Abstract
In Arabidopsis thaliana, the BEL1-like TALE homeodomain protein family consists of 13 members that form heterodimeric complexes with the Class 1 KNOX TALE homeodomain proteins, including SHOOTMERISTEMLESS (STM) and BREVIPEDICELLUS (BP). The BEL1-like protein BELLRINGER (BLR) functions together with STM and BP in the shoot apex to regulate meristem identity and function and to promote correct shoot architecture. We have characterized two additional BEL1-LIKE HOMEODOMAIN (BLH) proteins, SAWTOOTH1 (BLH2/SAW1) and SAWTOOTH2 (BLH4/SAW2) that, in contrast with BLR, are expressed in lateral organs and negatively regulate BP expression. saw1 and saw2 single mutants have no obvious phenotype, but the saw1 saw2 double mutant has increased leaf serrations and revolute margins, indicating that SAW1 and SAW2 act redundantly to limit leaf margin growth. Consistent with this hypothesis, overexpression of SAW1 suppresses overall growth of the plant shoot. BP is ectopically expressed in the leaf serrations of saw1 saw2 double mutants. Ectopic expression of Class 1 KNOX genes in leaves has been observed previously in loss-of-function mutants of ASYMMETRIC LEAVES (AS1). Overexpression of SAW1 in an as1 mutant suppresses the as1 leaf phenotype and reduces ectopic BP leaf expression. Taken together, our data suggest that BLH2/SAW1 and BLH4/SAW2 establish leaf shape by repressing growth in specific subdomains of the leaf at least in part by repressing expression of one or more of the KNOX genes.
INTRODUCTION
Morphogenesis in plants is achieved primarily through the regulation of differential growth in distinct domains of a developing organism. Such differential growth is specified by transcription factors expressed in specific domains in response to positional information. Among the transcription factors that regulate morphogenesis are members of the BEL1-LIKE HOMEODOMAIN (BLH) and KNOTTED1-LIKE HOMEOBOX (KNOX) TALE (for three amino acid loop extension) homeodomain protein families.
In Arabidopsis thaliana, two key members of the KNOX family whose functions have been defined are SHOOTMERISTEMLESS (STM), required for meristem maintenance and/or function (Barton and Poethig, 1993; Long et al., 1996), and BREVIPEDICELLUS (BP/KNAT1), required for regulation of inflorescence architecture (Douglas et al., 2002; Venglat et al., 2002). The Arabidopsis BLH proteins BELLRINGER (also called PENNYWISE, REPLUMLESS, VAAMANA [BLR/PNY/RPL/VAM]) and its paralog POUNDFOOLISH (PNF) play similar roles in shoot development and exhibit functional redundancy in these roles (Byrne et al., 2003; Smith and Hake, 2003; Bao et al., 2004; Bhatt et al., 2004; Smith et al., 2004).
KNOX proteins interact with specific members of BLH proteins in Arabidopsis (Bellaoui et al., 2001; Byrne et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004; Hackbusch et al., 2005; Kanrar et al., 2006). Such interactions have been shown to be required for site-specific DNA binding (Smith et al., 2002) and for nuclear localization of the transcription factor heterodimeric complex (Bhatt et al., 2004; Cole et al., 2006). Analogous BLH–KNOX interactions have also been reported in other plants, such as potato (Solanum tuberosum) and barley (Hordeum vulgare), indicating that these interactions are evolutionarily conserved and that the interaction is probably required for their biological functions (Muller et al., 2001; Chen et al., 2003).
The Arabidopsis BEL1 protein, required for ovule morphogenesis (Robinson-Beers et al., 1992; Modrusan et al., 1994), also interacts with BP and other KNOX proteins in yeast and in vitro (Bellaoui et al., 2001; Hackbusch et al., 2005). bel1 loss-of-function mutants have abnormal ovules due to defects in integument differentiation (Robinson-Beers et al., 1992; Modrusan et al., 1994). However, no KNOX-derived functions have been attributed to the mediation of BEL1-mediated ovule morphogenesis. Although the BEL1 gene is expressed in stems, leaves, sepals, inflorescence meristems, roots, and ovules (Reiser et al., 1995; Bellaoui et al., 2001; this study), no obvious phenotypes outside the ovule are visible in bel1 knockout lines. The lack of phenotypes in these other tissues might be attributable to the presence of other BLH proteins whose functions overlap with that of BEL1. To investigate this possibility, we have used a reverse genetics approach to begin to unravel the redundancy that we show exists in the BLH family.
In this study, we describe the characterization of two BLH proteins, SAWTOOTH1 (SAW1) and SAW2 (formerly called BLH2 and BLH4, respectively) that show significant amino acid sequence similarities with BEL1. The two SAW proteins are functionally redundant. The saw double mutants exhibit increased leaf serrations, giving the leaf margin a saw-like appearance. The leaf margins are also more revolute (abaxially curled) than the wild type. Overexpression of SAW1 impedes general growth of the plant. Molecular and genetic analyses reveal that the SAW proteins function in part by repressing BP (and other Class I KNOX genes) expression in the leaves.
RESULTS
BEL1, SAW1, and SAW2 Are Members of the BELL Family of Homeodomain Proteins
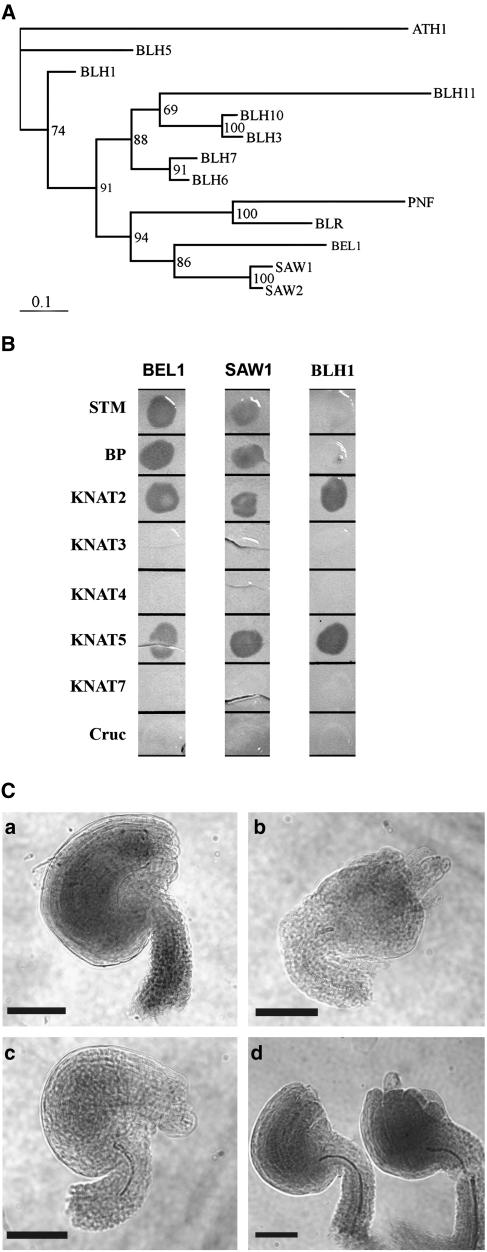
The loss of BEL1 gene function results in abnormal ovule growth and embryo sac arrest (Robinson-Beers et al., 1992; Modrusan et al., 1994). Consistent with its mutant phenotype, BEL1 is expressed in the developing ovule and in the inflorescence apex (Reiser et al., 1995; Bellaoui et al., 2001). However, RNA gel blot (Reiser et al., 1995) and in situ hybridization (Bellaoui et al., 2001) indicate that BEL1 is expressed in vegetative tissues where there is no obvious mutant phenotype. This lack of vegetative phenotype in a bel1 null mutant could be explained by genetic redundancy. In Arabidopsis, there are 12 TALE homeobox genes closely related in sequence to BEL1 that have been identified, including BLH1-7, BLH10, BLH11 (for BEL1-LIKE HOMEODOMAIN), BLR, PNF, and ATH1 (Quaedvlieg et al., 1995; Reiser et al., 1995; Bellaoui et al., 2001; Becker et al., 2002; Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Smith et al., 2004; Hackbusch et al., 2005; this study). These deduced BLH proteins share at least three regions of sequence similarity: the DNA binding homeodomain and the BELL and SKY domains involved in BEL1 interactions with KNOX TALE homeodomain proteins (Bellaoui et al., 2001; Becker et al., 2002). Phylogenetic analysis of this gene family (Figure 1A; Becker et al., 2002) indicates that the genes most closely related in sequence to BEL1 are two genes formerly referred to as BLH2 and BLH4 (Becker et al., 2002) that we designate SAW1 and SAW2, respectively, based on their loss-of-function leaf phenotype (see below). We focused on these two genes as the ones most likely to have overlapping functions with BEL1, similar to the situation observed with Arabidopsis MADS domain proteins (Pelaz et al., 2000). SAW1 and SAW2 proteins are 87% identical. The BEL1 gene is more closely related to BLH2/SAW1 and BLH4/SAW2 than to other BLH genes (Figure 1A; 73 and 72% protein sequence similarity, respectively). SAW2 and SAW1 are on chromosomes 2 and 4, respectively, and the chromosomal regions surrounding these genes share extensive sequence similarity (Table 1; see Supplemental Figure 1B online; Blanc et al., 2003), suggesting that they arose from segmental chromosomal duplication of one of the two loci. Since SAW1 and SAW2 exhibit sequence similarity, it is possible that the two genes share similar functions and exhibit similar expression patterns. Indeed, a comparison of the regions upstream to the translational start sites of SAW1 and SAW2 genes shows three conserved regions in the 5′ untranslated region (see Supplemental Figure 1C online), indicating that they might have common transcriptional and posttranscriptional regulatory elements. In support of this hypothesis, results of a PRIMe microarray coexpression gene search (http://prime.psc.riken.jp/?action=coexpression_index) with one of the two genes identifies the other as the best coexpression match, indicating that the two genes are expressed in similar domains.
Figure 1.
SAW and BEL1 Proteins Exhibit Sequence Similarity and Functional Redundancy.
(A) Phylogram of the 13 members of the BLH protein family. Bootstrap values are shown at the branch points.
(B) Yeast two-hybrid X-Gal filter assay showing interactions of BLH and KNOX proteins. The BLH proteins were used as baits and KNOX proteins as preys. Cruc, Cruciferin (negative control).
(C) Whole mounts of mature ovules showing complementation of the bel1 mutant by 35S:SAW1. (a) The wild type (b) bel1-3, (c) 35S:SAW1, and (d) 35S:SAW1 bel1-3. Bars = 50 μm.
Table 1.
Paralogous Genes Found in the Duplicated Region That Includes SAW1 and SAW2
| Gene (AGI Number) | Paralog | Description | Exp | Identity | Similarity |
|---|---|---|---|---|---|
| At2g23630 | At4g37160 | Pectinesterase-like protein | 3.8E-192 | 78% | 87% |
| At2g23680 | At4g37220 | Cold acclimation protein homolog | 1.0E-33 | 44% | 63% |
| At2g23690 | At4g37240 | Unknown/putative protein | 1.0E-28 | 59% | 66% |
| At2g23760 | At4g36870 | BEL1-like homeobox proteins (SAW2 and SAW1) | 2.9E-114 | 61% | 70% |
| At2g23790 | At4g36820 | Hypothetical/putative protein | 8.4E-100 | 66% | 80% |
| At2g23800 | At4g36810 | Geranylgeranyl pyrophosphate synthase | 1.2E-101 | 72% | 86% |
The genes were identified using the Web-based software available at http://wolfe.gen.tcd.ie/athal/index.html (Blanc et al., 2003). Various single copy genes occurring in the region due to deletions and chromosomal rearrangements have not been included. AGI, Arabidopsis Genome Initiative.
SAW1, but Not BLH1, Interacts with the Same KNOX Proteins as BEL1
We have previously reported that BEL1 selectively interacts with KNOX homeodomain proteins and that this interaction primarily involves the SKY and BELL domains (Bellaoui et al., 2001). Since these domains are highly conserved between BEL1, SAW1, and SAW2 proteins, it is possible that their ability to interact with KNOX proteins is also similar. To test this hypothesis, full-length cDNAs for BEL1, SAW1, and a more distantly related gene BLH1 were used in yeast two-hybrid assays as prey (TA) to test for interaction with KNAT homeodomain proteins and the seed storage protein Cruciferin (negative control) as bait (DB). As anticipated from previous results (Bellaoui et al., 2001), BEL1 was able to interact with STM, BP, KNAT2, and KNAT5 but not KNAT3, KNAT4, KNAT7, or Cruciferin. None of the BLH proteins in combination with DB-Cruciferin activated β-galactosidase, indicating that BLH proteins do not interact with GAL4 DB or nonspecifically with other proteins (Figure 1B). TA-SAW1 and TA-SAW2 promoted β-galactosidase activity in combination with DB-BP, KNAT2, KNAT5, and STM but not with DB-KNAT3, 4 or 7; an interaction profile identical to that of BEL1. However, TA-BLH1 promoted β-galactosidase activity only in combination with TA-KNAT2 and pTA-KNAT5. These data suggest that members of BLH family are generally capable of forming heterodimeric complexes with KNOX proteins but that the specific KNOX partners may vary.
SAW1, SAW2, and BEL1 Are Expressed in Partially Overlapping Expression Domains
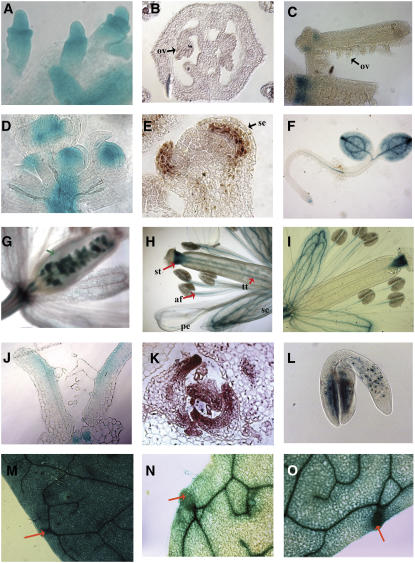
We investigated the expression domains of SAW1 and SAW2 in more detail using RT-PCR, RNA in situ hybridization (SAW1 and BEL1), and β-glucuronidase (GUS) reporter constructs fused to the SAW1, SAW2, and BEL1 promoter regions to provide clues to their function and determine the extent to which their expression domains overlap with each other and that of BEL1 (Figure 2; see Supplemental Figure 2 online). Unlike BEL1 (Reiser et al., 1995; Bellaoui et al., 2001), SAW1 and SAW2 were not expressed in developing ovules (Figures 2B and 2C) and therefore are unlikely to be redundant with BEL1 function in ovule development. These data are consistent with the fact that mutation of BEL1 results in an ovule mutant phenotype (Reiser et al., 1995). BEL1 is expressed in the inflorescence and floral meristems (Figure 2D; Bellaoui et al., 2001), but SAW1 and SAW2 are not (Figures 2E and 2F). However, in other parts of the plant, especially lateral organs, the expression domains of BEL1, SAW1, and SAW2 overlap extensively, indicating that these proteins might play a specialized role in the development of such lateral organs. In flowers, all three genes were expressed in the sepals. SAW1 and SAW2, but not BEL1, were expressed in the anther filament, style, and transmitting tract (Figures 2H and 2I), while only BEL1 was expressed in the ovary walls and at the base of the flower (receptacle; Figure 2G). SAW1 showed a unique, but faint, expression in the petals.
Figure 2.
Expression Analysis of SAW and BEL1 Genes.
(A) Immature ovules showing BEL1:GUS activity.
(B) SAW1 in situ hybridization of a cross section through the gynoecium showing immature ovules (ov).
(C) SAW2:GUS expression in whole-mount gynoecia. One of the gynoecia was split open to reveal the immature ovules.
(D) Whole mount of the inflorescence apex. BEL1:GUS expression is detected in the floral meristems of immature flowers.
(E) Longitudinal section of a stage 4 flower showing SAW1 expression (assayed by in situ hybridization) in the adaxial side of the developing sepal (se).
(F) SAW2:GUS expression in a 2-d-old seedling.
(G) to (I) Whole mounts of flowers showing BEL1:GUS, SAW1:GUS, and SAW2:GUS activity, respectively. st, style; tt, transmitting tract; af, anther filament; se, sepal; pe, petal.
(J) to (L) BEL1, SAW1, and SAW2 exhibit adaxial expression in developing lateral organs.
(J) A longitudinal section through the vegetative apex of a 7-d-old seedling showing adaxial expression of BEL1:GUS in the leaves.
(K) Cross section through the vegetative apex showing in situ localization of SAW1 in the adaxial side of developing leaves.
(L) SAW2:GUS expression in a developing embryo.
(M) to (O) Whole mounts of fifth leaves of 3-week-old plants showing BEL1:GUS, SAW1:GUS, and SAW2:GUS activity, respectively.
In developing leaves, BEL1:GUS was localized to the adaxial side (Figure 2J), similar to the adaxial expression observed for SAW1 in sepals and leaves by in situ hybridization experiments (Figures 2E and 2K) and for SAW2:GUS in the cotyledons of a developing embryo (Figure 2L), suggesting a role for these genes in the development of lateral organ asymmetry. SAW1, SAW2, and BEL1 were all expressed in mature leaves, with a more uniform adaxial/abaxial distribution. BEL1 expression is fairly uniform throughout the mature leaf; however, SAW1 and SAW2 show higher expression in vasculature and hydathodes in comparison to the mesophyll and epidermal cells (Figures 2M to 2O), and BEL1, SAW1, and SAW2 expression patterns also overlapped in the stem (cortex), pedicel, root, and embryo (data not shown).
35S:SAW1 Complements the bel1 Ovule Phenotype
Several lines of evidence suggest that SAW1 could be redundant with BEL1. To test this hypothesis directly, we introduced 35S:SAW1 into a bel1 mutant background and evaluated its ability to complement the bel1 mutant phenotype. A 35S:SAW1 gene construct was transformed into wild-type plants. A hemizygous 35S:SAW1 transformant was crossed to bel1-3 using bel1-3 as a pollen donor (the 35S:SAW1 plants are male sterile; see below) and the F1 progeny backcrossed to bel1-3. The bel1-3 homozygous testcross progeny were identified by PCR (see Methods) and the ovule phenotype examined. All 18 35S:SAW1 bel1-3/bel1-3 lines tested had ovules that appeared morphologically wild-type with both inner and outer integuments. Approximately 50% of the ovules in each silique were indistinguishable from the wild type in appearance, while the remainder had integuments that were shorter in length than the wild type (Figure 1C). Furthermore, all 35S:SAW1 bel1-3/bel1-3 lines that were cross-pollinated set seeds (data not shown), indicating that the ovules were fertile. Similar results were also obtained when 35S:SAW1 was crossed into the bel1-1 mutant (data not shown). These results demonstrate that 35S:SAW1 can complement the bel1-3 ovule defect.
saw1 and saw2 Affect Leaf Margin Development
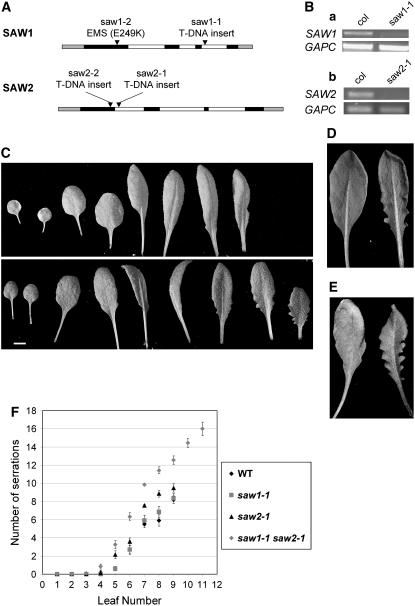
In an attempt to learn more about the function of SAW1 and SAW2 in planta, reverse genetic techniques were used to generate plant lines with loss-of-function SAW1 and SAW2 alleles. We examined saw1 and saw2 mutant alleles obtained through the Salk collection of T-DNA insertion lines (Alonso et al., 2003) and by screening ethyl methanesulfonate–mutagenized plants by TILLING (McCallum et al., 2000; Colbert et al., 2001; Till et al., 2004). Figure 3A shows the position of insertion/mutation in the gene in these alleles. A SAW1 TILLING allele (saw1-2) had a missense mutation that would be expected to result in a nonconservative amino acid substitution that could alter the function of the encoded protein. The saw1-1 allele has a T-DNA insertion in the third intron that separates the two exons that code for the homeodomain region. The saw1-1 transcript is severely reduced in abundance relative to SAW1 (Figure 3B), but transcript is still detectable. Thus, the saw1-1 mutant could be hypomorphic. The T-DNA insertional lines, saw2-1 and saw2-2, have insertions in the first intron and first exon, respectively, and have no detectable full-length transcripts (Figure 3B). All mutant alleles segregated in F2 populations as expected for single nuclear loci (data not shown). Under our normal growth conditions, no obvious morphological defects in the shoot segregated with any of the alleles examined.
Figure 3.
SAW Proteins Affect Leaf Margin Development.
(A) Graphical representations of the transcribed regions of SAW1 and SAW2 genes. Black, white, and gray boxes indicate exons, introns, and untranslated regions, respectively. The position of mutation/insertion in each mutant allele is marked by an arrowhead.
(B) RT-PCR analysis of SAW1 and SAW2 expression in wild-type (Col) and saw mutants. The fifth and sixth leaves were harvested from 4-week-old plants for RNA extraction and cDNA synthesis. (a) SAW1 expression is considerably reduced in saw1-1 compared with the wild type. Cytosolic-glyceraldehyde-3-phosphate dehydrogenase (GAPC) was used as a loading control. SAW1, 35 cycles; GAPC, 28 cycles. (b) There is no detectable SAW2 expression in the leaves of saw2-1 mutants, but SAW2 is detected in wild-type leaves. SAW2, 35 cycles; GAPC, 25 cycles.
(C) A comparison of the leaves of 4-week-old wild-type (top) and saw1-1 saw2-1 (bottom). saw1-1 saw2-1 double mutants had more serrations than the wild type from the seventh leaf onward. The fourth leaf of the double mutant was longer and more ovate than the fourth wild-type leaf.
(D) Leaf margins of saw 1-1 saw2-1 double mutants are more revolute than wild-type leaf margins, as shown by an abaxial view of the eighth leaf of 4-week-old wild-type (left) and saw1-1 saw2-1 (right) plants.
(E) The difference in the length of leaf serrations is more pronounced in older plants, as shown by an adaxial view of the ninth leaf of five-week-old wild-type (left) and saw1-1 saw2-1 plants (right).
(F) Leaves of both saw1-1 saw2-1 double mutants and saw2-1 single mutants had more serrations than wild-type leaves. Scatterplot showing the number of serrations for each leaf of 5-week-old wild-type, saw1-1, saw2-1, and saw1-1 saw2-1 plants. Serrations were counted only on the rosette leaves that had been initiated prior to bolting. Points represent means ± se. As can be seen from the chart, saw1-1 saw2-1 double mutants also initiate more leaves than the wild type prior to bolting.
Since SAW1 and SAW2 are products of chromosomal duplication (Table 1) and have very similar expression profiles, we suspected that the lack of visible defects in the single mutants might be because of functional redundancy and decided to examine saw1-1 saw2-1 double mutants. Plants from an F2 population segregating for both saw1-1 and saw2-1 were genotyped using molecular assays (see Methods). All saw1-1 saw2-1 double mutants, unlike the single mutants, had a distinct mutant phenotype characterized by increased serrations in leaf margins and revolute leaves that have margins curled abaxially (Figures 3C to 3E). Since in wild-type leaves SAW1 and SAW2 are specifically expressed in the adaxial side of developing leaves (Figures 2K and 2L) and in the hydathodes in the margins of older leaves (Figures 2N and 2O), these phenotypes could arise from loss of SAW function in these tissues. The saw2 single mutants also have slightly revolute margins, but the phenotype is not as obvious as in the double mutant. On closer examination, the single mutants also show a slight increase in serration length and numbers. The serrations observed were most prominent in the seventh and subsequent leaves initiated, and these leaves have a sawtooth appearance; hence, the genes were designated as SAW1 and SAW2. The double mutants had significantly more and deeper serrations than the wild type or the single mutants (Table 2). Also, the number of serrations increased with leaf number (Figure 3F). Plants heterozygous for either of the two mutations and homozygous for the other also exhibit serrations (less prominent than the double mutants), indicating that this phenotype is dosage dependant (data not shown). In addition to the leaf margin defects, saw1-1 saw2-1 double mutant leaves are darker green in color, and the third and fourth leaves of the double mutant are more elongated than the wild-type leaves of the same stage. The plants produce more leaves than the wild type and show a delay in flowering by 2 to 3 d in our continuous light growth conditions (Table 2).
Table 2.
Serration Length and Flowering Time in Wild-Type and saw Mutants
| Average Serration Length (mm) | Days to Bolting | Number of Rosette Leaves at Bolting | |
|---|---|---|---|
| Wild type | 0.54 ± 0.039a | 26.14 ± 0.27a | 7.95 ± 0.19a |
| saw1-1 | 0.75 ± 0.074a,b | 25.66 ± 0.2a | 7.92 ± 0.27a |
| saw2-1 | 0.86 ± 0.071b | 26.25 ± 0.45a | 8.08 ± 0.2a |
| saw1-1 saw2-1 | 1.31 ± 0.073c | 29.95 ± 0.35b | 10.36 ± 0.22b |
Values are mean ± se (n ≥ 9). The serrations were measured in the seventh leaf of 4-week-old plants. a, b, and c in each column denote significant differences (analysis of variance, P = 0.05). Means with the same letters are not significantly different.
We demonstrated that the observed phenotypes were due to disruption of saw1 and saw2 in three ways. First, all phenotypes cosegregated with the saw1-1 saw2-1 double mutant genotype. Second, saw double mutants carrying different saw1 and saw2 allele combinations (saw1-1 saw2-1; saw1-2 saw1-1 and saw1-1 saw2-2) had similar phenotypes (Table 3). Third, the saw1-1 saw2-1 double mutant phenotype was complemented by transformation with a SAW2 genomic DNA fragment (see Methods). Nineteen out of 28 transformants showed a significant reduction in the number of leaf serrations (see Supplemental Figure 3 online). Since complementation of the mutant phenotype looks to be dosage dependent, both the number of copies inserted and the position of insertion may influence the extent of complementation in the transformant.
Table 3.
Serrations in the Seventh Leaf of Different Allele Combinations of saw Mutants
| Wild Type | saw1-1 saw2-1 | saw1-2 saw2-1 | saw1-1 saw2-2 | |
|---|---|---|---|---|
| Average number of serrations | 5.5 ± 0.38a | 9.8 ± 0.37b | 9.7 ± 0.41b | 10.2 ± 0.24b |
Values are mean ± se (n = 12). a and b denote significant differences (analysis of variance, P = 0.05). Means with the same letters are not significantly different.
bel1 Does Not Affect the saw1 saw2 Double Mutant Phenotype
Since BEL1 and the SAW proteins are very similar in structure, expression, and interaction pattern, a triple mutant was generated with saw1-1, saw2-1, and bel1-1 to check for potential genetic redundancy or interaction. The progeny of an F2 plant that was homozygous for saw1-1 and saw2-1 and heterozygous for bel1-1 (bel1-1 is female sterile) were screened for triple mutants. The triple mutants identified showed an additive phenotype, including both bel1-1–like ovule defects and serrated leaves similar to those of the saw1-1 saw2-1 double mutant, indicating that BEL1 and SAW genes are not functionally redundant in tissues where their expression overlaps and/or as yet uncharacterized genes provide additional functional redundancy masking the affect of BEL1 loss.
SAW1 and the Class 1 KNOX Genes STM, BP, and KNAT2 Are Expressed in the Shoot Apex in Nearly Mutually Exclusive Domains
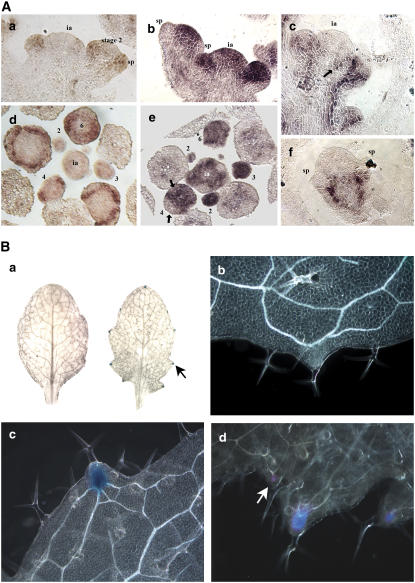
As shown above, SAW1 transcript in the shoot is found primarily in leafy organs and excluded from the meristematic region. Interestingly, although SAW1 has the ability to interact with Class 1 KNOX proteins STM, BP, and KNAT2, the available data for STM, BP, and KNAT2 gene expression has shown that all three genes are expressed in meristems, stem, and reproductive organs but not leaves, sepals, and petals (Lincoln et al., 1994; Long et al., 1996; Pautot et al., 2001). To verify the nonoverlap in expression pattern, we used in situ hybridization to compare SAW1 transcript distribution in the inflorescence with that for STM and BP. Our results (Figure 4A) clearly indicate that in these tissues, the SAW1 transcript distribution is adjacent to but does not significantly overlap with that of either STM or BP except in the stage 1 to 2 floral meristem. These data suggest that interaction between SAW1 and Class 1 KNOX proteins in the developing inflorescence either does not occur, occurs at early stages only to resolve to mutually exclusive domains, or occurs in cells along the common border of the different expression domains.
Figure 4.
SAW Genes Suppress BP Expression in Leaves.
(A) In situ hybridization showing SAW1, STM and BP expression patterns. (a) and (d) Probed with SAW1 antisense transcript. (b) and (e) Probed with STM antisense transcript. (c) and (f) Probed with BP antisense transcript. (a) to (c) Longitudinal sections; (d) and (e) transverse sections through an inflorescence apex. (f) Longitudinal section through a stage 3 flower. Numbers indicate the floral stages as described by Smyth et al. (1990). sp, sepal; ia, inflorescence apex.
(B) BP is misexpressed in leaves of saw double mutants. (a) Whole mounts of BP:GUS Col (left) and BP:GUS saw1-1 saw2-1 (right) leaves assayed for GUS activity. Arrow points to the BP:GUS expression localized to the hydathodes. (b) and (c) Higher magnification dark-field image of (a). (b) BP:GUS Col and (c) BP:GUS saw1-1 saw2-1. (d) Dark-field image of a whole mount of the ninth leaf of a 4-week-old BP:GUS saw1-1 saw2-1 plant assayed for GUS activity. Arrow points to BP:GUS expression in a secondary serration.
BP Is Ectopically Expressed in the Leaves of saw1 saw2 Double Mutants
The leaf serrations of the saw1-1 saw2-1 double mutant are similar to those found in the leaves of BP overexpression lines with weak phenotypes (Chuck et al., 1996). BP expression is not detected in wild-type Arabidopsis leaves (Byrne et al., 2000). To test if there is a change in BP expression pattern in the saw1-1 saw2-1 double mutants, we introduced a BP:GUS transgene (Ori et al., 2000) into a saw1-1 saw2-1 double mutant background by crossing the appropriate lines and screening the F2 population. BP:GUS expression was detected in the hydathodes (present at the tip of serrations) of the saw1-1 saw2-1 double mutants (Figure 4B) but not in the wild type. The intensity of GUS expression was roughly proportional to the size of the serration in the leaf margin (Figure 4B). BP:GUS expression was also observed in secondary serrations found in the ninth leaf and onwards (Figure 4B, d, arrow).
In addition to misexpression of BP in the leaf, 4-methylumbelliferyl β-d-glucuronide fluorometric assays (see Methods) revealed increased BP:GUS expression in the inflorescence and stem internodes of saw1 saw2 double mutants (see Supplemental Figure 4 online), indicating that BP (and perhaps other KNOX genes) may be upregulated in areas where SAW and BP expression overlap.
To further characterize the interactions of SAW and BP, saw1 saw2 bp triple mutants were generated. These plants had the characteristics of both saw1 saw2 double mutant (serrated leaves) and the bp phenotype (downward-pointing pedicels; see Supplemental Figure 6 online). The lack of a genetic interaction might be ascribed to the possible misexpression of other Class 1 KNOX genes, as was observed in the as1 and as2 mutants (Byrne et al., 2000; Ori et al., 2000).
In support, our RT-PCR data does indicate that besides BP, the KNOX genes KNAT6 and STM are also expressed in the leaves of saw1 saw2 but not in the wild type (see Supplemental Figure 7 online).
SAW1 and SAW2 Function in Parallel with AS1 to Regulate Leaf Margin Development
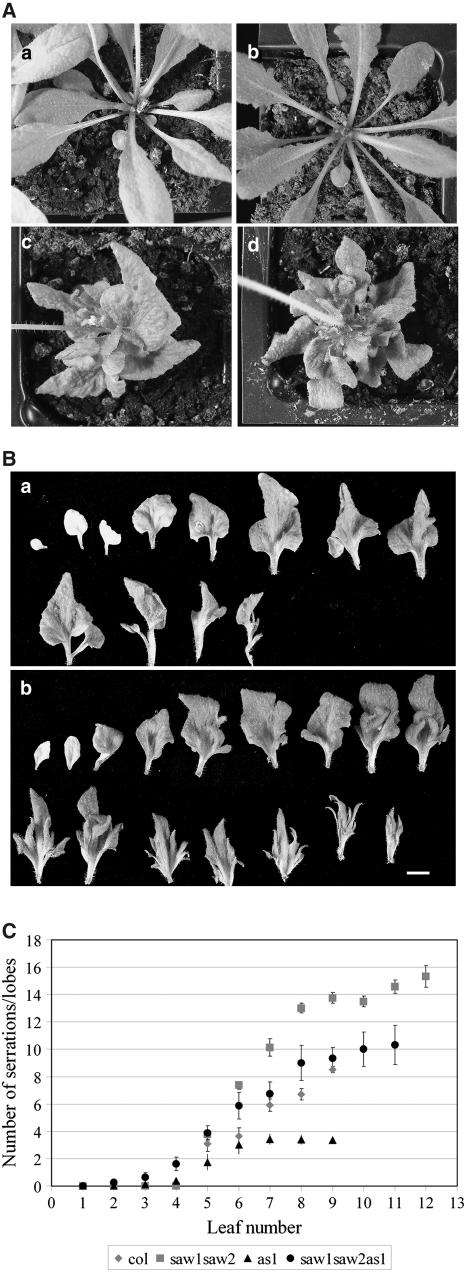
The as1 mutant of Arabidopsis exhibits lobing of the leaves and misexpression of KNOX genes, such as BP (Tsukaya and Uchimiya, 1997; Byrne et al., 2000; Semiarti et al., 2001). Like the saw1 saw2 double mutant serrations, the number of lobes in the as1 mutant increases with the leaf number (Ori et al., 2000). To investigate the genetic interaction between SAW1, SAW2, and AS1, we constructed the saw1 saw2 as1 triple mutant. The leaves (fifth leaf and higher) of saw1-1 saw2-1 as1-1 triple mutants exhibited more lobing/serrations than as1 and also showed deeper sinuses than either as1 or the saw1-1 saw2-1 double mutants (Figure 5). Although the triple mutant had more lobes than are found in as1 single mutants, the lobes were fewer in number than the serrations found in saw1-1 saw2-1 double mutants (Figure 5C). One possible explanation for this is that the as1 mutant has fewer hydathodes than the wild type and hence is unable to make as many serrations/lobes (Tsukaya and Uchimiya, 1997). Another possibility is that the longer serrations are formed at the expense of the serration number. Such an inverse correlation between serration length and serration number has been observed in a comparison of different species of Begonia (Mclellan and Dengler, 1995). In any case, these additive phenotypic effects observed in the double mutant suggest that the functions regulated by SAW1 and SAW2 are relatively independent of those regulated by AS1. Furthermore, RT-PCR analysis of SAW1 expression in wild-type and as1 leaves indicates that there is no difference in expression (see Supplemental Figure 5 online). Therefore, as1 phenotype is not a consequence of SAW1 downregulation. Taken together, these data indicate that AS1 and SAW1 are likely functioning in parallel pathways to repress BP expression.
Figure 5.
Phenotypes of saw1-1 saw2-1 as1 Triple Mutants.
(A) Rosette leaves of 5-week-old plants. (a) The wild type, (b) saw1-1 saw2-1, (c) as1, and (d) saw1-1 saw2-1 as1. The inflorescence has been removed from (b).
(B) A comparison of the leaves of 6-week-old as1 (top) and saw1-1 saw2-1 as1 (bottom). The top row of each panel has the rosette leaves formed prior to bolting. The bottom row rosette leaves are the ones formed after bolting (marked by the presence of secondary inflorescences in their axils). Bar = 5 mm.
(C) Scatterplot showing the number of serrations for each leaf of 5-week-old wild-type (Col), saw1-1 saw2-1, as1, and saw1-1 saw2-1 as1 plants. Serrations were counted only on the rosette leaves that had been initiated prior to bolting. Points represent means ± se.
Ectopic Expression of SAW1 Suppresses Shoot Growth
The saw1-1 saw2-1 loss-of-function phenotype suggests that the SAW genes function, at least in part, to suppress growth in developing leaves. To test this hypothesis, transgenic plant lines expressing SAW1 ectopically were generated by transforming Arabidopsis with a SAW1 gene under the control of a tandem cauliflower mosaic virus 35S promoter. Of 100 transformants recovered, 15 showed a similar set of morphological defects described below. All aspects of the phenotype segregated together with the T-DNA, and RNA gel blot analysis showed such plants to have a much higher level of SAW1 transcript than the wild type (see Supplemental Figure 9 online). Taken together, these data suggest that the phenotype is due to over/ectopic expression of SAW1.
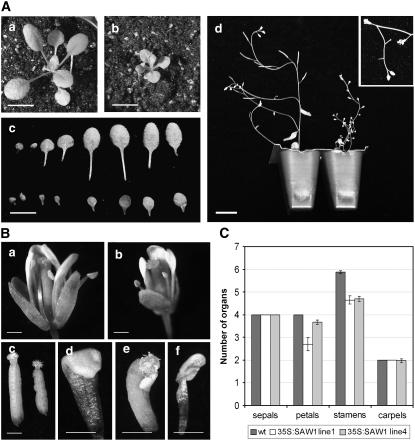
The 35S:SAW1 shoots were significantly smaller than that of the wild type (Figure 6A). Leaves were rounder in shape and either flat or curved in the dorsal-ventral axis toward the adaxial side unlike wild-type leaves that are slightly curved toward the abaxial side (Figure 6A, c) or the saw mutant, which has revolute leaves (Figure 3D). These data suggest defects in the abaxial-adaxial polarity of 35S:SAW1. Comparison of the cell types of the 35S:SAW1 abaxial and adaxial leaf surfaces with those of the wild type showed no obvious differences, indicating that the polarity defects were primarily due to differences in adaxial-abaxial growth rather than cell identity. Internodal length of the inflorescence stem was significantly shorter than the wild type, and measurements of stem epidermal cell length indicated that the growth defect was due to both a decrease in cell size and number (Figure 6A, d, Table 4).
Figure 6.
Phenotypes of 35S:SAW1.
(A) Shoot morphology. (a) and (b) 2.5-week-old plants: the wild type (a) and 35S:SAW1 (b) (bars = 5 mm). (c) A comparison of the leaves of 3-week-old wild type (top) and 35S:SAW (bar = 2 cm). (d) Four-week-old wild-type (left) and 35S:SAW1 (right) plants. Inset shows the fishbone-like growth defects observed in ∼5% of the inflorescences (including coflorescences) (bar = 2 cm).
(B) Organ defects in 35S:SAW1 flowers. (a) and (b) Stage 13 flowers of wild-type and 35S:SAW1 plants. Note that there are only three petals in this 35S:SAW1 flower. (c) Gynoecia of stage 13 flowers of wild-type (left) and 35S:SAW1 plants (right). (d) to (f) Floral organ defects observed in some of the 35S:SAW1 flowers. (d) Petal showing distal folding. (e) Stamen-carpel fusion. (f) Petal-stamen fusion. Bars = 1 mm.
(C) Floral organ numbers in the wild type and 35S:SAW1. Average number of sepals, petals, stamens, and carpels observed in the wild type and progeny of two independent transformed lines (lines 1 and 4). Error bars indicate se.
Table 4.
Cell Size and Number of Stem Epidermal Cells in the Wild Type and 35S:SAW1
| Average Cell Length (mm) | Length of Internode (mm) | Estimated Number of Cells in One Internode | |
|---|---|---|---|
| WT | 0.25 ± 0.006a | 18.9 ± 0.7a | 74 |
| 35S:SAW1 | 0.14 ± 0.003b | 8.5 ± 0.5b | 64 |
Values are mean ± se. a and b in each column denote significant differences (Student's t test, P = 0.05). Means with the same letters are not significantly different.
The flowers of 35S:SAW1 plants also showed morphological defects (Figure 6B). The number of organs in the second and third whorls was consistently less than in the wild type (Figures 6B and 6C), and organ fusions were not uncommon (Figure 6B, e and f). These phenotypes are similar to those described for the weak stm-2 allele, which also exhibits a reduction in organ number and organ fusions in the second and third whorls (Clark et al., 1996). Like the leaves, floral organs were smaller than the wild type. Petals were sometimes folded and pistils had an irregular bumpy appearance likely due to growth suppression in the silique valves (Figure 6B, c). Finally, 35S:SAW1 plants were sterile in part due to reduced pollen development in the anther and a decrease in stamen length that prevented self-fertilization. However, hand pollination of 35S:SAW1 pistils with their own pollen produced viable seed. Taken together, these data suggest that SAW1 can act as a negative regulator of growth throughout the plant.
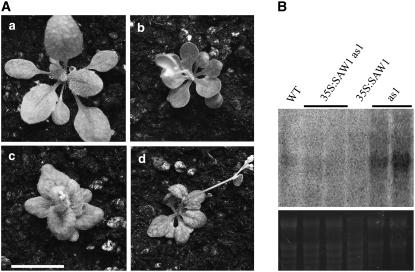
35S:SAW1 Can Suppress the Leaf Phenotype of as1 Mutants
As described earlier, the as1 mutant shows lobed leaves that are associated with ectopic expression of Class 1 KNOX genes. Since the SAW1 genes also suppress KNOX function in the leaf, we tested their ability to suppress the as1 mutant phenotype when expressed ectopically. The leaf phenotype of 35S:SAW1 as1 plants more closely resembled that of 35S:SAW1 alone, suggesting that SAW1 function can partially complement the as1 mutant phenotype (Figure 7A). To determine whether 35S:SAW1 has a direct effect on BP expression in as1 leaves, RNA gel blot analysis was used to compare BP expression in leaves of as1 and 35S:SAW1 as1 plants. 35S:SAW1 was sufficient to significantly repress BP expression in the leaves of as1 mutants (Figure 7B). Since the level of expression of SAW1 remained unchanged in an as1 mutant (see Supplemental Figure 5 online), it is quite possible that in wild-type leaves, AS1 and SAW1 repress BP expression in leaves by independent mechanisms, possibly in temporally and/or spatially distinct subdomains of the leaf.
Figure 7.
35S:SAW1 Suppresses the as1 Leaf Phenotype.
(A) Four-week-old plants. (a) The wild type, (b) 35S:SAW1, (c) as1, and (d) 35S:SAW1 as1. Bar = 10 mm.
(B) RNA gel blot showing BP expression in leaves. Ten micrograms of total RNA isolated from leaves was loaded and hybridized to a BP-specific probe. The bottom panel shows the ethidium bromide–stained gel (loading control).
DISCUSSION
During plant morphogenesis, shape is achieved primarily by establishing domains of differential growth within the various plant organs. In the meristem and stem, BLH proteins BLR and PNF and Class 1 KNOX TALE homeodomain proteins BP and STM interact to positively regulate growth (Byrne et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004; Kanrar et al., 2006). We have investigated the roles of two previously uncharacterized Arabidopsis BEL1-like TALE homeodomain genes, SAW1 and SAW2. Unlike BLR and PNF, the SAW genes contribute to morphogenesis of the leaf and perhaps other organs as well by reducing growth in specific domains. This negative regulation of growth by SAW is correlated with an inhibition of BP gene expression, suggesting a mechanism for growth suppression through inhibition of KNOX gene expression. Therefore, the SAW proteins play a critical role in plant morphogenesis and one that is in apparent direct opposition to their related homologs BLR and PNF.
SAW Proteins Are Negative Regulators of Growth
Loss- and gain-of-function mutant phenotypes suggest that a major role of SAW1 and SAW2 is to negatively regulate growth. SAW1 and SAW2 are expressed in the adaxial domain of the lateral organs early in development. saw1 saw2 loss-of-function lines have revolute/downward curling margins, suggesting an increase in the adaxial-to-abaxial growth of the leaf. Conversely, 35S:SAW1 leaves are either flat or slightly curved toward the adaxial side, suggesting a decrease in adaxial-to-abaxial growth. In addition, 35S:SAW1 leaves were substantially reduced in overall size with a phenotype similar to plants overexpressing ROTUNDIFOLIA4, a gene that regulates polar cell proliferation in leaves (Narita et al., 2004). Taken together, these data suggest that SAW function limits growth on the adaxial side, where it is expressed early in leaf development, perhaps to promote curvature of the leaf over the meristem until leaf blade expansion.
SAW1 and SAW2 are expressed in the hydathode regions of mature Arabidopsis leaves. Mild serrations occur naturally in wild-type Arabidopsis plants and typically occur at hydathodes, although a visible serration is not found at every hydathode (Tsukaya and Uchimiya, 1997; Candela et al., 1999). saw double mutants have increased numbers and sizes of serrations corresponding to the positions of the hydathodes in leaf margins of wild-type leaves, while 35S:SAW1 leaves have no obvious leaf serrations. These data suggest that one role of the SAW proteins is to limit growth at the hydathodes, thus limiting serration in the Arabidopsis leaf margin. That the serration could be due to increased growth at the hydathode is supported by the ectopic expression of the Class 1 KNOX gene BP at hydathodes in saw1 saw2 double mutants (see also below).
In addition to the specific effects of SAW function on leaves, constitutive expression of SAW1 appears to negatively regulate growth throughout the plant. The length and circumference of the stems and sizes of all lateral organs are reduced in 35S:SAW1 plants. 35S:SAW1 floral organ abnormalities involving a decrease in organ number and organ fusions suggest defects in maintaining appropriate growth within the meristem. It is noteworthy that a floral phenotype analogous to that of 35S:SAW1 observed in the weak stm-2 mutant has been ascribed to the reduced size of the floral meristem in this mutant (Clark et al., 1996). Given that SAW proteins negatively regulate BP expression, SAW1, when ectopically expressed in the floral meristem, may also interfere with STM function, leading to stm-2 like defects in floral patterning. Quantitative real-time PCR analysis of the expression of KNOX genes in the vegetative shoot apex did not reveal any significant reduction of expression in 35:SAW1 in comparison to the wild type (see Supplemental Figure 8 online). It is possible that the reduction of KNOX gene expression is localized, and this might be the reason why the disruption of meristem activity is not evident in all of the lateral organs.
Finally, SAW1 and/or SAW2 are expressed in a number of organs for which no obvious phenotype exists in the saw1 saw2 double mutant. These organs include petals, cotyledons, stems, styles, and roots. Indeed, the observed leaf phenotypes themselves are more subtle than the leaf expression pattern suggested. It is possible that redundancy with other BLH genes obscures SAW function in these organs. Alternatively, it is possible that both saw1 alleles obtained for this study are hypomorphic, and a stronger allele could have additional phenotypes. Thus, determination of the roles of the SAW genes in these organs awaits further genetic analysis.
SAW Is a Negative Regulator of BP
Several lines of evidence indicate that SAW proteins act at least in part by negatively regulating BP. First, BP is misexpressed in saw leaves at the regions marked by hydathodes (Figure 4B). Second, 35S:SAW1 is able to reverse the BP-associated leaf lobing observed in as1 mutants and decrease ectopic BP expression in as1 leaves (Figure 7). Third, SAW1 and SAW2 are expressed in dorsiventrally flattened organs, such as leaf and sepal, while BP and STM are excluded from these organs (Figures 2 and 3A). Arabidopsis BP protein has been associated with the promotion of growth. Loss of BP function in bp mutant plants results in a shortened inflorescence shoot and pedicels (Douglas et al., 2002; Venglat et al., 2002), indicating that BP is involved in proper elongation of stems. Arabidopsis plants transformed with 35S:BP exhibit leaf phenotypes of increased serrations and/or lobing as well as the establishment of ectopic meristems in the leaf margins (Lincoln et al., 1994; Chuck et al., 1996). Indeed, some of the weaker 35S:BP lines resemble saw1 saw2 mutants, suggesting that the ectopic expression of BP in the saw mutants may be sufficient to cause serration. However, the domain of BP expression in saw1 saw2 suggests a mechanism of serration growth under the region of expression in the leaf margin rather than a repression of growth in the sinuses, as observed in the 35S:BP plants (Lincoln et al., 1994; Chuck et al., 1996; Hay et al., 2003). Interestingly, a positive correlation between serration length and the size of the BP expression domain in the saw1 saw2 leaves is observed (Figure 4B, d). A more detailed analysis would be required to clarify this point. Nonetheless, it is possible that the SAW proteins regulate growth by suppressing KNOX expression in specific domains of lateral organs. In the absence of SAW function, localized BP expression might promote growth in regions below the site of expression, resulting in serrated, revolute leaves.
Interestingly, an STM-like protein in Cardamine hirsuta promotes compound leaf formation in wild-type plants by prolonging the duration of cell division in specific regions of the leaf primordia (Hay and Tsiantis, 2006). Since STM is closely related to BP, it is possible that BP-mediated cell divisions are the cause of leaf phenotypes in saw mutants. Alternatively, BP expression might be establishing a meristematic region where STM and other Class 1 KNOX genes are expressed. Since our RT-PCR results indicate STM misexpression in the leaves of saw mutants (see Supplemental Figure 7 online), either of these mechanisms may exist in the saw mutants.
The microRNA miR164A has been shown to regulate the extent of serration by regulating the levels of CUP-SHAPED COTYLEDON (CUC2) in the leaf sinuses (Nikovics et al., 2006). Absence of miR164A-mediated reduction of CUC2 transcripts results in increased leaf serrations, suggesting that CUC2 is possibly repressing growth in the sinuses, resulting in increased depths of serrations (Nikovics et al., 2006). It will be interesting to see if the CUC2 levels are changed in the saw1 saw2 mutants.
As observed for the saw1 saw2 double mutant, the BP gene is misexpressed in the leaves of the as1 mutant (Ori et al., 2000; Semiarti et al., 2001). AS1 is a MYB domain–containing transcriptional factor required for normal leaf development. as1 mutants have leaves with irregular lobed blades (Tsukaya and Uchimiya, 1997). This phenotype has been correlated with the misexpression of multiple Class 1 KNOX genes in the leaves (Byrne et al., 2000; Ori et al., 2000). Therefore, like SAW1 and SAW2, AS1 appears to establish a boundary between the meristem and the leaves in part by repressing expression of KNOX genes in the developing leaves. These data suggest that the functions of SAW and AS1 are related. Ectopic/overexpression of 35S:SAW1 is able to partially suppress the as1 mutant phenotype, and we found no evidence that one regulates the expression of the other, suggesting that SAW and AS1 function independently to repress expression of KNOX genes. However, the distinct phenotypes of the as1 single and saw1 saw2 double mutant and the additive effects observed in the saw1 saw2 as1 triple mutants suggest that AS1 and SAW functions are not identical. SAW and AS1 may be functioning in temporally/spatially separated domains in the leaf and/or might have distinct target gene sets.
AS1 works in the same regulatory pathway as AS2, a Leu zipper–containing protein (Byrne et al., 2002; Iwakawa et al., 2002; Xu et al., 2003). as2 mutants have phenotypes similar to those of as1 and also suppress KNOX gene expression in the leaf (Serrano-Cartagena et al., 1999; Semiarti et al., 2001). Therefore, it is likely that saw1 saw2 as2 interactions would be similar to those observed in saw1 saw2 as1 triple mutants.
SAW and KNOX Interactions
BLH and KNOX proteins are DNA binding transcription factors. Protein–protein interactions amongst members of the BLH and KNOX families have been well documented (Bellaoui et al., 2001; Muller et al., 2001; Smith et al., 2002; Smith and Hake, 2003; Hackbusch et al., 2005). Several lines of evidence suggest that heterodimerization amongst members of these families is required for DNA binding and nuclear localization (Bhatt et al., 2004; Cole et al., 2006). As described earlier, BP and the BLH protein BLR/PNY/RPL/VAN interact to positively regulate inflorescence growth (Smith and Hake, 2003; Bhatt et al., 2004). Similarly, BLR/PNY/RPL/VAN and the paralogous protein PNF positively interact with STM to regulate meristem function (Byrne et al., 2003; Kanrar et al., 2006). For the SAW proteins, it has been shown that both SAW1 and SAW2 interact with BP in yeast two-hybrid assays (Figure 1B; Hackbusch et al., 2005). However, genetic and molecular analyses have revealed that the SAW proteins negatively regulate BP expression and function (Figures 4B and 7). Furthermore, SAW and BP are mostly expressed in mutually exclusive domains (Figure 4A). These findings raise the question of what functions SAW–BP interactions could have. Based on our data, we hypothesize that SAW and BP interactions are required for negative regulation of BP function. Mechanistically, such negative regulation could occur if the SAW-BP heterodimer acts as a repressor of BP expression. Alternatively, the regulation might be a result of BP being sequestered by SAW proteins and hence unable to interact with BLR/PNY/VAN and PNF proteins. In the latter case, it must be assumed that BLR-BP complexes are activators of BP expression. Thus, the titers of competing BLH proteins in a given cell and/or tissue might decide the fate of BP-derived functions in the tissue. KNOX proteins have been shown to move across cell layers in the meristem (Kim et al., 2003, 2005). Long-range transport of KNOX transcripts via the phloem has also been reported (Kim et al., 2001, 2005). SAW proteins might repress the function of KNOX proteins trafficked into an inappropriate domain.
The fact that BP:GUS expression in stems of saw double mutants is more than double that found in wild-type stems suggests that SAW1 negatively regulates BP in some domain of the stem as well. However, unlike in leaves, BP, SAW1, and SAW2 are expressed in overlapping domains in the stem cortex (Venglat et al., 2002; R. Kumar and G.W. Haughn unpublished results). Further studies are required to clarify the role of any of the SAW proteins in the stem and its relationship with BP function.
Conservation of SAW Function in Dicotyledonous Plants
In this report, we have shown that SAW1 overexpression results in reduced organ size and flattened leaves. MDH1 (for Malus domestica Homeodomain protein 1) is a SAW1 ortholog from apple that shows higher amino acid sequence similarity to SAW1/SAW2 than to any other Arabidopsis BLH protein. Overexpression of MDH1 in Arabidopsis resulted in phenotypes (Dong et al., 2000) strikingly similar to the ones observed in the 35S:SAW1 plants. This suggests that SAW1 function is conserved in dicotyledonous plants and may represent a common regulatory mechanism of differential organ growth in dicots.
Redundancy between BEL1 and SAW Genes
The SAW proteins are most similar (in amino acid sequence) to the BEL1 protein, have overlapping expression patterns, and interact with a similar set of KNOX proteins. Moreover, the bel1 mutant phenotype can be complemented by 35S:SAW1 (Figure 1C). However, saw1 saw2 bel1 triple mutants have an additive phenotype. It is possible that there could be other BLH proteins that mask any functions common to SAW1, SAW2, and BEL1. Alternatively, it is also possible that BEL1 has diverged sufficiently from SAW1 and SAW2 to retain its ovule function but lost those needed for other tissues. In support of the latter hypothesis, we have found that unlike 35S:SAW1 plants, 35S:BEL1 does not have any observable changes outside the ovule (S.R. Hepworth and G.W. Haughn, unpublished results). A more complete understanding of BEL1 function outside the ovule must therefore await further analysis.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotypes Columbia-0 (Col-0), Col-2, Col with erecta mutation (Col er105), and Landsberg erecta were used as wild-type controls in phenotypic screens and other experiments for the mutants and transgenics, in accordance with their genetic backgrounds. BP:GUS was a gift from Sarah Hake (Ori et al., 2000). All Salk insertion lines, TILLING lines, and the as1-1 mutant were ordered from the ABRC (stock numbers: as1, CS3374; saw1, SALK_009120; saw1-2, CS87673; saw2-1, SALK_121117; and saw2-2, SALK_149402). The genotypes of the saw1 and saw2 mutants and any double mutants generated were confirmed by PCR-based genotyping. Plants were grown as previously described (Bellaoui et al., 2001; Hepworth et al., 2005) and transformed using the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998).
Sequence Analysis
The amino acid sequences for the BLH proteins were aligned using the ClustalX program (Thompson et al., 1994, 1997) with default alignment parameters except for the protein matrix, for which BLOSUM62 (Henikoff and Henikoff, 1992) was used. To construct the phylogram based on the maximum likelihood method (Felsenstein, 1981), the aligned sequences were imported into Bioedit for manual editing (Hall, 1999; http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Three highly conserved regions were identified (see Supplemental Figure 1A online), and these regions were selected for further analysis. The alignment of the conserved sequences was bootstrapped 1000 times using SEQBOOT (from PHYLIP version 3.6, distributed by J. Felsenstein, University of Washington, Seattle). The program Phyml (Guindon and Gascuel, 2003; Guindon et al., 2005) was then used to generate maximum likelihood–based trees for both bootstrapped and nonbootstrapped data sets. The trees were visualized using Treeview (R. Page; http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) and processed with IrfanView (I. Skiljan; http://www.irfanview.com/) and Photoshop (Adobe Systems), where the bootstrap values were converted to 100 and added to the tree branches.
RT-PCR and Quantitative Real-Time PCR
cDNA was synthesized by RT using Superscript II RT (Invitrogen) and 1 μg of total RNA isolated from required organs of the mutants or the wild type. PCR was performed using 1 μL of cDNA as template and Taq DNA polymerase (Invitrogen) according to the manufacturer's instructions. Primers were as follows: BEL1, p1 5′-CACAAGTCACCACAGCAACA-3′/p2 5′-TGCTTGAATCTGTCCCACAA-3′; SAW1, p1 5′-CAGCGGGAATCTCTTCTTCC-3′/p2 5′-TGGATTTTGTGCTCTTGTCG-3′; SAW2, p1 5′-CCATTGGAGGGATCTACACG-3′/p2 5′-ATCCCCTAGAAGCTCACAGC-3′; BP, p1 5′-CGATGTTGAAGCCATGAAGG-3′/p2 5′-GCTGTTGTCGAGCCTCAAAG-3′; KNAT2, p1 5′-CGCGTATTCGAAAAGCTGAG-3′/p2 5′-CATGGTTCTCTCGCTGAATCTC-3′; KNAT6, p1 5′-AAATCGCTTGTCATCCTTCG-3′/p2 5′-TCACTCTCCCGTTGAATCTCC-3′; STM, p1 5′-CAACCCTTGCTCCTCTTCC-3′/p2 5′-CCTGTTGGTCCCATAGATGC-3′. Either GAPC (Western et al., 2004) or ACTIN8 (An et al., 1996) was used as a loading control. These genes were amplified using the following primer pairs: GAPC p1 5′-TGGGGAGACATTCTTGCTG-3′/p2 5′-GATGGGCTTGTGTGTGTTTG-3′; ACT8 p1 5′-TGTGACAATGGTACTGGAATGG-3′/p2 5′-TTGGATTGTGCTTCATCACC-3′. All PCR reactions were repeated at least three times, and the PCR cycle numbers were adjusted to ensure that the amplification was in the logarithmic phase. The PCR products were resolved in a 1% agarose gel and stained with ethidium bromide. Quantitative real-time PCR was performed in the MJ MinipOticon real-time PCR system (Bio-Rad Laboratories) using iQ SYBR Green Supermix (Bio-Rad Laboratories). Each real-time PCR experiment was done in triplicate, and the graph was generated from the pooled data of two independent experiments.
GUS Analysis
Generation of Promoter:GUS Constructs
For all constructs made during this project, PCR fragments were amplified using Expand High-Fidelity polymerase (Roche; according to the manufacturer's protocol) from Col-2 genomic DNA isolated as described (Dellaporta et al., 1983). For the promoter-GUS constructs, the promoter regions of BEL1, SAW1, and SAW2, including the 5′ untranslated region, were amplified using the following gene-specific primers: BEL1, p1 5′-GAATTCCAATCTCTTTCACGTACTGTGCG-3′/p2 5′-GGATCCTGTCTCTCAAGAATTGAAAACCC-3′; SAW1, p1 5′-TTAGTCGACAAAGATTCCCACATGGTGTC-3′/p2 5′-TATCTCGAGGTTATTCCCATATCAATACTTCAATC-3′; SAW2, p1 5′-ATTATGAATTCTTGCAACCACCATTGAAGAG-3′/p2 5′-ATTATCTCGAGCAAAGCTCTTGGATCCTGTAAG-3′. The BEL1 promoter was cloned into pCR 2.1 blunt (Invitrogen) and then introduced into pBAR1 (a gift from Ben Holt and Doug Boyes, Dangl Lab, University of North Carolina) containing GUS that we subcloned from pBI101 (Clontech Laboratories). SAW1 and SAW2 promoters were cloned into pENTR1A (Invitrogen) and then subcloned into pGWB3 (a gift from Tsuyoshi Nakagawa, Research Institute of Molecular Genetics, Shimane University, Japan) by the Gateway LR recombination reaction (Invitrogen) according to the manufacturer's directions.
GUS Histochemical Assay
GUS histochemical assays were done on freshly isolated plant organs or whole seedlings using a protocol that was adapted for staining Arabidopsis (Sieburth and Meyerowitz, 1997). For whole mounts, stained tissues were destained by incubation in 70% ethanol overnight at room temperature. The destained samples were either directly viewed under a light microscope (Zeiss Axioskop microscope) or incubated in clearing solution (chloral hydrate:glycerol:water, 9:1:3 [w:v:v]; Mattsson et al., 2003) for 1 h to 4 d depending on the organ being cleared (ovules start clearing within an hour and leaves take 2 to 4 d) and photographed under the compound microscope or dissecting photomicroscope.
For resin embedding and sectioning, the stained samples were fixed with 3% glutaraldehyde (Canemco) in 0.5 M sodium phosphate buffer at 4°C overnight and then dehydrated through an ethanol series and embedded in Technovit 8100 resin based on the manufacturer's instructions (Heraeus Kulzer; Electron Microscopy Sciences, distributor). Embedded tissue was sectioned using a glass microtome. Sections were spread on glass slides and mounted with Entellan (Merck). The samples were then visualized under a light microscope and photographed using a SPOT digital camera (Diagnostic Instruments). The photographs were processed using Adobe Photoshop (Adobe Systems).
GUS Fluorometric Assay
GUS activity was quantified in crude protein extracts from stem internodes (the first two internodes from the base) and inflorescence apices (including the region from inflorescence meristem to the first open flower) from 5-week-old wild-type (Col-0), BP:GUS Col-0, and BP:GUS saw1 saw2 plants using the 4-methylumbelliferyl β-d-glucuronide fluorometric assay (Jefferson et al., 1987). Protein concentrations were quantified against a BSA standard curve using the Bio-Rad protein microassay according to the manufacturer's instructions. The concentration of the hydrolysis product, 4-methylumbelliferone (MU), in each aliquot was measured using a Finstruments Fluoroskan plate reader (Thermo Electron/Labsystems; excitation at 365 ± 7 nm; emission at 460 ± 15 nm) at 0 h, 30 min, and 1 h and reported as absolute fluorescence units. The amount of MU produced by each sample was determined in comparison with an MU standard curve.
Complementation of saw1 saw2 with a SAW2 Genomic Fragment
Expand high-fidelity polymerase (Roche) was used to amplify a wild-type genomic fragment containing the SAW2 coding sequence (including introns), as well as 3000 kb of the 5′ and 1000 kb of the 3′ sequence, using the primers 5′-TTCGCGGCCGCTGCTATTTCAAGGACGTGAGC-3′ and 5′-CTAGCGGCCGCATTGTGACTTATTGGCGCTTTCC-3′. Genomic DNA isolated by the method of Dellaporta et al. (1983) from leaves of wild-type plants was used as a template for PCR. The PCR-generated fragment was cloned directly into the NotI site of the pART27 binary vector (Gleave, 1992). This construct was used to transform it into saw1 saw2 double mutant plants. We then evaluated selected transformants for complementation of the mutant phenotypes.
Yeast Two-Hybrid Assays
A GAL4-based yeast two-hybrid system was used as described previously (Kohalmi et al., 1988). Plasmid constructs were generated by cloning full-length cDNAs into either the GAL4 DNA binding domain (DB) plasmid pBI-770 as the bait or the GAL4 transcriptional activation domain (TA) plasmid pBI-771 as the prey. Full-length SAW1 and BLH1 cDNAs were amplified from plasmids (obtained by screening flower-specific cDNA libraries) using the Expand high-fidelity PCR system (Invitrogen). The PCR product was digested with SalI and NotI and cloned into pBI-771 to create TA-SAW1 and TA-BLH1. Construction of TA-BEL1, all the DB-KNOX, and DB-Cruciferin has been described earlier (Bellaoui et al., 2001). The bait and prey constructs were transformed into the yeast strain YPB2 (Fields and Song, 1989), and interacting transformants were selected on medium lacking Leu, Trp, and His. The interactions were further visualized using X-Gal filter assays that were performed as described previously (Kohalmi et al., 1988).
In Situ Hybridization Analysis
Tissue fixation, sectioning, hybridization, and signal detection were performed as described previously (Samach et al., 1997). Full-length SAW1 cDNA was amplified from the TA-SAW1 construct (described earlier) and was used for probe preparation. Sections were viewed through a light microscope (Zeiss Axioskop II) and photographed under bright-field conditions using a SPOT digital camera (Diagnostic Instruments).
RNA Gel Blot Analysis
Approximately 10 μg of RNA isolated from leaves of as1, 35S:SAW1, and 35S:SAW1 as1 plants was resolved in a denaturing agarose gel and blotted onto a nylon membrane as previously described (Western et al., 2004). The membrane was then hybridized with a 32P-labeled BP probe and detected as previously described (Western et al., 2004), and the specific signal was detected with a Storm 860 PhosphorImager (Molecular Dynamics).
The 35S:SAW1 Construct
Full-length SAW1 cDNA was amplified by PCR from wild-type cDNA using the primers 5′-GCCTCTAGATGGAATAACTAAAACTTC-3′ and 5′-GCTCTAGACTAAAACCCCCAAACTC-3′. The resulting fragment was cut with XbaI and cloned into pGPTV pBAR (Becker et al., 1992) with a tandem 35S promoter with an AMV leader sequence (M. Bellaoui and G.W. Haughn, unpublished results) and transformed into Col-2 plants as described earlier.
Characterization of Mutant Phenotypes
All macroscopic images were either taken using a Nikon CoolPix camera or were scanned using an HP Scanjet 7400c (Hewlett Packard). Micrographs were taken using a dissection or compound microscope equipped with a Spot digital camera and Northern Eclipse software. Adobe Photoshop was used for image processing and the creation of montages.
To measure lengths of leaf serrations, leaves were scanned using a Canoscan 8400DF scanner (Canon). Scanned images were converted to gray scale, and threshold was adjusted to include only the leaves. Measurements were made using ImageJ software (National Institutes of Health; http://rsb.info.nih.gov/ij/) calibrated with a scanned ruler.
To evaluate cells of the 35S:SAW1 plants, epidermis was peeled from the bottom three internodes of stems of 5-week-old plants and mounted in 50% glycerol on glass slides. Cell counts and measurements were made using phase contrast optics of a compound light microscope.
Accession Numbers
The GenBank accession numbers and Arabidopsis Genome Initiative (AGI) locus identifiers of the SAW genes are as follows: BLH2/SAW1, AF173816 (GenBank), At4g36870 (AGI locus); BLH4/SAW2, AF353092 (GenBank), At2g23760 (AGI locus).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Conservation and Duplication in the BLH Proteins.
Supplemental Figure 2. RT-PCR Analysis of BEL1, SAW1, and SAW2.
Supplemental Figure 3. Complementation of saw1 saw2 Double Mutants by a SAW2 Genomic DNA Fragment.
Supplemental Figure 4. Increased BP:GUS Activity in Stems of saw1 saw2 Double Mutants.
Supplemental Figure 5. RT-PCR Analysis of SAW1.
Supplemental Figure 6. The saw1 saw2 bp Triple Mutant Phenotype.
Supplemental Figure 7. RT-PCR Analysis of Class 1 KNOX Gene Expression.
Supplemental Figure 8. Real-Time PCR Analysis of Class1 KNOX Genes.
Supplemental Figure 9. RNA Gel Blot Comparing SAW1 Expression in Wild-Type and 35S:SAW1 Plants.
Supplementary Material
Acknowledgments
We thank the Seattle TILLING Project for screening for saw1 mutants, the Salk Institute Genome Analysis Laboratory for T-DNA insertional lines, the ABRC at Ohio State University for providing us with the mutant seeds, Sarah Hake for the BP:GUS (KNAT1:GUS) line, Jeff Dangl for the pBAR1 plasmid, and Tsuyoshi Nakagawa for pGWB3 plasmid. We also thank Dmitry Belostotsky for providing necessary resources for the saw1 saw2 bp triple mutant screens, Tanya Hooker for critical reading of this manuscript, Juan Saldarriaga for comments on phylogenetic analysis, and Jin-Gui Chen for allowing us to use his MiniOpticon real-time PCR system. This work was supported by a Natural Sciences and Engineering Research Council (NSERC) Discovery grant to G.W.H., by NSERC postgraduate fellowships to M.S.P., and by a University of British Columbia Graduate Fellowship to R.K. and M.S.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: George W. Haughn (haughn@interchange.ubc.ca).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- An, Y.Q., McDowell, J.M., Huang, S.R., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10 107–121. [DOI] [PubMed] [Google Scholar]
- Bao, X., Franks, R.G., Levin, J.Z., and Liu, Z. (2004). Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Becker, A., Bey, M., Burglin, T.R., Saedler, H., and Theissen, G. (2002). Ancestry and diversity of BEL1-like homeobox genes revealed by gymnosperm (Gnetum gnemon) homologs. Dev. Genes Evol. 212 452–457. [DOI] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Etchells, J.P., Canales, C., Lagodienko, A., and Dickinson, H. (2004). VAAMANA–a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328 103–111. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Groover, A.T., Fontana, J.R., and Martienssen, R.A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130 3941–3950. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Candela, H., Martinez-Laborda, A., and Micol, J.L. (1999). Venation pattern formation in Arabidopsis thaliana vegetative leaves. Dev. Biol. 205 205–216. [DOI] [PubMed] [Google Scholar]
- Chen, H., Rosin, F.M., Prat, S., and Hannapel, D.J. (2003). Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 132 1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122 1567–1575. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colbert, T., Till, B.J., Tompa, R., Reynolds, S., Steine, M.N., Yeung, A.T., McCallum, C.M., Comai, L., and Henikoff, S. (2001). High-throughput screening for induced point mutations. Plant Physiol. 126 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, M., Nolte, C., and Werr, W. (2006). Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 34 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1 19–21. [Google Scholar]
- Dong, Y.H., Yao, J.L., Atkinson, R.G., Putterill, J.J., Morris, B.A., and Gardner, R.C. (2000). MDH1: An apple homeobox gene belonging to the BEL1 family. Plant Mol. Biol. 42 623–633. [DOI] [PubMed] [Google Scholar]
- Douglas, S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1981). Evolutionary trees from DNA sequences - A maximum-likelihood approach. J. Mol. Evol. 17 368–376. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O.K. (1989). A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Guindon, S., Lethiec, F., Duroux, P., and Gascuel, O. (2005). PHYML online - A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33 W557–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch, J., Richter, K., Muller, J., Salamini, F., and Uhrig, J.F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 102 4908–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hay, A., Jackson, D., Ori, N., and Hake, S. (2003). Analysis of the competence to respond to KNOTTED1 activity in Arabidopsis leaves using a steroid induction system. Plant Physiol. 131 1671–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., and Tsiantis, M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38 942–947. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Henikoff, J.G. (1992). Amino-acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth, S.R., Zhang, Y.L., Mckim, S., Li, X., and Haughn, G. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar, S., Onguka, O., and Smith, H.M. (2006). Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224 1163–1173. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., Rim, Y., Wang, L., and Jackson, D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19 788–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y., Yuan, Z., and Jackson, D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130 4351–4362. [DOI] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289. [DOI] [PubMed] [Google Scholar]
- Kohalmi, S.E., Reader, L.J.W., Samach, A., Nowak, J., Haughn, G.W., and Crosby, W.L. (1988). Identification and characterization of protein interactions using the yeast 2-hybrid system. In Plant Molecular Biology Manual M1, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–30.
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclellan, T., and Dengler, N. (1995). Pattern and form in repeated elements in the development of simple leaves of Begonia dregei. Int. J. Plant Sci. 156 581–589. [Google Scholar]
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fischer, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27 13–23. [DOI] [PubMed] [Google Scholar]
- Narita, N.N., Moore, S., Horiguchi, G., Kubo, M., Demura, T., Fukuda, H., Goodrich, J., and Tsukaya, H. (2004). Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 38 699–713. [DOI] [PubMed] [Google Scholar]
- Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., and Laufs, P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Pautot, V., Dockx, J., Hamant, O., Kronenberger, J., Grandjean, O., Jublot, D., and Traas, J. (2001). KNAT2: Evidence for a link between knotted-like genes and carpel development. Plant Cell 13 1719–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg, N., Dockx, J., Rook, F., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell 7 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83 735–742. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers, K., Pruitt, R.E., and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, A.H., Ferrandiz, C., and Yanofsky, M.F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13 1630–1635. [DOI] [PubMed] [Google Scholar]
- Samach, A., Kohalmi, S.E., Motte, P., Datla, R., and Haughn, G.W. (1997). Divergence of function and regulation of class B floral organ identity genes. Plant Cell 9 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena, J., Robles, P., Ponce, M.R., and Micol, J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261 725–739. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M., Campbell, B.C., and Hake, S. (2004). Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 14 812–817. [DOI] [PubMed] [Google Scholar]
- Smith, H.M., and Hake, S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B.J., Burtner, C., Comai, L., and Henikoff, S. (2004). Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 32 2632–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H., and Uchimiya, H. (1997). Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: Combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol. Gen. Genet. 256 231–238. [DOI] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Young, D.S., Dean, G.H., Tan, W.L., Samuels, A.L., and Haughn, G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.