Abstract
High-affinity sulfate transporters SULTR1;1 and SULTR1;2 are expressed at epidermis and cortex of Arabidopsis (Arabidopsis thaliana) roots during sulfur limitation. Here, we report that SULTR1;1 and SULTR1;2 are two essential components of the sulfate uptake system in roots and are regulated at posttranscriptional levels together with the previously reported transcriptional control. Double knockout of SULTR1;1 and SULTR1;2 by T-DNA insertion gene disruption resulted in complete lack of sulfate uptake capacity and severely affected plant growth under low-sulfur conditions. Expression of epitope-tagged proteins SULTR1;1mycHis and SULTR1;2mycHis, under the control of the cauliflower mosaic virus 35S promoter, rescued the uptake of sulfate and the growth of the sultr1;1 sultr1;2 double knockout mutant. The recovery of the double knockout phenotypes was attributable to the posttranscriptional accumulation of sulfate transporter proteins that derive from the epitope-tagged transgenic constructs. Both SULTR1;1mycHis and SUTLR1;2mycHis mRNAs were predominantly found in roots and slightly induced by long-term sulfur limitation. SULTR1;1mycHis and SULTR1;2mycHis proteins were found exclusively in roots, and significantly accumulated by sulfur limitation, correlating with the induction of sulfate uptake activities. In the time course of short-term sulfate starvation treatment, SULTR1;1mycHis and SULTR1;2mycHis proteins were significantly accumulated during the 8- to 72-h period, causing substantial induction of sulfate uptake activities, while their corresponding mRNAs were expressed constantly around the initial levels, except for the transient induction in the first 2 h. This study suggested the importance of root-specific and sulfur deficiency-inducible accumulation of SULTR1;1 and SULTR1;2 sulfate transporter proteins for the acquisition of sulfate from low-sulfur environment.
Sulfur is a macronutrient required for plant growth. Plants usually assimilate the oxidized form of sulfur, sulfate anion, to generate essential sulfur-containing amino acids (Leustek et al., 2000; Saito, 2004). The plasma membrane-bound sulfate transporters mediate the acquisition of sulfate at the root surface to initiate this assimilatory metabolic process (Takahashi et al., 2006).
Sulfate uptake and Cys synthesis are activated under low-sulfur conditions where plants require high demands for sulfur metabolites. Up to the present, numerous molecular studies have been performed to characterize the sulfur nutritional responses of metabolic genes in plant sulfur assimilatory pathways. Sulfate transporters adenosine 5′-phosphosulfate reductases and Ser acetyltransferases are the representatives showing significant increase in their mRNAs under sulfur-deficient conditions (Smith et al., 1995, 1997; Gutierrez-Marcos et al., 1996; Takahashi et al., 1997). More recent analyses of transcriptomes of Arabidopsis (Arabidopsis thaliana) plants indicated that sulfur-responsive genes, including sulfate transporters and metabolic enzymes, are coordinately regulated, responding to the changes in sulfur conditions (Hirai et al., 2003, 2004; Maruyama-Nakashita et al., 2003, 2005, 2006; Nikiforova et al., 2003, 2005).
Particularly for the sulfate acquisition process, two high-affinity sulfate transporter genes, SULTR1;1 and SULTR1;2, showed clear responses to sulfur limitation at epidermis and cortex of Arabidopsis roots (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). The SULTR1;1 and SULTR1;2 mRNA contents were elevated during sulfur limitation in parallel with the increase in sulfate uptake capacities of roots (Vidmar et al., 2000; Shibagaki et al., 2002). These two sulfate transporters exhibited slight differences in mRNA inducibilities; SULTR1;1 mRNA was predominantly detected under sulfur-deficient conditions, whereas SULTR1;2 was found abundantly even when plants were supplied with adequate sulfate and was less responsive to the fluctuation of sulfur status compared to SULTR1;1 (Yoshimoto et al., 2002). Recent studies on transgenic Arabidopsis plants expressing the promoter-reporter systems for SULTR1;1 and SULTR1;2 indicated that accumulation of their transcripts during sulfur limitation depends on their promoter activities (Maruyama-Nakashita et al., 2004, 2005). In SULTR1;1, the sulfur-responsive cis-acting element was present at the −2,777/−2,762 region of its promoter and served as an essential regulatory sequence conferring the sulfur responsiveness (Maruyama-Nakashita et al., 2005). Although the sulfur-responsive element in SULTR1;2 promoter region has not been verified, an EIL-family transcription factor, SLIM1, plays a key role in regulating the expression of this major sulfate uptake facilitator (Maruyama-Nakashita et al., 2006). In addition to the physiological importance of the demand-driven regulation of sulfate uptake processes, sulfate transporters that facilitate root-to-shoot transport (Takahashi et al., 1997; Kataoka et al., 2004a), phloem transport (Yoshimoto et al., 2003), and remobilization of vacuolar sulfate storage (Kataoka et al., 2004b) were generally responsive to sulfur-limitation stress. It is likely that all these internal transport systems are systematically controlled at mRNA levels to have an optimized balance of sulfate allocation between tissues and organelles showing varying sulfur requirements along with the changes in developmental stages and metabolic states.
Contrary to the cumulative information on transcriptional control, no reports (to our knowledge) have ever demonstrated posttranscriptional control of mRNAs and proteins of sulfate transporters in plants from the perspective of their responses to the changes in environmental sulfur conditions. In this study, we demonstrate that posttranscriptional regulation plays an important role in both sulfur-responsive and organ-specific expression of SULTR1;1 and SULTR1;2 in Arabidopsis roots. These additional regulatory mechanisms can work in parallel with the on-offs of transcription switches for SULTR1;1 and SULTR1;2 gene expression and may have importance in adjustment of sulfate influx under varying sulfur conditions in the environment.
RESULTS
sultr1;1 sultr1;2 Double Knockout Mutant Lacks High-Affinity Sulfate Uptake Activity
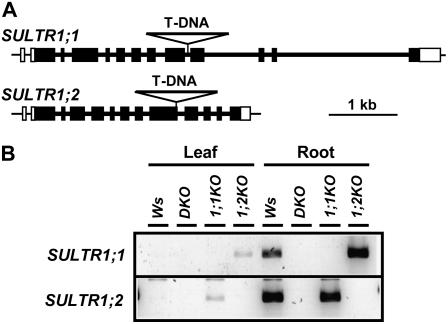
The physiological roles of SULTR1;1 and SULTR1;2 high-affinity sulfate transporters were confirmed by analyzing the phenotypes of knockout plants. We isolated transferred DNA (T-DNA) insertion lines (Arabidopsis Knockout Facility at the University of Wisconsin Biotech Center) by PCR-based reverse genetic strategy (Krysan et al., 1999). The mutant lines containing the T-DNA insertions in the ninth intron of SULTR1;1, and in the ninth exon of SULTR1;2, respectively, were used for the experiment (Fig. 1A). The sultr1;2 mutant used in this study is identical to the sel1-10 mutant that was previously reported (Maruyama-Nakashita et al., 2003). The sultr1;1 sultr1;2 double knockout mutant (DKO), which is deficient in both transport components, was generated by cross-fertilization of sultr1;1 (1;1KO) and sultr1;2 (1;2KO) knockout mutants. The mRNAs translatable to the functional SULTR1;1 and SULTR1;2 sulfate transporter proteins were completely absent in these mutants (Fig. 1B).
Figure 1.
Disruption of SULTR1;1 and SULTR1;2 genes by T-DNA insertion. A, Location of T-DNAs within the SULTR1;1 and SULTR1;2 genes. Thick bars and lines indicate exons and introns, respectively. White bars indicate the 5′- and 3′-untranslated regions. T-DNAs are not described in actual sizes. B, RT-PCR analysis of SULTR1;1 and SULTR1;2 in the Ws wild type and the DKO, 1;1KO, and 1;2KO mutants. RNA was extracted from leaves and roots of plants grown for 10 d on sulfur-sufficient medium containing 1,500 μm sulfate.
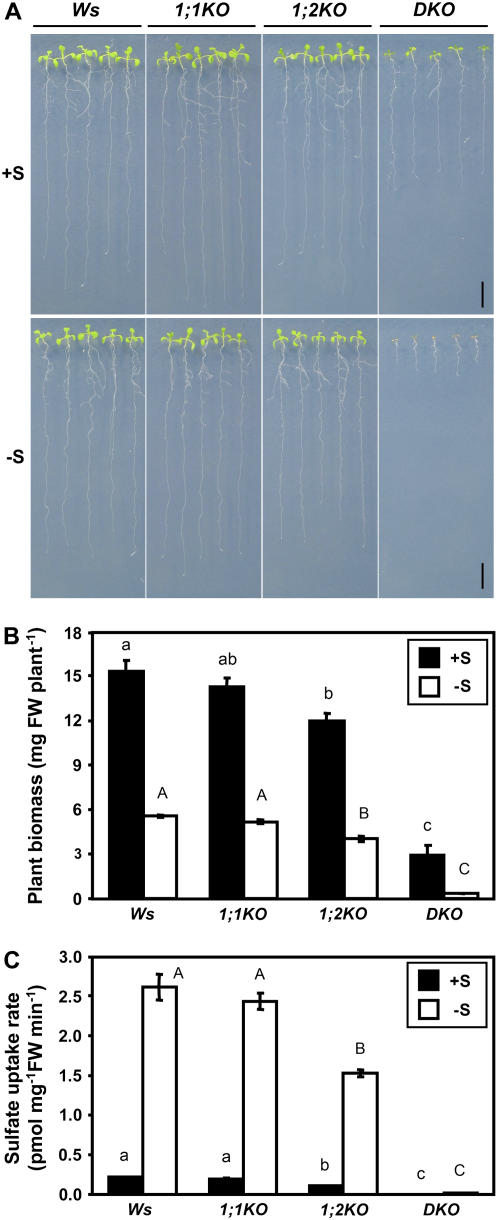
To analyze the effects of gene disruption of SULTR1;1 and SULTR1;2 on morphological phenotypes, Wassilewskija (Ws) wild-type, 1;1KO, 1;2KO, and DKO plants were grown vertically for 10 d on agar medium containing 1,500 or 5 μm sulfate as the sole sulfur source. The growth of DKO was inhibited under both sulfur-sufficient and -deficient conditions (Fig. 2A). In particular, the DKO plants were stunted and unable to expand their rosette leaves when grown with 5 μm sulfate (Fig. 2A). Ws and 1;1KO were visibly identical, and their plant biomasses were not significantly different from each other (Fig. 2B). However, 1;2KO plants showed slight growth retardation as compared to Ws, which resulted in a decrease in the plant biomass (Fig. 2B).
Figure 2.
Growth and sulfate uptake capacities of Ws, 1;1KO, 1;2KO, and DKO plants. A, Plants were grown for 10 d on agar medium containing 1,500 μm sulfate (+S) or 5 μm sulfate (−S) as the sole sulfur source. Scale bars = 1 cm. B, Biomass of plants. Plants were grown for 15 d on agar medium containing 1,500 μm sulfate (+S; black bars) or 5 μm sulfate (−S; white bars). Values are the means ± ses of four replicates of 20 seedlings. The fresh weights measured for 1;2KO and DKO were significantly different from Ws under both sulfur conditions. C, Sulfate uptake capacities of Ws, 1;1KO, 1;2KO, and DKO plants. Plants were grown for 15 d on agar medium containing 1,500 μm sulfate (+S; black bars) or 5 μm sulfate (−S; white bars), and incubated for 10 min in liquid medium containing 5 μm 35SO42−. Accumulation of radioactivities in the entire plants were determined (means ± ses, n = 6–8). Statistical differences among the genotypes were calculated for +S and −S conditions independently by one-way ANOVA. Different letters indicate means that were statistically different by Tukey's multiple-testing method (P < 0.05).
The activity of sulfate uptake was analyzed in Ws, 1;1KO, 1;2KO, and DKO plants (Fig. 2C). Plants were grown for 15 d on agar medium containing 1,500 or 5 μm sulfate, and sulfate uptake rates were measured in liquid medium containing 5 μm [35S]-labeled sulfate. DKO plants completely lacked the ability to take up sulfate (Fig. 2C), as expected from their growth defects on low-sulfate medium. The results demonstrate SULTR1;1 and SULTR1;2 are the two essential components of the sulfate uptake system that ultimately facilitates the absorption of sulfate externally supplied to the root system. In contrast to a complete lack of sulfate uptake in DKO under the low-sulfate condition, sulfate transport activities were substantially retained in 1;1KO and 1;2KO plants (Fig. 2C), indicating a single transport component, SULTR1;1 or SULTR1;2, is sufficient to facilitate the uptake of sulfate in Arabidopsis roots. The sulfate uptake rates of 1;2KO plants were 50% to 60% of those of the Ws wild-type plants (Fig. 2C). By contrast, 1;1KO plants showed no significant changes of sulfate uptake activities as compared with Ws plants, irrespective of the sulfur conditions of the preculture (Fig. 2C). However, contribution of SULTR1;1 to the uptake of sulfate was evident from the differences between DKO and 1;2KO.
Overexpression of SULTR1;1mycHis and SULTR1;2mycHis Epitope-Tagged Sulfate Transporters Rescues the Growth of DKO
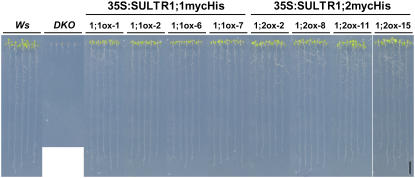
Using DKO as a parental line, we generated transgenic plants overexpressing epitope-tagged proteins of SULTR1;1 or SULTR1;2 under a constitutive promoter. The c-myc epitope and hexa-His tags were tandemly fused to the end of the coding region of SULTR1;1 and SULTR1;2 (SULTR1;1mycHis and SULTR1;2mycHis, respectively), and the fusion gene constructs were placed under the control of the cauliflower mosaic virus (CaMV) 35S RNA promoter. These fusion constructs were transformed to DKO plants, obtaining four independent, homozygous, single-insertion transgenic lines for each construct. Transgenic lines from both 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis constructs were able to rescue the growth of DKO parental plants on 5 μm sulfate medium (Fig. 3), indicating that both SULTR1;1mycHis and SULTR1;2mycHis can serve as functional sulfate transporters facilitating the uptake of trace amounts of sulfate (5 μm) in the culture medium. In particular, the growth of the aerial part was better improved with SULTR1;2mycHis than with SULTR1;1mycHis (Fig. 3).
Figure 3.
Overexpression of SULTR1;1mycHis and SULTR1;2mycHis rescues the growth of DKO plants. Ws wild type, DKO parental line, and 35S:SULTR1;1mycHis (1;1ox-1, 1;1ox-2, 1;1ox-6, and 1;1ox-7) and 35S:SULTR1;2mycHis (1;2ox-2, 1;2ox-8, 1;2ox-11, and 1;2ox-15) lines in DKO background were grown for 10 d on agar medium containing 5 μm sulfate as the sole sulfur source. Four independent transgenic lines from each construct were tested. Scale bar = 1 cm.
SULTR1;1mycHis and SULTR1;2mycHis Transcripts Are Predominantly Found in Roots
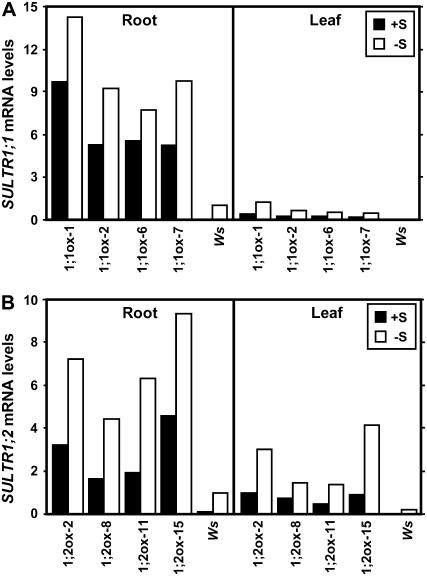
The SULTR1;1 and SULTR1;2 mRNA levels were analyzed in 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis transgenic plants and in Ws wild-type plants grown continuously for 15 d on agar medium containing 1,500 or 5 μm sulfate. The epitope-tagged and endogenous transcripts in transgenic and wild-type plants, respectively, were quantified by real-time reverse transcription (RT)-PCR using gene-specific primers for SULTR1;1 and SULTR1;2. In the roots of wild-type plants, mRNA levels of SULTR1;1 and SULTR1;2 in sulfur-starved plants (5 μm sulfate) were approximately 30 and 9 times higher, respectively, compared to those in the control plants (1,500 μm sulfate; Fig. 4). These sulfur responses were consistent with the results reported in previous studies (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002), which validates the experimental conditions where posttranscriptional regulation may take place in 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis transgenic lines.
Figure 4.
Accumulation of SULTR1;1 and SULTR1;2 transcripts by long-term sulfur limitation. A, SULTR1;1 mRNA levels in 35S:SULTR1;1mycHis lines (1;1ox-1, 1;1ox-2, 1;1ox-6, and 1;1ox-7) and Ws wild-type plants. B, SULTR1;2 mRNA levels in 35S:SULTR1;2mycHis lines (1;2ox-2, 1;2ox-8, 1;2ox-11, and 1;2ox-15) and Ws wild-type plants. Plants were continuously grown for 15 d on agar medium containing 1,500 μm sulfate (+S; black bars) or 5 μm sulfate (−S; white bars). The mRNA levels were determined by quantitative real-time RT-PCR using gene-specific primers for SULTR1;1 (A) and SULTR1;2 (B), and normalized using UBQ2 as described in the “Materials and Methods.” The mRNA levels are indicated as relative values calculated by comparison with the value of −S (5 μm sulfate) Ws root sample as 1.
CaMV 35S promoter is suggested to be constitutively active irrespective of plant organs; however, the accumulation of both SULTR1;1mycHis and SULTR1;2mycHis mRNAs occurred predominantly in root tissues (Fig. 4). SULTR1;1mycHis mRNA was detected mainly in the roots of all four 35S:SULTR1;1mycHis transgenic lines, showing 1.5- to 2-fold higher levels of mRNA accumulation in sulfur-starved plants than in the control plants grown with adequate sulfur supply (Fig. 4A). This small increase was statistically significant by paired Student's t test (P = 0.007); however, the rate of induction was rather limited as compared to the strong induction of the endogenous SULTR1;1 mRNA in the wild type. SULTR1;2mycHis mRNA was also regulated posttranscriptionally in 35S:SULTR1;2mycHis plants (Fig. 4B). The SULTR1;2mycHis mRNA was predominantly accumulated in the roots of all four transgenic lines, although the transcript was detected in both roots and leaves (Fig. 4B). As for the sulfur responsiveness, the SULTR1;2mycHis mRNA levels in roots were 2- to 3.5-fold higher in sulfur-starved plants than in the control plants (P = 0.003, paired Student's t test; Fig. 4B). The results indicated that SULTR1;1mycHis and SULTR1;2mycHis mRNAs are strictly regulated to accumulate in roots and are increased by long-term sulfur limitation, despite the fusion constructs were driven by constitutive CaMV 35S promoters.
SULTR1;1mycHis and SULTR1;2mycHis Proteins Are Accumulated Exclusively in the Roots of Sulfur-Starved Plants
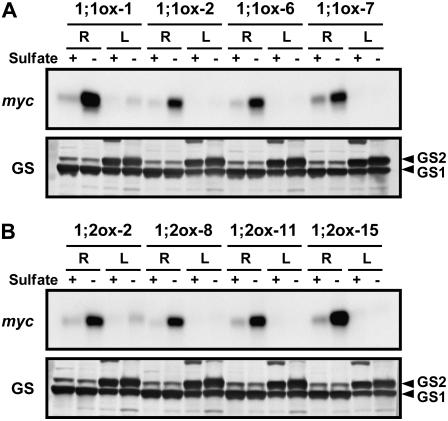
We further analyzed the effects of sulfur deprivation on protein levels of SULTR1;1mycHis and SULTR1;2mycHis. 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis transgenic lines were grown for 15 d on agar medium containing 1,500 or 5 μm sulfate, and used for preparation of crude protein extracts and subsequent western-blot analysis. Anti-myc antibody detected a specific band at approximately 59 kD in the extracts from 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis lines (Fig. 5, A and B), but not in the extract from Ws wild-type plants (data not shown). These specific bands migrated to somewhat lower positions than the predicted molecular masses of the SULTR1;1mycHis and SULTR1,2mycHis proteins (72.6 and 73.7 kD, respectively). High hydrophobicities of these membrane proteins may have resulted in faster migration on the gel.
Figure 5.
SULTR1;1mycHis and SULTR1;2mycHis proteins are exclusively accumulated in the roots of sulfur-starved plants. Protein-blot analysis was performed on crude protein extracts from roots (R) and leaves (L) of 35S:SULTR1;1mycHis lines (1;1ox-1, 1;1ox-2, 1;1ox-6, and 1;1ox-7; A) and 35S:SULTR1;2mycHis lines (1;2ox-2, 1;2ox-8, 1;2ox-11, and 1;2ox-15; B). Plants were grown on agar medium containing 1,500 μm sulfate (+) or 5 μm sulfate (−) for 15 d (same as in Fig. 4). The presence of SULTR1;1mycHis (A) and SULTR1;2mycHis (B) proteins was detected using anti-myc antibody. Each lane contains 5 μg of crude protein extract. Gln synthetases GS1 (lower band) and GS2 (upper band) serve as the loading controls.
Both SULTR1;1mycHis and SULTR1;2mycHis proteins were accumulated significantly in the roots under low-sulfate conditions (Fig. 5). Sulfur starvation caused drastic accumulation of SULTR1;1mycHis protein (Fig. 5A), which cannot be explained by the moderate increase of SULTR1;1mycHis mRNA (Fig. 4A). The results indicated SULTR1;1mycHis is regulated predominantly at the level of protein accumulation upon sulfur limitation. SULTR1;2mycHis protein was also accumulated exclusively in the roots of sulfur-starved plants (Fig. 5B). Although substantial quantities of SULTR1;2mycHis mRNAs were found in leaves as well as in roots of sulfur-starved plants (Fig. 4B), the signals for SULTR1;2mycHis proteins were hardly detectable in the leaves (Fig. 5B). In addition, the results indicated that accumulation of SULTR1;2mycHis protein by sulfur limitation (Fig. 5B) was significantly greater than the increase of SULTR1;2mycHis mRNA in roots (Fig. 4B).
Sulfur-Dependent Control of Sulfate Uptake in SULTR1;1mycHis and SULTR1;2mycHis Overexpressors
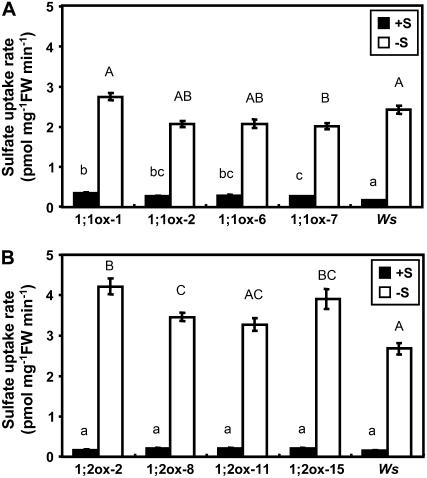
Sulfate uptake rates of transgenic and wild-type plants were measured to determine whether the sulfur-dependent posttranscriptional regulation of SULTR1;1mycHis and SULTR1;2mycHis causes the changes in the capacities of sulfate uptake. Plants were precultured for 15 d under sulfur-sufficient (1,500 μm sulfate) or -deficient (5 μm sulfate) conditions, and the uptake rates were measured in liquid medium containing 5 μm [35S]-labeled sulfate (Fig. 6).
Figure 6.
Sulfur-dependent regulation of the sulfate uptake activities of 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis plants. Sulfate uptake activities were measured in 5 μm 35SO42− liquid medium as described in Figure 2C using plants precultured for 15 d on 1,500 μm sulfate (+S; black bars) or 5 μm sulfate (−S; white bars; means ± ses, n = 6–8). The transgenic lines and culture conditions were the same as in Figures 4 and 5. A, Sulfate uptake rates of 35S:SULTR1;1mycHis lines (1;1ox-1, 1;1ox-2, 1;1ox-6, and 1;1ox-7) and Ws wild-type plants. B, Sulfate uptake rates of 35S:SULTR1;2mycHis lines (1;2ox-2, 1;2ox-8, 1;2ox-11, and 1;2ox-15) and Ws wild-type plants. Statistical differences among the wild-type and transgenic lines were calculated for +S and −S conditions independently by one-way ANOVA. Different letters indicate means that were statistically different by Tukey's multiple-testing method (P < 0.05).
When plants were precultured under sulfur-sufficient conditions, 35S:SULTR1;1mycHis transgenic lines showed 1.6 to 2.1 times greater capacities of sulfate uptake compared with the Ws wild-type plants (Fig. 6A, black bars). The sulfate uptake capacities of 35S:SULTR1;1mycHis transgenic lines increased about 7-fold by preculturing the plants under sulfur-deficient conditions. The same low-sulfate culture of Ws plants caused approximately 14-fold increase in sulfate uptake (Fig. 6A). Consequently, the 35S:SULTR1;1mycHis transgenic lines and Ws showed comparable levels of sulfate uptake when plants were precultured under 5 μm sulfate (Fig. 6A). The induction of sulfate uptake by sulfur limitation was fairly consistent with the accumulation of SULTR1;1mycHis proteins in transgenic lines (Fig. 5A); however, the case in Ws wild-type plants may also include the effects of the induction of SULTR1;1 and SULTR1;2 transcripts under the native promoters.
The SULTR1;2mycHis overexpressors showed slightly different responses. The sulfate uptake rates in 35S:SULTR1;2mycHis lines were around the same levels as in Ws wild-type plants when plants were precultured under sulfur-sufficient conditions. In three out of four 35S:SULTR1;2mycHis lines, the sulfate uptake rates were elevated by sulfur limitation more significantly than in Ws (Fig. 6B). The induction of sulfate uptake by sulfur limitation was 16- to 24-fold in 35S:SULTR1;2mycHis plants (Fig. 6B), which was higher than in the case of 35S:SULTR1;1mycHis plants (Fig. 6A). The induction of sulfate uptake was generally consistent with the accumulation of SULTR1;2mycHis proteins in roots (Fig. 5B).
Time-Course Induction of SULTR1;1mycHis and SULTR1;2mycHis by Sulfur Starvation
To examine the kinetics of the induction of SULTR1;1mycHis and SULTR1;2mycHis, a time-course experiment was performed. Plants were grown for 12 d on sulfur-replete agar medium containing 1,500 μm sulfate, and then transferred onto agar medium containing no sulfur source, harvesting root samples 0, 2, 8, 24, and 72 h after the transfer. The mRNA and protein levels of SULTR1;1mycHis or SULTR1;2mycHis were determined during this time course.
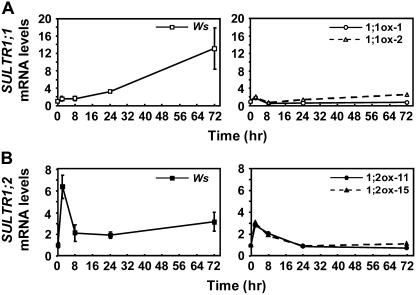
In Ws wild-type plants, SULTR1;1 mRNA increased continuously during the time course of sulfate starvation, resulting in 13-fold induction in 72 h (Fig. 7A, left). On the other hand, SULTR1;2 mRNA showed a transient increase in the first 2 h but was resumed at 8 h, maintaining its level around 2- to 3-fold of the initial level during the remaining period of sulfur starvation (Fig. 7B, left). Except for this transient induction of SULTR1;2 at 2 h, the accumulation of SULTR1;1 and SULTR1;2 mRNAs during the time course of sulfur limitation from 8 to 72 h was generally consistent with the results of previous reports (Takahashi et al., 2000; Yoshimoto et al., 2002).
Figure 7.
Time-course analysis of SULTR1;1 and SULTR1;2 transcript levels during short-term sulfate starvation. A, SULTR1;1 mRNA levels in Ws wild-type plants (left) and 35S:SULTR1;1mycHis lines (1;1ox-1 and 1;1ox-2; right). B, SULTR1;2 mRNA levels in Ws wild-type plants (left) and 35S:SULTR1;2mycHis lines (1;2ox-11 and 1;2ox-15; right). Plants were grown for 12 d on agar medium containing 1,500 μm sulfate, and then incubated for 0, 2, 8, 24, and 72 h on agar medium containing no sulfur source. Total RNA was extracted from the root tissues, and quantitative real-time RT-PCR was performed using gene-specific primers for SULTR1;1 (A) and SULTR1;2 (B). The mRNA levels were determined by fitting Ct values to standard curves and normalized by UBQ2 as described in the “Materials and Methods.” Means of independent triplicate RNA samples and sd values are shown for Ws wild-type plants. For 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis plants, two independent transgenic lines from each construct were analyzed to confirm the reproducibility of the results. The mRNA levels are indicated as relative values calculated by comparison with the value of the 0-h sample in each line as 1.
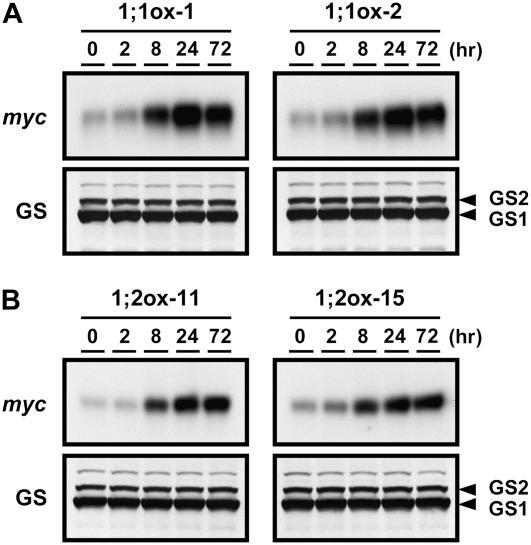
In 35S:SULTR1;1mycHis and 35S:SUTLR1;2mycHis transgenic plants, SULTR1;1mycHis and SULTR1;2mycHis transcripts were transiently accumulated (2- to 3-fold), peaking 2 h after the transfer (Fig. 7, A and B, right); however, this temporal induction was not reflected to protein accumulation (Fig. 8, A and B). During the period from 8 to 72 h, the transcript levels of both SULTR1;1mycHis and SULTR1;2mycHis shifted constantly around the initial level. The results were consistent between the two independent transgenic lines of each construct (Fig. 7, A and B). Contrary to this static behavior of mRNA accumulation during the 8- to 72-h period, the abundance of both SULTR1;1mycHis and SULTR1;2mycHis proteins drastically increased at 8 h and reached the maximum at 24 h after the withdrawal of sulfate (Fig. 8, A and B). It was evident that the accumulation of SULTR1;1mycHis and SULTR1;2mycHis proteins in the overexpressor plants occurs rapidly and effectively by short-term sulfate starvation, although the transgenes were expressed under the constitutive CaMV 35S promoters.
Figure 8.
SULTR1;1mycHis and SULTR1;2mycHis proteins are accumulated by short-term sulfate starvation. The transgenic lines and culture conditions were the same as in Figure 7. A, SULTR1;1mycHis proteins in 35S:SULTR1;1mycHis lines (1;1ox-1 and 1;1ox-2). B, SULTR1;2mycHis proteins in 35S:SULTR1;2mycHis lines (1;2ox-11 and 1;2ox-15). One microgram (1;1ox-1) or 5 μg (1;1ox-2, 1;2ox-11, and 1;2ox-15) of crude proteins from root tissues were loaded to each lane of an SDS-polyacrylamide gel, and the accumulation of SULTR1;1mycHis or SULTR1;2mycHis proteins was detected using anti-myc antibody. Gln synthetases GS1 (lower band) and GS2 (upper band) serve as the loading controls.
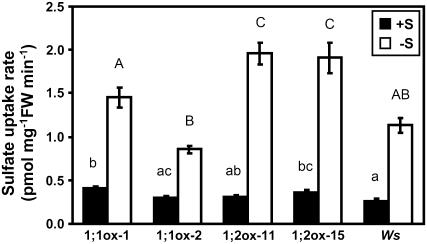
The physiological relevance of protein accumulation was confirmed by measurements of sulfate uptake rates in 35S:SULTR1;1mycHis and 35S:SUTLR1;2mycHis transgenic plants. Transgenic and Ws wild-type plants were precultured for 12 d on agar medium containing 1,500 μm sulfate and subjected to 24-h sulfur starvation. Plants that were not subjected to this short-term sulfur-starvation treatment were used as controls. Both the wild-type and transgenic plants showed nearly the same levels of sulfate uptake under the control condition (Fig. 9, +S). Under sulfur-starved conditions (Fig. 9, −S), two independent lines of 35S:SULTR1;1mycHis plants, 1;1ox-1 and 1;1ox-2, showed 3.6- and 2.8-fold increase in sulfate uptake rates, respectively. Those were comparable to the induction of sulfate uptake observed in wild-type plants. In 35S:SULTR1;2mycHis plants, the induction of sulfate uptake by 24-h sulfur starvation was significantly higher than in the wild type. The induction of sulfate uptake rates were 6.4- and 5.3-fold for line 1;2ox-11 and line 1;2ox-15, respectively.
Figure 9.
Sulfate uptake activities are increased by short-term sulfur starvation in 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis plants. The experiments were carried out in the same transgenic lines used for the RT-PCR (Fig. 7) and protein-blot analysis (Fig. 8). 35S:SULTR1;1mycHis lines (1;1ox-1 and 1;1ox-2), 35S:SULTR1;2mycHis lines (1;2ox-11 and 1;2ox-15), and Ws wild-type plants were grown for 12 d on agar medium containing 1,500 μm sulfate (+S; black bars), and then incubated for 24 h on agar medium without sulfur source (−S; white bars). Sulfate uptake activities were measured in 5 μm 35SO42− liquid medium as described in Figure 2C (means ± ses, n = 10). Induction of sulfate uptake by 24-h sulfur starvation was statistically significant (P < 0.00001, Student's t test) in all transgenic lines and wild-type plants. Statistical differences among the wild type and transgenic lines were calculated for +S and −S conditions independently by one-way ANOVA. Different letters indicate means that were statistically different by Tukey's multiple-testing method (P < 0.05).
In both SULTR1;1mycHis and SULTR1;2mycHis overexpressors, drastic increase of sulfate uptake by sulfur limitation occurred even with constitutive CaMV 35S promoters (Figs. 6 and 9). The induction of sulfate uptake was generally consistent with the increase in protein levels in both long-term (Fig. 5) and short-term (Fig. 8) sulfur-starvation treatments.
DISCUSSION
Induction of sulfate uptake is the primary response to sulfur-starvation stress. Previously we have reported that SULTR1;1 and SULTR1;2 high-affinity sulfate transporters are expressed at the cortical and epidermal cell layers of Arabidopsis roots (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002), and their transcripts are strictly regulated by sulfur status under the control of sulfur-responsive sequences in the promoter regions (Maruyama-Nakashita et al., 2004, 2005). In this report, we proved that SULTR1;1 and SULTR1;2 substantially function as two essential components of the high-affinity sulfate uptake system necessary for the acquisition of sulfate from the environment. We further demonstrated that both SULTR1;1 and SULTR1;2 are controlled posttranscriptionally mainly at the levels of protein accumulation in roots responding to the changes in environmental sulfur conditions. This study provides new regulatory schemes of plant sulfur assimilation in addition to the previously reported transcriptional regulation of the sulfate uptake systems.
We first clarified the functional significance of SULTR1;1 and SULTR1;2 for the acquisition of sulfate by analyzing the sulfate uptake capacities of the knockout mutants. DKO plants failed to absorb sulfate from low-sulfate medium (Fig. 2), demonstrating that the high-affinity sulfate transporters SULTR1;1 and SULTR1;2 are essentially required for the uptake of micromolar sulfate from the rhizosphere. In contrast to the severe growth defects and complete loss of sulfate uptake in DKO, both 1;1KO and 1;2KO were viable and retained substantial capacities to take up sulfate from low-sulfate medium (Fig. 2). These results suggested that SULTR1;1 and SULTR1;2 may have overlapping functions and act independently for the uptake of sulfate when either of them is deleted. In fact, overexpression of SULTR1;1mycHis or SULTR1;2mycHis alone conferred sulfate uptake activity in DKO plants (Figs. 3 and 6), suggesting a single transporter component is sufficient for the acquisition of sulfate. Comparison of 1;1KO and 1;2KO suggested that the contribution of SULTR1;2 to total sulfate uptake predominates over the function of SULTR1;1 (Fig. 2C), reflecting the differences of their transcript abundance. Furthermore, we found that overexpression of SULTR1;1mycHis and SULTR1;2mycHis show slight differences in rescuing the sulfate uptake capacities of DKO; the 35S:SULTR1;2mycHis plants were able to absorb sulfate more efficiently compared to the 35S:SULTR1;1mycHis plants during sulfur starvation (Figs. 6 and 9). Consistent with the differences in sulfate uptake rates, recovery of growth was more substantial in 35S:SULTR1;2mycHis than in 35S:SULTR1;1mycHis plants (Fig. 3). It is suggested that SULTR1;2 protein may have a higher capacity for sulfate uptake as compared with SULTR1;1.
The posttranscriptional regulation of SULTR1;1 and SULTR1;2 was determined in transgenic plants overexpressing the epitope-tagged proteins SULTR1;1mycHis and SULTR1;2mycHis in DKO background. Although the epitope-tagged transporters were expressed under the constitutive CaMV 35S promoters, SULTR1;1mycHis and SULTR1;2mycHis mRNAs were predominantly found in roots (Fig. 4). Furthermore, the mRNA levels were slightly increased by a long-term continuous low-sulfur culture (15 d from germination). It was previously reported that the levels of CaMV 35S-driven GFP are unaffected by long-term sulfur limitation (Ohkama et al., 2002). Therefore, the increase of SULTR1;1mycHis and SULTR1;2mycHis mRNAs on long-term sulfur limitation (Fig. 4) may have some relevance to up-regulation of sulfate uptake in response to sulfur starvation, although the effects are suggested to be limited. In fact, the increase of transcripts was rather moderate as compared with the drastic changes in protein accumulation (Fig. 5), which largely contributed to the induction of sulfate uptake capacities (Fig. 6).
We further performed time-course sulfur-starvation experiments to demonstrate the significance of posttranscriptional regulation of sulfate transporters in short time periods. For the time-course experiment, plants were cultured for 12 d with adequate sulfur supply before transfer to sulfur-less medium. Therefore, the results indicated here represent the comparison of plants at the same developmental stages. SULTR1;1mycHis and SULTR1;2mycHis proteins started to accumulate no later than 8 h after the transfer and reached the maximum levels in 24 h, whereas the levels of their corresponding mRNAs showed no significant increase under the same conditions (Figs. 7 and 8). It is suggested that accumulation of SULTR1;1mycHis and SULTR1;2mycHis proteins during sulfur starvation can be caused by increased translation or by changes in protein turnover/stability. By contrast, accumulation of SULTR1;1mycHis and SULTR1;2mycHis mRNAs occurred under prolonged low-sulfate stress (Fig. 4) or transiently after the replacement of culture medium (Fig. 7, 2 h), the latter of which had no relevance to protein accumulation (Fig. 8). The time-course experiments further indicated that the accumulation of SULTR1;1mycHis or SULTR1;2mycHis proteins in transgenic plants occurs rapidly and even more significantly (Fig. 8) compared with the up-regulation of SULTR1;1 or SULTR1;2 transcripts in Ws wild-type plants (Fig. 7, A and B, left). These results suggest that control of protein level is a dominant mechanism to optimize the uptake of sulfate in response to sulfur deficiency. The mechanism proposed here controls the amounts of SULTR1;1 and SULTR1;2 sulfate transporters and may have close linkage to the regulation of the sulfate uptake process in roots.
The physiological significance of this regulatory mechanism was manifested by the induction of sulfate uptake (Figs. 6 and 9) that occurred in parallel with the increase in SULTR1;1mycHis and SULTR1;2mycHis proteins in sulfur-starved plants (Figs. 5 and 8). The conditions where SULTR1;1mycHis and SULTR1;2mycHis proteins accumulated in overexpressors were identical with those necessary for the promoter-dependent induction of the endogenous SULTR1;1 and SULTR1;2 transcripts (Maruyama-Nakashita et al., 2004, 2005). Under these conditions, Arabidopsis plants strictly control the expression of SULTR1;1 and SULTR1;2, increasing the transcript levels to induce de novo synthesis of functional sulfate transporter proteins that facilitate the uptake of sulfate. The results presented in this study clearly suggested the existence of an additional and efficient mechanism that drives protein accumulation of SULTR1;1 and SULTR1;2, fulfilling the demands to elevate the sulfate uptake capacities upon sulfur deficiency.
Sulfate transporters contain a conserved region named STAS (sulfate transporter and anti-sigma factor antagonist) domain at their C terminus. The STAS domain shares significant similarity with the Bacillus sp. anti-sigma factor, and is suggested to function as a domain for protein-protein interaction, which may play a role in regulating the activity and/or stability of sulfate transporters. The role of the STAS domain in the control of both activity and biosynthesis/stability of SULTR1;2 has been studied using yeast cells that express SULTR1;2 proteins containing no or mutated STAS domains (Rouached et al., 2005; Shibagaki and Grossman, 2004, 2006). Mutation of the Thr residue in the STAS domain resulted in complete reduction of sulfate transport activity of SUTLR1;2, which may suggest that phosphorylation of the STAS domain is a key factor for modulation of SULTR1;2 activity (Rouached et al., 2005). Functional interaction between multiple sulfate transporter isoforms might be another possible posttranscriptional regulatory mechanism that may modulate sulfate transport activities. Arabidopsis SULTR3;5 sulfate transporter, which has no sulfate transport activity itself, significantly up-regulated the sulfate transport capacity of SULTR2;1 low-affinity sulfate transporter when both were coexpressed in yeast (Kataoka et al., 2004a). It is suggested that SULTR3;5 might play roles in stabilizing or activating SULTR2;1, presumably by forming a heteromeric sulfate transporter complex. The sulfate uptake process can be regulated additionally by these posttranslational mechanisms. However, the physiological relevance of these mechanisms is still unverified.
Besides the regulation of sulfate transporters demonstrated in this study, multiplicity of regulatory mechanisms has been reported for other nutrient uptake systems. The root-specific and iron deficiency-inducible expression of Arabidopsis IRT1, the major transporter for high-affinity iron uptake, was controlled at the levels of both transcription and protein accumulation (Connolly et al., 2002). For nitrate acquisition, the presence of ammonium-dependent decrease or inactivation of NRT2.1 high-affinity nitrate transporter was suggested from the analysis of transgenic Nicotiana plumbaginifolia plants overexpressing NRT2.1 under control of the constitutive promoter (Fraisier et al., 2000). This mechanism seems to work in parallel with down-regulation of endogenous NRT2.1 transcript under ammonium-excess conditions. The posttranscriptional mechanisms might be a general phenomenon in plant nutrient response.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown on agar medium (Fujiwara et al., 1992) at 22°C under 16-h-light/8-h-dark cycles. Sulfate-deficient medium was prepared by replacing sulfate salts contained in the medium with equivalent chloride salts as described previously (Hirai et al., 1995).
The Arabidopsis sultr1;1 mutant (Wisconsin T-DNA insertion lines, Ws accession, available from the Arabidopsis Biological Resource Center) was isolated by PCR screening as described by Krysan et al. (1999). The integration site of the T-DNA in SULTR1;1 was determined by sequencing the PCR product amplified with the SULTR1;1-specific primer and T-DNA border primer. The sultr1;1 sulr1;2 double mutant was generated by cross-fertilization of sultr1;1 and sultr1;2 (Maruyama-Nakashita et al., 2003) mutants.
RT-PCR
Total RNA was extracted using the RNeasy plant mini kit (Qiagen) and treated with DNase I (Invitrogen). RT was carried out using OmniScript reverse transcriptase (Qiagen) and oligo-d(T)12-18. Knockouts of SULTR1;1 and SULTR1;2 (Fig. 1) were confirmed by RT-PCR using the primer pairs described by Yoshimoto et al. (2002). The PCR products were separated in agarose gels and stained with SYBR green (Takara). Signals were detected using FluorImager 595 (Molecular Dynamics) with a 515- to 545-nm band-pass filter.
Real-time RT-PCR was performed for the quantification of SULTR1;1 and SULTR1;2 transcript levels (Figs. 4 and 7). First-strand cDNA was synthesized from 1 μg of total RNA using oligo-d(T)12-18 and OmniScript reverse transcriptase (Qiagen). The first-strand cDNA that derives from 10 ng of total RNA was used for real-time PCR. The reaction was performed using SYBR Premix Ex Taq (Takara), and the signals were detected with 7500 Fast Real-Time PCR system (Applied Biosystems). The SULTR1;1mycHis and SULTR1;2mycHis mRNA levels were determined using the gene-specific primer pairs for SULTR1;1, SULTR1;2, and UBIQUITIN2 (UBQ2), reported previously by Maruyama-Nakashita et al. (2004). Plasmid DNAs harboring SULTR1;1, SULTR1;2, and UBQ2 cDNAs, respectively, were used to generate the standard curves of Ct values versus the amounts of target cDNAs. The standard plasmid DNAs ranging from 0.1 fg to 10 pg were used as templates to generate the standard curves. The amounts of SULTR1;1, SULTR1;2, and UBQ2 mRNAs in each sample were calculated from these standard curves. The mRNA levels of SULTR1;1 and SULTR1;2 were normalized by those of UBQ2 as an internal control.
Transgenic Plants Expressing SULTR1;1mycHis and SULTR1;2mycHis
Molecular biological experiments were carried out according to the standard protocols (Sambrook et al., 1989). The fusion gene of CaMV 35S RNA promoter and SULTR1;1mycHis (35S:SULTR1;1mycHis) was constructed as follows. The 3′ end of the coding region of SULTR1;1 cDNA was tagged with myc-epitope and amplified by PCR using oligonucleotide primers 1;1-SalI (5′-GTCGACATGTCCGGGACTATTAATCCCCCGGA-3′) and 1;1-myc-R (5′-CAGGTCCTCCTCTGAGATCAGCTTCTGCTCAGTTTGTTGCTCAGCCACT-3′). The SULTR1;1myc fragment, the product of first PCR, was subsequently tagged with six His residues and reamplified by PCR using oligonucleotides 1;1-SalI and SacI-His-R (5′-GAGCTCTCAATGGTGATGGTGATGATGCAGGTCCTCCTCTGAGATC-3′). The underlined sequences in the 1;1-SalI and SacI-His-R primers correspond to the SalI and SacI restriction sites used for fusion construction. The resultant PCR-amplified fragment of SULTR1;1mycHis was cloned into pCR-BluntII-TOPO (Invitrogen) and fully sequenced. The HindIII-SalI fragment of CaMV 35S promoter from pTH2 vector (Chiu et al., 1996) and the SalI-SacI fragment of SULTR1;1mycHis were ligated to obtain the HindIII-SacI fragment of the 35S:SULTR1;1mycHis fusion cassette.
The 35S:SULTR1;2mycHis fusion was constructed as follows. CaMV 35S promoter fragment from pTH2 vector (Chiu et al., 1996) and the coding region of SULTR1;2 cDNA was fused by adaptamer-mediated PCR. The 3′ end of the CaMV 35S promoter and the 5′ end of the SULTR1;2 coding region were fused using a set of oligonucleotide primers, 35Somega-1;2-F (5′-CAGTCGACTCTAGAGGATCCATGTCGTCAAGAGCTCACCCTGTGGACG-3′) and 35Somega-1;2-R (5′-TGAGCTCTTGACGACATGGATCCTCTAGAGTCGACTGTAATTGTAAATA-3′), followed by amplification of 35S:SULTR1;2 fusion. The 3′ end of the 35S:SULTR1;2 fusion gene was tagged with myc-epitope and amplified by PCR using oligonucleotide primers pBI-35Somega-F (5′-AAGCTTGCATGCCTGCAGATCCCCCCTCAGAAGACC-3′) and 1;2-myc-R (5′-CAGGTCCTCCTCTGAGATCAGCTTCTGCTCGACCTCGTTGGAGAG-3′). The 35S:SULTR1;2myc fusion was subsequently tagged with six His residues and reamplified by PCR using the pBI-35Somega-F and SacI-His-R primers. pBI-35Somega-F and SacI-His-R contain HindIII and SacI restriction sites (underlined), respectively, for the fusion construction. The final PCR product that corresponded to the 35S:SULTR1;2mycHis fusion cassette was cloned into pCR-BluntII-TOPO (Invitrogen) and fully sequenced.
The kanamycin selection marker in pBI101 (CLONTECH) was replaced with the hygromycin resistance gene. The NheI-HindIII fragment covering the 3′-end region of nopaline synthase promoter, hygromycin phosphotransferase coding region, and polyadenylation signal of Arabidopsis RbcS-2B gene was cut out from the pSMAH vector (provided by Dr. H. Ichikawa, National Institute of Agrobiological Sciences, Japan), and inserted between the NheI and HindIII sites in pBI101 (CLONTECH) for reconstruction of nopaline synthase promoter at the NheI site and for complete replacement of neomycin phosphotransferase II and nopaline synthase terminator with hygromycin phosphotransferase and RbcS-2B terminator. This hygromycin-resistant binary vector was used for plant transformation. The HindIII-SacI fragments of the 35S:SULTR1;1mycHis and 35S:SULTR1;2mycHis fusion cassettes were inserted between the HindIII and SacI sites of this binary vector, replacing the GUS gene.
The resulting binary plasmids were transferred to Agrobacterium tumefaciens GV3101(pMP90) (Koncz and Schell, 1986) by the freeze-thaw method (Höfgen and Willmitzer, 1988). Arabidopsis sultr1;1 sultr1;2 double mutant plants were transformed according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on agar medium containing half-strength Murashige and Skoog salts (Murashige and Skoog, 1962), 1% (w/v) Suc, and 25 mg L−1 hygromycin B. All experiments were conducted using homozygous T3 transgenic segregants containing a single T-DNA insertion.
Protein Extraction and Immunoblot Analysis
Total protein was prepared from the roots and leaves of plants grown vertically on agar medium containing 1,500 or 5 μm sulfate as the sole sulfur source. Tissues were ground under liquid N2 and homogenized in the extraction buffer [50 mm Tris-MES, pH 7.5, 300 mm Suc, 150 mm NaCl, 10 mm CH3COOK, 5 mm EDTA, 20 mm leupeptin, 10 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 200 mm phenylmethylsulfonyl fluoride]. After centrifugation at 9,000g for 10 min to remove cell debris, the supernatant was used as a total protein fraction. Protein content was determined by the Coomassie Blue dye-binding method (Bio-Rad) using bovine serum albumin as a standard.
To detect SULTR1;1mycHis and SULTR1;2mycHis proteins, total protein was mixed with the SDS-PAGE sample buffer and incubated for 30 min at 4°C, and applied to the 7.5% (w/v) polyacrylamide gel. The amount of total protein used for the analysis is described in the figure legends. Protein was separated in the gel and transferred to Hybond-P (Amersham Biosciences). The blot was incubated with anti-myc mouse monoclonal antibody (Invitrogen), followed by the incubation with goat anti-mouse IgG conjugated to alkaline phosphatase (Promega). As a control for equal loading, two different isozymes of Gln synthetase, GS1 and GS2, were detected using anti-GS1 polyclonal antibody and goat anti-rabbit IgG alkaline phosphatase conjugate (Promega) as described previously by Ishiyama et al. (2004). GS1 (40 kD) is constitutively expressed in leaves and roots, whereas GS2 (44 kD) is present predominantly in leaves. The presence of immunoreactive protein was detected by ECL chemiluminescence (Amersham Bioscience).
35SO42− Influx Assay
Experiments were carried out according to the method described by Kataoka et al. (2004b) and Maruyama-Nakashita et al. (2004) with minor modifications. For the experiments of long-term continuous sulfur deficiency (Fig. 6), plants were germinated on nylon mesh and grown vertically for 15 d on agar medium containing 1,500 or 5 μm sulfate as the sole sulfur source. For the experiments of short-time sulfur limitation (Fig. 9), plants were grown for 12 d on the sulfur-replete agar medium containing 1,500 μm sulfate, and then transferred on agar medium without any sulfur source and incubated for 24 h before the analysis. Labeling was started by submerging the roots into liquid nutrient medium (Hirai et al., 1995) containing 5 μm Na235SO4 (Amersham Bioscience). After 10 min of incubation, plants were rinsed twice in nonlabeled liquid medium and left 1 h in the nonlabeled liquid medium to wash out the radioactivities in the apoplasm. Plants were weighed in scintillation vials and covered with 0.1 n HCl. The vials were left overnight for extraction. Scintillation cocktail (Perkin-Elmer) was added to the extract, and the incorporated radioactivity was determined in a scintillation counter (Aloka).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB018695 (SULTR1;1), AB042322 (SULTR1;2), and J05508 (UBQ2).
Acknowledgments
We thank the Arabidopsis Biological Resource Center and the Arabidopsis Knockout Facility of the University of Wisconsin Biotech Center for providing T-DNA insertion mutants. We thank Dr. Hiroaki Ichikawa (National Institute of Agrobiological Sciences, Japan) for providing the pSMAH vector, and Dr. Yasuo Niwa (University of Shizuoka, Japan) for providing the pTH2 vector. We also thank Dr. Soichi Kojima (RIKEN Plant Science Center) for useful discussions on statistical analysis.
This work was supported in part by Special Postdoctoral Fellowship of RIKEN (to N.Y.) and Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hideki Takahashi (hideki@riken.jp).
Open Access articles can be viewed online without a subscription.
References
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6 325–330 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisier V, Gojon A, Tillard P, Daniel-Vedele F (2000) Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J 23 489–496 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Hirai MY, Chino M, Naito S (1992) Effects of sulfate nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol 99 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL (1996) Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA 93 13377–13382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Chino M, Naito S (1995) Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol 36 1331–1339 [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama K, Inoue E, Watanabe-Takahashi A, Obara M, Yamaya T, Takahashi H (2004) Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J Biol Chem 279 16598–16605 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004. a) Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H (2004. b) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of T-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick J, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51 141–166 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42 305–314 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H (2004) A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J 38 779–789 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R (2005) Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J Exp Bot 56 1887–1896 [DOI] [PubMed] [Google Scholar]
- Ohkama N, Goto DB, Fujiwara T, Naito S (2002) Differential tissue-specific response to sulfate and methionine of a soybean seed storage protein promoter region in transgenic Arabidopsis. Plant Cell Physiol 43 1266–1275 [DOI] [PubMed] [Google Scholar]
- Rouached H, Berthomieu P, El Kassis E, Cathala N, Catherinot V, Labesse G, Davidian JC, Fourcroy P (2005) Structural and functional analysis of the C-terminal STAS (sulfate transporter and anti-sigma antagonist) domain of the Arabidopsis thaliana sulfate transporter SULTR1.2. J Biol Chem 280 15976–15983 [DOI] [PubMed] [Google Scholar]
- Saito K (2004) Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol 136 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning. A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Shibagaki N, Grossman AR (2004) Probing the function of STAS domains of the Arabidopsis sulfate transporters. J Biol Chem 279 30791–30799 [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Grossman AR (2006) The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J Biol Chem 281 22964–22973 [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29 475–486 [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, Vanden Berg PJ, Belcher AR, Warrilow AGS (1997) Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J 12 875–884 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J 23 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, de Almeida Engler J, Engler G, Van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yoshimoto N, Saito K (2006) Anionic nutrient transport in plants: the molecular bases of sulfate transporter gene family. In JK Setlow, ed, Genetic Engineering, Principles and Methods, Vol 27. Springer, New York, pp 67–80 [DOI] [PubMed]
- Vidmar JJ, Tagmount A, Cathala N, Touraine B, Davidian JE (2000) Cloning and characterization of a root specific high-affinity sulfate transporter from Arabidopsis thaliana. FEBS Lett 475 65–69 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H (2003) Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol 131 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K (2002) Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J 29 465–473 [DOI] [PubMed] [Google Scholar]