To study gene function, generation of loss-of-function mutants by RNA interference (RNAi) approaches and T-DNA insertional mutagenesis are widely used in plant science and have proven to be very successful in determining gene functions. AGAMOUS-LIKE18 (AGL18) is a MADS-box gene that is expressed during Arabidopsis (Arabidopsis thaliana) pollen development (Alvarez-Buylla et al., 2000; Pina et al., 2005). To reveal a function for AGL18, different transgenic AGL18 RNAi populations were produced and a T-DNA insertion line from the Salk collection (Alonso et al., 2003) was analyzed in parallel. Surprisingly, a pollen lethality phenotype found in many RNAi lines was not detectable in the AGL18 T-DNA knockout mutant. To investigate these contradicting results, we made a series of transgenic lines using different vectors, promoters, and reporter genes. Here, we present data proving that the abnormal pollen development observed in RNAi AGL18 plants is not caused by loss of AGL18 function but is a common feature occurring in transgenic RNAi populations.
Three different promoters, depicted in Figure 1A, were fused to the AGL18 RNAi cassette, namely, the cauliflower mosaic virus 35S promoter, the AGL18 promoter (Lehti-Shiu et al., 2005) and the pollen-specific DEFH125 promoter (Lauri et al., 2006). These RNAi constructs were transformed into Arabidopsis wild-type plants using Agrobacterium strain GV3101. Among the 300 T1 plants examined from each transgenic population, we found that more than 20% of the plants failed to produce normal, wild type-like pollen. Instead, they developed anthers in which approximately 20% to 50% of the pollen was aborted by the time the anthers dehisced (Fig. 2; Table I). Except for the pollen defects (Fig. 2), these plants revealed a normal vegetative and reproductive development, producing wild type-like siliques under our growth conditions (data not shown). To determine whether the phenotype observed in these RNAi lines correlates with a reduced AGL18 expression, we performed semiquantitative reverse transcription-PCRs with six transgenic T1 plants from one AGL18∷AGL18 RNAi population, all producing 50% aborted pollen (Fig. 1A). As shown in Figure 1D, AGL18 transcription varied among the different individuals, and wild type-like as well as reduced AGL18 expression levels were observed. No AGL18 transcript was detectable in the agl18-3 T-DNA mutant (Fig. 1, B and C), strongly suggesting that it represents a loss-of-function mutant. However, in contrast to the RNAi lines, agl18-3 anthers produce wild type-like pollen, similar to two other recently analyzed agl18 knockout lines (Lehti-Shiu et al., 2005). Taken together, these data indicate that the pollen lethality phenotype observed in AGL18 RNAi plants is likely not related to loss of AGL18 function.
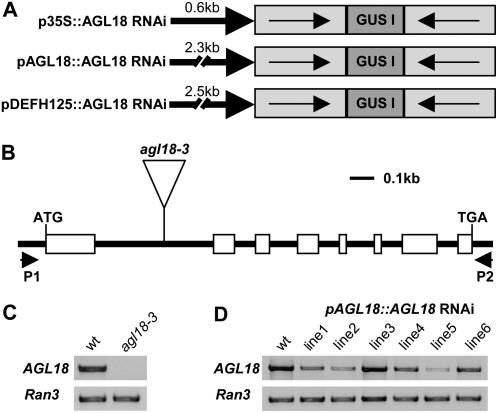
Figure 1.
Gene structure and expression analysis. A, AGL18 RNAi constructs (derivatives of the binary RNAi vector pGSA1252; http://chromdb.org/rnai). AGL18 cDNA (without MADS box, gray boxes) in sense (→) and antisense (←) orientation, separated by a 370-bp GUS intron (GUS I, green box), was cloned into the RNAi cassette driven by three different promoters (bold arrows, promoter length indicated above). B, AGL18 gene structure and T-DNA insertion site of the agl18-3 (NASC ID, N875156) allele. P1 (5′-TAATCTCTTCTCTCTATATCTCTTCTC-3′) and P2 (5′-AGATGAAATAAAGCAAAAGAACAGCCAG-3′) are primers used for reverse transcription-PCR in C and D. C and D, Comparison of AGL18 transcript level between wild-type Columbia plants, agl18-3 T-DNA, and six different RNAi T1 mutants, harboring the AGL18∷AGL18 RNAi construct. Total RNA was isolated from inflorescences of 6-week-old plants grown under long-day conditions (16 h light/8 h dark, 22°C). cDNA was produced from 4 μg of total RNA, treated with RNase-free DNase I (Roche) and using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. PCRs were performed with 31 cycles. Five microliters from 25-μL reactions were loaded on ethidium bromide-stained 1.0% (w/v) agarose gels. Images were scanned with the phosphor imager Typhoon 8600 (Amersham Biosciences). As a control, Ran3 (At5g55190) was amplified using the primers 5′-ACCAGCAAACCGTGGATTACCCTAGC-3′ and 5′-ATTCCACAAAGTGAAGATTAGCGTCC-3′.
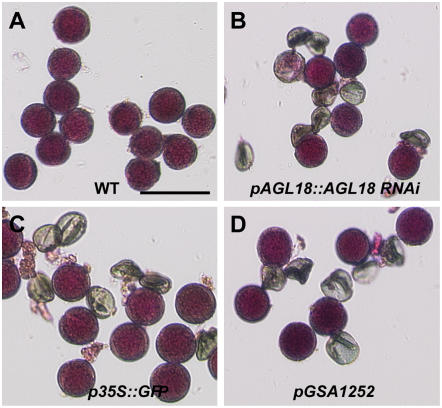
Figure 2.
Lethal pollen phenotypes in representative transgenic plants harboring RNAi, reporter gene constructs, and empty vectors, respectively. Pollen was isolated from anthers after dehiscence and examined by Alexander staining (Alexander, 1969). Five hundred pollen grains were counted from each T1 plant. A, Wild-type (Columbia) pollen grains. B, Pollen grains from an AGL18∷AGL18 RNAi plant. C, Pollen grains from a 35S∷GFP plant. D, Pollen grains from a transgenic plant harboring the empty vector pGSA1252. Bar = 50 μm. [See online article for color version of this figure.]
Table I.
Percentage of plants showing the pollen lethality phenotype in transgenic Arabidopsis populations
Pollen lethality phenotype is defined as the production of 20% to 50% aborted pollen per anther. Percentage of plants exhibiting this phenotype in each transgenic population was determined by three independent scorings, each comprising 100 transgenic T1 plants. Information on vectors is available upon request. Values are depicted as means ± sd.
| Genotype | Percentage of Plants with Pollen Lethality Phenotype |
|---|---|
| Wild type | 0.0 ± 0.0 |
| AGL18∷AGL18 RNAi | 28.3 ± 2.5 |
| DEFH125∷AGL18 RNAi | 25.3 ± 2.2 |
| DEFH125∷GFP RNAi | 23.8 ± 2.0 |
| DEFH125∷DEFH125 RNAi | 22.5 ± 2.0 |
| 35S∷ROXY1 RNAi | 22.1 ± 2.0 |
| 35S∷AGL18 RNAi | 21.6 ± 1.9 |
| 35S∷GFP | 14.6 ± 1.7 |
| ROXY1∷GUS | 14.2 ± 1.5 |
| AGL17∷GUS | 13.5 ± 1.5 |
| pBarA | 13.3 ± 1.5 |
| 35S∷AGL18 antisense | 11.7 ± 1.3 |
| 35S∷DEFH125 | 11.4 ± 1.3 |
| pGSA1252 | 11.1 ± 1.0 |
To test if pollen development is generally affected by the RNAi mechanism, two genes were selected for which no function during pollen development has been reported. The heterologous fluorescent reporter GFP and ROXY1, a glutaredoxin involved in Arabidopsis petal development (Xing et al., 2005), were cloned into an RNAi cassette and expressed under the control of the pollen-specific DEFH125 promoter and the cauliflower mosaic virus 35S promoter, respectively. Strikingly, more than 20% of both transgenic T1 plants displayed the pollen lethality phenotype, with 20% to 50% of the pollen being aborted (Table I). As these data were generated with the binary vector pGSA1252, a transgenic DEFH125∷DEFH125 RNAi population was produced using another binary vector, a derivative of pGPTV (Lauri et al., 2006). Again, 22.5% of the transgenic plants showed the pollen lethality phenotype (Table I). Thus, these data demonstrate that all different RNAi T1 populations tested here comprise a large proportion, about 20% to 30%, of T1 plants that produce 20% to 50% nonviable pollen (Table I), irrespective of the gene and binary vector type used for RNAi vector construction.
Next, we examined whether not only RNAi constructs, but also transgenes in general, such as antisense constructs (35S∷AGL18 antisense), reporter gene constructs like GUS (ROXY1∷GUS; AGL17∷GUS) and GFP (35S∷GFP), as well as overexpression constructs (35S∷DEFH125) or even empty vectors (pBarA; pGSA1252; pGSA1285), also disturb normal pollen development. We observed that in all different T1 populations, more than 10% of the transgenic plants showed the pollen lethality phenotype (Table I). To conclude, occurrence of the pollen lethality phenotype seems to be a general, common phenomenon among transgenic plant populations. However, the number of plants with this phenotype was significantly increased in all investigated transgenic RNAi populations.
Then, we determined at which stage pollen development is affected in the RNAi lines. To make pollen abortion analysis easier, three selected AGL18∷AGL18 RNAi lines exhibiting 50% pollen lethality were crossed into a qrt1-2 mutant background, where the four meiosis products of the pollen mother cells do not separate but stick together as tetrads (Francis et al., 2006). In the three F2 populations, plants showed identical pollen lethality phenotypes and pollen development was further analyzed. Until the end of meiosis, no obvious morphological differences among haploid uninucleate microspores were detectable (data not shown). Onset of abnormal pollen development was determined to occur at a late uninucleate microspore stage. Normally, big vacuoles are formed, moving the nuclei to the microspore wall (Fig. 3, B and C). However, microspores lacking formation of a big vacuole degenerate their nuclei and cytoplasm and get aborted (Fig. 3, C–H). Occasionally, pollen quartets containing four wild type-like pollen grains were formed (Fig. 3J). Also, other deviations from a 2:2 ratio of aborted pollen to nonaborted pollen were observed in the quartets (data not shown), inferring that pollen defects in these RNAi plants might result from sporophytic effects (Johnson-Brousseau and McCormick, 2004).
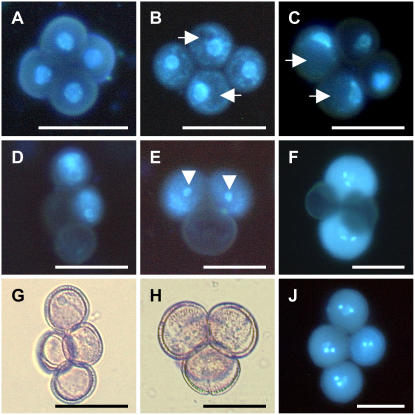
Figure 3.
Pollen development analysis of T1 AGL18∷AGL18 RNAi plants forming 50% nonviable pollen in a qrt1-2 background. Representative microspores, isolated from anthers at different uninucleate and binucleate stages and stained with 4′,6-diamidine-2′-phenylindole dihydrochloride (1 μg/mL dissolved in PBS), are shown. A, Early uninucleate microspore stage. Due to the qrt1-2 background, microspores still stick together, revealing that wild type-like nuclei are located in the center of the cells. B and C, At the late uninucleate microspore stage, formation of big vacuoles (arrows) in two microspores moves nuclei from the center to the cell wall. The two other microspores are lacking big vacuoles and likely degenerated later. D, Nuclei disappeared in two microspores; others just completed the first mitotic division. E, At the two-cell stage, two microspores proceeded further in development and generative cells are visible (arrowheads). F, Only wild type-like microspores underwent second mitosis and contain two sperm cells and one big vegetative cell; the two other pollen grains were aborted. G and H, Bright-field images of Figure 3, D and F (respectively). J, Four wild type-like pollen grains in the quartet at a mature stage. Bar = 25 μm. [See online article for color version of this figure.]
Finally, we tested if the pollen lethality phenotype is heritable. Six T1 plants from different transgenic populations (two from AGL18∷AGL18 RNAi; two from 35S∷GFP; two from pGSA1252) were selfed over five generations and the pollen lethality phenotype was still detectable in the progeny. Given the observation that no plant could be scored producing more than 50% lethal pollen, this suggests that the aborted pollen contains the transgene and the lethal pollen phenotype might be transmitted through the female. To test this idea, pollen from the investigated transgenic plants showing 50% pollen lethality was used to pollinate wild-type plants. Of the 334 F1 plants analyzed, none showed the pollen lethality phenotype. However, when transgenic plants were fertilized with wild-type pollen, pollen abortion was still detectable in about 50% of the F1 progeny (146/300 analyzed plants). To conclude, these data indicate that the observed pollen lethality phenotype in the different transgenic plants is inheritable and transmitted through the female.
DISCUSSION
Our data demonstrate that at least 10% of transgenic Arabidopsis plants produce 20% to 50% nonviable pollen in anthers and this phenomenon is independent of the construct types used for transformation. However, the number of plants with this pollen phenotype was 2 to 3 times higher when the RNAi constructs were used for generation of transgenic plants (Table I). This pollen lethality phenotype will be easily overlooked if the focus is not on investigating pollen development, as transgenic plants do not display other obvious deviations from wild-type development.
Why so many transgenic lines show this pollen phenotype, particularly in RNAi transgenic populations, is still a puzzle. One possible explanation is the positional effect of the T-DNA integration. T-DNA insertions can cause chromosome rearrangements and deletions that can lead to pollen developmental defects (Nacry et al., 1998; Tax and Vernon, 2001; Oh et al., 2003). It has been estimated that about 3,500 genes are expressed in the anther (Scott et al., 2004), and random targeting of these genes by T-DNA insertions might cause, directly or indirectly, pollen developmental defects. Alternatively, off-target effects might be involved; these have been reported from animal studies and require as few as 11 bp of sequence identity to direct silencing of nontargeted transcripts (Jackson et al., 2003; Birmingham et al., 2006; Moffat et al., 2007). Using GFP and AGL18 coding region sequences as well as binary vector sequences to blast the whole Arabidopsis genome or Arabidopsis Genome Initiative transcripts (www.arabidopsis.org) easily identifies a number of matching 16 to 22 nucleotides that may mediate, via an off-targeting mechanism, a higher frequency of lethal pollen formation in the RNAi populations.
Currently, RNAi and T-DNA mutant analysis are broadly and successfully applied approaches to study gene functions. However, pollen lethality is a by-product affecting about 10% of transgenic plants regardless of the construct type used, and the phenomenon is enhanced to more than 20% if RNAi technology is applied to analyze pollen development. Given our observations, it is highly recommendable to combine RNAi data with independent T-DNA mutant analysis and complementation studies. This seems to be particularly crucial for analysis of genes in species such as Antirrhinum majus and Zea mays, where raising larger T1 populations is hampered by laborious transformation techniques and where pollen sterility, obtained in only a few transgenic T1 plants, could lead to misinterpretation of observed pollen lethality phenotypes.
Acknowledgments
We thank Peter Huijser and Andrea Busch for helpful comments on the manuscript. S.Z. is grateful to Heinz Saedler for ongoing support. This work has been initiated in the Arabidopsis Functional Genomics Network, supported by the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sabine Zachgo (szachgo@mpiz-koeln.mpg.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF (2000) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J 24 457–466 [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al (2006) 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3 199–204 [DOI] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21 635–637 [DOI] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39 761–775 [DOI] [PubMed] [Google Scholar]
- Lauri A, Xing S, Heidmann I, Saedler H, Zachgo S (2006) The pollen-specific DEFH125 promoter from Antirrhinum is bound in vivo by the MADS-box proteins DEFICIENS and GLOBOSA. Planta 224 61–71 [DOI] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Adamczyk BJ, Fernandez DE (2005) Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol Biol 58 89–107 [DOI] [PubMed] [Google Scholar]
- Moffat J, Reiling JH, Sabatini DM (2007) Off-target effects associated with long dsRNAs in Drosophila RNAi screens. Trends Pharmacol Sci 28 149–151 [DOI] [PubMed] [Google Scholar]
- Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D (1998) Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Park SK, Jang I, Howden R, Moore JM, Grossniklaus U, Twell D (2003) halfman, an Arabidopsis male gametophytic mutant associated with a 150 kb chromosomal deletion adjacent to an introduced Ds transposable element. Sex Plant Reprod 16 99–102 [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16 S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax FE, Vernon DM (2001) T-DNA-associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol 126 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132 1555–1565 [DOI] [PubMed] [Google Scholar]