Abstract
MicroRNAs and small interfering RNAs (siRNAs) are two classes of small regulatory RNAs derived from different types of precursors and processed by distinct Dicer or Dicer-like (DCL) proteins. During evolution, four Arabidopsis thaliana DCLs and six rice (Oryza sativa) DCLs (Os DCLs) appear to have acquired specialized functions. The Arabidopsis DCLs are well characterized, but those in rice remain largely unstudied. Here, we show that both knockdown and loss of function of rice DCL4, the homolog of Arabidopsis DCL4, lead to vegetative growth abnormalities and severe developmental defects in spikelet identity. These phenotypic alterations appear to be distinct from those observed in Arabidopsis dcl4 mutants, which exhibit accelerated vegetative phase change. The difference in phenotype between rice and Arabidopsis dcl4 mutants suggests that siRNA processing by DCL4 has a broader role in rice development than in Arabidopsis. Biochemical and genetic analyses indicate that Os DCL4 is the major Dicer responsible for the 21-nucleotide siRNAs associated with inverted repeat transgenes and for trans-acting siRNA (ta-siRNA) from the endogenous TRANS-ACTING siRNA3 (TAS3) gene. We show that the biogenesis mechanism of TAS3 ta-siRNA is conserved but that putative direct targets of Os DCL4 appear to be differentially regulated between monocots and dicots. Our results reveal a critical role of Os DCL4-mediated ta-siRNA biogenesis in rice development.
INTRODUCTION
MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are two types of noncoding RNAs that play fundamental roles in developmental regulation, epigenetic modifications, and viral defense (Kidner and Martienssen, 2005; Vaucheret, 2006). miRNAs and siRNAs are very similar in their structural and functional features. Both are 21 to 24 nucleotides in length, with a 5′ phosphate and a 3′ OH, and both serve as sequence-specific regulators to guide target gene repression (Bartel, 2004). However, they are produced from very different types of precursors (Bartel, 2004; Jones-Rhoades et al., 2006). miRNAs originate from single RNA molecules that form imperfect local hairpin-like secondary structures (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001; Reinhart et al., 2002). siRNAs are produced from either endogenous or exogenous long double-stranded RNAs (dsRNAs) with perfect complementarity (Zamore et al., 2000; Elbashir et al., 2001).
Key components in the miRNA and siRNA biogenesis pathways are the Dicer or Dicer-like (DCL) proteins. Arabidopsis thaliana encodes four DCL proteins, and the rice (Oryza sativa) genome has six putative DCL proteins. Biochemical analysis has shown that Dicer activities producing 21- to 24-nucleotide siRNAs were present in a wheat germ system (Tang et al., 2003). Further biochemical studies also revealed that DCL1 and DCL3 produce 21- and 24-nucleotide siRNAs, respectively, in Arabidopsis silencing pathways (Qi et al., 2005). Although molecular and genetic analysis demonstrated that At DCL4 was predominantly involved in the biogenesis of 21-nucleotide trans-acting siRNAs (ta-siRNAs), virus-induced siRNAs, and transgenic siRNAs, direct biochemical evidence is lacking (Vazquez et al., 2004; Allen et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). Genetic analysis in Arabidopsis has revealed both specialized and overlapping functions of Dicer proteins (Fahlgren et al., 2006; Henderson et al., 2006).
DCL1 is required for the maturation of miRNAs, and dcl1 loss-of-function mutants show pleiotropic developmental defects (Park et al., 2002; Reinhart et al., 2002; Schauer et al., 2002; Kurihara and Watanabe, 2004). All four DCLs appear to be involved in siRNA biogenesis either with specific, redundant or partially compensatory and antagonistic roles reflecting the complexity of siRNA production (Xie et al., 2004, 2005; Gasciolli et al., 2005; Yoshikawa et al., 2005; Bouche et al., 2006; Henderson et al., 2006; Margis et al., 2006).
DCL2 is involved in the processing of 22-nucleotide siRNAs derived from Turnip crinkle virus (Xie et al., 2004) and 24-nucleotide siRNAs derived from natural antisense transcript pairs under salt stress (Borsani et al., 2005). DCL3 is involved in the biogenesis of 24-nucleotide siRNAs that are derived from endogenous repeated sequences associated with heterochromatin or silenced transgenes (Xie et al., 2004). Neither dcl2 nor dcl3 shows developmental defects in Arabidopsis.
DCL4 is responsible for the processing of 21-nucleotide ta-siRNAs that are required for normal plant development (Vazquez et al., 2004; Allen et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). ta-siRNAs are endogenous and a plant-specific class of siRNAs. The biogenesis of ta-siRNAs requires both DCL1 and DCL4 activities in Arabidopsis (Vazquez et al., 2004; Yoshikawa et al., 2005). Transcripts from TRANS-ACTING siRNA (TAS) genes are cleaved by DCL1-processed miRNAs, and the cleavage products are stabilized by SUPPRESSOR OF GENE SILENCING3 (SGS3) and then amplified by RNA-DEPENDENT RNA POLYMERASE6 (RDR6) to generate dsRNAs that are further processed by DCL4 into ta-siRNAs (Allen et al., 2005; Yoshikawa et al., 2005; Axtell et al., 2006).
Mutations in DCL4 result in slightly elongated, downward-curled rosette leaves and accelerated juvenile-to-adult vegetative phase change in Arabidopsis, suggesting a specific role of the DCL4-mediated ta-siRNA pathway (Gasciolli et al., 2005; Xie et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006). Several TAS gene families in Arabidopsis have been identified (Vazquez et al., 2004; Rajagopalan et al., 2006; Howell et al., 2007). However, the juvenile-to-adult phase transition is only suppressed by At TAS3 ta-siRNAs through negative regulation of Auxin Response Factor3 (ETT/ARF3) and ARF4 in Arabidopsis (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). Moreover, the weak dcl4 phenotype can be enhanced by overexpression of nontargeted ARF3 (ARF3mut) in the rdr6 mutant, resulting in further acceleration of phase change and severe developmental defects of leaves and floral organs (Fahlgren et al., 2006).
By contrast with the well-characterized DCL proteins in Arabidopsis, little is known about their action in any other plants. Previously, we demonstrated that knockdown of Os DCL1, like Arabidopsis dcl1, caused pleiotropic phenotypes in rice due to a failure of miRNA metabolism (Schauer et al., 2002; Liu et al., 2005). However, phylogenetic analysis suggested that the functional diversification of DCLs occurred before the divergence of monocots and dicots ∼200 million years ago (Henderson et al., 2006; Margis et al., 2006). Therefore, it is possible that during evolution, rice and Arabidopsis DCLs might have acquired distinct functions in small RNA biogenesis and/or plant development for each species.
Here, we analyze the role of DCL4 in rice and compare it with that of DCL4 in Arabidopsis. Biochemical and genetic analyses demonstrate that Os DCL4 generates 21-nucleotide siRNAs in vitro and is responsible for 21-nucleotide siRNA production derived from inverted repeat transgenes and TAS3 ta-siRNA biogenesis, indicating the fundamental similarity of the ta-siRNA pathways between rice and Arabidopsis. By contrast, loss of function of Os DCL4 leads to vegetative developmental defects and severe disruption of spikelet organ identity, resulting in sterility, revealing a much broader role for Os DCL4 in development than for its Arabidopsis counterpart. Furthermore, we identify an endogenous mRNA with a long fold-back structure that could be processed by Os DCL4 into 21-nucleotide siRNAs, supporting the hypothesis that miRNA genes are evolved from inverted duplication of target genes (Allen et al., 2004), thus providing further evidence for an evolutionary link between siRNA and miRNA.
RESULTS
Knockdown of Os DCL4 Results in Abnormal Spikelet Morphology
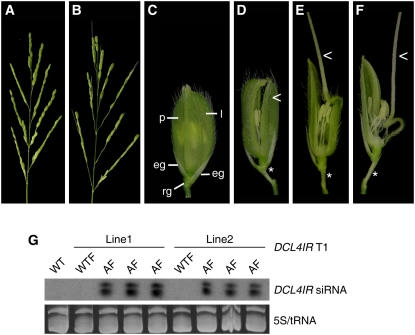
In other organisms, RNA interference (RNAi) has been used successfully to study the components involved in the RNAi pathway (Dudley et al., 2002; Kim et al., 2005). To determine the role of Os DCL4 encoded by Os 04g43050 in small RNA biogenesis and rice development, 15 individual transgenic plants expressing an inverted repeat of Os DCL4 (DCL4IR) were generated (Liu et al., 2005). During reproductive development, the basic branching architecture of the panicle in DCL4IR transformants was normal except for the absence of certain spikelets in the lower part of the rachis (Figures 1A and 1B). A spikelet, the basic morphological unit of the rice inflorescence, contains a pair of empty glumes and a pair of bract-like structures named the lemma and the palea, which function like sepals of other plants (Schmidt and Ambrose, 1998; Ferrario et al., 2004; Prasad et al., 2005) (Figure 1C). Compared with wild-type plants (Figure 1C), spikelet organ identity was greatly disrupted in the DCL4IR transformants (Figures 1D to 1F). Three typical phenotypes from weak to strong were observed, including a slight opening between the lemma and the palea (Figure 1D) and the lemma being partially or completely degenerated to the awn (Figures 1E and 1F). Moreover, the empty glumes and the rudimentary glumes were separated in DCL4IR transformants (Figures 1D to 1F) instead of being close together, as in the wild-type control (Figure 1C). As a result, the strong loss-of-function transgenic plants (Figure 1F) were sterile and only two weak loss-of-function plants could produce seeds for further genetic analysis (Figure 1D). The offspring from the weak loss-of-function plants (Figure 1D) showed the expected 1:3 segregation of normal to abnormal phenotype. When 41 T1 plants from two individual transgenic lines were examined, the abnormal phenotype cosegregated with the DCL4IR transgenes (Figure 1G). Previously, we showed that Os DCL4 mRNA levels were reduced but those of the other DCLs were not in DCL4IR transformants (Liu et al., 2005). Therefore, we conclude that the significant reduction or loss of function of Os DCL4 caused abnormal spikelet morphology in rice and that the phenotypic variations could be due to variable RNAi efficiency within individual spikelets.
Figure 1.
Knockdown of Os DCL4 Causes Abnormal Morphology in Rice.
(A) An inflorescence of wild-type Nipponbare rice.
(B) An inflorescence of DCL4IR transgenic rice.
(C) A spikelet of wild-type rice. The palea (p), lemma (l), a pair of empty glumes (eg), and a pair of rudimentary glumes (rg) are indicated.
(D) to (F) The spikelets of DCL4IR transformants showing weak (D), intermediate (E), and strong (F) phenotypes. Asterisks represent the space between the empty glume and the rudimentary glume; carets indicate the impaired lemma.
(G) Small RNA gel blot analysis of DCL4IR siRNA levels (indicated at right) in wild-type and DCL4IR transgenic progeny (WTF and AF, respectively). Line 1 and line 2 are two independent T1 generation lines of DCL4IR transformants. AF, transgenic plants displaying abnormal flowering; WTF, wild-type phenotypic progeny. 5S/tRNA stained with ethidium bromide was used as a control.
Identification of an Os dcl4 Recessive Mutant
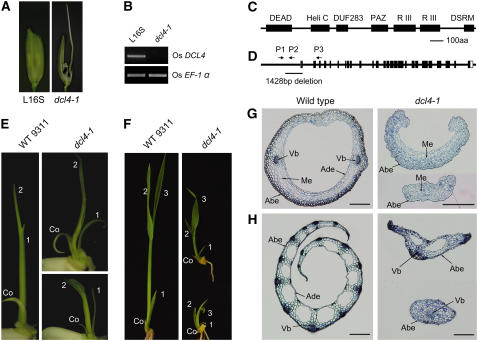
By searching for mutants with similar phenotypes to the strong DCL4IR loss-of-function transformants, we isolated a natural mutant from rice strain L16S of the indica variety background (Figure 2A). In this mutant, full-length DCL4 mRNA was not detectable by RT-PCR (Figure 2B). Sequence analysis showed that DCL4 mRNA is composed of 26 exons and encodes a polypeptide with 1657 amino acids (Figures 2C and 2D). DNA sequencing of Os DCL4 from the natural mutant revealed a 1428-bp deletion corresponding to the promoter, the 5′ untranslated region, and the leading 72 bp in the first exon of the gene (Figure 2D). We thus named the natural mutant dcl4-1.
Figure 2.
Identification of a Loss-of-Function dcl4 Mutant.
(A) The spikelet phenotype of dcl4-1 (right) and its parent L16S (left).
(B) RT-PCR analysis of Os DCL4 expression levels in wild-type L16S and dcl4-1. Os EF-1α was used as an internal control.
(C) The domain structure of Os DCL4 protein. Conserved domains are shown as black boxes. DEAD, DEAD box helicase; Heli C, helicase conserved C-terminal domain; DUF283, domain of unknown function 283; PAZ, the PAZ domain, which is named after the proteins Piwi, Argonaut, and Zwille; R III, ribonuclease III domain; DSRM, dsRNA binding motif. Bar = 100 amino acids (aa).
(D) Schematic representation of the Os DCL4 gene and the location of the dcl4-1mutation. Exons are shown as black boxes and introns are shown as lines. The 3′ untranslated region is shown as an open box. The black bar indicates the 1428-bp deletion in the promoter and the 72-bp deletion in the first exon of Os DCL4 in dcl4-1. P1, forward primer; P2, reverse primer located within the deletion region; P3, reverse primer. Primers are used for genotyping of the dcl4-1mutation.
(E) and (F) Phenotypes of dcl4-1 in the juvenile vegetative stage. Seedlings of the wild type (left panels) and dcl4-1 (right panels) at 3.5 DAG (E) and 8 DAG (F). Co, coleoptile; 1 to 3, first, second, and third leaves.
(G) and (H) Anatomy of wild-type and dcl4-1 coleoptiles (G) and first leaf sheaths (H) at 3.5 DAG. In the right panels of dcl4-1, the top and bottom plants represent the majority and some thread-like leaves of dcl4-1 plants, respectively. Abe, abaxial epidermis; Ade, adaxial epidermis; Me, mesophyll; Vb, vascular bundle. Bars = 200 μm.
To confirm that the abnormal spikelet phenotype in dcl4-1 was caused by loss of function of Os DCL4, we introgressed dcl4-1 into indica variety 9311 with two backcrosses. The F1 plants displayed a normal phenotype, and in the F2 progeny, segregation at a ratio of ∼1:3 (660:2175) between mutant and wild-type plants was observed. The abnormal phenotypes cosegregated with dcl4-1 homozygotes when 660 F2 plants were examined, indicating that the lesion causing abnormal spikelet phenotypes was tightly linked to the Os DCL4 locus.
Os dcl4 Mutants Undergo Abnormal Juvenile and Adult Vegetative Development
Vegetative development in rice consists of three stages: juvenile (one to two leaves), intermediate (three to five leaves), and adult (six leaves to flowering) (Itoh et al., 2005). The transitions between these stages are gradual (Asai et al., 2002). During the juvenile stage, dcl4-1 mutants displayed abnormal coleoptiles and leaf phenotypes compared with wild-type plants (Figure 2E). In the wild type, the first two leaves were rolled up inside the coleoptile or the first leaf sheath to form a cone (Figure 2E, left panel). However, dcl4-1 mutants produced radialized coleoptiles, and the first and second leaves were not enclosed by the coleoptile or the first leaf, respectively (Figure 2E, right panel, top). With lower (5 to 10%) frequency, severe phenotypes were observed, including development of the first leaf into a thread-like structure (Figure 2E, right panel, bottom). By the intermediate stage, nearly normal leaf phenotypes of dcl4-1 plants were observed, except that leaves were shorter than those of the wild type (Figure 2F). In the mature stage, dcl4-1 mutant plants appeared semidwarfed in stature (see Supplemental Figure 1 online).
For further anatomical analyses of the coleoptiles and the first leaf sheaths, we made transverse sections of the wild type and dcl4-1 at 3.5 d after germination (DAG). In the wild type, the coleoptile formed a circle with the two vascular bundles opposite each other on either side (Figure 2G, left panel). In addition, wild-type coleoptiles displayed adaxial/abaxial polarization, with ∼10 layers of differentiated mesophyll cells between the adaxial and abaxial epidermises (Figure 2G, left panel). In contrast with the coleoptiles of wild-type plants, the coleoptiles of dcl4-1 plants lacked the two vascular bundles (Figure 2G, right panel, top). In addition, the shapes of the sheath and margins of the mature first leaves were different between the wild type and dcl4-1. In the wild type, the sheath is fully developed with multiple vascular bundles, and the margin of the sheath is pointed and membranous (Itoh et al., 2005). The first leaves of dcl4-1 mutants displayed a reduced number of vascular bundles and abnormally thick margins (Figure 2H, right panel, top). In some severe dcl4-1 mutants, single vascular bundles were observed from thread-like leaves (Figure 2H, right panel, bottom).
Moreover, transverse sectioning analysis also revealed the adaxial/abaxial polarity alteration in both coleoptiles and the first leaves in dcl4-1 (Figures 2G and 2H). In wild-type coleoptiles, abaxial and adaxial surfaces had distinct epidermal cells. The abaxial epidermal cells were relatively small and arranged densely, whereas the adaxial epidermal cells were rectangular in shape and arranged into a smooth surface (Figure 2G, left panel). By contrast, the adaxial epidermis of dcl4-1 developed into an irregular shape (Figure 2G, right panel, top), and part of the adaxial epidermis displayed abaxial characteristics, especially in those severe mutants (Figure 2G, right panel, bottom). In addition, typical adaxial epidermal cells of the first leaves are present in the wild type; however, the adaxial epidermis of dcl4-1 mutants partial or fully transformed into densely arranged abaxial epidermis (Figure 2H, right panel). These observations indicate that Os DCL4 plays an important role in the adaxial/abaxial polarization of the coleoptiles and the first leaves.
Lemma Polarity Is Disrupted in Os dcl4 Mutants
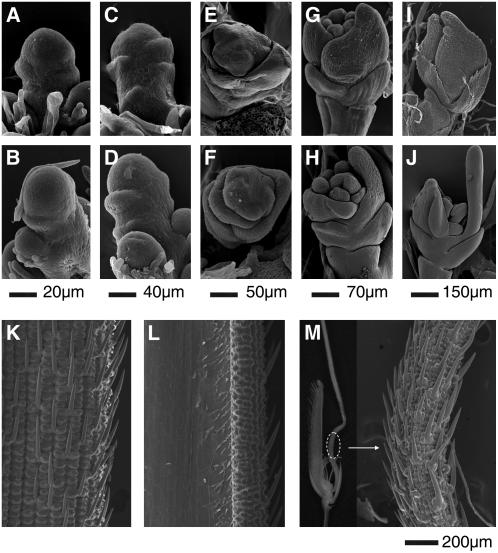
In the reproductive stage, the lemma and palea of the rice DCL4IR transformants and dcl4-1 mutants showed severe abnormalities. We determined the stage of onset of these abnormalities using scanning electron microscopy (Itoh et al., 2005) (Figure 3). We did not observe clear developmental differences between DCL4IR transformants and wild-type plants during stages Sp1 to Sp6 (Figures 3A to 3F; data not shown). The lemma of DCL4IR transformants elongated ectopically during Sp7 (the stage at which the carpel forms and stamens differentiate into filaments and anthers) (Figures 3G and 3H). During Sp8, the apical part of the lemma in the DCL4IR transformants tended to become a needle-like awn and the inner reproductive organs were not enclosed by the lemma and palea as they were in wild-type plants (Figures 3I and 3J).
Figure 3.
Scanning Electron Microscopy Images of Spikelet Organs.
(A) to (J) Top panels, wild-type Nipponbare; bottom panels, DCL4IR transformants.
(A) and (B) Stage Sp1 to Sp2, formation of a pair of primordia of rudimentary glume and empty glume, respectively.
(C) and (D) Stage Sp2 to Sp3, formation of lemma primordium.
(E) and (F) Stage Sp5 to Sp6, formation of lodicule and stamen primordia, respectively.
G) and (H) Stage Sp7, formation of carpel primordium.
(I) and (J) Stage Sp8, ovule and pollen formation.
(K) Scanning electron microscopy image of wild-type 9311 lemma abaxial epidermis.
(L) Scanning electron microscopy image of wild-type 9311 lemma adaxial epidermis.
(M) A spikelet of dcl4-1 (left panel) and scanning electron microscopy image of its lemma epidermis (right panel).
Scale bars are shown below each stage.
During the heading stage, the wild-type lemma showed shaggy abaxial (Figure 3K) and smooth adaxial (Figure 3L) epidermis. By contrast, the lemma of dcl4-1 developed into a radial abaxialized awn (Figure 3M, left panel), with the whole epidermis displaying abaxial characteristics (Figure 3M, right panel). This observation indicated that the polarity of the lemma was completely disrupted in dcl4-1.
Os DCL4 Provides the Main Dicer Activity for Generating 21-Nucleotide siRNAs
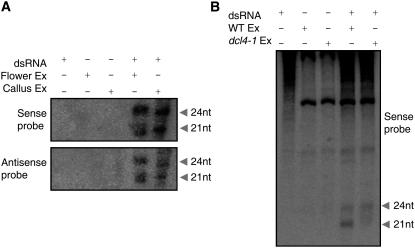
The unique phenotype of the dcl4-1mutant prompted us to analyze the role of Os DCL4 in small RNA biogenesis. Previously, we showed that knockdown of Os DCL4 in DCL4IR plants did not affect miRNA biogenesis (Liu et al., 2005). This result was confirmed by analysis of rice carrying the dcl4-1 null allele. Therefore, the abnormal phenotype is not due to defects in miRNA accumulation (see Supplemental Figure 2 online). Because direct biochemical evidence for DCL4 protein activity in plants is lacking, we performed a biochemical analysis for DCL4 activity in rice. Using protein extracts from young flowers or 30-d-old callus as enzyme sources and the dsRNA of the β-glucuronidase (GUS) gene as substrate, both 21- and 24-nucleotide siRNAs could be detected when hybridized with sense (Figure 4A, top panel) and antisense (Figure 4A, bottom panel) RNA probes, indicating that Dicer-like activities generating 21- and 24-nucleotide siRNAs exist in both rice tissues tested. By comparing the Dicer-like activities in the extracts from young flower tissues of wild-type and dcl4-1 plants, we found that extract from dcl4-1 could produce 24-nucleotide siRNAs with reduced 21-nucleotide siRNAs (Figure 4B). This result provides direct biochemical evidence that efficient 21-nucleotide siRNA production in rice requires DCL4.
Figure 4.
Dicer-Like Activity Assay in Rice.
(A) A 568-bp dsRNA, flower extract (Flower Ex), or callus extract (Callus Ex) were mixed as shown at top. The siRNAs originating from dsRNA of the GUS gene were detected using the sense strand (top panel) or the antisense strand (bottom panel) as a probe. nt, nucleotides.
(B) The dsRNA was incubated with wild-type or dcl4-1 extract as shown at top. siRNAs were detected by sense strand 32P-labeled RNA probe. The positions of the produced 21- and 24-nucleotide siRNAs are indicated.
Os DCL4 Is Required for the Production of 21-Nucleotide siRNAs
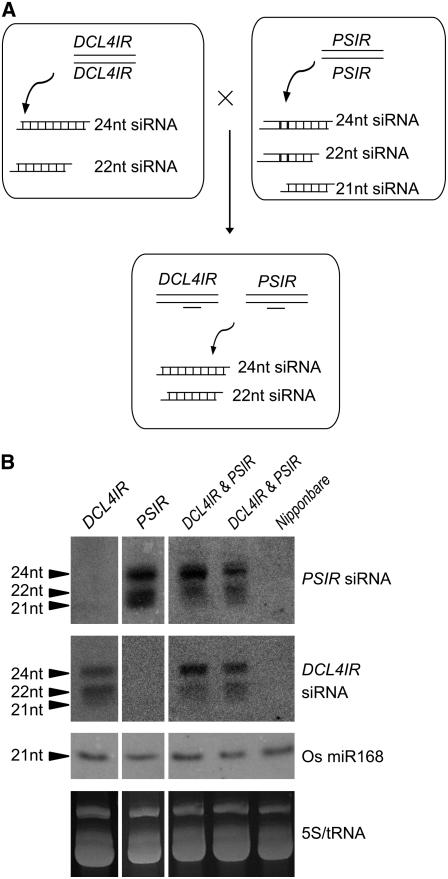
It is well established that overexpression of an inverted repeat often leads to transgene silencing associated with the production of 21-, 22-, and 24-nucleotide siRNAs (Mette et al., 2000). To examine the specific role of Os DCL4 in siRNA biogenesis, we studied small RNAs corresponding to the inverted repeat region of the Os DCL4IR transgene (Figure 5A, top, left panel). The 22- and 24-nucleotide siRNAs, but not the 21-nucleotide siRNAs, were easily detected in the DCL4IR transformants (Figure 5B, left panel). To determine whether the absence of 21-nucleotide siRNAs in the DCL4IR transformants is a general phenotype, a transgenic line containing an inverted repeat of the PHYTOCHROMOBILIN SYNTHASE (PSIR) gene was crossed into DCL4IR transformants (Figure 5A). The resulting F1 plants containing both DCL4IR and PSIR transgenes were examined. siRNAs derived from the PSIR inverted repeat with lengths of 21, 22, and 24 nucleotides were all detected in the PSIR lines (Figure 5B, middle panel). The 21-nucleotide PSIR siRNAs, but not the 22- and 24-nucleotide PSIR siRNAs, were almost eliminated in the PSIR lines carrying DCL4IR (Figure 5B, right panel). We conclude that the production of the 21-nucleotide siRNAs in rice requires Os DCL4 function (Figure 5).
Figure 5.
Os DCL4 Acts in the Biogenesis of 21-Nucleotide siRNAs Associated with the Transgene.
(A) Diagrammatic representation of the biogenesis of 21-, 22-, and 24-nucleotide (nt) siRNAs in DCL4IR transformants, PSIR transformants, and their crossed offspring.
(B) Small RNA gel blot analysis of inverted repeat–associated siRNAs. Samples from DCL4IR transformants (DCL4IR), PSIR transformants (PSIR), and two double mutants (DCL4IR and PSIR) are listed at top. The wild type (Nipponbare) was used as a negative control. Os miR168 and 5S/tRNA stained with ethidium bromide were used as loading controls (bottom two panels).
Conserved Biogenesis Mechanism of TAS3 ta-siRNAs between Monocots and Eudicots
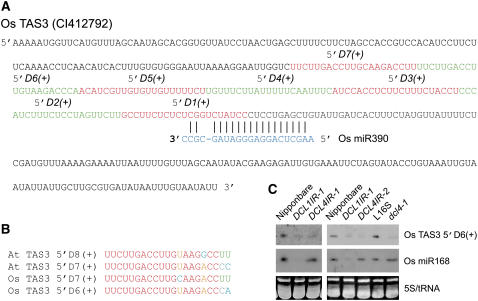
In pursuing the candidate target genes downstream of Os DCL4, we took a whole genome approach to compare transcript profiles in the young inflorescences of wild-type Nipponbare and DCL4IR transformants (Ma et al., 2005) (see Supplemental Table 1 online). An Auxin Response Factor gene from rice (Os ARF, Os 01g48060) showed upregulation in the DCL4IR transformants. The closest homologous gene, ARF3 in Arabidopsis, is a direct target of TAS3 ta-siRNAs (Allen et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). Phylogenetic analysis indicated that TAS3 is conserved between Arabidopsis and rice (Allen et al., 2005; Williams et al., 2005). In order to test whether Os DCL4 plays a role in ta-siRNA biogenesis in rice, we first analyzed the sequences of Os TAS3 (CI412792) to confirm that Os TAS3 ta-siRNAs exist in rice (Figure 6A). By comparing consecutive 21-nucleotide-long sequences 5′ to the miR390 directed cleavage point, we were able to identify two pairs of highly homologous 21-nucleotide-long sequences between Arabidopsis and rice, the At TAS3 5′D7(+) versus the Os TAS3 5′D6(+) and the At TAS3 5′D8(+) versus the Os TAS3 5′D7(+), which indicated that Os TAS3 5′D6(+) and 5′D7(+) are bona fide ta-siRNAs (Figures 6A and 6B) (Allen et al., 2005).
Figure 6.
Characterization of the Biogenesis and Function of Os TAS3 ta-siRNAs.
(A) The diagram represents the biogenesis of predicted Os TAS3 ta-siRNAs from Os TAS3 transcript directed by Os miR390 (blue). Putative ta-siRNAs are shown alternately in red and green.
(B) Alignment of At TAS3 ta-siRNAs and Os TAS3 ta-siRNAs. Os TAS3 5′D6(+) is homologous with At TAS3 5′D7(+), and Os TAS3 5′D7(+) is homologous with At TAS3 5′D8(+).
(C) Accumulation of Os TAS3 ta-siRNAs in DCL1IR, DCL4IR transformants, and dcl4-1 as revealed by RNA gel blot analysis. Nipponbare was used as a control for DCL1IR and DCL4IR transformants; L16S was the wild-type control for dcl4-1. Os miR168 and 5S/tRNA stained with ethidium bromide were used as loading controls in the middle and bottom panels, respectively.
To determine whether any DCLs play a role in ta-siRNA biogenesis in rice, we compared the expression levels of Os TAS3 ta-siRNAs between DCL1IR and DCL4IR transformants and wild-type Nipponbare controls. Levels of Os TAS3 5′D6(+) ta-siRNAs were substantially reduced in both the DCL1IR and DCL4IR transformants (Figure 6C, left panel). Consistent with impaired Os TAS3 5′D6(+) ta-siRNA biogenesis in the DCL4IR transformants, dcl4-1 also displayed decreased expression of Os TAS3 ta-siRNAs (Figure 6C, right panel), indicating that the Os DCL4 activity is required for the biogenesis of ta-siRNAs in rice and that the ta-siRNA biogenesis mechanism is conserved between monocots and eudicots.
Os DCL4 Regulates Multiple Os ARF Genes
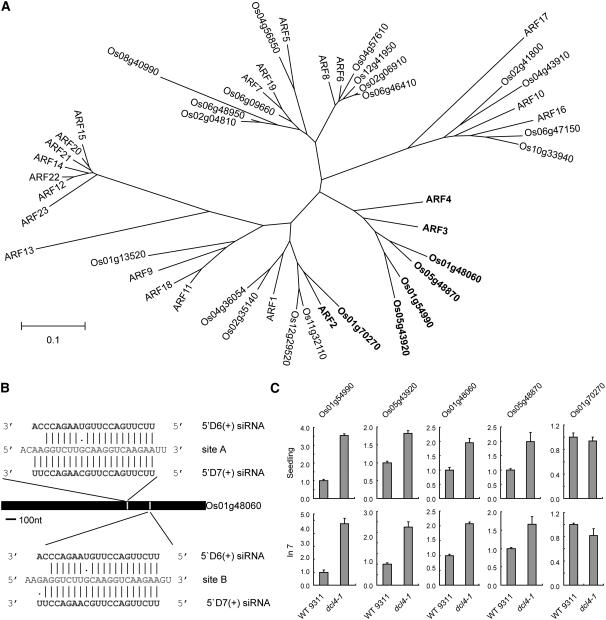
Computational and experimental methods have identified and validated ARF2, ARF3, and ARF4 as targets of TAS3 ta-siRNA regulation in Arabidopsis (Allen et al., 2005; Williams et al., 2005). Sequence comparison analysis identified four close homologs of Arabidopsis ARF3 and one homolog of Arabidopsis ARF2 in rice (Williams et al., 2005) (Figure 7A). Among these, the four ARF3 homologs contained two putative target sites for both Os TAS3 5′D6 (+) and Os TAS3 5′D7(+) ta-siRNAs (Figure 7B, Os 01g48060 as an example), while the ARF2 homolog (Os 01g70270) had only one target site for Os TAS3 5′D6(+) and Os TAS3 5′D7(+) ta-siRNAs.
Figure 7.
Target Genes of Os TAS3 ta-siRNAs.
(A) Phylogenetic relationships of ARF proteins in Arabidopsis and rice. The predicted targets of TAS3 ta-siRNAs are indicated in boldface. Bootstrap values (1000 replicates) for this tree are shown in Supplemental Figure 3 online. The bar indicates substitutions per site.
(B) The diagram represents a detailed example of Os TAS3 ta-siRNAs and their target gene Os 01g48060.
(C) Quantitative RT-PCR analysis of the expression of the five putative target genes of Os TAS3 ta-siRNAs in wild-type 9311 rice and dcl4-1. The expression levels of the assayed genes were normalized according to the expression value of Os EF-1α, and the expression from wild-type 9311 was set to 1.0. Error bars indicate sd. Seedling, the aerial part of plants at 6 DAG; In7, inflorescence in the floral organ differentiation stage. Os 01g54990, Os 05g43920, Os 01g48060, Os 05g48870, and Os 01g70270 were previously named ARF3-likeB, ARF3-likeA, ARF3-like2, ARF3-like1, and ARF2-like, respectively (Williams et al., 2005).
Transcript levels of the five Os ARF genes in dcl4-1 and wild-type 9311 were compared at 6 DAG and at inflorescence stage 7 (the floral organ differentiation stage) (Itoh et al., 2005). The expression level of Os 01g54990 was upregulated by about fourfold in seedlings and inflorescences of dcl4-1(Figure 7C, left panel). The transcripts of Os 05g43920, Os 01g48060, and Os 05g48870 were upregulated by about twofold in seedlings and inflorescences of dcl4-1 (Figure 7C, middle three panels). The expression of the Arabidopsis ARF2 homolog Os 01g70270 was not changed significantly (Figure 7C, right panel). The differences in expression between individual Os ARF genes regulated by Os TAS3 ta-siRNAs may be related to the developmental regulation of Os DCL4 function.
Endogenous 21-Nucleotide siRNAs Derived from an Imperfect Inverted Repeat of AK120922 Are Processed by Os DCL4
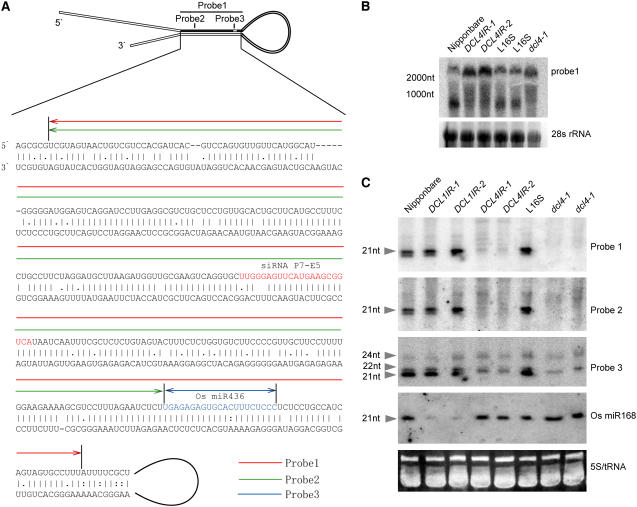
In our microarray analysis, a unique transcript (AK120922) (Kikuchi et al., 2003) showed threefold higher expression in DCL4IR transformants compared with that of the wild type. AK120922 is a rice-specific sequence that has no conserved homolog in currently available genome sequences of any other species. The transcript of AK120922 can form a hairpin-like structure with a number of mismatches, which is structurally similar to known miRNA precursors or putative transitional RNA genes (containing a long fold-back structure with very few mismatches) (Figure 8A) (Allen et al., 2004).
Figure 8.
Expression Pattern of AK120922 Transcript and Its Derived siRNAs in Different Mutants.
(A) Diagrammatic representation of the fold-back structure of AK120922 RNA and the sequence alignment of the fold-back double-stranded sequences. Probes used in the RNA gel blot hybridization are underlined in different colors: red for probe 1, green for probe 2, and blue for probe 3. Probe 1 covers probe 2 and probe 3. Probe 3 represents the region previously identified as miR436. The previously identified siRNA P7-E5 is located within probe 2 and labeled in red.
(B) RNA gel blot analysis of the expression levels of AK120922 in DCL4IR transformants and dcl4-1 using probe 1. nt, nucleotides.
(C) AK120922-related siRNAs in DCL1IR transformants, DCL4IR transformants, and dcl4-1. Probe 1 and probe 2 were 32P-labeled transcripts, and probe 3 was a specific LNA nucleotide probe.
To confirm that the increased transcript levels of AK120922 resulted from loss of function of Os DCL4, RNA gel blot analysis was performed using a probe corresponding to the entire top arm of the inverted repeat region of AK120922 (Figures 8A and 8B). As expected, the level of the full-length AK120922 was increased in DCL4IR and dcl4-1 plants compared with the wild-type controls, indicating that AK120922 was indeed processed by Os DCL4 in wild-type plants.
As shown in Figure 8A, 21- and 22-nucleotide small RNAs that have been cloned previously and identified as Os miR436 (Sunkar et al., 2005a) and P7-E5 siRNA (Sunkar et al., 2005b), respectively, may originate from the AK120922 fold-back region. Data from the small RNA Massively Parallel Signature Sequencing database (http://mpss.udel.edu/rice/) revealed that large amounts of small RNAs were generated from the AK120922 transcript (see Supplemental Figure 4 online) (Nobuta et al., 2007). In order to determine whether the small RNAs derived from AK120922 were miRNAs or siRNAs, small RNA gel blots were used to examine their accumulation in DCL1IR and DCL4IR transformants and in the dcl4-1 mutant (Figure 8C). We found that expression of 21-nucleotide small RNAs corresponding to the top arm of the inverted repeat region was eliminated in DCL4IR transformants and dcl4-1 but not in the wild-type Nipponbare, the DCL1IR transformants, or the L16S rice strain (the wild-type control for dcl4-1). Similar results were also obtained when a different probe for that region was used (the top arm of the inverted repeat region that does not overlap the Os miR346 region). Interestingly, when a more sensitive lock-modified DNA (LNA) probe corresponding to the small RNA of Os miR436 was used, 21-nucleotide siRNAs appeared to be reduced in DCL4IR and dcl4, while the levels of 22- and 24-nucleotide siRNAs were not changed (Figure 8C). This result demonstrates the specificity of Os DCL4 in the production of 21- but not 22- or 24-nucleotide siRNAs. By contrast, the expression level of Os miR168 was reduced in DCL1IR but not DCL4IR transformants. Our results suggest that the small RNAs derived from AK120922, including the previously reported Os miR436, are siRNAs, unless they are redefined as special miRNAs produced by Os DCL4 (Rajagopalan et al., 2006).
DISCUSSION
The role of RNA silencing in plant development has been the focus of much recent research activity. miRNAs generated by DCL1 are known to regulate the expression of numerous key developmental genes (Jones-Rhoades et al., 2006). Our previous work indicates that Os DCL1 is exclusively responsible for miRNA biogenesis, and lesions in Os DCL1 cause pleiotropic developmental defects similar to Arabidopsis dcl1 mutants (Liu et al., 2005). ta-siRNAs processed by DCL1 and DCL4 activities are involved in leaf development in Arabidopsis. This study describes the role of Os DCL4 in rice, showing its biochemical functions to be conserved with Arabidopsis, but with a much broader role in developmental regulation.
Os DCL4 Loss-of-Function Rice Mutants Display Phenotypes Distinct from Their Arabidopsis Counterparts
Here, we demonstrate that Os DCL4 is the main, if not sole, enzyme that generates 21-nucleotide siRNAs in vivo. We also prove that Os DCL4 is responsible for the production of the 21-nucleotide siRNAs derived from inverted repeat transgenes and endogenous ta-siRNAs. Although the ta-siRNA biogenesis mechanism is conserved between monocots and eudicots, rice DCL4IR transformants and dcl4-1 mutants display severe lateral organ polarity defects in contrast with Arabidopsis dcl4 mutants.
In Arabidopsis, dcl4 mutants display inconspicuous leaf changes, including downward-curled rosette leaves and increased ratio of length to width, as well as accelerated juvenile-to-adult vegetative phase changes indicated by the appearance of trichomes on the abaxial epidermis in leaves (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006). By contrast, the rice dcl4-1 mutants display abnormal coleoptiles in which the two vascular bundles are undeveloped. Furthermore, the first leaves of dcl4-1 plants have a reduced number of vascular bundles and abnormally thick margins and even form thread-like leaves with abaxialized epidermis and single vascular bundles (Figure 2H). These phenotypes of dcl4-1 resemble phenotypes caused by the ectopic expression of KANADI, a member of the GARP-type transcription factor in Arabidopsis, which results in abaxialized cotyledons and inhibits the formation of vascular bundles (Eshed et al., 2001; Kerstetter et al., 2001). In the reproductive stage, the lemmas of dcl4-1 mutants develop into awn-like organs with abaxialized epidermis and spikelet organ identity is disrupted, resulting in sterility. While other regulatory pathways, in addition to that including TAS3, may be involved in regulating leaf polarity, the extent to which the different pathways are used may vary between different organs. The most obvious defects of lemma in dcl4-1 indicate that the TAS3-containing pathway plays a critical role in the determination of lemma polarity. Recently, Nagasaki et al. (2007) have independently reported that the Os DCL4 (also known as SHOOT ORGANIZATION1)–mediated ta-siRNA pathway regulates the critical step of shoot apical meristem formation during rice embryogenesis. In conclusion, loss-of-function mutations of Os DCL4 cause severe developmental defects in rice but not in Arabidopsis. The striking phenotypes of dcl4-1 rice mutants indicate the essential role of Os DCL4 in siRNA biogenesis for rice development, especially for normal lateral organ polarity.
Involvement of the siRNA Pathway in Leaf Polarity Control in Different Plant Species
In Arabidopsis, the protein-coding genes ASYMMETRIC LEAVES1 (AS1) and AS2 and the ta-siRNA–encoding TAS3 gene redundantly regulate leaf polarity (Garcia et al., 2006; Kidner and Timmermans, 2007). Mutations in neither TAS3 nor AS1 show obvious defects. Only double mutants between as1 and the ta-siRNA pathway show enhanced leaf-patterning defects (Garcia et al., 2006). Here, we demonstrate that dcl4 single mutants develop severe lateral organ polarity defects, implying that the ta-siRNA pathway plays a critical role in the development of adaxial/abaxial leaf polarity in rice. Similarly, leafbladeless (lbl1) is the maize (Zea mays) homolog of Arabidopsis SGS3, and severe lbl1 mutants also develop radially symmetric, thread-like leaves and exhibit a completely abaxialized epidermis (Timmermans et al., 1998; Juarez et al., 2004; Nogueira et al., 2007). The comparison of obvious phenotypes of dcl4 in rice and lbl1 in maize with their Arabidopsis counterpart reinforces the idea that polarity pathways vary in their importance in different plant species (Kidner and Timmermans, 2007). Although we do not know whether the rice AS1 homolog (Os AS1) acts in proximal/distal patterning, like ROUGH SHEATH2 in maize, or in adaxial/abaxial patterning, like AS1 in Arabidopsis, from the distinct phenotypes of Os dcl4 and At dcl4, we would predict that the ta-siRNA pathway may not be directly coupled with Os AS1 to redundantly regulate lateral organ polarity. Therefore, loss of function of Os DCL4, the major component ta-siRNA pathway, leads to severe developmental defects in rice.
TAS3 ta-siRNA, ARF3/4-LIKE Genes, and Lateral Organ Polarity
ARFs such as the ta-siRNA targets that are negatively regulated by TAS3 ta-siRNAs play a role in lateral organ polarity determination in Arabidopsis (Pekker et al., 2005). Overexpression of a modified ETT cDNA in which TAS3 ta-siRNA target sites were mutated (35S:ETTAB) produced a higher frequency of aberrant phenotypes than did overexpression of a wild-type ETT cDNA (35S:ETT), indicating that a higher ectopic expression level of the TAS3 ta-siRNA target correlates with the severity of the abnormal phenotypes (Hunter et al., 2006). In maize, four ARF3/ARF4-like target genes of TAS3 ta-siRNA have also been identified, although only ARF3a has been experimentally validated to be a target (Nogueira et al., 2007). Phylogenetic analysis reveals that ARF3/ARF4-like genes have been duplicated, resulting in four Os ARF genes highly related to ARF3/ARF4 in rice (Williams et al., 2005). In this work, we demonstrate that all four Os ARF genes are also upregulated in rice dcl4, similar to the regulation of their homologs in Arabidopsis, whereas the distinct phenotypes of dcl4 mutants hint that the functional divergence of ARFs and/or variation in their downstream genes would be another possible explanation for the abnormal defects in Os dcl4.
Relationship between Endogenous siRNAs Derived from Inverted Repeats and miRNAs in Plants
In this study, we identified an endogenous RNA, AK120922, that was upregulated in loss-of-function rice dcl4 mutants. The AK120922 transcript can form an inverted repeat structure similar to miRNA precursors. It has been proposed that miRNA genes evolved from the inverted duplication of target genes (Allen et al., 2004). During evolution, the duplicated genes would evolve from the original inverted repeat with a perfect match to transitional genes (containing a long fold-back structure with very few mismatches) and miRNA genes (Allen et al., 2004). Recently, this hypothesis was experimentally validated in Arabidopsis (Rajagopalan et al., 2006; Fahlgren et al., 2007). In rice, AK120922 contains all of the features of transitional genes, and the 21-nucleotide siRNAs generated from the double-stranded region of AK120922 are dependent on Os DCL4 but not Os DCL1. This discovery supports the existence of a third class of small RNAs either transitional between siRNA and miRNA or a novel miRNA that depends on DCL4.
Unlike miRNA genes, these loci with long inverted repeats are not conserved in plants (Lu et al., 2006). The lack of conservation at the sequence level and the diversity of these loci in plants may partially explain why loss of function of DCL4 in Arabidopsis and rice resulted in distinct phenotypes.
METHODS
Plant Materials
Rice (Oryza sativa) subsp japonica (Nipponbare) and indica (L16S, 9311) plants were used in this study. The DCL4IR and DCL1IR transformants were in the Nipponbare background (Liu et al., 2005). Nipponbare was used as the wild-type control of DCL4IR and DCL1IR transformants. The dcl4-1 mutant was identified from an indica variety, L16S, and introgressed into 9311 by backcross. 9311 was used as the wild-type control of dcl4-1. The molecular markers used to PCR genotype the dcl4-1 mutation were as follows: P1 (CX1240, 5′-CAGACGGTGTATGTAACACC-3′) and P2 (CX1794, 5′-CGGACTCCAAGACGCAATATGT-3′) for the wild type, with a predicted size of 637 bp; and P1 and P3 (CX1335, 5′-TTGCGATCACCACAGCTTGC-3′) for dcl4-1 and the wild-type, with predicted sizes of 1.6 and 3 kb, respectively (Figure 2D). To map the dcl4-1 mutant, every 10 plants with mutant phenotypes were pooled, and 66 pools were obtained from 660 F2 populations. Genomic DNA was extracted from each pool as a template for PCR-based genotyping.
Tissue Collection
Young inflorescences enclosed in sheaths were collected from plants grown in the field, frozen in liquid nitrogen immediately after harvest, and preserved in a −80°C refrigerator for RT-PCR, RNA gel blot analysis, and microarray hybridization. For biochemical analysis, inflorescences of ∼4 cm in length were collected from the field. Callus tissues were induced from embryogenic calli on Murashige and Skoog medium with 2 mg/L 2,4-D for 30 d before harvesting. Seedlings were grown on Murashige and Skoog medium with a daylength of 16 h and a night of 8 h at 25°C.
Histological Analysis
The coleoptiles and first leaves of 3.5-DAG seedlings were fixed overnight with 2% glutaraldehyde and 1% paraformaldehyde. Tissues were dehydrated in a graded ethanol series, then transferred to propylene oxide and embedded in Spurr's resin (SPI-CHEM). Microtome sections at 3 μm were stained with 0.5% toluidine blue and observed with a light microscope (Olympus BX51 plus DP70).
Rice Oligonucleotide Array
The rice 70-mer oligonucleotide microarray was used in this study as described previously (Jiao et al., 2005; Ma et al., 2005).
RNA Isolation, Probe Labeling, and Microarray Hybridization
Rice inflorescences (∼4 to 8 cm in length) were frozen in liquid nitrogen, and total RNA was isolated using RNAwiz reagent (Ambion) and purified with the RNeasy kit (Qiagen). For each sample, 100 μg of total RNA was labeled with aminoallyl-dUTP (Sigma-Aldrich) by reverse transcription. The aminoallyl-dUTP–labeled cDNAs were purified using a Microcon YM-30 filter (Millipore) and resuspended in 0.1 M NaHCO3. The purified cDNAs were further fluorescently labeled by conjugating monofunctional Cy3 or Cy5 dye (Amersham) to the aminoallyl functional groups. Two independent biological replicates were performed with a dye swap. After coupling at room temperature for 1 h, the labeling reaction was stopped by ethanolamine. The labeled probes were separated from unincorporated dye using the QIAquick PCR purification kit (Qiagen) and concentrated with a Microcon YM-30 filter. The protocols for microarray hybridization, microarray slide washing, and array scanning were as described previously (Ma et al., 2005). Hybridized slides were scanned with a GenePix 4000B scanner (Axon), and independent TIFF images for Cy3 and Cy5 channels were used for subsequent analysis.
Microarray Data Processing
After manual removal of spots with aberrant morphology, microarray spot intensity signals were acquired using the Axon GenePix Pro 5.0 software package without correction for background, and each slide included two groups of intensity data corresponding to Cy5 and Cy3 channels. We first removed the dye effect on each slide using the LOWESS normalization method, which was applied to log2-transformed mutant versus wild-type expression values with two-sample hypothesis and equal variation assumptions. Subsequently, quartile normalization was applied to the LOWESS-normalized microarray data to remove biases among slides. For detection of differentially expressed spots between mutant and wild-type rice panicles, the normalized data were log2-transformed and fitted into a mixed-effect analysis of variance model with the software MAANOVA under an R environment. After multiple testing between pair-wise comparisons, spots with false discovery rate–corrected P < 0.05 were regarded as differentially expressed genes.
dsRNA Synthesis
A 568-bp PCR fragment was produced from the GUS gene using the sense primer CX1415 (5′-GAAGATCTGGTATCAGCGCGAAGTCT-3′) and the antisense primer CX1416 (5′-CCGCTCGAGTTCATAGAGATAACC-3′). The PCR fragment was cloned into pGEM-T Easy vector (Promega) for in vitro transcription. Both sense and antisense RNA transcripts produced by T7 RNA polymerase were equally mixed and denatured at 100°C for 5 min and annealed at room temperature for 10 min. dsRNAs were purified from a 1% agarose gel to remove single-stranded RNAs.
Preparation of Protein Extract
The total protein extract was prepared as described previously (Qi et al., 2005), and the protein concentration was adjusted to 2 mg/mL with exaction buffer (20 mM Tris-HCl, pH 7.5, 4 mM MgCl2, 5 mM DTT, and 1 tablet/10 mL protease inhibitor cocktail from Roche) for the Dicer activity assay.
Dicer Activity Analysis
dsRNA (1 ng) was added to 30 μL of protein extract and 8 μL of 5× Dicer buffer (0.5 M NaCl, 5 mM ATP, 1 mM GTP, 6 mM MgCl2, 125 mM creatine phosphate, 150 μg/mL creatine kinase, and 2 units of RNasin RNase inhibitor from Promega) in a total volume of 40 μL (Qi et al., 2005). The mixture was incubated at 30°C for 2 h. The reaction was stopped by adding Trizol solution and precipitated with 4 volumes of ethanol. The precipitate was resolved on a 15% denaturing PAGE gel and transferred to a Bio-Rad Zeta-Probe GT nylon membrane for RNA gel blot analysis. The produced siRNAs were detected with 32P-labeled RNA probe.
Phylogenetic Analysis
Full-length protein sequences were used for phylogenetic analyses. The sequences derived from 29 known or predicted Os ARF genes were found in The Institute for Genomic Research rice genome annotation database (http://www.tigr.org/tdb/e2k1/osa1/GeneNameSearch.shtml) using “auxin response factor” as the search query. The 23 highly related Os ARF protein sequences were used for alignments. The protein sequences of 23 ARF genes from ARF1 to ARF23 were downloaded from The Arabidopsis Information Resource database (http://www.Arabidopsis.org/index.jsp), referring to the locus numbers as listed previously (Remington et al., 2004). Alignments of protein sequences were performed using ClustalX version 1.81 with default parameters (gap opening, 10.00; gap extension, 0.20; delay divergent sequences, 30%; DNA transition weight, 0.50) (see Supplemental Figure 5 online). A bootstrapping phylogenetic tree was constructed by the MEGA2 program with the Unweighted Pair Group Method and the Arithmetic Mean method. The number of bootstrap replicates was 1000.
RT-PCR and Real-Time PCR Analysis of Gene Expression
RT-PCR was performed as described previously to detect the expression of Os DCL4 (Liu et al., 2005) using primer pairs CX0758 (5′-GGACTAGTTACACGAACGTCCTCTTCTTTTGGTAGGT-3′) and CX0857 (5′-CGATGAGAGAACTTCGAGAGCT-3′). The Os EF-1α gene was used as an internal loading control with the following primer pairs: CX1597 (5′-GCACGCTCTTCTTGCTTTCACTCT-3′) and CX1598 (5′-AAAGGTCACCACCATACCAGGCTT-3′).
Real-time PCR analysis was performed to measure transcript levels using the system reported previously (Deng et al., 2007). PCR was performed using hot-start Taq DNA polymerase (TaKaRa Taq Hot Start version; Code DR007B). For each sample, quantifications were made in triplicate. Melt curves were read at the end of each amplification by steps of 0.3°C from 65 to 95°C to ensure that the quantifications were derived from real PCR products and not primer dimers. Specific gene expression was normalized to the internal control gene Os EF-1α using the primers just described; the gene expression value of the wild type was used as a control and set at 1.0. For each target gene, primer pairs were as follows: for Os 05g48870, CX1548 (5′-GTGATACAGACCCTATGTGGCAT-3′) and CX1549 (5′-TTACTCGCATCGCTGGAGCAACT-3′); for Os 01g48060, CX1540 (5′-TTGGAAGCAGAGAGGCAGATCCAA-3′) and CX1541 (5′-AGGAAAACCCAGGGTTTCCACTT-3′); for Os 01g54990, CX2259 (5′-AAAGGCTTCAACACCTGGGA-3′) and CX2260 (5′-GGCTCATAACTAGGTCTAGA-3′); for Os 05g43920, CX2261 (5′-TGAAGAGCCTGAACCATCCA-3′) and CX2262 (5′-TTGGTTCGCTTAGACCCAGA-3′); and for Os 01g70270, CX2263 (5′-TCGATGGTGAATTGGTGTCT-3′) and CX2264 (5′-AGACTCCATGCGATCTACCT-3′).
RNA Gel Blot Analysis
Total RNAs were extracted using Trizol solution from the young panicles of rice and dissolved in RNase-free water. Small RNAs were enriched by adding the same volume of 8 M LiCl and centrifuging at 12,000 rpm for 30 min at 4°C. RNA filter hybridizations were performed as described previously (Liu et al., 2005). The templates for transcript probes 1 and 2 were amplified from rice genomic DNA by PCR with the following primers: for probe 1, CX0761 (5′-TCGTAGTAACTGTCGTCCACGA-3′) and CX0762 (5′-AAAGGCACTACTGATGGCAGGA-3′); for probe 2, CX0761 (5′-TCGTAGTAACTGTCGTCCACGA-3′) and CX1443 (5′-AGAGATTCTAAAGGACGCT-3′). The PCR products were then cloned into T-Easy vector under the control of the T7 promoter, and the templates for transcription of the RNA probes were linearized by PCR with the following primers: for probe 1, M13 (5′-GTAAAACGACGGCCAG-3′) and CX0762; for probe 2, M13 and CX1443. Probe 3 was end-labeled with LNA (5′-GGAGAAAGTGcAcTCTcTCA-3′; lowercase letters represent LNAs) (TaKaRa) (Valoczi et al., 2004). Probe for Os TAS3 ta-siRNA was end-labeled with LNA (5′-GTGGGTCTTACAaGgTcAaGAA-3′; lowercase letters represent LNAs). To prepare RNA probe for PSIR, the template was amplified from the RNAi vector using primer CX1947 (5′-CCCTCCCAGGAAAAGTTCA-3′) and primer CX1948 (5′-CCAAGTTCAGGGATATGAGCA-3′). DCL4IR probe was prepared as described previously (Liu et al., 2005). End-labeled oligo DNA probe (5′-GTCCCGATCTGCACCAAGCGA-3′) was used to detect Os miR168.
Accession Number
The cDNA sequence for Os DCL4 was deposited in the GenBank data library under accession number EU009924.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mature Plants of dcl4-1.
Supplemental Figure 2. Accumulation of Os miRNAs as Revealed by RNA Filter Hybridization with End-Labeled Oligo DNA Probes.
Supplemental Figure 3. Neighbor-Joining Phylogenetic Tree of Protein Sequences with Bootstrap Values.
Supplemental Figure 4. Seventeen-Nucleotide Small RNA Massively Parallel Signature Sequencing Signatures Generated from the Fold-Back Region of AK120922 Transcript.
Supplemental Figure 5. Multiple alignments of the ARF Protein Sequences by ClustalX.
Supplemental Table 1. Expression Data for Genes That Were Significantly Changed between the Inflorescences of Wild-Type and DCL4IR Transgenic Rice.
Supplementary Material
Acknowledgments
We thank Catherine Kidner from the Royal Botanic Garden Edinburgh and the University of Edinburgh for helpful comments and English editing of the manuscript. We also thank Yonghong Wang from Jiayang Li's laboratory, Xia Wang, and Ying Lan for assistance with microscopy for morphologic analysis. We thank Lihuang Zhu for providing the dcl4-1 allele. This work was supported by the National Natural Science Foundation of China (Grants 30621001 and 30325015 to X.C.), the State High-Tech Project (Grant 2006AA10A101 to Y.X.), the National Basic Research Program of China (Grants 2005CB120806 and 2005CB522400), and the Chinese Academy of Sciences (Grant CXTD-S2005-2 to X.C.). The authors gratefully acknowledge the support of the K.C. Wong Education Foundation, Hong Kong.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xiaofeng Cao (xfcao@genetics.ac.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36 1282–1290. [DOI] [PubMed] [Google Scholar]
- Asai, K., Satoh, N., Sasaki, H., Satoh, H., and Nagato, Y. (2002). A rice heterochronic mutant, mori1, is defective in the juvenile-adult phase change. Development 129 265–273. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., Jan, C., Rajagopalan, R., and Bartel, D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127 565–577. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R., and Zhu, J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche, N., Lauressergues, D., Gasciolli, V., and Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W., Liu, C., Pei, Y., Deng, X., Niu, L., and Cao, X. (2007). Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol. 143 1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, N.R., Labbe, J.C., and Goldstein, B. (2002). Using RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. USA 99 4191–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Fahlgren, N., Howell, M.D., Kasschau, K.D., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., Law, T.F., Grant, S.R., Dangl, J.L., and Carrington, J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2 e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Ferrario, S., Immink, R.G., and Angenent, G.C. (2004). Conservation and diversity in flower land. Curr. Opin. Plant Biol. 7 84–91. [DOI] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Gasciolli, V., Mallory, A.C., Bartel, D.P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15 1494–1500. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., and Jacobsen, S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38 721–725. [DOI] [PubMed] [Google Scholar]
- Howell, M.D., Fahlgren, N., Chapman, E.J., Cumbie, J.S., Sullivan, C.M., Givan, S.A., Kasschau, K.D., and Carrington, J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, C., Willmann, M.R., Wu, G., Yoshikawa, M., de la Luz Gutierrez-Nava, M., and Poethig, S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., Kitano, H., and Nagato, Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46 23–47. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., et al. (2005). A tiling microarray expression analysis of rice chromosome 4 suggests a chromosome-level regulation of transcription. Plant Cell 17 1641–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., Bartel, D.P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57 19–53. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Twigg, R.W., and Timmermans, M.C. (2004). Specification of adaxial cell fate during maize leaf development. Development 131 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2005). The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8 38–44. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Timmermans, M.C. (2007). Mixing and matching pathways in leaf polarity. Curr. Opin. Plant Biol. 10 13–20. [DOI] [PubMed] [Google Scholar]
- Kikuchi, S., et al. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379. [DOI] [PubMed] [Google Scholar]
- Kim, J.K., Gabel, H.W., Kamath, R.S., Tewari, M., Pasquinelli, A., Rual, J.F., Kennedy, S., Dybbs, M., Bertin, N., Kaplan, J.M., Vidal, M., and Ruvkun, G. (2005). Functional genomic analysis of RNA interference in C. elegans. Science 308 1164–1167. [DOI] [PubMed] [Google Scholar]
- Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294 853–858. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., and Ambros, V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294 862–864. [DOI] [PubMed] [Google Scholar]
- Liu, B., Li, P., Li, X., Liu, C., Cao, S., Chu, C., and Cao, X. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Kulkarni, K., Souret, F.F., MuthuValliappan, R., Tej, S.S., Poethig, R.S., Henderson, I.R., Jacobsen, S.E., Wang, W., Green, P.J., and Meyers, B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., et al. (2005). A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 15 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margis, R., Fusaro, A.F., Smith, N.A., Curtin, S.J., Watson, J.M., Finnegan, E.J., and Waterhouse, P.M. (2006). The evolution and diversification of Dicers in plants. FEBS Lett. 580 2442–2450. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki, H., Itoh, J., Hayashi, K., Hibara, K., Satoh-Nagasawa, N., Nosaka, M., Mukouhata, M., Ashikari, M., Kitano, H., Matsuoka, M., Nagato, Y., and Sato, Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta, K., Venu, R.C., Lu, C., Belo, A., Vemaraju, K., Kulkarni, K., Wang, W., Pillay, M., Green, P.J., Wang, G.L., and Meyers, B.C. (2007). An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 25 473–477. [DOI] [PubMed] [Google Scholar]
- Nogueira, F.T., Madi, S., Chitwood, D.H., Juarez, M.T., and Timmermans, M.C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker, I., Alvarez, J.P., and Eshed, Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, K., Parameswaran, S., and Vijayraghavan, U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43 915–928. [DOI] [PubMed] [Google Scholar]
- Qi, Y., Denli, A.M., and Hannon, G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, D.L., Vision, T.J., Guilfoyle, T.J., and Reed, J.W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7 487–491. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., and Ambrose, B.A. (1998). The blooming of grass flower development. Curr. Opin. Plant Biol. 1 60–67. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Girke, T., Jain, P.K., and Zhu, J.K. (2005. a). Cloning and characterization of microRNAs from rice. Plant Cell 17 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., Girke, T., and Zhu, J.K. (2005. b). Identification and characterization of endogenous small interfering RNAs from rice. Nucleic Acids Res. 33 4443–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C., Schultes, N.P., Jankovsky, J.P., and Nelson, T. (1998). Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125 2813–2823. [DOI] [PubMed] [Google Scholar]
- Valoczi, A., Hornyik, C., Varga, N., Burgyan, J., Kauppinen, S., and Havelda, Z. (2004). Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32 e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16 69–79. [DOI] [PubMed] [Google Scholar]
- Williams, L., Carles, C.C., Osmont, K.S., and Fletcher, J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Wilken, A., and Carrington, J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101 25–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.