Abstract
The ruvA, ruvB, and ruvC genes of Escherichia coli provide activities that catalyze branch migration and resolution of Holliday junction intermediates in recombination. Mutation of any one of these genes interferes with recombination and reduces the ability of the cell to repair damage to DNA. A suppressor of ruv mutations was identified on the basis of its ability to restore resistance to mitomycin and UV light and to allow normal levels of recombination in a recBC sbcBC strain carrying a Tn10 insertion in ruvA. The mutation responsible was located at 12.5 min on the genetic map and defines a new locus which has been designated rus. The rus suppressor works just as well in recBC sbcA and rec+ sbc+ backgrounds and is not allele specific. Mutations in ruvB and ruvC are suppressed to an intermediate level, except when ruvA is also inactive, in which case suppression is complete. In all cases, suppression depends on RecG protein, a DNA-dependent ATPase that catalyzes branch migration of Holliday junctions. The rus mutation activates an additional factor that probably works with RecG to process Holliday junction intermediates independently of the RuvAB and RuvC proteins. The possibility that this additional factor is a junction-specific resolvase is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
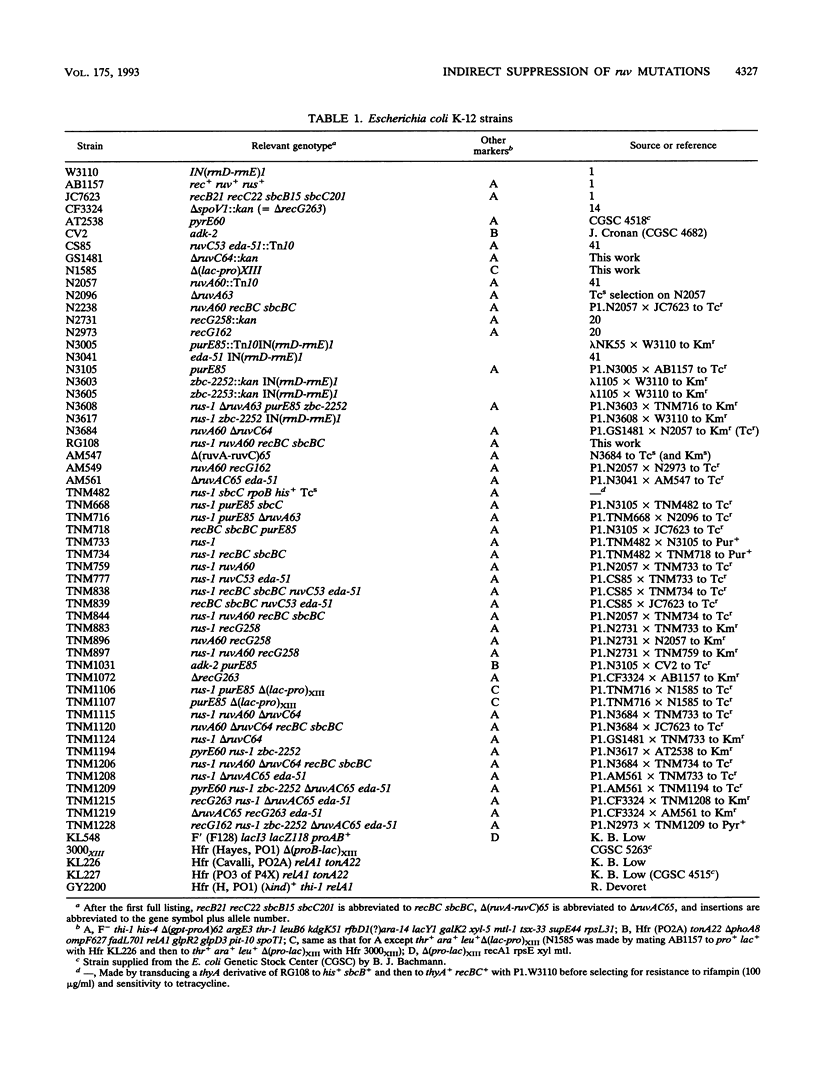
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson F. E., Illing G. T., Sharples G. J., Lloyd R. G. Nucleotide sequencing of the ruv region of Escherichia coli K-12 reveals a LexA regulated operon encoding two genes. Nucleic Acids Res. 1988 Feb 25;16(4):1541–1549. doi: 10.1093/nar/16.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson F., Collier S., Lloyd R. G. Evidence of abortive recombination in ruv mutants of Escherichia coli K12. Mol Gen Genet. 1991 Feb;225(2):266–272. doi: 10.1007/BF00269858. [DOI] [PubMed] [Google Scholar]
- Connolly B., Parsons C. A., Benson F. E., Dunderdale H. J., Sharples G. J., Lloyd R. G., West S. C. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunderdale H. J., Benson F. E., Parsons C. A., Sharples G. J., Lloyd R. G., West S. C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991 Dec 19;354(6354):506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., Theriot L., Stedeford J. B. Some properties of excision-defective recombination-deficient mutants of Escherichia coli K-12. J Bacteriol. 1969 Mar;97(3):1134–1141. doi: 10.1128/jb.97.3.1134-1141.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Flanders P., West S. C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984 May 17;309(5965):215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Shiba T., Makino K., Nakata A., Shinagawa H. Overproduction, purification, and ATPase activity of the Escherichia coli RuvB protein involved in DNA repair. J Bacteriol. 1989 Oct;171(10):5276–5280. doi: 10.1128/jb.171.10.5276-5280.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Nakata A., Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992 Nov;6(11):2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991 Dec;10(13):4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman M., Murphy H., Cashel M. The nucleotide sequence of recG, the distal spo operon gene in Escherichia coli K-12. Gene. 1992 Jan 2;110(1):95–99. doi: 10.1016/0378-1119(92)90449-y. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Bender J., Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
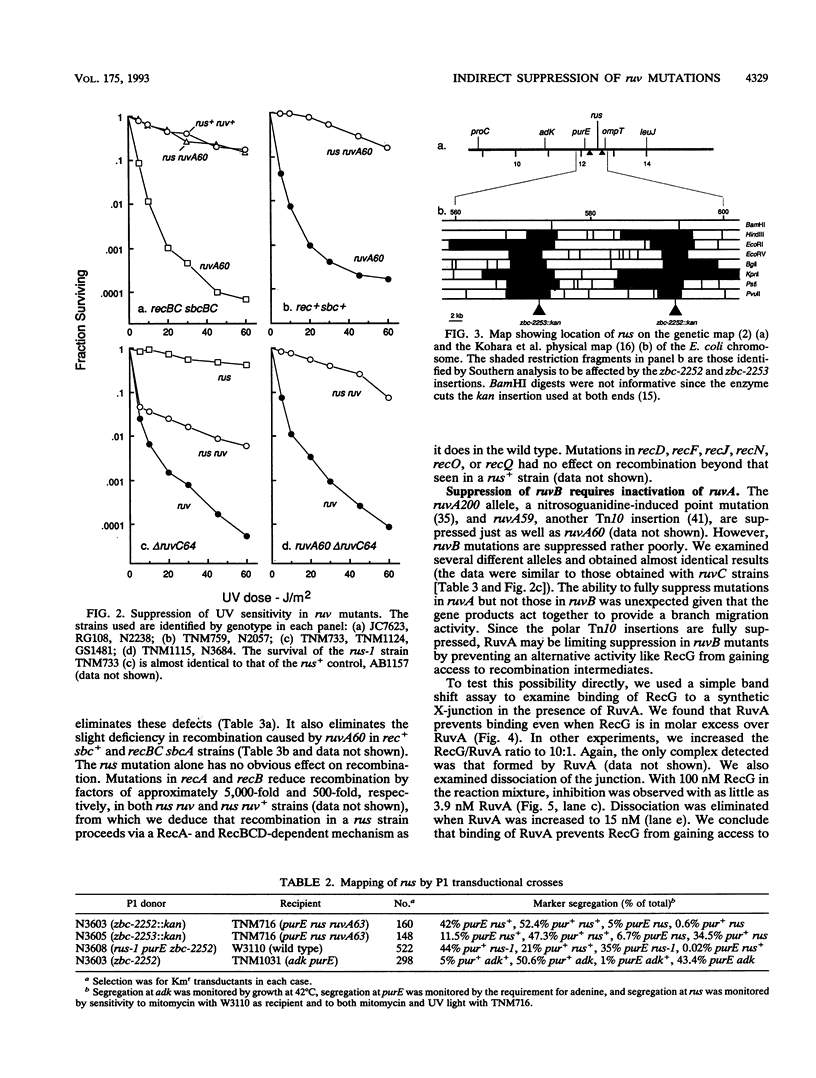
- Lloyd R. G., Benson F. E., Shurvinton C. E. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol Gen Genet. 1984;194(1-2):303–309. doi: 10.1007/BF00383532. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991 Feb;173(3):1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985 Nov;164(2):836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991 Sep;173(17):5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Evans N. P., Buckman C. Formation of recombinant lacZ+ DNA in conjugational crosses with a recB mutant of Escherichia coli K12 depends on recF, recJ, and recO. Mol Gen Genet. 1987 Aug;209(1):135–141. doi: 10.1007/BF00329848. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Low B., Godson G. N., Birge E. A. Isolation and characterization of an Escherichia coli K-12 mutant with a temperature-sensitive recA- phenotype. J Bacteriol. 1974 Oct;120(1):407–415. doi: 10.1128/jb.120.1.407-415.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Porton M. C., Buckman C. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol Gen Genet. 1988 May;212(2):317–324. doi: 10.1007/BF00334702. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993 Jan;12(1):17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Molecular organization and nucleotide sequence of the recG locus of Escherichia coli K-12. J Bacteriol. 1991 Nov;173(21):6837–6843. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res. 1993 Apr 25;21(8):1719–1725. doi: 10.1093/nar/21.8.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
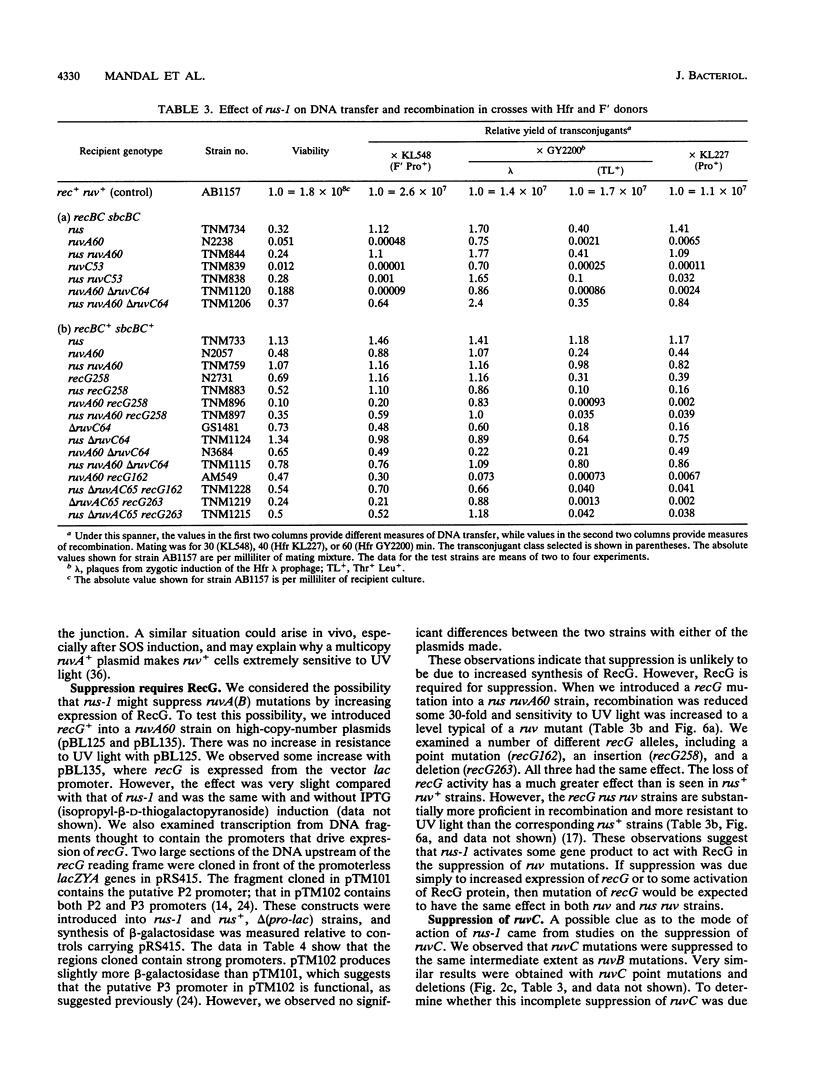
- Oden K. L., DeVeaux L. C., Vibat C. R., Cronan J. E., Jr, Gennis R. B. Genomic replacement in Escherichia coli K-12 using covalently closed circular plasmid DNA. Gene. 1990 Nov 30;96(1):29–36. doi: 10.1016/0378-1119(90)90337-q. [DOI] [PubMed] [Google Scholar]
- Otsuji N., Iyehara H., Hideshima Y. Isolation and characterization of an Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J Bacteriol. 1974 Feb;117(2):337–344. doi: 10.1128/jb.117.2.337-344.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons C. A., Kemper B., West S. C. Interaction of a four-way junction in DNA with T4 endonuclease VII. J Biol Chem. 1990 Jun 5;265(16):9285–9289. [PubMed] [Google Scholar]
- Parsons C. A., Tsaneva I., Lloyd R. G., West S. C. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W. D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968 Jan 28;31(2):291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- Rupp W. D., Wilde C. E., 3rd, Reno D. L., Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971 Oct 14;61(1):25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- Sargentini N. J., Smith K. C. Role of ruvAB genes in UV- and gamma-radiation and chemical mutagenesis in Escherichia coli. Mutat Res. 1989 Nov;215(1):115–129. doi: 10.1016/0027-5107(89)90224-8. [DOI] [PubMed] [Google Scholar]
- Sharples G. J., Benson F. E., Illing G. T., Lloyd R. G. Molecular and functional analysis of the ruv region of Escherichia coli K-12 reveals three genes involved in DNA repair and recombination. Mol Gen Genet. 1990 Apr;221(2):219–226. doi: 10.1007/BF00261724. [DOI] [PubMed] [Google Scholar]
- Sharples G. J., Lloyd R. G. Resolution of Holliday junctions in Escherichia coli: identification of the ruvC gene product as a 19-kilodalton protein. J Bacteriol. 1991 Dec;173(23):7711–7715. doi: 10.1128/jb.173.23.7711-7715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T., Iwasaki H., Nakata A., Shinagawa H. SOS-inducible DNA repair proteins, RuvA and RuvB, of Escherichia coli: functional interactions between RuvA and RuvB for ATP hydrolysis and renaturation of the cruciform structure in supercoiled DNA. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8445–8449. doi: 10.1073/pnas.88.19.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H., Makino K., Amemura M., Kimura S., Iwasaki H., Nakata A. Structure and regulation of the Escherichia coli ruv operon involved in DNA repair and recombination. J Bacteriol. 1988 Sep;170(9):4322–4329. doi: 10.1128/jb.170.9.4322-4329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C. E., Lloyd R. G., Benson F. E., Attfield P. V. Genetic analysis and molecular cloning of the Escherichia coli ruv gene. Mol Gen Genet. 1984;194(1-2):322–329. doi: 10.1007/BF00383535. [DOI] [PubMed] [Google Scholar]
- Shurvinton C. E., Lloyd R. G. Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol Gen Genet. 1982;185(2):352–355. doi: 10.1007/BF00330811. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Tsaneva I. R., Müller B., West S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992 Jun 26;69(7):1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990 Mar;54(1):18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- West S. C., Cassuto E., Howard-Flanders P. Homologous pairing can occur before DNA strand separation in general genetic recombination. Nature. 1981 Mar 5;290(5801):29–33. doi: 10.1038/290029a0. [DOI] [PubMed] [Google Scholar]
- West S. C., Cassuto E., Howard-Flanders P. Mechanism of E. coli RecA protein directed strand exchanges in post-replication repair of DNA. Nature. 1981 Dec 17;294(5842):659–662. doi: 10.1038/294659a0. [DOI] [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]