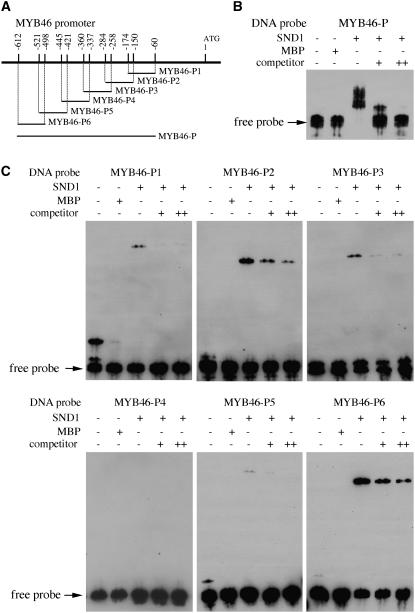
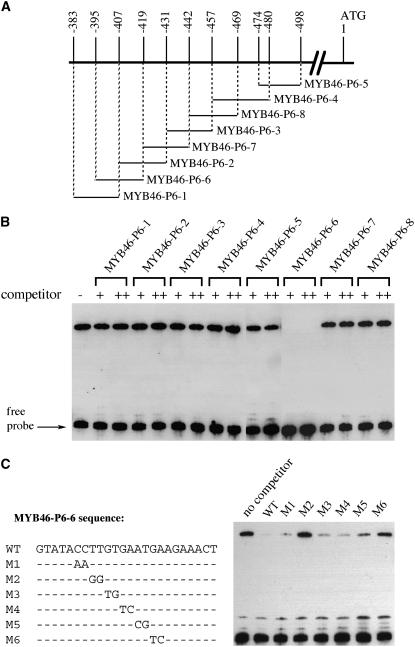
To investigate whether SND1 binds to the MYB46 promoter, we applied electrophoretic mobility shift assay (EMSA) using recombinant maltose binding protein (MBP)-SND1 fusion protein and a 553-bp MYB46 promoter fragment that was shown to be activated by SND1 in the transactivation analysis. It was found that the recombinant SND1 protein was able to bind to the MYB46 promoter fragment and caused a mobility shift (Figures 4A and 4B). The observations that addition of unlabeled MYB46 promoter fragment competed with the binding in a dose-dependent manner and the mobility shift was not seen when the MYB46 promoter fragment was incubated with MBP alone (Figure 4B) indicate that the binding of SND1 to the MYB46 promoter is specific. Further EMSAs using six ∼100-bp overlapping fragments of the MYB46 promoter sequence demonstrated that two regions, MYB46-P2 and MYB46-P6, exhibited very strong binding by SND1, and two other fragments, MYB46-P1 and MYB46-P3, appeared to show weak binding (Figures 4A and 4C). These results indicate that there are at least two strong SND1 binding sites in the MYB46 promoter.
Figure 4.
EMSA of SND1 Binding to the Promoter Sequence of MYB46 Gene.
The NAC domain of SND1 fused with MBP was expressed in E. coli, and the purified recombinant protein was incubated with biotin-labeled MYB46 promoter fragments. The samples were subjected to EMSA by polyacrylamide gel electrophoresis. The biotin-labeled DNA fragments were detected with the chemiluminiscent method.
(A) Diagram of the MYB46 promoter showing the DNA fragments used for EMSA. The positions of individual fragments relative to the start codon are shown above the line.
(B) EMSA of SND1 binding to a 553-bp fragment (located between −60 and −612 relative to the start codon) of the MYB46 promoter. MBP was used as a control protein. For competition analysis, unlabeled DNA fragments (competitors) in 30-fold (+) or 60-fold (++) molar excess relative to the labeled probes were included in the reactions.
(C) EMSA of SND1 binding to different regions of the MYB46 promoter. The locations of DNA fragments used for EMSA are shown in (A). The MYB46-P2 and MYB46-P6 fragments exhibited strong binding by SND1.
We next used overlapping oligonucleotides within the MYB46-P6 fragment to define the sequence that was bound by SND1 (Figure 5). Competition analysis demonstrated that a 24-bp oligonucleotide, MYB46-P6-6, was able to compete with the MYB46-P6 fragment for binding to SND1 (Figure 5B). The MYB46-P6-1 and MYB46-P6-2 oligonucleotides, which overlapped with the 5′ 12-bp and 3′ 12-bp sequences, respectively, of MYB46-P6-6, did not show any competition with MYB46-P6. Further competition analysis with mutated MYB46-P6-6 oligonucleotides revealed that mutations in M2 and M6 resulted in a loss of their ability to compete with MYB46-P6 for binding to SND1 (Figure 5C), suggesting that the nucleotides mutated in M2 and M6 were critical for SND1 binding. These results demonstrate that SND1 binds to the 24-bp MYB46-P6-6 sequence.
Figure 5.
SND1 Binds to a 24-bp Sequence in the MYB46-P6 Fragment.
(A) Diagram of the overlapping DNA oligonucleotides in the MYB46-P6 fragment used in EMSA. The positions of individual oligonucleotides relative to the start codon are shown.
(B) Competition analysis of SND1 binding to MYB46-P6 by the overlapping DNA oligonucleotides shown in (A). Unlabeled double-stranded DNA oligonucleotides (competitors) in 30-fold (+) or 60-fold (++) molar excess relative to the labeled MYB46-P6 probe were used for competition in EMSA. A 24-bp oligonucleotide, MYB46-P6-6, was found to compete with MYB46-P6 for binding to SND1.
(C) Competition analysis of SND1 binding to MYB46-P6 by mutated MYB46-P6-6 oligonucleotides. The wild-type and mutated (M1 to M6) MYB46-P6-6 sequences are shown in the left panel. Dashes in M1 to M6 denote nucleotides that are identical to the wild-type sequence. Unlabeled wild-type and mutated MYB46-P6-6 oligonucleotides in 30-fold molar excess relative to the labeled MYB46-P6 probe were used as competitors in EMSA (right panel).
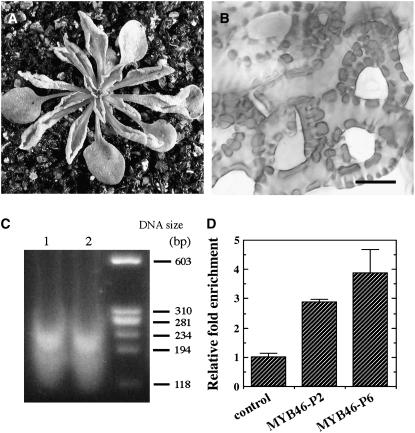
To further substantiate the results from the EMSA, we used the chromatin immunoprecipitation approach to examine whether SND1 binds to the MYB46 promoter in vivo. To do this, we overexpressed GFP-tagged SND1 in Arabidopsis. Overexpression of GFP-tagged SND1 led to ectopic secondary wall thickening in leaf epidermal and mesophyll cells (Figure 6B) and a concomitant curling of leaves (Figure 6A), the same phenotypes as those caused by overexpression of SND1 (Zhong et al., 2006). This indicates that the SND1-GFP fusion protein acts the same as SND1 in the activation of secondary wall biosynthetic pathways. Therefore, transgenic plants overexpressing SND1-GFP can be used for analysis of SND1 binding sequences. Formaldehyde cross-linked chromatin from SND1-GFP overexpressors was fragmented into 100 to 250 bp in size (Figure 6C) and immunoprecipitated with the GFP antibody for enrichment of SND1-GFP bound DNA fragments. The immunoprecipitated DNA fragments were then used as templates in quantitative PCR detection of MYB46 promoter sequences. It was found that the two fragments in the MYB46 promoter, MYB46-P2 and MYB46-P6, that were strongly bound by SND1 in the EMSA were enriched threefold to fivefold compared with control DNA (Figure 6D). The results from both in vitro and in vivo binding analyses demonstrate that SND1 directly binds to the promoter of the MYB46 gene to regulate its expression.
Figure 6.
Chromatin Immunoprecipitation Analysis of SND1 Binding to the MYB46 Promoter Sequence in Vivo.
Chromatin isolated from transgenic Arabidopsis seedlings expressing SND1-GFP was immunoprecipitated with the GFP antibody. The DNA before (input) and after (bound) immunoprecipitation was used for analysis of enrichment of the MYB46 promoter sequence.
(A) Overexpression of SND1-GFP fusion protein in transgenic Arabidopsis plants caused the same curly leaf phenotype as SND1 overexpression.
(B) Overexpression of SND1-GFP results in ectopic deposition of thick secondary walls in the epidermal cells of leaves. Bar = 8 μm.
(C) Agarose gel separation of sonicated chromatin isolated from SND-GFP overexpressors showing the DNA fragments in the range of 100 to 250 bp. Lanes 1 and 2 are DNA fragments from two independent chromatin preparations. The DNA size markers are indicated at right.
(D) Real-time quantitative PCR analysis showing the enrichment of the MYB46 promoter sequence after chromatin immunoprecipitation. The values of bound over input for MYB46-P2 and MYB46-P6 promoter fragments were normalized against that of the control DNA. Error bars represent se of three replicates of two biological samples.
Dominant Repression of MYB46 Causes a Drastic Reduction in Secondary Wall Thickening
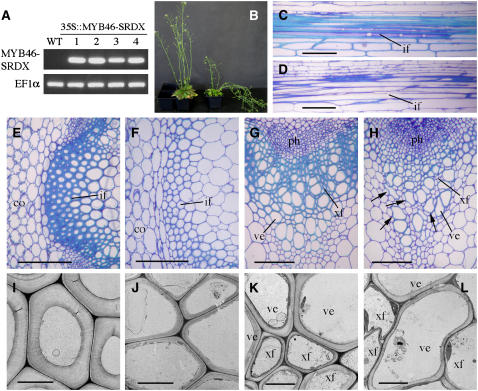
Because the MYB46 gene is directly regulated by SND1, we reasoned that it is potentially involved in the regulation of secondary wall biosynthesis. We next investigated whether repression of MYB46 functions could lead to inhibition of secondary wall thickening in fibers and vessels. It has been demonstrated that the functions of several secondary wall–associated transcription factors, such as VND6, VND7, NST1, NST2, and SND1, could be inhibited by overexpression of these transcription factors fused with the EAR-dominant repression domain (Kubo et al., 2005; Mitsuda et al., 2005; Zhong et al., 2006). To repress the functions of endogenous MYB46, the EAR repression domain was fused to MYB46 and expressed under the control of the CaMV 35S promoter in transgenic Arabidopsis plants. Histological examination of fibers in the stems showed that among 42 transgenic plants in the first generation, 14 of them had an apparent reduction in the secondary wall thickening of interfascicular fibers and a concomitant weak stem phenotype (Figure 7B). RT-PCR analysis confirmed the expression of the MYB46 repressor transcripts in these plants (Figure 7A). Three transgenic lines showing the strongest reduction in secondary wall thickness were chosen for further analyses, and the results from one representative line are shown hereafter. Cross sections of the basal part of stems revealed that the wall thickness of interfascicular fibers was reduced to 26% of that of the wild type by MYB46-dominant repression (Figures 7E, 7F, 7I, and 7J, Table 1). The wall thickness of vessels and xylary fibers was decreased to 67 and 24%, respectively, of that of the wild type (Figures 7G, 7H, 7K, and 7L, Table 1). Some of the vessels in the MYB46 repressors exhibited a deformed morphology (Figure 7H), probably due to the reduced wall thickness that rendered vessels unable to resist the negative pressure caused by transpiration. The length of interfascicular fiber cells was not affected by MYB46 repression (Figures 7C and 7D). These results demonstrate that dominant repression of MYB46 reduces secondary wall deposition in both fibers and vessels. It should be noted that the phenotypes caused by dominant repression experiments may not directly reflect those of knockout mutants. Dominant repressors inhibit not only the functions of the transcription factors targeted for repression but also the functions of their homologs by competing with their binding to the same cis-elements or interacting proteins. The dominant repression approach has been successfully used to facilitate the analysis of functionally redundant transcription factors (Hiratsu et al., 2004). For example, it has been shown that dominant repression of either NST1 or NST2 causes the same secondary wall defects in endothecium as in the double knockout mutant of NST1 and NST2, whereas the single knockout mutants of NST1 or NST2 do not show any phenotypes (Mitsuda et al., 2005). No phenotypes were observed in the T-DNA knockout lines of MYB46 (data not shown), probably due to the functional redundancy of its homologs.
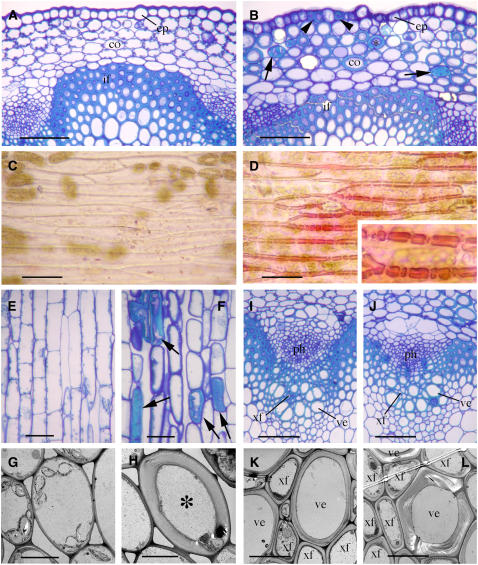
Figure 7.
Reduction of Secondary Wall Thickening in Fibers and Vessels by Dominant Repression of MYB46.
The full-length MYB46 cDNA fused in frame with the dominant EAR repression sequence was expressed in transgenic Arabidopsis plants. The bottom internodes of 8-week-old plants were used for examination of secondary walls in fibers and vessels. co, cortex; if, interfascicular fiber; ph, phloem; ve, vessel; xf, xylary fiber. Bars = 103 μm in (C) to (F), 40 μm in (G) and (H), and 6 μm in (I) to (L).
(A) RT-PCR analysis showing the presence of the MYB46 repressor (MYB46-SRDX) transcript in the stems of four representative transgenic lines. The expression level of EF1α was used as a control.
(B) Wild-type plant (left) and a transgenic Arabidopsis plant expressing the MYB46 repressor (right).
(C) and (D) Longitudinal sections of the interfascicular regions of stems showing the similar length of fiber cells in the wild type (C) and the MYB46 repressors (D).
(E) and (F) Cross sections of the interfascicular regions showing a reduction in the wall thickness of fibers in the MYB46 repressors (F) compared with the wild type (E).
(G) and (H) Cross sections of vascular bundles showing mildly collapsed vessels (arrows) and an apparent reduction in the wall thickness of xylary fibers in the MYB46 repressors (H) compared with the wild type (G).
(I) and (J) Transmission electron micrographs of interfascicular fiber walls of the wild type (I) and the MYB46 repressors (J).
(K) and (L) Transmission electron micrographs of xylem cells showing reduced wall thickness in both xylary fibers and vessels in the MYB46 repressors (L) compared with the wild type (K).
Table 1.
Wall Thickness of Vessels and Fibers in the Stems of Wild-Type, MYB46 Repressors, and MYB46 Overexpressors
| Plant | Interfascicular Fibers | Vessels | Xylary Fibers |
|---|---|---|---|
| Wild type | 2.27 ± 0.27 | 0.92 ± 0.15 | 0.59 ± 0.08 |
| MYB46 repressors | 0.58 ± 0.08 | 0.62 ± 0.10 | 0.14 ± 0.04 |
| MYB46 overexpressors | 1.72 ± 0.23 | 1.56 ± 0.31 | 0.60 ± 0.07 |
Wall thickness was measured from transmission electron micrographs of fibers and vessels. Data are means (μm) ± se from 15 cells.
Overexpression of MYB46 Activates Secondary Wall Biosynthetic Pathways and Results in Ectopic Deposition of Secondary Walls
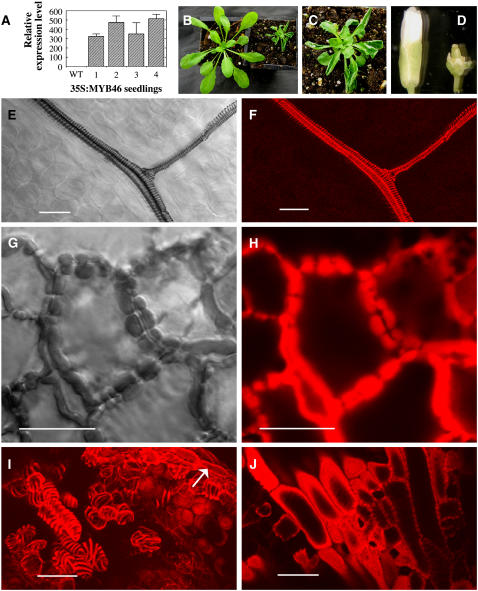
To investigate whether MYB46 is sufficient to activate secondary wall biosynthesis, we overexpressed MYB46 in wild-type Arabidopsis (Figure 8A). Among 64 transgenic plants, 47 of them had small rosette size due to the stunted growth of leaves with severely upward-curling blades (Figures 8B and 8C). Examination of leaf epidermis and mesophyll cells revealed ectopic deposition of secondary walls. In wild-type leaves, lignified secondary wall thickening was only observed in the xylem strands of veins but absent in epidermis and mesophyll cells (Figures 8E and 8F). By contrast, in the leaves of MYB46 overexpressors, band-like lignified secondary walls were deposited in many epidermal (Figures 8G and 8H) and mesophyll cells (data not shown). It appeared that the thick-walled epidermal cells lack the typical sinuous morphology (Figure 8G) as seen in the wild type, indicating that the ectopic secondary wall thickening occurred before the cells reached their full expansion.
Figure 8.
Overexpression of MYB46 Leads to Ectopic Deposition of Secondary Walls in the Epidermis of Leaves and Parenchyma Cells of Floral Organs.
The full-length MYB46 cDNA driven by the CaMV 35S promoter was expressed in transgenic Arabidopsis plants. Leaves from 3-week-old plants were examined for ectopic deposition of lignified secondary walls by collecting differential interference contrast (DIC) and lignin autofluorescence images. Bars = 24 μm in (E) to (J).
(A) Real-time quantitative PCR analysis showing MYB46 overexpression in the seedlings of four representative transgenic lines. Error bars represent se of three replicates.
(B) Three-week-old seedlings of the wild type (left) and an MYB46 overexpressor (right) showing a reduction in the rosette size.
(C) High magnification of a MYB46 overexpressor showing upward-curling leaves.
(D) Flowers of the wild type (left) and a MYB46 overexpressor (right) showing the reduced size of floral organs.
(E) and (F) DIC (E) and lignin autofluorescence (F) images of a wild-type leaf showing the helical secondary wall thickening in veins.
(G) and (H) DIC (G) and lignin autofluorescence (H) images of leaf epidermis of MYB46 overexpressors showing ectopic deposition of lignified secondary walls with a banded pattern.
(I) and (J) Lignin autofluorescence images of the anther (I) and carpel (J) of MYB46 overexpressors showing ectopic deposition of lignified secondary walls with a helical pattern in parenchyma cells. Arrow in (I) indicates the normal secondary wall thickening in the endothecium. In wild-type anthers and carpels, lignified secondary walls are only present in endothecium and vascular strands, respectively (data not shown).
Overexpression of MYB46 also led to ectopic secondary wall thickening in floral organs and inflorescence stems. The flowers of MYB46 overexpressors were often sterile and had severely shortened sepals, petals, stamens, and carpels (Figure 8D). In these organs, many cells that are normally parenchymatous in the wild type deposited helical or band-like lignified secondary walls (Figures 8I and 8J). Similarly, secondary walls were ectopically laid down in many epidermal and cortical cells in the inflorescence stems (Figures 9A to 9D, 9G, and 9H). The ectopic secondary walls in the cortical cells of stems were deposited in a reticulated or pitted pattern (Figure 9B), whereas those in the epidermal cells formed thick bands (Figure 9D). It was evident that the ectopic secondary walls of some cortical cells were even thicker than that of normal fiber cells (Figure 9B). The length of the cortical cells with ectopic secondary walls appeared to be comparable with that of the wild type (Figures 9E and 9F). In addition, overexpression of MYB46 led to an apparent increase in the secondary wall thickness of vessels (Figures 9K and 9L, Table 1). By contrast, the wall thickness of interfascicular fibers was reduced (Table 1).
Figure 9.
MYB46 Overexpression Causes Ectopic Deposition of Secondary Walls in the Parenchyma Cells of Inflorescence Stems and Results in an Increased Secondary Wall Thickening in Vessels.
Stems of 8-week-old plants were used for examination of secondary wall thickening by toluidine blue or phloroglucinol-HCl staining. co, cortex; ep, epidermis; if, interfascicular fiber; ph, phloem; ve, vessel; xf, xylary fiber. Bars = 66 μm in (A), (B), (E), (F), (I), and (J), 39 μm in (C) and (D), and 8 μm in (G), (H), (K), and (L).
(A) and (B) Cross sections of stems showing ectopic deposition of thick secondary walls in cortical cells (arrows and asterisk) and epidermis (arrowheads) in the MYB46 overexpressors (B) compared with the wild type (A). Note that some cortical cells showed reticulated secondary wall thickening (arrows), and the walls of some cortical cells (asterisk) are thicker than those of interfascicular fibers.
(C) and (D) Phloroglucinol-HCl staining of the epidermal peels of stems showing the ectopic deposition of lignified secondary walls (inset) in the MYB46 overexpressors (D) compared with the wild type (C).
(E) and (F) Longitudinal sections of cortical regions showing that the thick-walled cortical cells (arrows) in the MYB46 overexpressors (F) had similar length as those of the wild type (E).
(G) and (H) Transmission electron micrographs of cortical cells showing their thick walls (asterisk) in the MYB46 overexpressors (H) compared with the wild type (G).
(I) and (J) Cross sections of vascular bundles of the wild type (I) and MYB46 overexpressors (J).
(K) and (L) Transmission electron micrographs of xylem showing an increase in secondary wall thickening in the vessels of MYB46 overexpressors (L) compared with the wild type (K).
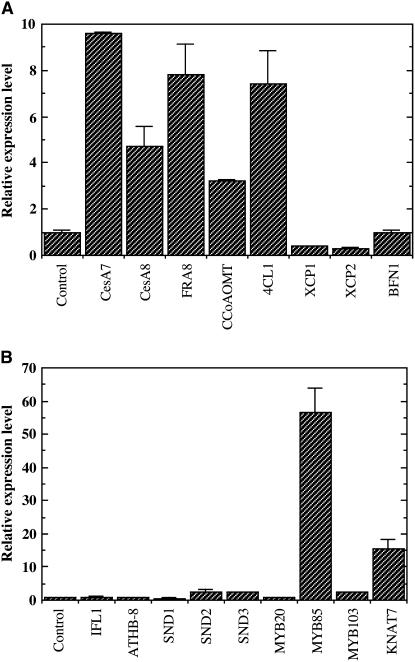
To investigate whether the ectopic secondary wall thickening consists of typical secondary wall components, we examined the presence of cellulose, xylan, and lignin in the ectopically deposited secondary walls in the stems of MYB46 overexpressors. Histological staining of cellulose and lignin and immunostaining of xylan revealed the presence of all three major secondary wall components, including cellulose, xylan, and lignin (Figure 10). Quantitative PCR analysis (Figure 11A) showed that overexpression of MYB46 resulted in a drastic induction in the expression of cellulose synthase genes CesA7 and CesA8 (Taylor et al., 2004), the xylan biosynthetic gene FRAGILE FIBER8 (FRA8) (Zhong et al., 2005; Pena et al., 2007), and the lignin biosynthetic genes 4CL1 (for hydroxycinnamate CoA ligase) and CCoAOMT (for caffeoyl CoA O-methyltransferase) (Boerjan et al., 2003). These results suggest that overexpression of MYB46 activates the biosynthetic pathways of all three major secondary wall components, thus leading to ectopic deposition of secondary walls. The expression of programmed cell death–associated genes, including XYLEM CYSTEINE PEPTIDASE1 (XCP1), XCP2, and BIFUNCTIONAL NUCLEASE1 (BFN1) (Funk et al., 2002; Ito and Fukuda, 2002), was not affected by MYB46 overexpression.
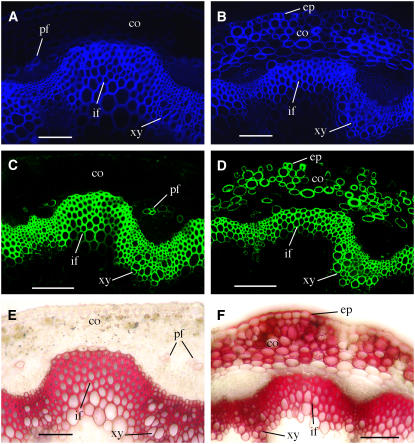
Figure 10.
Deposition of Cellulose, Xylan, and Lignin in the Ectopic Secondary Walls in MYB46 Overexpressors.
The stem sections were stained with Calcofluor White or phloroglucinol-HCl for detection of cellulose and lignin, respectively. The LM10 xylan monoclonal antibody was used for immunodetection of xylan. co, cortex; ep, epidermis; if, interfascicular fiber; pf, phloem fiber; xy, xylem. Bars = 105 μm.
(A) and (B) Calcofluor White staining of stem sections showing intensive cellulose staining in the walls of cortical cells and epidermis in addition to interfascicular fibers and xylem cells in MYB46 overexpressors (B) compared with the wild type (A).
(C) and (D) Stem sections probed with the LM10 xylan monoclonal antibody showing intensive xylan staining in the walls of cortical cells and epidermis in addition to interfascicular fibers and xylem cells in MYB46 overexpressors (D) compared with the wild type (C).
(E) and (F) Phloroglucinol-HCl staining of stem sections showing intensive lignin staining in the walls of cortical cells and epidermis in addition to interfascicular fibers and xylem cell walls in MYB46 overexpressors (F) compared with the wild type (E).
Figure 11.
Induction of Secondary Wall Biosynthetic Genes and Transcription Factors by MYB46 Overexpression.
(A) Real-time quantitative PCR analysis of the expression of secondary wall biosynthetic genes in 3-week-old seedlings of MYB46 overexpressors compared with the wild type. The expression levels of genes involved in the biosynthesis of cellulose (CesA7 and CesA8), xylan (FRA8), and lignin (CCoAOMT and 4CL1) and genes involved in programmed cell death (XCP1, XCP2, and BFN1) were examined. Error bars represent se of three replicates. Note the drastic induction in the expression of secondary wall biosynthetic genes but not cell death–associated genes.
(B) Real-time quantitative PCR analysis of the expression levels of secondary wall–associated transcription factors in 3-week-old seedlings of MYB46 overexpressors. Note the dramatic induction in the expression of MYB85 and KNAT7. The expression levels of two non-secondary wall–associated transcription factors, IFL1 and ATHB-8, which are associated with the differentiation of interfascicular fibers and xylem, respectively, are shown for comparison.
Overexpression of MYB46 Upregulates the Expression of Two Secondary Wall–Associated Transcription Factors
It has been previously shown that overexpression of SND1 induces the expression of six secondary wall–associated transcription factors, which led to the proposal that SND1 is a key regulator in coordinating the expression of these transcription factors, which in turn activate different sets of genes involved in secondary wall biosynthesis (Zhong et al., 2006). The demonstration that MYB46 is a direct target of SND1 prompted us to investigate whether MYB46 also induces the expression of the same sets of transcription factors as does SND1. Quantitative PCR analysis of the six transcription factors upregulated by SND1 demonstrated that the expression of MYB85 (At4g22680) and KNAT7 (At1g62990) were induced by MYB46 overexpression (Figure 11B). The expression of MYB103 and two NAC genes, SND2 (At4g28500) and SND3 (At1g28470), was only slightly induced. The expression of SND1 was not affected (Figure 11B). In addition, MYB46 overexpression did not affect the expression of the IFL1 gene, which is involved in the initiation of fiber differentiation (Zhong and Ye, 1999) or that of the ATHB8 gene, which was proposed to be involved in the early stage of xylem differentiation (Baima et al., 2001). These results demonstrate that MYB46 upregulates the expression of MYB85 and KNAT7, two secondary wall–associated transcription factors that are also induced by SND1.
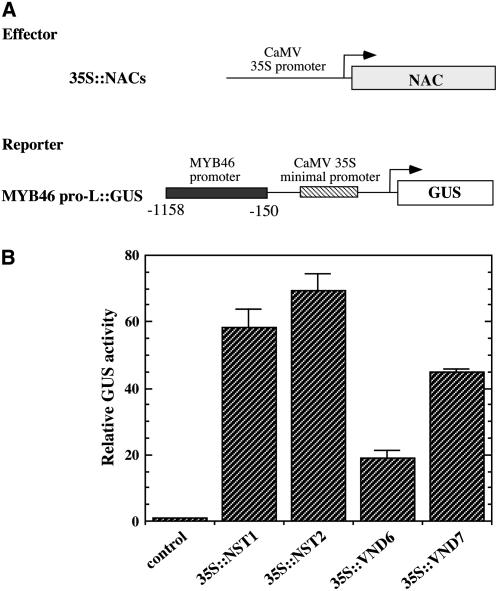
The MYB46 Promoter Can Be Activated by the SND1 Homologs NST1, NST2, VND6, and VND7
It was previously shown that SND1 and its close homolog, NST1, act redundantly in the regulation of secondary wall biosynthesis in fibers (Zhong et al., 2006, 2007; Mitsuda et al., 2007). In addition, three other SND1 homologs, NST2, VND6, and VND7, have been demonstrated to be important for secondary wall thickening in endothecium or vessels (Kubo et al., 2005; Mitsuda et al., 2005). We investigated whether these secondary wall–associated NACs could also activate the MYB46 promoter by transactivation analysis. Cotransfection of Arabidopsis protoplasts with the expression constructs of NACs and the MYB46 promoter-driven GUS reporter gene construct revealed that NST1, NST2, VND6, and VND7 were able to activate the expression of the GUS reporter gene (Figure 12), suggesting that MYB46 might be a common target of these secondary wall–associated NACs.
Figure 12.
Activation of the MYB46 Promoter by NST1, NST2, VND6, and VND7.
(A) Diagrams of the effector and reporter constructs used for transactivation analysis. The effector constructs contain the NST1, NST2, VND6, or VND7 cDNA driven by the CaMV 35S promoter. The reporter construct consists of the GUS reporter gene driven by the MYB46 promoter linked with the CaMV 35S minimal promoter sequence.
(B) Transactivation analysis showing that NST1, NST2, VND6, and VND7 activate the MYB46 promoter-driven expression of GUS reporter gene. Error bars represent se of three biological replicates.
DISCUSSION
The biosynthesis of secondary walls in fibers and tracheary elements during wood formation requires the coordinated activation of the biosynthetic pathways of cellulose, xylan, and lignin. Because wood, the most abundant biomass produced by plants, is an important raw material for traditional forest products and potentially for biofuel production, it is imperative to understand the molecular mechanisms underlying the regulation of secondary wall biosynthesis. Recent studies have demonstrated that two NAC domain transcription factors, SND1 and NST1, function redundantly in the activation of secondary wall biosynthesis and in the regulation of expression of several secondary wall–associated transcription factors, which opens a new avenue to study how secondary wall biosynthesis is regulated during wood formation. Our current findings that MYB46 is a direct target of SND1 and itself is also essential for secondary wall thickening mark another important step toward molecular dissection of the transcriptional network regulating secondary wall biosynthesis.
MYB46 Is a Direct Target of SND1
We have demonstrated that MYB46 is a direct target of SND1 in regulating secondary wall biosynthesis based on the following lines of evidence. First, MYB46 is highly induced by overexpression of SND1 but not vice versa. Second, MYB46 is downregulated by simultaneous inhibition of SND1 and NST1. Third, SND1 is able to activate the expression of the MYB46 promoter-driven GUS reporter gene in leaf protoplasts. Fourth, SND1 directly binds to the MYB46 promoter as demonstrated in the EMSA and the chromatin immunoprecipitation analysis. These results demonstrate that SND1 activates the expression of the MYB46 gene by directly binding to its promoter. They further suggest that MYB46 is an important player in the SND1-mediated transcriptional regulation of secondary wall biosynthesis.
The demonstration that MYB46 is a direct target of SND1 is consistent with their expression patterns. Both SND1 and MYB46 are expressed during secondary wall biosynthesis in interfascicular fibers and xylary fibers, indicating that they function in the same cell types. However, MYB46 was also found to be expressed in vessels. Because SND1 is not expressed in vessels, it is likely that other SND1 homologs are involved in the activation of the MYB46 gene in vessels. Possible candidates are the VND proteins that belong to the same subgroup as SND1 (Zhong et al., 2006). Indeed, two of the VND genes, VND6 and VND7, which are key regulators for vessel development (Kubo et al., 2005), were found to be able to activate the MYB46 promoter (Figure 12). It should be noted that although MYB46 is expressed in vessels and essential for their secondary wall thickening, it appears not to be involved in the regulation of genes involved in programmed cell death during vessel development. This suggests that the transcriptional controls leading to secondary wall thickening and programmed cell death in vessels are not coupled by MYB46.
MYB46 Regulates Secondary Wall Biosynthesis in Both Fibers and Vessels
The available evidence suggests that MYB46 is a key player in the SND1-mediated induction of secondary wall biosynthesis. The MYB46 gene is predominantly expressed in cells undergoing secondary wall thickening in stems, and dominant repression of MYB46 causes a reduction in secondary wall thickening in both fibers and vessels. Overexpression of MYB46 activates the biosynthetic pathways of cellulose, xylan, and lignin and concomitantly leads to ectopic deposition of secondary walls in cells that are normally parenchymatous. These results indicate that MYB46 is another key switch regulating the overall secondary wall biosynthesis. Several MYB genes have been demonstrated to be associated with the regulation of monolignol biosynthetic pathway (Tamagnone et al., 1998; Patzlaff et al., 2003; Karpinska et al., 2004; Goicoechea et al., 2005). For example, the Eucalyptus Eg MYB2 protein, which is a close homolog of MYB46, has been shown to bind to the promoters of lignin biosynthetic genes, and when overexpressed in tobacco (Nicotiana tabacum), causes upregulation of lignin biosynthetic genes and a concomitant increase in the secondary wall thickness (Goicoechea et al., 2005). It is not known whether Eg MYB2 overexpression also affects the biosynthesis of other secondary wall components in addition to lignin. So far, only two members in the MYB family, MYB46 and MYB26, have been conclusively demonstrated to be involved in regulation of the biosynthetic pathways of all three major secondary wall components, albeit in different cell types.
It has been observed that overexpression of either SND1 or MYB46 leads to ectopic secondary wall deposition in many different cell types and organs, including epidermis and mesophyll cells of leaves, epidermis of stems, and floral organs. This indicates that these cell types are competent for induction of secondary wall thickening by SND1 and MYB46. It is interesting to note that although overexpression of SND1 rarely induces secondary wall thickening in the cortical cells of stems, MYB46 overexpression results in ectopic deposition of secondary walls in these cells. This could be due to differential responses of the cortical cells to SND1 and MYB46. Alternatively, the cortical cells in MYB46 overexpressors might accumulate a higher level of MYB46 protein than those of SND1 overexpressors, which is needed to turn on the secondary wall biosynthetic pathways. The overexpression studies of transcription factors from this and several other reports (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006; Yang et al., 2007) demonstrate that most parenchyma cells remain competent to enter the secondary wall biosynthetic program and that whether a particular cell type is parenchymatous or sclerenchymatous may be determined by the expression of these key transcriptional regulators.
Transcriptional Network Regulating Secondary Wall Biosynthesis
In addition to MYB46, SND1 regulates the expression of several other secondary wall–associated transcription factors, including MYB20, MYB85, MYB103, SND2, SND3, and KNAT7 (Zhong et al., 2006), suggesting that they also might be downstream targets of SND1.
Based on the findings that MYB46 is a direct target of SND1 and a key player in activating secondary wall biosynthesis, we propose that a transcriptional hierarchy is involved in the regulation of secondary wall biosynthesis in fibers and vessels. In this scenario, SND1 coordinately activates a transcriptional cascade in which a set of transcription factors is induced by SND1 and they then activate a subset of transcription factors involved in turning on secondary wall biosynthetic genes. This hypothesis is supported by the fact that two SND1-upregulated transcription factors, MYB85 and KNAT7, are also induced by MYB46, indicating that they may be targets of MYB46. It is also consistent with previous findings that some MYB genes are specifically involved in the regulation of the monolignol biosynthetic pathway (Tamagnone et al., 1998; Patzlaff et al., 2003; Karpinska et al., 2004; Goicoechea et al., 2005) through direct activation of monolignol biosynthetic genes via binding to their promoters (Patzlaff et al., 2003; Goicoechea et al., 2005). A similar transcriptional cascade may also exist for the regulation of secondary wall thickening in endothecium of anthers, in which MYB26 upregulates the expression of two secondary wall–promoting NAC genes, NST1 and NST2 (Yang et al., 2007).
It is intriguing to find that the SND1 close homologs, NST1, NST2, VND6, and VND7, can also activate the MYB46 promoter. This finding indicates that these secondary wall–associated NACs are functional homologs of SND1, and they may activate the same downstream targets, thereby regulating secondary wall biosynthesis. Since these NAC genes have been shown to be required for secondary wall deposition in different cell types (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2007), it is conceivable that they control secondary wall biosynthesis in a cell type–specific manner.
In conclusion, we have demonstrated that MYB46 is an important player in the SND1-mediated transcriptional network regulating secondary wall biosynthesis. Our findings provide an additional framework for further dissecting the cascade of transcription factors and their roles in the regulation of secondary wall biosynthetic genes. It will be interesting to find out which transcription factors directly control the biosynthetic pathways of cellulose, xylan, and lignin. It is likely that different transcription factors are involved in the activation of different biosynthetic pathways of secondary wall components. Because secondary walls constitute the bulk part of wood, which is the most abundant plant biomass widely used for pulping and construction as well as potentially for biofuel production, further studies of the transcriptional network regulating secondary wall biosynthesis will likely enable us to genetically alter the biosynthetic pathways of individual secondary wall components. Knowledge gained from such studies may potentially lead to better strategies for genetic manipulation of fiber or wood quality and quantity.
METHODS
Gene Expression Analysis
For analysis of gene expression in different cell types, interfascicular fiber cells, xylem cells, and pith cells from inflorescence stems of 6-week-old Arabidopsis thaliana plants were isolated using the PALM microlaser system (PALM Microlaser Technologies), and the isolated cells were used for RNA isolation and amplification as described (Zhong et al., 2006).
For analysis of gene expression in different organs, total RNA from Arabidopsis seedlings, leaves, roots, stems, and apices was isolated using a Qiagen RNA isolation kit. The seedlings used were 2 weeks old. Mature leaves were from 6-week-old plants. Inflorescence apices and mature roots were from 8-week-old plants. Stems I and II were from 4- and 8-week-old plants, respectively.
For quantitative PCR analysis, total RNA was treated with DNase I and used for first-strand cDNA synthesis. The first-strand cDNA was used as template for real-time quantitative PCR analysis with the QuantiTect SYBR Green PCR kit (Clontech). The relative mRNA levels were determined by normalizing the PCR threshold cycle number of each gene with that of the EF1α reference gene. The expression level of each gene in the wild-type control or in the sample with the lowest expression level was set to 1, and the data were the average of three replicates.
The developmental expression pattern of the MYB46 gene was studied using the GUS reporter gene. A 6.9-kb genomic DNA fragment containing a 3-kb 5′ upstream sequence, the entire MYB46 exon and intron region, and a 1.9-kb 3′ downstream sequence were used. The GUS reporter gene was inserted in frame right before the stop codon of the MYB46 gene and then cloned into the binary vector pBI101 (Clontech) to create the MYB46:GUS construct. The construct was transformed into wild-type Arabidopsis plants by the Agrobacterium tumefaciens–mediated transformation procedure (Bechtold and Bouchez, 1994). Transgenic plants were selected on kanamycin and used for expression analysis of the GUS reporter gene as described previously (Zhong et al., 2005).
Subcellular Localization of MYB46
The full-length MYB46 cDNA was fused in frame with the YFP cDNA and ligated between the CaMV 35S promoter and the nopaline synthase terminator in pBI221 (Clontech). The MYB46-YFP construct was introduced into Arabidopsis leaf protoplasts by polyethylene glycol–mediated transfection according to Sheen (2001). After 20 h of incubation, the transfected protoplasts were examined for yellow fluorescence signals using a Leica TCs SP2 spectral confocal microscope (Leica Microsystems). Images were saved and processed with Adobe Photoshop Version 7.0 (Adobe Systems).
Transcriptional Activation Analysis
For testing the ability of MYB46 to activate transcription, the MYB46 R2R3 domain or the MYB46 with deletion of the R2 motif was fused in frame with the GAL4 DNA binding domain and ligated between the CaMV 35S promoter and the nopline synthase terminator in the pBI221 vector (Clontech) to create the effector constructs. The reporter construct was created by placing the GAL4 upstream activation sequence linked with the CaMV 35S minimal promoter sequence (from −1 to −46 bp) in front of the GUS reporter gene. Arabidopsis leaf protoplasts were cotransfected with the reporter and effector constructs together with a construct containing the firefly luciferase gene driven by the CaMV 35S promoter for determination of the transfection efficiency. After 20 h of incubation, transfected protoplasts were lysed, and the soluble extracts were used for analysis of GUS and luciferase activities as described (Gampala et al., 2001). The GUS activity was normalized against the luciferase activity in each transfection, and the data were the average of three biological replicates.
For the transactivation analysis of the MYB46 promoter by SND1 and its close homologs, an effector construct was created by placing the full-length cDNA of SND1 or its homologs between the CaMV 35S promoter and the nopline synthase terminator in pBI221. The MYB46 promoter sequence between −150 and −1158 or −150 to −612 relative to the start codon was linked with the CaMV 35S minimal promoter, which was ligated upstream of the GUS reporter gene to create the reporter constructs. The reporter and effector constructs together with a luciferase construct were transfected into Arabidopsis leaf protoplasts, and the activities of GUS and luciferase were measured. The data were the average of three biological replicates.
EMSA
The NAC domain of SND1 was fused in frame with MBP and expressed in Escherichia coli. The recombinant SND1-MBP protein was purified using amylose resin and then used for EMSA with the MYB46 promoter fragments. The MYB46 promoter fragments were PCR amplified and biotin labeled at the 3′ end using the Biotin 3′ End DNA labeling kit (Pierce). The biotin-labeled DNA fragments were incubated for 30 min with 100 ng of SND1-MBP in binding buffer [10 mM Tris, pH 7.5, 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 0.05% Nonidet P-40, and 100 ng/μL poly(dI-dC)]. The SND1-bound DNA fragments were separated from the unbound ones by polyacrylamide gel electrophoresis. The DNA was electroblotted onto nitrocellulose membrane and detected by the chemiluminescent method.
Chromatin Immunoprecipitation Analysis
The full-length SND1 cDNA was fused in frame with the GFP cDNA (ABRC; developed by S.J. Davis and R.D. Vierstra) and ligated downstream of the CaMV 35S promoter in pBI121 (Clontech). The construct was introduced into wild-type Arabidopsis plants by Agrobacterium-mediated transformation, and transgenic plants overexpressing SND1-GFP were used for chromatin immunoprecipitation analysis as described (Gendrel et al., 2002). Transgenic seedlings exhibiting ectopic secondary wall deposition in leaves were cross-linked in 1% formaldehyde for 15 min under vacuum and quenched in 0.125 M glycine for 5 min. The seedlings were washed twice with deionized water and then ground in liquid nitrogen into a fine powder for extraction of chromatin.
To extract chromatin, the powder was resuspended in 30 mL of extraction buffer 1 (10 mM Tris-HCl, pH 8.0, 0.4 M sucrose, 5 mM β-mercaptoethanol, 1 mM PMSF, 10 μg/mL aprotinin, and 10 μg/mL leupeptin) and filtered through two layers of Miracloth before centrifugation at 2000g for 20 min. The pellet was resuspended in 1 mL of extraction buffer 2 (10 mM Tris-HCl, pH 8.0, 0.25 M sucrose, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 1 mM PMSF, 10 μg/mL aprotinin, and 10 μg/mL leupeptin) and centrifuged at 14,000g for 10 min. The pellet was resuspended in 300 μL of extraction buffer 3 (10 mM Tris-HCl, pH 8.0, 1.7 M sucrose, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, 1 mM PMSF, 10 μg/mL aprotinin, and 10 μg/mL leupeptin) and then layered on top of a cushion of 300 μL of extraction buffer 3 in an Eppendorf tube before centrifugation at 14,000g for 1 h at 4°C. The chromatin pellet was resuspended in 500 μL of nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, 1 mM PMSF, 10 μg/mL aprotinin, and 10 μg/mL leupeptin) and sonicated to small fragments with an average size of 100 to 250 bp.
The sonicated chromatin was diluted 1:10 in chromatin immunoprecipitation dilution buffer (16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.2 mM EDTA, and 1.1% Triton X-100) and precleared by incubation with Protein A agarose beads blocked with salmon sperm DNA (Upstate Biotechnology) for 1 h. The precleared chromatin was then incubated with 1 μg of GFP antibody (Clontech) overnight at 4°C. The SND1-GFP–bound chromatin was purified by incubation with Protein A agarose beads blocked with salmon sperm DNA for 1 h and subsequent washing for 5 min each in low-salt wash buffer (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 0.5% Triton X-100, 0.2% SDS, and 150 mM NaCl), high-salt wash buffer (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 0.5% Triton X-100, 0.2% SDS, and 500 mM NaCl), LiCl wash buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% Na deoxycholate, and 0.25 M LiCl), and TE buffer. The purified chromatin was incubated with 10% Chelex (Bio-Rad) and boiled for 10 min to reverse cross-linking. Chromatin DNA was further purified by incubation with proteinase K (0.2 mg/mL) for 30 min to remove proteins before used for quantitative PCR analysis. The enrichment of the MYB46 promoter sequence was examined by real-time quantitative PCR analysis of chromatin samples before (input) and after (bound) immunoprecipitation. The promoter sequence of the IFL1 gene, which has been shown not to be a downstream target of SND1 (Zhong et al., 2006), was used as an internal control for nonspecific binding of genomic DNA. The values of bound over input for MYB46-P2 and MYB46-P6 promoter fragments were normalized against that of the control DNA. The experiments were repeated twice, and identical results were obtained.
Dominant Repression and Overexpression of MYB46
The MYB46 dominant repression construct (35S:MYB46-SRDX) was created by fusing the full-length MYB46 cDNA in frame with the dominant EAR repression sequence (Hiratsu et al., 2004), which was ligated downstream of the CaMV 35S promoter in pBI121 (Clontech). The MYB46 overexpression construct was made by ligating the full-length MYB46 cDNA downstream of the CaMV 35S promoter in pBI121. The MYB46 dominant repression and overexpression constructs were introduced into wild-type Arabidopsis plants by Agrobacterium-mediated transformation. Transgenic plants were selected on kanamycin, and the first generation was used for examination of phenotypes.
Microscopy
Tissues were fixed in 2% formaldehyde and embedded in low viscosity (Spurr's) resin (Electron Microscopy Sciences) as described (Burk et al., 2006). Sections (1 μm thick) were cut with a microtome and stained with toluidine blue for light microscopy. For transmission electron microscopy, 85-nm-thick sections were cut, poststained with uranyl acetate and lead citrate, and observed using a Zeiss EM 902A transmission electron microscope (Carl Zeiss).
Histological Detection of Cellulose and Lignin
Sections (1 μm thick) of stems were stained for cellulose with 0.01% Calcoflour White and observed with a UV fluorescent microscope as described (Hughes and McCully, 1975). Under the conditions used, only secondary walls exhibited brilliant fluorescence. Sections (50 μm thick) of stems were stained with phloroglucinol-HCl for lignin, which was shown as bright red color. For visualization of lignified secondary walls in leaves and flowers, methanol-cleared leaves and flowers were used for examination of lignin autofluorescence and collection of DIC images under a microscope.
Immunolocalization of Xylan
For examination of xylan in secondary walls, 1-μm-thick sections of stems were probed with the LM10 monoclonal antibody, which binds to 4-O-methylglucuronoxylan (McCartney et al., 2005), and detected with fluorescein isothiocyanate–conjugated secondary antibodies. The fluorescence-labeled xylan signals were visualized with a confocal microscope.
Statistical Analysis
The data in the experiments of quantitative PCR, transcriptional activation of the GUS reporter gene, and measurement of cell wall thickness were subjected to statistical analysis using the Student's t test program (http://www.graphpad.com/quickcalcs/ttest1.cfm). The quantitative differences between two groups of data for comparison in all these experiments were shown to be statistically significant (P < 0.001).
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for genes investigated in this study are MYB46 (At5g12870), SND1 (At1g32770), SND2 (At4g28500), SND3 (At1g28470), MYB20 (At1g66230), MYB85 (At4g22680), MYB103 (At1g63910), KNAT7 (At1g62990), IFL1 (At5g60690), ATHB-8 (At4g32880), NST1 (At2g46770), NST2 (At3g61910), VND6 (At5g62380), VND7 (At1g71930), CesA7 (At5g17420), CesA8 (At4g18780), FRA8 (At2g28110), CCoAOMT (At4g34050), 4CL1 (At1g51680), XCP1 (At4g35350), XCP2 (At1g20850), and BFN1 (At1g11190).
Acknowledgments
We thank the editor and the reviewers for their constructive suggestions and comments and Ryan L. McCarthy for critical reading of the manuscript. This work was supported by a grant from the U.S. Department of Energy–Bioscience Division (DE-FG02-03ER15415).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zheng-Hua Ye (zhye@plantbio.uga.edu).
References
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., Zhong, R., Morrison III, W.H., and Ye, Z.-H. (2006). Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J. Integr. Plant Biol. 48 85–98. [Google Scholar]
- Demura, T., and Fukuda, H. (2007). Transcriptional regulation in wood formation. Trends Plant Sci. 12 64–70. [DOI] [PubMed] [Google Scholar]
- Funk, V., Kositsup, B., Zhao, C., and Beers, E.P. (2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128 84–94. [PMC free article] [PubMed] [Google Scholar]
- Gampala, S.S., Hagenbeek, D., and Rock, C.D. (2001). Functional interactions of lanthanum and phospholipase D with the abscisic acid signaling effectors VP1 and ABI1-1 in rice protoplasts. J. Biol. Chem. 276 9855–9860. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., Lapierre, C., Pollet, B., Verhaegen, D., Chaubet-Gigot, N., and Grima-Pettenati, J. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43 553–567. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Mitsuda, N., Matsui, K., and Ohme-Takagi, M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321 172–178. [DOI] [PubMed] [Google Scholar]
- Hughes, J., and McCully, M.E. (1975). The use of an optical brightener in the study of plant structure. Stain Technol. 50 319–329. [DOI] [PubMed] [Google Scholar]
- Ito, J., and Fukuda, H. (2002). ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinska, B., Karlsson, M., Srivastava, M., Stenberg, A., Schrader, J., Sterky, F., Bhalerao, R., and Wingsle, G. (2004). MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol. Biol. 56 255–270. [DOI] [PubMed] [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, L., Marcus, S.E., and Knox, J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53 543–546. [DOI] [PubMed] [Google Scholar]
- Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulates secondary wall thickening and are required for anther dehiscence. Plant Cell 17 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L.J., Smith, C., Bevan, M.W., Mansfield, S., Whetten, R.W., Sederoff, R.R., and Campbell, M.M. (2003). Characterization of a pine MYB that regulates lignification. Plant J. 36 743–754. [DOI] [PubMed] [Google Scholar]
- Pena, M.J., Zhong, R., Zhou, G.-K., Richardson, E.A., O'Neill, M.A., Darvill, A.G., York, W.S., and Ye, Z.-H. (2007). Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven, P.H., Evert, R.F., and Eichhorn, S.E. (1999). Biology of Plants, 6th ed. (New York: W.H. Freeman and Company).
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Steiner-Lange, S., Unte, U.S., Eckstein, L., Yang, C., Wilson, Z.A., Schmelzer, E., Dekker, K., and Saedler, H. (2003). Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 34 519–528. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F.A., Roberts, K., and Martin, C. (1998). The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Gardiner, J.C., Whiteman, R., and Turner, S.R. (2004). Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11 329–338. [Google Scholar]
- Yang, C., Xu, Z., Song, J., Conner, K., Vizcay-Barrena, G., and Wilson, Z.A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z.-H., Freshour, G., Hahn, M.G., Burk, D.H., and Zhong, R. (2002). Vascular development in Arabidopsis. Int. Rev. Cytol. 220 225–256. [DOI] [PubMed] [Google Scholar]
- Ye, Z.-H., York, W.S., and Darvill, A.G. (2006). Important new players in secondary wall synthesis. Trends Plant Sci. 11 162–164. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., and Ye, Z.-H. (2001). Fibers. A model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol. 126 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Demura, T., and Ye, Z.-H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Pena, M.J., Zhou, G.-K., Nairn, C.J., Wood-Jones, A., Richardson, E.A., Morrison, W.H., Darvill, A.G., York, W.S., and Ye, Z.-H. (2005). Arabidopsis Fragile Fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Richardson, E.A., and Ye, Z.-H. (2007). Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225 1603–1611. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]