Abstract
Regulated RNA metabolism appears to be a critical component of molecular mechanisms directing flowering initiation in plants. A group of RNA binding proteins exerts their roles through the autonomous flowering pathway. Posttranscriptional mechanisms regulated by microRNAs (miRNAs) also play a key role in flowering-time control. Here, we demonstrate that the GIGANTEA (GI)-regulated miR172 defines a unique genetic pathway that regulates photoperiodic flowering by inducing FLOWERING LOCUS T (FT) independent of CONSTANS (CO). A late-flowering mutant in which a miR172 target gene, TARGET OF EAT1, is constitutively activated by the nearby insertion of the cauliflower mosaic virus 35S enhancer normally responded to vernalization and gibberellic acid treatments. By contrast, its response to daylength changes was severely disrupted. In the mutant, FT was significantly repressed, but other flowering genes were unaffected. Notably, miR172 abundance is regulated by photoperiod via GI-mediated miRNA processing. Accordingly, miR172-overproducing plants exhibit early flowering under both long days and short days, even in the absence of functional CO, indicating that miR172 promotes photoperiodic flowering through a CO-independent genetic pathway. Therefore, it appears that GI-mediated photoperiodic flowering is governed by the coordinated interaction of two distinct genetic pathways: one mediated via CO and the other mediated via miR172 and its targets.
INTRODUCTION
Seasonal control of flowering is essential for reproductive success in plants and therefore regulated by coordinated interactions between various endogenous signals and environmental cues (Mouradov et al., 2002). Light is one of the most important environmental factors that substantially affect seasonal flowering. Environmental light is actually a complex factor consisting of diverse components, such as intensity, quality (wavelength), photoperiod, and directionality, among which effects of photoperiod and light quality are most prominent in flowering-time control (Cerdan and Chory, 2003; Hayama and Coupland, 2003).
Plants sense seasonal progression by monitoring the daily variation of photoperiod lengths through circadian rhythms that are generated by the clock system. The intimate relationship between circadian rhythms and photoperiodic flowering has been studied extensively (Samach and Coupland, 2000; Hayama and Coupland, 2003). Most mutants with disrupted clock function exhibit altered flowering time, depending on photoperiod lengths. Accordingly, many Arabidopsis thaliana genes that regulate photoperiodic flowering show circadian regulation of expression patterns. Photoreceptors, including phytochromes, cryptochromes, and ZEITLUPE blue light receptors, mediate the input light pathway to synchronize the clock (Lin, 2000; Nelson et al., 2000; Somers et al., 2000; Putterill, 2001). The circadian clock then generates a series of rhythmic output signals in the regulation of photoperiodic flowering, which are principally mediated by GIGANTEA (GI) and CONSTANS (CO) and regulate floral integrators, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF EXPRESSION OF CO1 (SOC1) (Yanovsky and Kay, 2003; Mizoguchi et al., 2005).
CO promotes flowering in inductive light conditions by activating FT transcription. The role of CO is regulated at the transcriptional level by GI-mediated circadian rhythms as well as at the posttranscriptional level by the photoreceptors (Valverde et al., 2004; Mizoguchi et al., 2005). It has been known that light stabilizes the CO protein in the evening under long days (LDs) when CO mRNA abundance reaches its peak. This indicates that light and the internal clock act on the CO protein to sense daylength, supporting the coincidence of an internal rhythm and a light-sensitive phase as a molecular mechanism for floral promotion.
Regulated RNA metabolism is an important molecular scheme functioning in flowering-time control in plants. A group of genes encoding RNA binding proteins, including FPA, FCA, and FLK, regulates flowering initiation through the autonomous flowering pathway (Macknight et al., 1997; Schomburg et al., 2001; Lim et al., 2004; Quesada et al., 2005). FCA and FPA encode RNA recognition motif–containing proteins, whereas FLK encodes a K homology domain–containing protein. FY is apparently a polyadenylation factor that mediates alternative polyadenylation of FCA pre-mRNA and is required for FCA autoregulation. Additional proteins, including PIE1, CBP80, ABSCISIC ACID–HYPERSENSITIVE1, HUA2, and EARLY FLOWERING, have also been shown to function in FLOWERING LOCUS C (FLC) mRNA accumulation and RNA processing (Hugouvieux et al., 2001; Noh and Amasino, 2003; Bezerra et al., 2004; He et al., 2004; Doyle et al., 2005). Interestingly, it was recently reported that FCA is an abscisic acid receptor (Razem et al., 2006). Abscisic acid directly affects the FCA-mediated processing of pre-mRNA, supporting a role for RNA metabolism in mediating the interactions of intrinsic and environmental signals in flowering-time control.
Of particular interest are microRNAs (miRNAs). Three miRNAs have been functionally characterized in flowering-time control: miR156 (Schwab et al., 2005), miR159 (Achard et al., 2004), and miR172 (Aukerman and Sakai, 2003; Chen, 2003). A group of SQUAMOSA PROMOTER BINDING PROTEIN–LIKE (SPL) genes, such as SPL3, SPL4, and SPL5, which promote flowering, are targets of miR156. Gibberellic acid (GA) upregulates miR159 abundance, and overproduction of miR159 causes repression of the floral integrator LEAFY (LFY), resulting in late flowering specifically under short days (SDs). The most extensively studied is miR172. miR172 promotes flowering primarily by posttranscriptionally repressing a set of APETALA2 (AP2)-like genes, including TARGET OF EAT1 (TOE1), TOE2, and TOE3 (Aukerman and Sakai, 2003). Overexpression of TOE1 causes late flowering, whereas miR172-overexpressing plants exhibit early flowering under both LDs and SDs. A global analysis of Arabidopsis gene expression recently identified additional miR172 targets: SCHLAFMÜTZE (SMZ) and SCHNARCHZAPFEN (SNZ) (Schmid et al., 2003). They repress flowering but are downregulated upon floral initiation under LDs.
It has been observed that miR172 is regulated by daylength and that the daylength effect on miR172 is disrupted in the co and ft mutants (Schmid et al., 2003), suggesting a role for miR172 in photoperiodic flowering. However, it was later suggested that miR172 abundance is not affected by daylength and CO (Aukerman and Sakai, 2003). As a result, miR172 has not been assigned to particular flowering pathways, and the underlying molecular mechanisms have not been determined due to the controversial results on the effects of daylength and CO on miR172 abundance.
In this work, we show that miR172 mediates photoperiodic flowering by regulating FT. The functional role of miR172 depends on GI but does not require functional CO, demonstrating that miR172 belongs to a unique photoperiod flowering pathway. Our data indicate that GI-mediated photoperiodic flowering is coordinately regulated by at least two distinct molecular schemes: one mediated by CO (Suárez-López et al., 2001; Valverde et al., 2004) and the other mediated by miR172 and its targets. In addition, our data support the notion that GI regulates miR172 abundance at the miRNA processing step rather than at the transcriptional level.
RESULTS
toe1-2D Is Late Flowering Due to TOE1 Overexpression
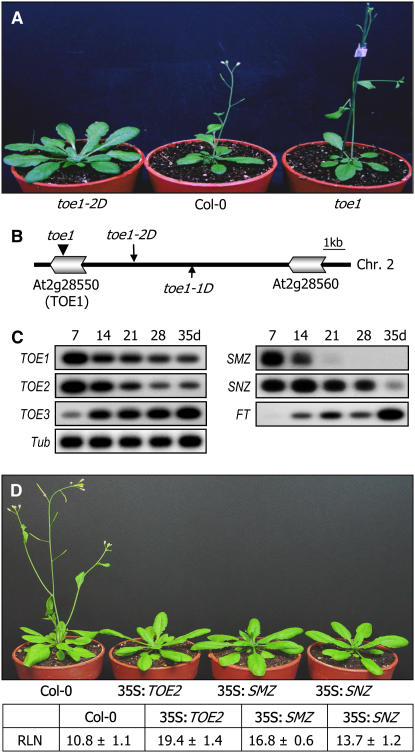
We isolated a late-flowering mutant, toe1-2D, by screening an Arabidopsis activation-tagged mutant pool generated by randomly inserting the cauliflower mosaic virus (CaMV) 35S enhancer into the genome of ecotype Columbia-0 (Col-0) (Weigel et al., 2000). Except for the late-flowering phenotype, other growth and developmental aspects, such as growth rate and organ morphologies, were essentially normal in the mutant (Figure 1A). The toe1-2D mutant initiated flowering at a rosette leaf number (RLN) of 22.6 ± 1.2 under LDs, whereas control plants flowered at a RLN of 11.6 ± 1.3 under the same growth conditions.
Figure 1.
Flowering Phenotype and Mapping of the T-DNA Insertion Site in toe1-2D.
(A) Flowering phenotype of toe1-2D. toe1 is a T-DNA insertional knockout mutant (SALK 069677).
(B) T-DNA insertion site in toe1-2D. A single copy of the T-DNA enhancer element is inserted near the TOE1 gene locus in the toe1-2D genome (long arrow). The toe1-1D mutant, which has been characterized by Aukerman and Sakai (2003), is marked by a short arrow. The T-DNA insertional knockout mutant toe1 is marked by an arrowhead.
(C) Growth stage–dependent transcription of TOE1 and other miR172 targets. Transcript levels were examined by RT-PCR–based DNA gel blot analysis. d, days after germination. A tubulin gene (Tub) was included as a control for constitutive expression.
(D) Flowering times of transgenic plants with altered expression of other miR172 target genes. Thirty plants were measured and averaged. Values in the table are means ± se.
A three-step thermal asymmetric interlaced–PCR analysis revealed that TOE1 encoding an AP2 domain–containing transcription factor was activated by the nearby insertion of the 35S enhancer in the toe1-2D genome (Figure 1B). Genomic DNA gel blot analysis confirmed that there was a single T-DNA insertion event occurring in the mutant (see Supplemental Figure 1A online). The toe1-2D mutant is allelic to toe1-1D, a late-flowering mutant that was isolated previously by a similar approach (Aukerman and Sakai, 2003). By contrast, a T-DNA insertional knockout mutant, toe1, showed slightly early flowering (Figure 1A), as has been reported previously (Aukerman and Sakai, 2003), demonstrating that TOE1 overexpression underlies the late-flowering phenotype of toe1-2D (see Supplemental Figure 1B online).
TOE1 possesses two AP2 domains in similar positions as those identified in the homeotic protein AP2, but sequence similarities are entirely restricted to the AP2 domains (see Supplemental Figure 2 online) (Okamuro et al., 1997). This structural distinction certainly reflects the functional diversity between AP2 and TOE1.
TOE1 transcript level was high in very young seedlings but gradually decreased throughout the life cycle (Figure 1C). A similar pattern was also observed with TOE2 and SNZ. Notably, SMZ transcript level decreased more rapidly and was barely detectable in 3-week-old or older plants. By contrast, TOE3 exhibited a reversed expression pattern. Its expression was relatively low in young seedlings but increased rapidly as plants grew until flowering. The diverse expression patterns suggest that TOE3 may be functionally distinct from other AP2-like genes. To further examine the physiological roles of TOE1 and related genes targeted by miR172, transgenic plants overexpressing individual genes and T-DNA insertional mutants were obtained and their flowering phenotypes were measured. Whereas overexpression of TOE2, SNZ, or SMZ caused late flowering as in TOE1 (Figure 1D), TOE3 overexpression did not exhibit late flowering. The toe2, toe3, smz, and snz mutants were phenotypically indistinguishable from control plants and apparently normal in flowering phenotypes, unlike the toe1 mutant, suggesting a functional redundancy among the miR172 target genes. Consistent with this notion, the toe1 toe2 double mutant flowered earlier than the toe1 mutant (see Figure 6C below). These observations indicate that the miR172 target genes are differentially regulated and may play distinct as well as overlapping roles in flowering promotion and other developmental processes.
Figure 6.
FT Expression in 172OX Plants Grown under Either LDs or SDs, and Flowering Phenotypes of toe1 and toe1 toe2 under Different Daylengths.
Two-week-old plants grown under LDs were further grown for 5 d under either LDs or SDs before harvesting plant materials. Transcript levels were measured by RT-PCR–based DNA gel blot hybridization.
(A) and (B) FT transcript levels in 172OX plants grown either under LDs (A) or under SDs (B).
(C) and (D) Flowering phenotypes of toe1 and toe1 toe2 under SDs. RLNs were counted at flowering. Thirty plants were measured and averaged for each plant group. Error bars denote se. Statistical significance was determined using Student's t test (P < 0.01).
(E) Expression of CO and FT in toe1 and toe1 toe2. Transcription levels were examined by RT-PCR.
Levels of miR172 and TOE1 Transcript Are Mutually Complementary
It has been shown that miR172 abundance increases as plants grow, and it regulates its target mRNAs primarily through translational repression (Aukerman and Sakai, 2003; Lauter et al., 2005). However, it was later shown that TOE1 mRNA is also cleaved by miR172, although the steady state mRNA level is unchanged due to feedback regulation (Schwab et al., 2005).
Transcript levels of four target genes, TOE1, TOE2, SMZ, and SNZ, were steadily reduced as plants grew. By contrast, miR172 abundance increased rapidly until flowering (Figure 2A), as has been observed (Aukerman and Sakai, 2003). Analysis of the primary transcript levels of MIR172 genes revealed that transcription of at least three MIR172 genes, MIR172a-1, MIR172a-2, and MIR172b-1, was elevated steadily throughout the plant growth stages. However, expression of genes mediating miRNA metabolism, such as Dicer-like1 (DCL1) (Schauer et al., 2002) and SERRATE (SE) (Yang et al., 2006), was unaltered, indicating that the steady increase of miR172 abundance is governed mainly at the transcriptional level rather than by miRNA metabolism. The transcript levels of MIR172b-2 and MIR172c were very low and did not exhibit any temporal regulation.
Figure 2.
TOE1 mRNA Cleavage by miR172 in N. benthamiana Leaf Cells.
(A) Growth stage–dependent abundance of miR172 and its target gene transcripts. DCL1 and SE were also included for comparison in the assays.
(B) Abundances of TOE1 mRNA and miR172 in plant tissues. The 5S rRNA served as a loading control. CL, cauline leaves; F, flowers; RL, rosette leaves; FB, floral buds; R, roots; S, stems.
(C) Expression constructs of TOE1 genes and the MIR172a-2 gene. They were subcloned under the control of the CaMV 35S promoter.
(D) A truncated form (ΔTOE1) of the TOE1 gene used in the mRNA cleavage assays. It was used instead of a full-size one, since the miR172 cleavage site is located near the 3′ end. ORF, open reading frame; UTR, untranslated region.
(E) TOE1 mRNA cleavage by miR172 in N. benthamiana leaves. Cleavage products were examined by RNA gel blot hybridization using a TOE1-specific probe. Green fluorescent protein (GFP) transcription was also assayed to verify infiltration efficiencies. The arrow indicates the cleavage product derived from the 5′ side of the miR172 cleavage site.
To obtain further insights into how miR172 regulates its targets, miR172 abundance and TOE1 transcript level were compared in plant tissues. We observed that levels of miR172 and TOE1 transcript exhibited a mutual complementarity in plant tissues, which is consistent with the proposed threshold model for miR172 function (Aukerman and Sakai, 2003). The relative levels of miR172 and TOE1 transcript were inversely related to each other in most plant tissues. For example, miR172 abundance was relatively high but TOE1 mRNA level was low in flowers, floral buds, and stem tissues (Figure 2B). In addition, miR172 was very low in root tissue in which TOE1 mRNA level was very high. These complementarities are likely to be caused by miR172-directed TOE1 mRNA cleavage occurring at least in these plant tissues. However, the mutual complementarity was not observed in leaf tissue. This may be due to different expression domains of miR172 and its targets or to feedback regulatory mechanisms in this tissue (Schwab et al., 2005). Notably, other miR172 target genes showed distinct spatial expression patterns. Transcript levels of TOE2, TOE3, and SMZ as well as AP2 were relatively high in flowers and floral buds but low in leaves and stems (see Supplemental Figure 3 online). Tissue-specific expression patterns of SNZ were most similar to those of TOE1, with comparatively high expression in leaves and stems, although the level was very low in roots. Together, these observations suggest that temporal and spatial regulation of the MIR172 genes and their targets would be important for their functions in flowering promotion and floral architecture.
Our data supported that at least TOE1 mRNA might be cleaved by miR172 (Figures 1C, 2A, and 2B). To more directly examine whether TOE1 mRNA is cleaved by miR172, we employed the Nicotiana benthamiana transient expression system (Llave et al., 2002). To ensure the miR172-mediated cleavage reactions, a miR172-resistant TOE1 mutant (ΔmTOE1) was generated (Figure 2C; see Supplemental Figure 4 online). Since the putative cleavage site is located near the 3′ end of the TOE1 open reading frame, partial constructs (ΔTOE1 or ΔmTOE1) consisting of ∼400 bases were used instead of full-size gene sequences to more clearly discriminate the cleavage products (Figure 2D). The TOE1 gene constructs were then subcloned into the Gateway plant expression vector system (pB2GW7; Invitrogen) under the control of the 35S promoter (Figure 2C). The results showed that TOE1 mRNA is evidently, although not very efficiently, cleaved by miR172 (Figure 2E). A cleavage product of ∼300 bp was detected only in the presence of miR172. However, it was not detected from ΔmTOE1 mRNA, indicating that miR172 specifically cleaves TOE1 mRNA. The basal level of cleavage product in the absence of miR172 might be due to intrinsic miR172 activity existing in N. benthamiana cells.
FT and Its Downstream Genes Are Downregulated in toe1-2D
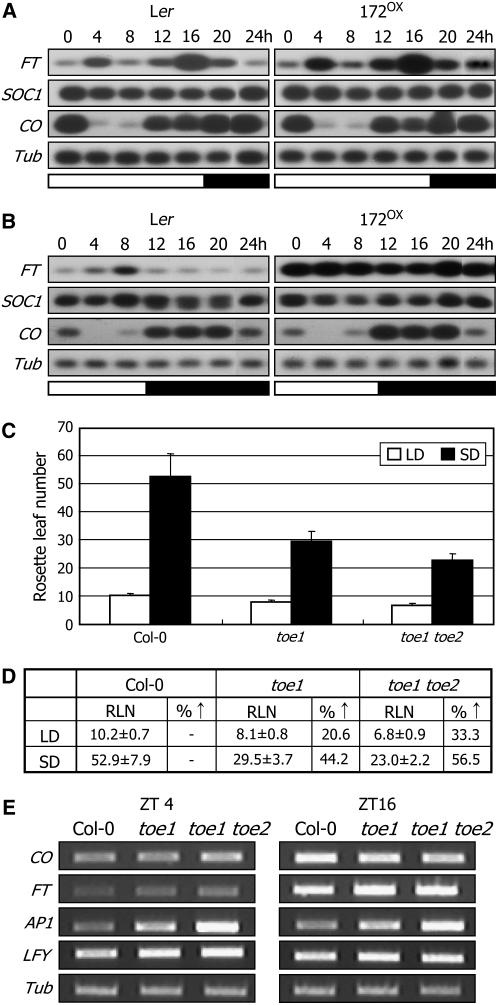
In order to explore the molecular mechanisms by which miR172 and TOE1 regulate flowering initiation, the expression of diverse flowering-time genes was examined in toe1-2D and toe1. Whereas FT and floral homeotic genes, such as AP1 and LFY, were repressed significantly, expression of CO, FLC, and other genes functioning in the autonomous and GA pathways was unaltered (Figure 3A). These results indicate that miR172 regulation of TOE1 exerts its role by inducing FT through a genetic pathway other than the autonomous and GA pathways.
Figure 3.
Expression Patterns of Flowering-Time Genes in toe1-2D.
(A) Expression of flowering-time genes in toe1-2D and toe1 examined by RT-PCR–based DNA gel blot hybridization. Ten-day-old plants grown under normal growth conditions (23°C, LD) were harvested at either ZT4 or ZT16 for total RNA extraction. A tubulin gene (Tub) was included as a control for constitutive expression. ZT, Zeitgeber time.
(B) Levels of pre-miR172 and mature miR172 in floral buds and open flowers. A FT-deficient mutant (ft-10) was included in the assays (Yoo et al., 2005). The 5S rRNA served as a loading control.
(C) Flowering times of primary ft-10 transformants overproducing miR172 under LDs (n = 92). The p172a-2 expression construct (Figure 2C) was transformed into ft-10. The arrowhead indicates the range of flowering time of the transgenic plants overproducing miR172 (35S:172a-2), and the arrow marks the range of flowering time of ft-10.
(D) Localized TOE1 expression in rosette leaves compared with that of FT. Promoter sequences of ∼4.5 kb were transcriptionally fused to a β-glucuronidase (GUS)–coding sequence, and the fusion constructs were transformed into Arabidopsis.
The next question concerned the hierarchic relationship between miR172 and FT. It has been suggested that miR172 functions downstream of FT (Schmid et al., 2003). However, such FT dependence of miR172 abundance was not observed in another study (Aukerman and Sakai, 2003). To resolve this uncertainty, levels of pre-miR172 and mature miR172 were examined in the knockout ft-10 mutant (Yoo et al., 2005). We used floral tissues, since miR172 levels are highest in the adult reproductive stage (Figure 2A). The levels of pre-miR172 and miR172 were identical in the floral buds and fully open flowers of control and ft-10 plants (Figure 3B). It was anticipated that analysis using floral tissues would be confounded by the potential effects of different developmental stages. Therefore, we examined the levels of miR172 in young seedlings. Again, the miR172 levels were identical in control plants, 35S∷FT transgenic plants, and ft mutants (see Supplemental Figure 5 online). Together with FT repression in toe1-2D and induction in toe1 (Figure 3A), these results indicate that FT functions downstream of miR172 and TOE1. Consistent with this view, ft-10 mutants overproducing miR172 still showed late flowering (Figure 3C), comparable with the parental mutant. Furthermore, the expression domains of FT and TOE1 were mutually complementary (Figure 3D; see Supplemental Figure 6 online). While GUS activity was primarily distributed in the distal leaf area in FT promoter–GUS transgenic plants, it was detected mainly in the proximal leaf area in TOE1 promoter–GUS transgenic plants, further supporting the functional relationship between FT and TOE1.
miR172 Abundance Is Altered in gi-2 and Photoreceptor Mutants
To determine the genetic pathway by which TOE1 and its upstream regulator miR172 regulate flowering time, the toe1-2D and toe1 mutants were grown until flowering under different daylengths or subjected to vernalization and GA treatments. toe1-2D flowering normally responded to vernalization, a prolonged treatment at 4°C for 6 weeks, and GA treatments. The late-flowering phenotype was efficiently rescued by both treatments (see Supplemental Figure 7 online). By contrast, its response to daylength changes was obviously disrupted. toe1-2D was late flowering under LDs (Figure 4A). However, it did not exhibit delayed flowering under SDs. Meanwhile, it has been shown that miR172 abundance is unchanged in a CO mutant (Aukerman and Sakai, 2003). We observed that CO is also unaffected in toe1-2D (Figure 3A). Together, these observations suggest that miR172 and its target TOE1 regulate photoperiodic flowering via FT, but possibly independent of CO.
Figure 4.
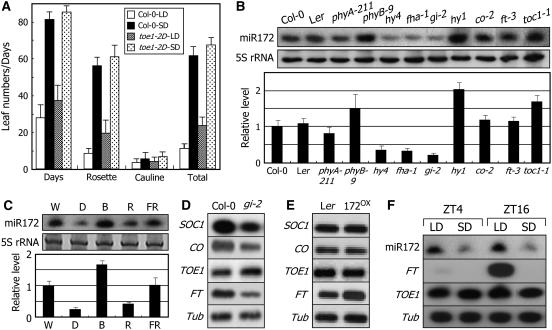
Photoperiodic Responses of toe1-2D and Effects of Light Wavelengths and Daylengths on miR172 Abundance.
A tubulin gene (Tub) or 5S RNA was included as a loading control. Transcript levels were examined by RT-PCR–based DNA gel blot hybridization, and miR172 abundance was examined by RNA gel blot analysis.
(A) Photoperiodic responses of toe1-2D. Thirty plants were measured and averaged for each plant group. Error bars denote se. Statistical significance was determined using Student's t test (P < 0.01).
(B) miR172 abundance in flowering-time mutants. Photoreceptor mutants and a chromophore-deficient mutant (hy1) were also included in the assays. Ler, Landsberg erecta.
(C) miR172 abundance under different light wavelengths. Two-week-old plants were further grown under white light (W), dark (D), red light (R), blue light (B), or far-red light (FR) for an additional 1 d before harvesting plant materials.
(D) and (E) TOE1 expression in gi-2 (D) and 172OX (E) plants.
(F) miR172 abundance under different daylengths. Two-week-old plants grown under LDs or SDs were harvested at either ZT4 or ZT16.
To obtain insights into how miR172 and TOE1 regulate photoperiodic flowering, miR172 abundance was examined in the co-2, gi-2, and various photoreceptor mutants. Interestingly, miR172 abundance was substantially reduced in gi-2 but unaffected in co-2 (Figure 4B). By contrast, it was greatly upregulated in toc1-1, a SD-specific early-flowering mutant (Somers et al., 1998). The level of miR172 was also altered in the photoreceptor mutants. Whereas it was reduced markedly in the cryptochrome mutants and slightly in phyA-211, it increased detectably in phyB-9. Furthermore, a dramatic increase was observed in hy1, a mutant that lacks functional phytochromes (Muramoto et al., 1999). These results are well consistent with the antagonistic roles of phyB and the cryptochromes in regulating photoperiodic flowering (Lin, 2000; El-Din El-Assal et al., 2003; Mockler et al., 2003). Consistent with the altered miR172 abundances in the photoreceptor mutants, it increased significantly after blue light illumination but decreased after red light illumination (Figure 4C). These observations support the notion that miR172 is regulated by GI but independent of CO through the photoperiod flowering pathway. However, TOE1 transcript level was unaltered in gi-2 (Figure 4D) as well as in co-2 and photoreceptor mutants (see Supplemental Figure 8 online), which is consistent with the fact that the miR172 regulation of target mRNAs occurs primarily at the level of translation.
We additionally found that miR172 abundance was unaltered in ft-3 (Figure 4B). By contrast, FT transcription increased drastically in 172OX transgenic plants overexpressing MIR172a-1 (Figure 4E). Furthermore, miR172 abundance was much higher in plants grown under LDs but lower in plants grown under SDs (Figure 4F). Therefore, it is evident that miR172 functions upstream of FT in response to daylength changes.
miR172 Abundance Is Regulated by GI through miRNA Metabolism
GI regulates photoperiodic flowering, at least in part, by inducing CO (Fowler et al., 1999; Park et al., 1999). In addition, GI and its downstream genes, such as CO and FT, are regulated by the circadian clock, and their transcript levels cycle with a diurnal rhythm of 24 h (Suárez-López et al., 2001). Therefore, it was anticipated that miR172 abundance might exhibit a daily oscillation. However, it did not oscillate with a diurnal rhythm under both LDs and SDs (Figure 5A). It was constantly higher under LDs but lower under SDs. Together with the reduced miR172 abundance in the gi-2 mutant grown under LDs (Figure 5B), these observations suggest that miR172 may be regulated by a clock-independent GI pathway (Mizoguchi et al., 2005; Oliverio et al., 2007).
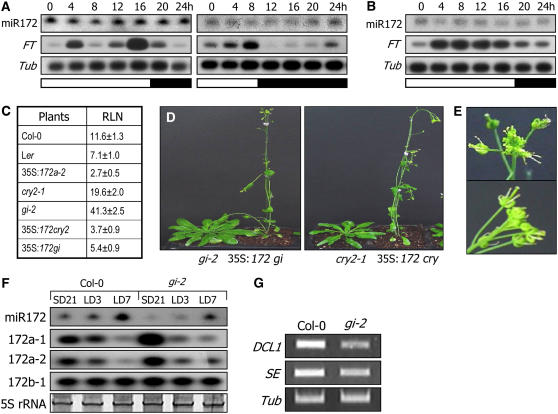
Figure 5.
Regulation of miR172 Abundance by GI.
A tubulin gene (Tub) or 5S rRNA was included as a loading control. Transcript levels of FT and MIR172 genes were examined by RT-PCR–based DNA gel blot analysis, and miR172 abundance was examined by RNA gel blot analysis. Expression of DCL1 and SE was examined by RT-PCR.
(A) miR172 abundance in Col-0 grown under LDs (left panel) or under SDs (right panel). Plants were grown for 2 weeks before harvesting plant materials.
(B) miR172 abundance in gi-2 grown under LDs.
(C) and (D) Flowering phenotypes of gi-2 or cry2-1 overproducing miR172 (35S∷172 gi or 35S:172 cry, respectively). The mutant plants were transformed with the binary pBA002 plasmid containing the MIR172a-2 gene sequence under the control of the CaMV 35S promoter. Thirty plants were measured and averaged (C). The cry2-1 (Col-0) mutant was also included in the assays for comparison. Representative plants were photographed (D).
(E) Altered floral structures. Flowers of the transgenic plants exhibited altered structures, indicative of miR172 overproduction.
(F) Levels of primary miR172 transcripts in gi-2. The pre-miR172 levels were examined by RT-PCR–based DNA gel blot hybridization.
(G) Transcript levels of DCL1 and SE in gi-2. Transcript levels were examined by RT-PCR.
To further examine the functional relationship between GI and miR172, the MIR172a-2 gene was overexpressed under the control of the CaMV 35S promoter in the gi-2 mutant. The resultant 35S∷172 gi plants flowered at a RLN of 5.4 ± 0.9 (Figures 5C and 5D), demonstrating that miR172 functions downstream of GI, which is also consistent with the reduced miR172 abundance in the gi-2 mutant (Figure 5B). In addition, the miR172-overproducing fha-1 mutant (35S∷172 cry2) also exhibited early flowering, initiating flowering at a RLN of 3.7 ± 0.9 (Figures 5C and 5D). Floral architecture of the transgenic plants was disrupted (Figure 5E), indicative of miR172 overproduction in the transgenic plants.
miR172 abundance was reduced significantly in the gi-2 mutant (Figure 5B) and regulated by daylength (Figure 4F). A critical question, therefore, was how GI regulates miR172 abundance. To obtain insights into the molecular mechanisms by which GI regulates miR172 abundance, control and gi-2 plants grown for 3 weeks under SDs were transferred to LDs and further grown for 3 to 7 d, and levels of miR172 and MIR172 transcripts were examined. miR172 abundance increased upon transfer to LDs in both control and gi-2 plants, but with a considerably reduced rate in the latter (Figure 5F). Interestingly, MIR172 transcript levels were higher in the gi-2 mutant than in control plants when grown under SDs and were reduced slightly in both plants upon exposure to LDs. Therefore, it is likely that GI regulates miR172 maturation (processing) rather than MIR172 transcription. In accordance with this view, genes encoding miRNA processing enzymes, such as DCL1 and SE, were obviously reduced in the gi-2 mutant (Figure 5G).
miR172 Regulates the Amplitude of FT Expression but Not Its Daily Rhythm
The 172OX plants flowered extremely early at RLNs of ∼2 and ∼6 under LDs and SDs, respectively (Aukerman and Sakai, 2003). We observed that the amplitude and daily rhythm of CO transcription were unaltered in the 172OX plants grown under both LDs and SDs (Figures 6A and 6B), further supporting the CO independence of miR172 function. SOC1 expression was also unchanged in the same growth conditions. By contrast, FT expression was detectably upregulated in the 172OX plants grown under LDs, while the daily rhythm was maintained (Figure 6A). Interestingly, the level of FT transcript was very high, but with a disrupted daily rhythm in the 172OX plants grown under SDs (Figure 6B). Together, these observations indicate that miR172 regulates the amplitude of FT expression but not its daily rhythm. Furthermore, this view is not inconsistent with the role of CO in regulating the daily rhythm of FT expression (Suárez-López et al., 2001).
Our data indicate that miR172 abundance is regulated by daylength. It was relatively very low under SDs (Figures 4F and 5A), suggesting that the TOE1 activity would be higher under SDs. Therefore, it was assumed that the early-flowering phenotypes of toe1 would be more evident under SDs than under LDs. To examine this hypothesis, control and toe1 mutant plants were grown under LDs and SDs and their flowering times was compared. A toe1 toe2 double mutant was also included in the assays. While toe1 flowering was accelerated by 20.6% under LDs, it was accelerated by 44.2% under SDs compared with control plants (Figures 6C and 6D). The flowering acceleration was more evident in the toe1 toe2 double mutant. It was accelerated by 33.3% under LDs and by 56.5% under SDs. These results further support the regulation of miR172 by daylength and the role of TOE1 in flowering-time control. Consistent with the early-flowering phenotype of toe1 and toe1 toe2, transcript levels of FT, LFY, and AP1 were proportionally higher in the mutants (Figure 6E).
The miR172-Mediated Flowering Pathway Is Independent of CO
Our data support the notion that miR172 is independent of CO. To confirm this idea, a null mutant, co-2, was transformed with the MIR172a-2 gene and the flowering phenotype of the transformants was analyzed. The 35S∷172 co plants flowered at a RLN of 7.2 ± 0.8 under LDs (Figure 7A), similar to the previous observation in which 35S-EAT plants overexpressing MIR172a-2 flowered at a RLN of 6.1 ± 1.2 under SDs (Aukerman and Sakai, 2003). However, 35S∷172 co flowering under LDs was slightly later than that of 35S-EAT plants grown under LDs. These results demonstrate that miR172 functions independent of CO.
Figure 7.
CO-Independent miR172 Function in Photoperiodic Flowering.
(A) Measurements of flowering times in 35S∷172 co plants. Plants grown for 30 d were measured and averaged. Values in the table are means ± se.
(B) FT expression in 35S∷172 co plants grown under either LDs or SDs. Transcript levels were examined by RT-PCR, and miR172 abundance was examined by RNA gel blot hybridization.
(C) A schematic working model for miR172 function in photoperiodic flowering. miR172 mediates light signals from GI and promotes flowering by inducing FT independent of CO.
It is also envisioned that the flowering initiation of the 35S∷172 co plants at a RLN of 7.2 ± 0.8 may reflect a maximal capacity of the miR172 pathway in promoting flowering in the absence of functional CO. Full functioning of FT may require both the CO- and miR172-mediated signals in photoperiodic flowering. This reasoning also explains why the 172OX flowering was delayed under SDs, when the CO-mediated FT induction became negligible, in the previous report (Aukerman and Sakai, 2003). Consistent with the early-flowering phenotype of the 35S∷172 co plants both under LDs and SDs (Figure 7A), the transcript levels of FT were upregulated in the 35S∷172 co plants regardless of daylength changes (Figure 7B), further confirming that miR172 promotes flowering by inducing FT in a CO-independent manner. Together, our data demonstrate that miR172 mediates clock-independent GI signals in regulating photoperiodic flowering via FT (Figure 7C). In this view, GI plays a dual role in photoperiodic flowering: circadian rhythmic regulation of CO expression and regulation of miR172 metabolism, both of which are required for FT function.
miR172 Abundance Is Influenced by Autonomous Pathway Mutations
Flowering of the photoperiod pathway mutants, including ft and co, is accelerated by vernalization, but to a lesser degree compared with that of the autonomous pathway mutants (Koornneef et al., 1991). We observed that toe1-2D flowering was substantially accelerated by vernalization (see Supplemental Figure 7A online), suggesting that miR172 and its targets might also be affected by other genetic pathways in addition to the photoperiodic flowering pathway.
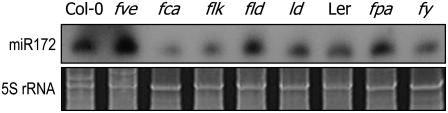
To examine this possibility, miR172 abundance was examined in a series of mutants that belong to other flowering pathways. Although we found no discernible changes in miR172 abundance in the mutants functioning in the vernalization and GA pathways, it was significantly reduced in a few autonomous pathway mutants. While it was at least threefold lower in fca and flk and twofold lower in fy, a visible increase was observed in fve (Figure 8). Other autonomous pathway mutations did not influence miR172 abundance to a detectable level, suggesting that there might be more than one signaling cascade in the autonomous pathway, as has been proposed previously (Simpson et al., 2003). These observations suggest that the miR172-mediated flowering pathway is somewhat influenced by the autonomous pathway. This notion may be related to the growth stage–dependent regulation of miR172 abundance (Aukerman and Sakai, 2003; Lauter et al., 2005) (see Discussion).
Figure 8.
miR172 Abundance in the Autonomous Pathway Mutants.
miR172 abundance was examined by RNA gel blot hybridization. The 5S rRNA was included as a loading control.
DISCUSSION
miR172 in Photoperiodic Flowering
miR172 is unique among the characterized miRNAs mediating flowering initiation in that it regulates both floral architecture and flowering time by controlling a group of AP2 domain–containing transcription factors. It regulates a floral organ identity gene, AP2, and a group of related genes, such as TOE1, TOE2, SMZ, and SNZ, through mRNA cleavage and translational repression (Schwab et al., 2005). Whereas plants overexpressing TOE1 are late-flowering with normal floral morphologies, mutations in AP2 exhibit altered floral structures without affecting flowering time (Okamuro et al., 1997). In addition, although TOE3 is a target of miR172, transgenic plants overexpressing TOE3 are not late-flowering. Furthermore, its expression profile is different from those of other targets (Figure 1C). It seems that miR172 exerts its role through multiple signaling pathways.
miR172 regulation of flowering time has also been demonstrated in N. benthamiana (Mlotshwa et al., 2006). Transgenic N. benthamiana lines overexpressing the Arabidopsis MIR172a-1 gene exhibit early flowering. In maize (Zea mays), miR172 activity has not been examined directly in flowering-time control. However, it has been shown that transgenic maize overexpressing an AP2 homolog, glossy15, exhibits delayed reproductive development (Lauter et al., 2005). In addition, a maize mutant, Corngrass1, that overproduces miR156 exhibits late flowering, and miR172 level is lowered in the mutant (Chuck et al., 2007). These observations suggest that miR172 activity is conserved at least in these plant species.
There are five MIR172 genes in Arabidopsis. Whereas the expression of MIR172a-1, MIR172a-2, and MIR172b-1 is elevated as plants reach the reproductive growth stage, that of MIR172b-2 and MIR172c is very low and does not respond to growth stages, supporting the notion that the growth stage–dependent increase of miR172 abundance is regulated primarily at the transcriptional level. Furthermore, its target genes also show differential expression patterns spatially and temporally. It will be interesting to investigate transgenic plants or mutants with altered expression of individual genes or of multiple genes in terms of detailed aspects of flowering time and floral architecture, as well as other developmental aspects, such as seed germination.
miR172 was once suggested to function via the photoperiodic flowering pathway. It has been shown that the target genes are suppressed upon flowering and that this downregulation is dependent on CO and FT (Schmid et al., 2003). In addition, expression of MIR172a-2 was affected by daylength changes, as judged by RT-PCR of its primary transcript, and this effect was dependent on CO and FT, suggesting that miR172 may function downstream of CO and FT. However, the pattern of steady increase of miR172 abundance throughout plant growth was unaffected either by daylength changes or by mutations in CO. Consequently, assignment of miR172 and its target genes to particular flowering pathways was unclear. It was also poorly understood how miR172 regulates flowering initiation at the molecular level.
In this work, we analyzed the flowering phenotypes of diverse mutants and transgenic plants with altered miR172 abundance or with altered target gene expression. We observed that an activation-tagging mutant overexpressing the miR172 target TOE1 was late-flowering only under LDs but responded normally to vernalization and GA treatments, indicating that TOE1 belongs to the photoperiodic flowering pathway. miR172 abundance was regulated by daylength changes via GI. However, it was independent of CO. Based on our observations, we concluded that miR172 and its targets constitute a unique genetic pathway that regulates flowering time by mediating FT expression in response to daylength changes. In a previous report (Schmid et al., 2003), the level of the primary transcript of MIR172a-2 was examined by RT-PCR and found to be altered in the ft and co mutations. Through miRNA RNA gel blot analysis, we confirmed that miR172 abundance was unaltered in these mutants. Different plant materials used for miRNA extraction may explain the discrepancy between the previous findings and our observations.
Our data suggest that GI regulates miR172 processing rather than transcription of the MIR172 genes. This view is also supported by the downregulation of DCL1 and SE in the gi-2 mutant. These results suggest that processing of additional miRNAs would also be regulated by GI. This notion would also explain the pleiotropic phenotypes associated with the gi mutants (Fowler et al., 1999), which may involve other miRNAs as well. Analysis of levels of pre-miRNAs and mature miRNAs in the gi mutants would clarify this.
Physiological Role of miR172 in Flowering Initiation
Our data illustrate a unique photoperiodic flowering pathway in Arabidopsis, in which miR172 promotes flowering by upregulating FT. The miR172 pathway is responsive to daylength changes but not linked to the well-established CO-mediated pathway, which is similar to the role of the Ehd1 response regulator in rice (Oryza sativa) (Doi et al., 2004). The CO- and miR172-mediated flowering-promotive pathways apparently converge at FT. Furthermore, both pathways are essential for photoperiodic flowering, since disruption of either one of the two pathways causes late flowering under LDs.
Photoperiodic flowering requires diverse aspects of light signals, including photosensory signals that regulate CO protein stability and those resetting the circadian clock (Imaizumi and Kay, 2006). The interrelationships of these signaling components have been described by two alternative models: the internal coincidence model and the external coincidence model (Davis, 2002; Yanovsky and Kay, 2002). Recent studies strongly support the latter, which states that photoperiodic flowering is promoted when the illuminated part of the day and a photoinducible phase of the circadian clock coincide with each other (Yanovsky and Kay, 2003; Hayama and Coupland, 2004; Putterill et al., 2004). Based on our data and previous observations (Koornneef et al., 1991; Michaels and Amasino, 1999), we propose a revised external coincidence model. In the revised model, photoperiodic flowering is promoted by the coincidence of the light stabilization of CO, the GI-mediated circadian regulation of CO expression, and the GI-mediated miR172 signal for FT induction (Figure 7C). This view may adopt both the external and internal coincidence models (Valverde et al., 2004).
It has been observed that miR172 expression is temporally regulated (Aukerman and Sakai, 2003). We also observed that miR172 was barely detectable in young seedlings but gradually increased as plants approach the reproductive phase. In addition, this temporal pattern of miR172 abundance is proportional to the gradual decrease of the transcript levels of TOE1 and TOE2 and to the gradual increase of FT transcript level throughout plant growth (Figure 1C). These observations suggest that the miR172-mediated photoperiodic pathway may measure plant developmental age and promote flowering when the level of TOE floral repressors decreases below a critical threshold. However, it is unlikely that miR172 simply measures plant age, because miR172 abundance is relatively lower in plants grown under SDs than in those grown under LDs. We observed that miR172 abundance also gradually increased as plants grew under SDs, although the rate of increase was much slower compared with that in plants grown under LDs. miR172 would monitor developmental age rather than growth stages. It will be interesting to examine whether there is a correlation between miR172 abundance and developmental traits, such as trichome formation and distribution, during the transition from the vegetative to the reproductive phase.
METHODS
Plant Materials and Growth Conditions
The hy4, fha-1, hy1, ft-3, and co-2 mutants and the miR172-overproducing line (172OX) overexpressing MIR172a-1 under the control of the CaMV 35S promoter were in the Landsberg erecta ecotype. All other Arabidopsis thaliana lines used were in the Col-0 ecotype. The 172OX seeds were kindly provided by X. Chen (Rutgers University). Plants were grown in a controlled culture room set at 23°C with a RH of 60%. The photoperiods were 16 h of light and 8 h of dark for the LD conditions and 8 h of light and 16 h of dark for the SD conditions, with white light illumination (120 μmol·m−2·s−1) provided by FLR40D/A fluorescent tubes (Osram). For light wavelength treatments, plants were grown in darkness or under red, far-red, or blue light (12, 8, and 15 μmol·m−2·s−1, respectively) in a VS-940L-DUAL incubator (Vision) equipped with red, far-red, or blue light–emitting diodes.
A knockout mutant, toe1 (SALK 069677), which has a single T-DNA insertion into the sixth exon of TOE1, was isolated from a pool of T-DNA insertion lines (ABRC, Ohio State University). The absence of TOE1 expression in toe1 was confirmed by RT-PCR. The knockout mutant is identical to the toe1 mutant characterized by Aukerman and Sakai (2003). Other knockout mutants used were toe2 (SALK 065370; insertion in the first exon) and smz1 (SALK 135576; insertion in the first intron), isolated from a mutant pool of T-DNA insertion lines (ABRC), and snz1 (SM_3_2856; insertion in the seventh exon), isolated from a pool of John Innes Centre SM lines deposited at the Nottingham Arabidopsis Stock Centre. Absence of gene expression in each mutant was verified by RT-PCR.
Generation of miR172-Overproducing Plants and Arabidopsis Transformation
To generate transgenic Arabidopsis plants overproducing miR172, a MIR172a-2 gene sequence covering a 1.4-kb transcription unit was amplified from Col-0 genomic DNA using a pair of primers that had been designed in a similar way as those described by Aukerman and Sakai (2003). The primers used were 172a2f (5′-GACTACTCGAGCACCTCTCACTCCCTTTCTCTAAC-3′) and 172a2r (5′-GACTAACTAGTGTTCTCAAGTTGAGCACTTGAAAAC-3′), with XhoI (underlined) and SpeI (underlined and boldface) sites, respectively, for cloning purposes. The genomic PCR product was double-digested with XhoI and SpeI and subcloned into the binary pBA002 Ti plasmid under the control of the CaMV 35S promoter. Agrobacterium tumefaciens–mediated transformation of Arabidopsis plants was performed by a modified floral dip method as described previously (Clough and Bent, 1998), and homozygotic lines were obtained through selection for three consecutive generations.
For transient expression of MIR172 genes and their targets in Nicotiana benthamiana leaf cells, the gene constructs were subcloned into the pB2GW7 Gateway vector (Invitrogen). Agrobacterial cells containing the vector constructs were injected directly into the leaves. miRNA was extracted from the leaves at 4 d after injection as described and analyzed on a 1.2% formaldehyde–agarose gel (Kim et al., 2005).
Isolation of the toe1-2D Mutant
Ecotype Col-0 was transformed with the activation-tagging vector pSKI015 that contains the CaMV 35S enhancer element. The pSKI015 vector was kindly provided by D. Weigel and used as described previously (Weigel et al., 2000). To select herbicide-resistant transformants, T0 seeds were collected, sown in soil, and sprayed twice a week with a 1:1000 dilution (in water) of Finale (AgrEvo) containing 5.78% Basta. The herbicide-resistant plants were then screened for the late-flowering phenotype. The mutants were further examined through two additional generations. Among the homozygotic T3 plants that exhibited late flowering, a mutant (toe1-2D) was chosen for further analysis.
The single T-DNA insertion event in toe1-2D was verified by genomic DNA gel blot hybridization using the 35S enhancer sequence as a probe, followed by analysis of segregation ratios. The sequences flanking the T-DNA insertion site in toe1-2D were determined by three-step thermal asymmetric interlaced–PCR (Liu et al., 1995) and DNA sequencing.
Flowering-Time Measurements
Plants were grown on soil under either LDs or SDs. Flowering time was measured both by counting the total number of rosette and cauline leaves at flowering initiation and by days from sowing to floral bud formation. Thirty to 35 plants were measured and averaged for each plant group. Statistical significance was determined using Student's t test.
Vernalization and GA Treatments
For vernalization treatments, plants were germinated and grown on soil at 4°C for 6 weeks under SDs, transferred to normal growth conditions (23°C, LDs), and grown until flowering. To examine GA effects on flowering initiation, plants were grown on soil at 23°C under LDs, and a GA solution of 20 μM was sprayed twice a week until flowering. Thirty to 35 plants were measured and averaged for each treatment. Statistical significance was determined using Student's t test.
RNA Gel Blot Hybridization and DNA Gel Blot Hybridization of RT-PCR Products
Transcript levels were determined either by RNA gel blot hybridization or by DNA gel blot hybridization of PCR products. For RNA gel blot hybridization, total RNA samples were extracted from plant materials using the RNeasy plant total RNA isolation kit (Qiagen). Routinely, 20 μg of each RNA sample was denatured in denaturation buffer (20 mM MOPS, 8 mM sodium acetate, and 1 mM EDTA) supplemented with 50% (v/v) formamide and 2.2 M formaldehyde by incubating at 65°C for 10 min, resolved on a 1.0% denaturing agarose gel, and transferred to a Hybond-N+ nylon membrane (Amersham-Pharmacia). The blotted membranes were hybridized with gene-specific probes labeled with digoxigenin-UTP using the DIG RNA labeling kit (Roche Applied Science).
Prior to RT-PCR, total RNA samples were extensively pretreated with RNase-free DNaseI to eliminate any contaminating genomic DNA. To determine the linear amplification range for the RT-PCR of each gene, a certain amount (0.5 to 2 μg) of each total RNA sample was used for RT-PCR runs with different numbers of cycles (15 to 35 cycles). The RT-PCR products were subjected to DNA gel blot hybridization, and densitometric scores were plotted against the numbers of PCR cycles. RT-PCR runs were performed for 20 to 35 cycles, depending on the linear range of PCR amplification for each gene.
Each PCR cycle included incubations at 94°C for 1 min, 55°C for 1 min, and 72°C for 5 min. One additional cycle at 72°C for 10 min was run after the last cycle to allow trimming of incomplete polymerizations. Whenever possible, positive and negative control genes were included in the reactions to ensure the feasibility of the assay conditions.
For RT-PCR–based DNA gel blot hybridization, <10 ng of PCR products was analyzed on a 1% agarose gel and transferred to a Hybond-N+ nylon membrane (Amersham-Pharmacia). The membrane was hybridized with gene-specific probes labeled with [32P]dCTP using the Megaprime DNA labeling system (Amersham-Pharmacia). The primers used for the RT-PCR are listed in Supplemental Table 1 online. Quantitation of transcript levels was determined by densitometry of images using the Labwork image acquisition and analysis program (Media Cybernetics).
miRNA Extraction and RNA Gel Blot Hybridization
Plants were grown for 2 weeks before harvesting plant materials, unless specified otherwise. For miRNA RNA gel blot hybridization, total RNA samples were extracted from plant materials using the Trizol reagent (Invitrogen) according to the procedure described previously (Pfeffer et al., 2003), but with a few modifications. After isopropanol precipitation, the Eppendorf tube containing the RNA pellet was briefly centrifuged without rinsing with ethanol, and the remaining isopropanol was completely removed by pipetting. The RNA pellet was then dissolved in 95% formamide and 25 mM EDTA without rinsing with 70% ethanol. We found that these modified steps greatly improve both the yield and the solubility of miRNA in total RNA preparations. We also found that the high-salt condition (1.2 M NaCl and 0.8 M Na citrate), which is required for RNA preparations used for subsequent enzymatic reactions, should be eliminated for maximal yields.
miRNA RNA gel blot hybridizations were performed using the ULTRA-Hyb Oligo solution according to the procedure supplied by the manufacturer (Ambion). The oligonucleotide probes were 5′ end–labeled using [32P]γATP and T4 polynucleotide kinase. An oligonucleotide complementary to 5S rRNA was also processed in the same way and used as a quality control for RNA preparations.
For the detection and measurement of primary MIR172 transcripts, DNA gel blot hybridization of RT-PCR products was performed using the primer pairs described in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TOE1 Activation by a Nearby Insertion of the 35S Enhancer in the toe1-2D Mutant.
Supplemental Figure 2. TOE1 Protein Structure.
Supplemental Figure 3. Expression Patterns of the miR172 Target Genes.
Supplemental Figure 4. Sequence Comparison of miR172s and Their Target Genes.
Supplemental Figure 5. miR172 Abundance in ft Mutants and 35S:FT Transgenic Plants.
Supplemental Figure 6. Tissue-Specific Expression Patterns of the TOE1 Gene.
Supplemental Figure 7. Flowering Responses of the toe1-2D Mutant to Vernalization and GA Treatments.
Supplemental Figure 8. TOE1 Expression in Flowering-Time Mutants.
Supplemental Table 1. PCR Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank X. Chen for providing the 172OX line and S.K. Kang for transformation of gi-2 and co-2 with the MIR172a-2 gene construct. This work was supported by the BK21, Biogreen21 (Grant 20050301034456), and National Research Laboratory (Grant 2005-01039) programs and by grants from the Plant Signaling Network Research Center and the Korea Research Foundation (Grant 2005-070-C00129).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Chung-Mo Park (cmpark@snu.ac.kr).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra, I.C., Michaels, S.D., Schomburg, F.M., and Amasino, R.M. (2004). Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 40 112–119. [DOI] [PubMed] [Google Scholar]
- Cerdan, P.D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423 881–885. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2003). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Cigan, A.M., Saeteurn, K., and Hake, S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39 544–549. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Davis, S.J. (2002). Photoperiodism: The coincidental perception of the season. Curr. Biol. 12 R841–R843. [DOI] [PubMed] [Google Scholar]
- Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., Yano, M., and Yoshimura, A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, M.R., Bizzell, C.M., Keller, M.R., Michaels, S.D., Song, J., Noh, Y.S., and Amasino, R.M. (2005). HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J. 41 376–385. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal, S., Alonso-Blanco, C., Peeters, A.J., Wagemaker, C., Weller, J.L., and Koornneef, M. (2003). The role of Cryptochrome 2 in flowering in Arabidopsis. Plant Physiol. 133 1504–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2003). Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 6 13–19. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Doyle, M.R., and Amasino, R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., and Kay, S.A. (2006). Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 11 550–558. [DOI] [PubMed] [Google Scholar]
- Kim, J., Jung, J.-H., Reyes, J.L., Kim, Y.-S., Kim, S.-Y., Chung, K.-S., Kim, J.-A., Lee, M., Lee, Y., Kim, V.N., Chua, N.-H., and Park, C.-M. (2005). MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stem. Plant J. 42 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229 57–66. [DOI] [PubMed] [Google Scholar]
- Lauter, N., Kampani, A., Carlson, S., Goebel, M., and Moose, S.P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 102 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, M.H., Kim, J., Kim, Y.S., Chung, K.S., Seo, Y.H., Lee, I., Kim, J., Hong, C.B., Kim, H.J., and Park, C.M. (2004). A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. (2000). Photoreceptors and regulation of flowering time. Plant Physiol. 123 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89 737–745. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S., Yang, Z., Kim, Y., and Chen, X. (2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol. Biol. 61 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y.C., Dolan, S., and Lin, C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.): S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto, T., Kohchi, T., Yokota, A., Hwang, I., and Goodman, H.M. (1999). The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101 331–340. [DOI] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio, K.A., Crepy, M., Martin-Tryon, E.L., Milich, R., Harmer, S.L., Putterill, J., Yanovsky, M.J., and Casal, J.J. (2007). GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 144 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., Lagos-Quintana, M., and Tuschl, T. (2003). Cloning of small RNA molecules. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidmann, J.A. Smith, and K. Struhl, eds (New York: John Wiley & Sons), p. 26. [DOI] [PubMed]
- Putterill, J. (2001). Flowering in time: Genes controlling photoperiodic flowering in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356 1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., Laurie, R., and Macknight, R. (2004). It's time to flower: The genetic control of flowering time. Bioessays 26 363–373. [DOI] [PubMed] [Google Scholar]
- Quesada, V., Dean, C., and Simpson, G.G. (2005). Regulated RNA processing in the control of Arabidopsis flowering. Int. J. Dev. Biol. 49 773–780. [DOI] [PubMed] [Google Scholar]
- Razem, F.A., El-Kereamy, A., Abrams, S.R., and Hill, R.D. (2006). The RNA-binding protein FCA is an abscisic acid receptor. Nature 439 290–294. [DOI] [PubMed] [Google Scholar]
- Samach, A., and Coupland, G. (2000). Time measurement and the control of flowering in plants. Bioessays 22 38–47. [DOI] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7 487–491. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schomburg, F.M., Patton, D.A., Meinke, D.W., and Amasino, R.M. (2001). FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8 517–527. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., Dijkwel, P.P., Quesada, V., Henderson, I., and Dean, C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113 777–787. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A., Pearson, M., and Kay, S.A. (1998). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125 485–494. [DOI] [PubMed] [Google Scholar]
- Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Liu, Z., Lu, F., Dong, A., and Huang, H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47 841–850. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2003). Living by the calendar: How plants know when to flower. Nat. Rev. Mol. Cell Biol. 4 265–275. [DOI] [PubMed] [Google Scholar]
- Yoo, S.K., Chung, K.S., Kim, J., Lee, J.H., Hong, S.M., Yoo, S.J., Yoo, S.Y., Lee, J.S., and Ahn, J.H. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.