Abstract
S-adenosylmethionine (SAM) is the substrate used in the methylation of homogalacturonan (HGA) in the Golgi apparatus. SAM is synthesized in the cytosol, but it is not currently known how it is then transported into the Golgi. In this study, we find that HGA methyltransferase is present in Golgi-enriched fractions and that its catalytic domain faces the lumen of this organelle. This suggests that SAM must be imported into the Golgi. We performed uptake experiments using [methyl-14C]SAM and found that SAM is incorporated into the Golgi vesicles, resulting in the methylation of polymers that are sensitive to pectinase and pectin methylesterase but not to proteases. To avoid detecting the transfer reaction, we also used [carboxyl-14C]SAM, the uptake of which into Golgi vesicles was found to be sensitive to temperature, detergents, and osmotic changes, and to be saturable with a Km of 33 μm. Double-label uptake experiments using [methyl-3H]SAM and [carboxyl-14C]SAM also revealed a time-dependent increase in the 3H to 14C ratio, suggesting that upon transfer of the methyl group, the resulting S-adenosylhomocysteine is not accumulated in the Golgi. SAM incorporation was also found to be inhibited by S-adenosylhomocysteine, whereas UDP-GalA, UDP-GlcA, and acetyl-CoA had no effect. DIDS, a compound that inhibits nucleotide sugar transporters, also had little effect upon SAM incorporation. Interestingly, the combination of UDP-GalA + acetyl-CoA or UDP-GlcA + acetyl-CoA produced a slight increase in the uptake of SAM. These results support the idea that a SAM transporter is required for HGA biosynthesis.
S-adenosylmethionine (SAM) is the donor substrate for a number of methylation reactions within the cell, including DNA and protein methylation (Fontecave et al., 2004). In plants, SAM is also involved in other metabolic pathways, such as ethylene biosynthesis (Kende, 1993) and the methylation of pectins (Kauss and Hassid, 1967; Vannier et al., 1992; Boulard et al., 1997; Goubet et al., 1998). The pectins are a group of complex polysaccharides present in the primary cell wall in plants and are composed of homogalacturonan (HGA), rhamnogalacturonan I, and rhamnogalacturonan II (O'Neill et al., 1990; Albersheim et al., 1996).
Previous studies using monoclonal antibodies against pectins and also involving immunoelectronmicroscopy have suggested that the pectins are synthesized in the Golgi apparatus (Zhang and Staehelin, 1992). In addition, Goubet and Mohnen (1999a) have described a Golgi-localized HGA methyltransferase (HGA-MT) in tobacco (Nicotiana tabacum) cells, thus indicating that the methylation of HGA also takes place in the Golgi. Furthermore, they also found that by treating Golgi fractions with proteinase K, the HGA-MT activity was decreased only in permeabilized membranes, suggesting that this enzyme is located in the lumen of the Golgi apparatus.
The reported evidence to date indicates that SAM biosynthesis occurs in the cytosol (Wallsgrove et al., 1983; Ravanel et al., 1998). However, since HGA-MT utilizes SAM in the lumen of the Golgi apparatus, this substrate must necessarily be imported into this subcellular compartment. However, to our knowledge no evidence for the transport of SAM into the Golgi apparatus has so far been provided. Hence, to establish whether SAM is indeed transported into the Golgi and used in the methylation of pectins, we have analyzed the incorporation of SAM into Golgi vesicles obtained from etiolated pea (Pisum sativum) epicotyls in this study. Our results suggest that SAM is indeed transported into the lumen of the Golgi and used in the methylation of pectins. Moreover, this importation process shows features that differ from other Golgi-localized transporters, such as the nucleotide sugar transporters (NSTs), suggesting that a new class of Golgi transporter is involved in polysaccharide biosynthesis.
RESULTS
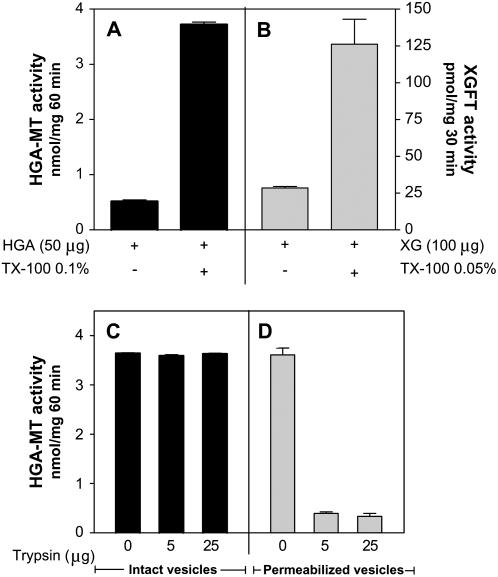
To determine the mechanisms underlying the importation of SAM into the Golgi, we purified Golgi vesicles from etiolated pea epicotyls, an experimental model plant that has been utilized previously to analyze the topology of enzymes involved in the biosynthesis of polysaccharides, as well as the transport of metabolites and ions (Muñoz et al., 1996; Neckelmann and Orellana, 1998; Wulff et al., 2000; Sterling et al., 2001; Ordenes et al., 2002). We first determined the orientation of the catalytic domain of HGA-MT in our Golgi vesicle preparations by measuring its enzymatic activity in the presence of exogenous HGA as the acceptor for the methylation reaction. This polymer cannot access the lumen of the Golgi vesicles unless the integrity of the membrane is altered, and we thus measured HGA-MT activity both in the presence and absence of 0.1% Triton X-100.
A 6-fold increase in the activity of HGA-MT was observed when the Golgi vesicle membranes were permeabilized with detergent (Fig. 1A). This activation level was also similar to those observed for xyloglucan (XG)-fucosyltransferase (FT) in the presence of 0.1% Triton X-100 (Fig. 1B). XG-FT is a type II membrane protein with a catalytic site that faces the lumen of the Golgi apparatus (Perrin et al., 1999; Wulff et al., 2000). Since HGA-MT and XG-FT were measured in the presence of acceptors that cannot cross the membrane, the increases we detected in their activity following the permeabilization of the Golgi vesicle membranes could be explained by the luminal localization of the active site of both enzymes. To confirm whether the active site of HGA-MT indeed faces the lumen of the Golgi, we treated both intact and permeabilized Golgi vesicles with trypsin to disrupt both the cytosolic and lumenal domains of the membrane proteins and also any Golgi soluble proteins. Following these trypsin treatments, the intact vesicles were also permeabilized and the total remaining HGA-MT activity was measured using exogenous HGA (Fig. 1, C and D). The results show that trypsin has no effect on the HGA-MT activity in intact vesicles, but almost completely abolishes this activity in already Triton X-100-treated Golgi vesicles. These results strongly suggest that the catalytic domain of HGA-MT faces the lumen of the Golgi apparatus in etiolated peas. These results are also in agreement with the previously reported findings of Goubet and Mohnen (1999a) in tobacco cells.
Figure 1.
HGA-MT activity is found in the lumen of Golgi vesicles. A, HGA-MT activity was measured for 1 h on intact or permeabilized Golgi vesicles (50 μg of protein) in the presence (+) or absence (−) of 50 μg of HGA. Vesicles were treated with or without 0.1% (v/v) Triton X-100. B, XG-FT was measured for 30 min on intact or permeabilized Golgi vesicles (100 μg of protein) in the presence (+) or absence (−) of 100 μg of tamarind XG. Vesicles were permeabilized using 0.05% (v/v) Triton X-100. C and D, Golgi vesicles (50 μg of protein) were treated for 10 min with 5 or 25 μg of trypsin, in the absence (see C) or presence (see D) of 0.02% (v/v) Triton X-100. Trypsin was inactivated using trypsin inhibitor as described in “Materials and Methods.” The trypsin-treated vesicles were then used to assess the HGA-MT activity. The total activity present in each sample was measured in the presence of HGA (50 μg), [methyl-14C]SAM, and 0.1% (v/v) Triton X-100 as described in “Materials and Methods.” HGA was precipitated in 20% TCA and the radioactivity associated to the pellet was measured using a liquid scintillation counter.
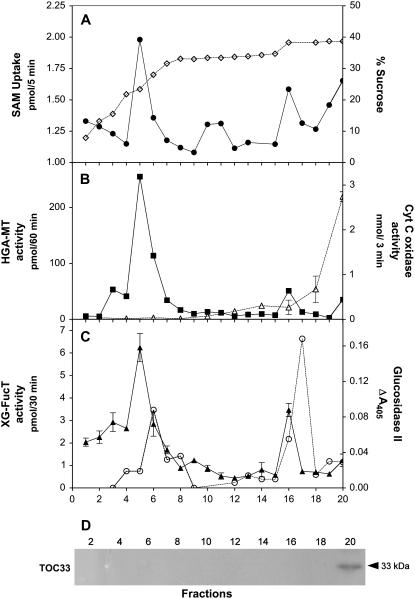
Since the newly synthesized HGA and HGA-MT are located in the lumen of the Golgi, we postulated that SAM would also need to be incorporated into this compartment. To test this, we performed subcellular fractionation of etiolated pea stem organelles and measured the uptake of SAM into these different subcellular fractions. We observed that the uptake of SAM was at the highest levels in fractions 4, 5, and 6 (Fig. 2A), which also contained high HGA-MT and XG-FT activity. We also detected SAM uptake in other subcellular fractions that contain endoplasmic reticulum markers and toward the bottom of the gradient at the mitochondrial and plastid sedimentation points, suggesting that SAM is also incorporated into these organelles in pea epicotyls. No detectable activity of mitochondrial or plastid markers was detected in the Golgi fraction, indicating that SAM uptake observed in the Golgi fraction was not due to mitochondria or plastid present in that fraction.
Figure 2.
Uptake of SAM and HGA-MT activity in subcellular fractions of etiolated pea epicotyls. Pea epicotyls were homogenized and the organelles separated in a discontinuous Suc gradient. Aliquots were collected and utilized in the measurement of different activities. A, SAM uptake measured in the presence of 3 μm [methyl-14C]SAM for 5 min (•). Suc concentration was determined by refractometry (⋄). B, HGA-MT activity measured in the presence of 4 μm [methyl-14C]SAM and 6 μm cold SAM, 0.1% Triton X-100, and 50 μg of polygalacturonic acid (HGA) in STM (Suc-Tris-magnesium; ▪); cytochrome c oxidase activity (mitochondrial marker; ▵). C, Glucosidase II activity (endoplasmic reticulum marker; ○); XG-FT activity (Golgi marker; ▴). D, Immunoblot using anti-TOC33 (plastid marker). The measurements were done in duplicate and the average activity is plotted.
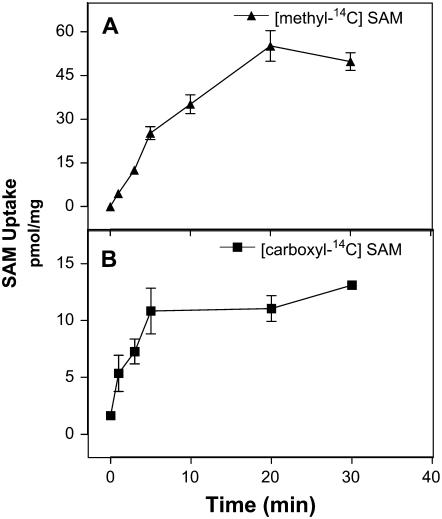
To further characterize the import of SAM into the Golgi, we measured the uptake of two different radiolabeled forms of this substrate using enriched Golgi vesicle fractions. One of these, [methyl-14C]SAM, is radiolabeled on the methyl group that is transferred onto HGA by HGA-MT, whereas the other, [carboxyl-14C]SAM, is radiolabeled on the carboxyl group that remains in the S-adenosylhomocysteine (SAH) molecule following the methylation reaction. The results shown in Figure 3 indicate that both substrates were incorporated into Golgi vesicles but that the uptake of [carboxyl-14C]SAM plateaued at 5 min, whereas the uptake of [methyl-14C]SAM required 20 min to reach this peak. Upon reaching a plateau, the incorporation of [methyl-14C]SAM was found to be between 5- and 6-fold higher than [carboxyl-14C]SAM, suggesting that when SAM is metabolized, the radiolabeled methyl and carboxyl groups have a different fate within the Golgi lumen.
Figure 3.
Time dependence of SAM uptake in Golgi vesicles. Golgi vesicles (100 μg of protein) were incubated with 3 μm [methyl-14C]SAM (A) or [carboxyl-14C]SAM (B) in a final volume of 100 μL for the times indicated. The reaction was terminated by 10-fold dilution of the reaction and filtering the incubation medium through 0.45-μm cellulose ester filters. The filters were washed with cold buffer, dried, and the radioactivity associated with them estimated in a liquid scintillation counter.
To further analyze the differences observed during the incorporation of the methyl- and carboxyl-radiolabeled SAM substrates, we carried out a double-labeling experiment by incubating Golgi vesicles in the presence of both [carboxyl-14C]SAM and [methyl-3H]SAM. As shown in Table I, both radiolabeled substrates were incorporated into the Golgi vesicles but the methyl group clearly accumulated at higher levels than the carboxyl group, leading to a time-dependent increase in the 3H to 14C ratio. These results suggest that SAM is transported into the lumen and that the methyl group is then rapidly transferred onto endogenous acceptors. It is therefore likely that the transfer of the methyl group leads to the release of SAH, and our results further suggest that SAH is not retained within the Golgi lumen since the 3H to 14C ratio does not remain constant but increases with time.
Table I.
Ratio of 14C to 3H found in Golgi vesicles after incorporation of a mixture of [carboxyl-14C]SAM + [methyl-3H]SAM
Vesicles (100 μg) were incubated with 3 μm SAM containing a mixture of [carboxyl-14C]SAM + [methyl-3H]SAM. After 30 s, 1 min, or 3 min of incubation, the reaction was stopped and filtered, and the radioactivity associated with the filters was determined. The numbers in parentheses are pmoles of SAM and are the average of triplicate measurements. The sd was less than 10%.
| Medium | 0 s | 30 s | 1 min | 3 min |
|---|---|---|---|---|
| 1.00 | 1.14 | 1.71 | 2.22 | 2.45 |
| (300/300) | (1.64/1.43) | (3.01/1.76) | (5.24/2.36) | (5.34/2.38) |
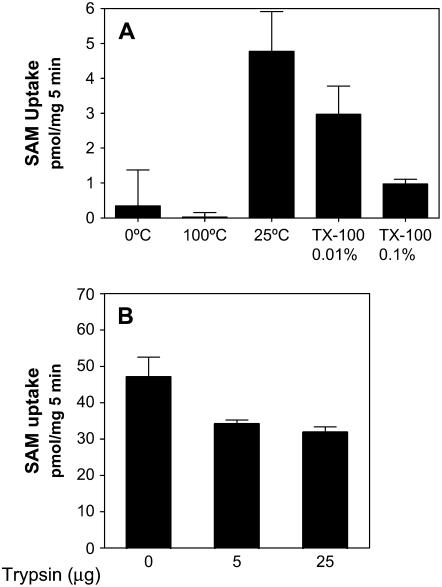
The use of methyl-radiolabeled SAM in incorporation assays makes it difficult to distinguish between the transport of SAM and the transfer of its donor methyl group. To therefore characterize the transport process alone, we performed experiments using only [carboxyl-14C]SAM as the substrate to assess whether SAM is truly transported into the Golgi lumen. We evaluated the importance of the membrane integrity during this importation process by treating the vesicles with 0.01% and 0.1% Triton X-100. This treatment led to a strong decrease in the uptake of [carboxyl-14C]SAM (Fig. 4A), which was also found to be sensitive to temperature since it showed a 10-fold higher activity at 25°C compared with 0°C. Boiling of the vesicles prior to the assay completely inactivated the incorporation of SAM (Fig. 4A).
Figure 4.
Effect of temperature, detergent, and proteases in the incorporation of SAM into Golgi vesicles. A, Golgi vesicles (100 μg of protein) were incubated with 3 μm [carboxyl-14C]SAM for 5 min under different conditions: 25°C; TX-100, incubation in the presence of 0.01% or 0.1% (v/v) Triton X-100; 0°C, incubation in an ice bath; and 100°C, vesicles boiled for 5 min prior to the assay. Incorporation was measured by a filtration assay. B, Golgi vesicles (50 μg of protein) were treated for 10 min with 5 or 25 μg of trypsin, which was then blocked using a specific inhibitor as described in “Materials and Methods.” The protease-treated vesicles were then used to measure the uptake of SAM as described in Figure 4 by incubation with 3 μm [methyl-14C]SAM for 5 min.
To obtain further evidence that a protein is involved in this process, we treated the Golgi vesicles with trypsin and then measured the uptake of SAM. The results of this experiment (Fig. 4B) showed that trypsinization produces a decrease in the uptake of SAM (25%–30%). The use of the same amounts of trypsin produced a larger decrease (90%) in the HGA-MT activity levels (Fig. 1C). These findings support the idea that a SAM transporter is involved in the import of this substrate into the Golgi and also indicates that the transport and methyltransferase activities associated with SAM in the Golgi are separate events.
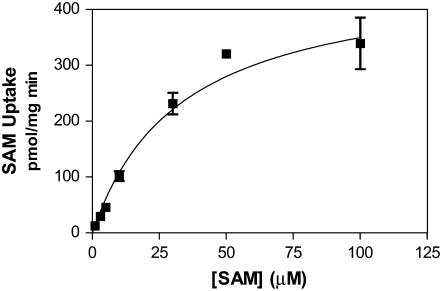
To further characterize the transport of SAM, we measured the substrate dependence of this process. In addition, to minimize the contribution of the transfer reaction to the measurement of radioactivity during the uptake of [carboxyl-14C]SAM, the assay was performed after a 30-s incubation only. Our results show that the uptake of SAM was saturable with an apparent Km of 33 μm (Fig. 5). We also measured the sensitivity of the uptake of [carboxyl-14C]SAM into Golgi vesicles to osmotic changes as these will affect the vesicle volumes. The uptake of SAM was found to be lower in the presence of a higher Suc concentration (Supplemental Table S1), a condition under which the volume of the Golgi vesicles decreases. These results strongly suggest that SAM is imported into the Golgi by an active transporter.
Figure 5.
Initial rates of SAM uptake as a function of external [carboxyl-14C]SAM concentrations in Golgi vesicles. Golgi vesicles (100 μg) were incubated with various concentrations of [carboxyl-14C]SAM and incorporation into Golgi vesicles was estimated by a filtration assay. The curve was fitted with the Michaelis-Menten equation, thus giving Km and Vmax values of 33 μm and 227 pmol/mg 30 s.
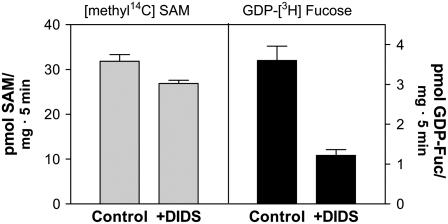
The chemical structure of SAM resembles that of the nucleotide sugars (NSs), and, since different NSTs have been shown previously to be localized in the Golgi apparatus in plants (Baldwin et al., 2001; Bakker et al., 2005; Norambuena et al., 2005; Rollwitz et al., 2006), we wanted to evaluate whether the transport of SAM is also mediated by a NST-like transporter. We thus performed our SAM import assay in the presence of DIDS, a general inhibitor of the NSTs (Wulff et al., 2000). Under conditions where DIDS produced a 10% to 15% decrease in the uptake of SAM, the incorporation of GDP-Fuc was found to be decreased by 70% (Fig. 6). This result indicates that at the biochemical level, the Golgi SAM transporter does not behave as a NST.
Figure 6.
SAM uptake has low sensitivity to DIDS. Golgi vesicles (100 μg) were incubated with 3 μm [methyl-14C]SAM or 1 μm GDP-[3H]Fuc by 5 min at 25°C in the presence or absence of 50 μm DIDS. The SAM uptake was measured using a filtration assay.
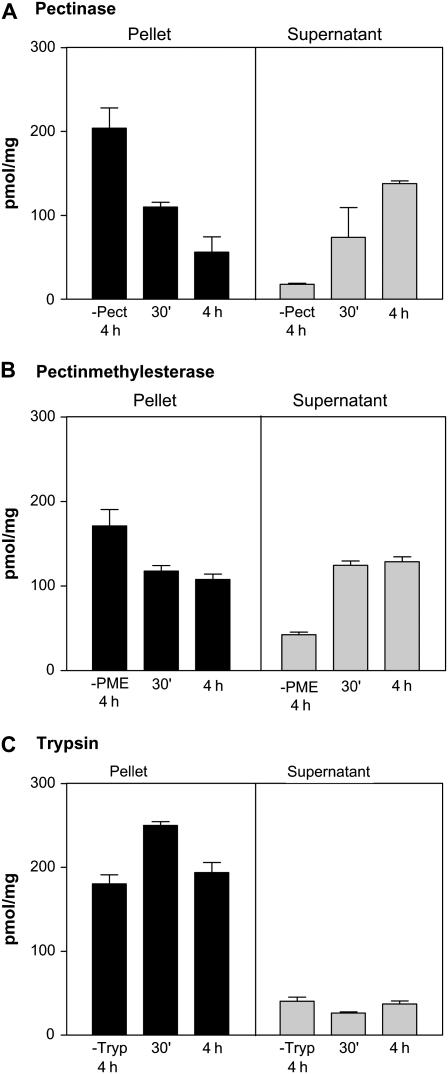
Because our data indicate that the methyl group of SAM is transferred onto endogenous acceptors in the lumen of the Golgi, we postulated that HGA was likely to be one such acceptor. To test this possibility, we incubated Golgi vesicles with [methyl-14C]SAM and then adjusted the incubation media with TCA to a final concentration of 10%, a treatment that sediments both proteins and pectic polysaccharides (Goubet and Mohnen, 1999b). The results showed that radioactive material was indeed precipitated under these conditions (Fig. 7). Additionally, this radioactive material was also precipitated by 70% ethanol, suggesting that the methyl group is transferred onto polysaccharides (data not shown). To assess whether the TCA-precipitated radiolabeled material corresponded to pectin, we separately treated this precipitate with pectinase and pectin methylesterase (PME) followed by a reprecipitation with 10% TCA. As shown in Figure 7, treatment with both enzymes decreased the amount of radiolabeled material that could again be precipitated with 10% TCA and at the same time increased the amount of soluble radiolabeled material that was released into the supernatant. Since TCA also precipitates proteins, we determined whether the TCA-precipitable material was sensitive to trypsin but found that treatment with this protease did not result in the release of any radiolabeled material (Fig. 7). These results thus suggest that SAM molecules that are incorporated into Golgi vesicles are used as donor substrates for the transfer of methyl groups onto pectin but not protein acceptors.
Figure 7.
Methylated products obtained upon incubation with SAM are sensitive to enzymes that degrade pectins. Golgi vesicles (100 μg of protein) were incubated with 3 μm [methyl-14C]SAM for 60 min. Immediately after, polysaccharides and proteins were precipitated by adding 1 volume of 20% TCA. The precipitated fraction was in buffers with different pH (described in “Materials and Methods”) and treated with pectinase (pH 4.0; A), PME (pH 7.5; B), or trypsin (pH 7.5; C) for 30 min and 4 h. Then, the polysaccharides and proteins were reprecipitated with a volume of 20% TCA, and the radioactivity remaining both in the supernatant and pellet was determined by scintillation counting. Experiments were done in triplicate.
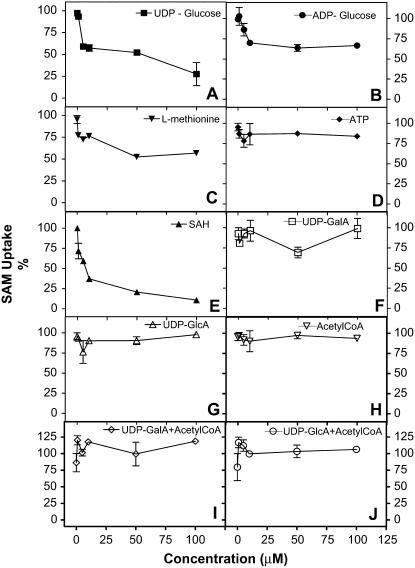
Since many substrates involved in polysaccharide biosynthesis are known to be transported into the Golgi apparatus, we analyzed whether a combination of these biosynthetic precursors had any effect in the uptake of SAM. UDP-Glc had an inhibitory effect at high concentrations, whereas ADP-Glc and l-Met showed only minor inhibitory effects. In contrast, ATP, acetyl-CoA, UDP-GalA, and UDP-GlcA showed no effects upon the uptake of SAM. Interestingly, different combinations of substrates likely to be involved in HGA biosynthesis, such as UDP-GalA + acetyl-CoA or UDP-GlcA + acetyl-CoA, appeared to cause slight increases in the uptake of SAM (Fig. 8). Significantly, SAH, the principal by-product of the HGA-MT/SAM methyltransferase reaction, showed the strongest inhibitory effects upon the uptake of SAM into Golgi vesicles. This inhibitory effect of SAH has also been observed in the transport of SAM into chloroplasts and mitochondria. However, SAH also produced similar inhibitory effects upon HGA-MT activity (Supplemental Fig. S1), and, since the transport of SAM and the transfer of the methyl group are tightly associated, we cannot rule out the possibility that the effects of this molecule on the uptake of SAM are a consequence of its inhibition of HGA-MT.
Figure 8.
Effect of different substrates in the uptake of SAM by Golgi vesicles. Golgi vesicles (100 μg of protein) were incubated for 10 min with increasing concentrations (0, 5, 10, 50, 100 μm) of different compounds. Then, the vesicles were incubated with 3 μm [methyl-14C]SAM for 5 min in urea final volume of 100 μL. The incorporation of SAM was measured using a filtration assay. The values are expressed as a percentage of the SAM uptake activity determined in the absence of any of the compounds used in the experiment. The highest value on each case corresponds to (mean ± se): A, 35.92 ± 0.97 pmol mg−1 5 min−1; B, 30.23 ± 0.19 pmol mg−1 5 min−1; C, 42.73 ± 2.10 pmol mg−1 5 min−1; D, 42.73 ± 2.10 pmol mg−1 5 min−1; E, 41.38 ± 1.78 pmol mg−1 5 min−1; F, 32.89 ± 2.61 pmol mg−1 5 min−1; G, 44.11 ± 2.02 pmol mg−1 5 min−1; H, 34.73 ± 1.66 pmol mg−1 5 min−1; I, 38.44 ± 4.44 pmol mg−1 5 min−1; and J, 36.99 ± 6.93 pmol mg−1 5 min−1.
DISCUSSION
The methylation of HGA occurs in the Golgi apparatus, and biochemical analyses have indicated that the catalytic domains of the methyltransferases involved in this process face the lumen of this organelle (Goubet and Mohnen, 1999a; this study). Proteomic analyses of cellular organelles have identified a number of putative Golgi methyltransferases (GMTs), two of which (GMT1 and GMT2) have now been confirmed to be localized in the Golgi apparatus (Dunkley et al., 2006). Hydropathy analysis of these proteins has further indicated that they are type II membrane-bound proteins, supporting the idea that most of the enzyme, including its active site, faces the lumen of the Golgi. Although none of the candidate GMTs has yet been functionally tested, it is likely that some of them are involved in the methylation of HGA in the lumen of the Golgi. Recently, Mouille et al. (2007) showed that the QUA2 locus from Arabidopsis (Arabidopsis thaliana) encodes a putative methyltransferase that is Golgi localized. A QUA2 mutant exhibited a decrease in HGA, and the corresponding gene is coexpressed with GAUT1, a galacturonosyltransferase involved in HGA biosynthesis (Sterling et al., 2006). In addition, the QUA2 gene encodes a putative type II membrane protein that contains a methyltransferase domain that would necessarily face the lumen of the Golgi. These findings strongly indicate that the methyltransferases involved in HGA biosynthesis harbor a catalytic domain that faces the lumen of the Golgi apparatus and that the methylation of these polysaccharides takes place in this compartment.
SAM is a methyltransferase substrate and it is widely accepted that the enzymes involved in its biosynthesis are exclusively located in the cytosol (Schröder et al., 1997; Ravanel et al., 1998; Hanson and Roje, 2001). Hence, for the methylation of HGA, SAM must be imported into the lumen of the Golgi. In this study, we provide different lines of evidence indicating that SAM is indeed transported into the lumen of the Golgi. These include data showing the following: (1) by subcellular fractionation analysis, strong levels of SAM uptake occur in the Golgi fraction; (2) the uptake of SAM is dependent upon the membrane integrity of the Golgi vesicles as treatments with detergent inhibit this process; (3) SAM uptake into the Golgi is saturable with an apparent Km of 33 μm; (4) SAM uptake is sensitive to both temperature and osmotic changes that alter the volume of the Golgi vesicles; and (5) trypsin treatments inhibit SAM uptake. Taken together, these data strongly support our conclusion that SAM is transported into the Golgi lumen and that a SAM transporter is localized at the membrane of this organelle.
SAM transport activity has been detected previously in mitochondria and plastids (Horne, et al., 1997; Ravanel et al., 2004; Bouvier et al., 2006). The Km for the SAM transporter from plastids is 38 μm, which is very similar to the Km value we obtained in our present analysis for the uptake of SAM by the Golgi vesicles (33 μm). We do not yet know what the free cytosolic concentration of SAM is, and it is thus difficult to predict whether its transporter is working under saturating conditions. This is an important issue to be elucidated in the future because, depending on the concentration of SAM in the cytosol, this transporter may be a limiting step in the methylation process.
The uptake of SAM was found to be inhibited by SAH, which had also been found to be the case for other SAM transporters. However, we found that the activity of HGA-MT was also inhibited by SAH, and therefore cannot yet exclude the possibility that the inhibitory effects of this molecule are not due to a direct effect upon the transporter but are in fact caused by a failure in the transfer of methyl groups that ultimately leads to a decrease in the transport of SAM. UDP-Glc was also found to have inhibitory effects upon SAM uptake at high concentrations, but none of the other NSs that we tested has this effect. We do not yet know the reason for the inhibition of this process by UDP-Glc, but it does not seem to be a general effect of UDP-sugars since none of the other NSs inhibited the import of SAM into the Golgi. Whether this effect is due to some negative interaction between glucosylation reactions in the Golgi and the incorporation of SAM remains to be determined.
Neither UDP-GalUA nor UDP-GlcUA, both substrates of the pectin biosynthesis pathway, affected the incorporation of SAM, but the addition of acetyl-CoA in conjunction with either UDP-GalA or UDP-GlcA marginally stimulated SAM uptake. This result suggests that a coordinated uptake of the substrates involved in the biosynthesis of pectins may occur in the Golgi apparatus.
With regards to the putative existence of a SAM transporter, another family of Golgi-localized transporters has already been described. These are the NSTs that are responsible for the import of glycosyltransferase substrates, and the catalytic domains for these enzymes also face the Golgi lumen. It is noteworthy that the chemical structure of SAM shows some similarity with NSs, particularly at the nitrogenated base that is present in both structures. Hence, one of the questions that arises is whether SAM may in fact be transported by a protein related to NSTs. However, we tested the effect of DIDS, an inhibitor of anionic transporters that has been shown to inhibit NSTs (Wulff et al., 2000), and found that it showed little effect upon the uptake of SAM in Golgi vesicles.
The genes encoding SAM transporters have now been cloned in yeast, human, and Arabidopsis (Marobbio et al., 2003; Agrimi et al., 2004; Bouvier et al., 2006; Palmieri et al., 2006), and belong to the mitochondrial carrier family. One of the interesting features of these carriers is that they are inhibited by SAH. We also found that SAH was a strong inhibitor of SAM uptake into Golgi vesicles. In addition, these characterized transporters work as antiporters; therefore, it is likely that the Golgi-localized transporter may also work as an antiporter, taking the SAH produced in the lumen of the organelle back to the cytosol. Hence, it is likely that a similarity may exist between the putative SAM Golgi-localized transporter and those located in mitochondria and plastids. Significantly in this regard, phylogenetic analyses of the SAM transporters and the NSTs show that they form different clusters, suggesting that the putative Golgi-localized SAM transporter may be encoded by a member of the mitochondrial carrier family. Future studies should lead us to the identification of this gene.
MATERIALS AND METHODS
Chemicals
The chloride salt of SAM, the ammonium salt of UDP-GalA, SAH, acetyl-CoA, ATP, l-Met, Triton X-100, trypsin and trypsin inhibitor, polygalacturonic acid (referred to as HGA in this article), pectinase from Rhizobium sp. (crude powder), and PME from orange peel were each purchased from Sigma. [Methyl-14C]SAM (specific activity 52.7 mCi/mmol) and GDP-[3H]Fuc (specific activity 20.0 Ci/mmol) were purchased from New England Nuclear. [Carboxyl-14C]SAM (specific activity 62 mCi/mmol) and [methyl-3H]SAM (specific activity 84.0 Ci/mmol) were obtained from Amersham Pharmacia. Cellulose ester filters (0.45 μm) were purchased from Millipore.
Plant Material
Pea (Pisum sativum var. Alaska) seeds were obtained from ANASAC and grown in moist vermiculite for 7 or 8 d in the dark at 25°C. Stem segments (1 cm) were excised from the elongating region of the epicotyls and kept on ice until homogenization.
Subcellular Fractionation and Enrichment for Golgi Vesicles
Vesicles were obtained essentially as described previously by Muñoz et al. (1996). Briefly, pea stems were homogenized using razor blades in the presence of 1 volume of 0.5 m Suc, 0.1 m KH2PO4, pH 6.65, 5 mm MgCl2. The homogenate was then filtered through Miracloth (Calbiochem) and centrifuged at 1,000g for 5 min. The supernatant was loaded onto a 1.3 m Suc cushion and centrifuged at 100,000g for 90 min. The upper phase was then removed without disturbing the interface fraction. A discontinuous gradient was then formed on top of the interface fraction by the addition of 1.1 and 0.25 m Suc cushions, followed by centrifugation for 100 min at 100,000g. The interface 0.25/1.1 m Suc fraction was then collected, diluted in 1 volume of distilled water, and centrifuged at 100,000g for 50 min. The pellet was resuspended using a Dounce homogenizer in buffer containing 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5 (STM buffer). The vesicles were kept at −80°C until use. All of these procedures were performed on ice or at 4°C. The UDPase latency of the vesicles varied between 75% and 90% for different preparations.
Enzymatic Assays and Organelle Markers
XG-FT activity was measured at 25°C for 30 min in a final volume of 100 μL, as described by Wulff et al. (2000). Briefly, the reaction contained 1 μm GDP-[3H]Fuc, 100 μg of tamarind XG, 10 mm HEPES/KOH, pH 7.0, 10 mm MnCl2, and 0.05% (v/v) Triton X-100 and was terminated after 30 min by adding 250 mL of 95% (v/v) ethanol. The samples were kept on ice for 30 min, and then filtered through 0.45-μm cellulose ester filters using a filtration system (model FH225V; Hoefer). The filters were then washed three times with 1 mL of 70% (v/v) ethanol containing 1 mm EDTA and dried. The radioactivity was estimated by liquid scintillation counting. To estimate the XG-FT activity in permeabilized vesicles, we added 0.05% (v/v) Triton X-100 to the incubation medium.
The HGA-MT assay was modified from the method described by Goubet et al. (1998). Briefly, Golgi vesicles (50 μg of protein) or gradient aliquots were incubated in a final volume of 50 μL containing 6 μm [methyl-14C]SAM (final concentration), 0.25 m Suc, 10 mm Tris-HCl, pH 7.5, 1 mm MgCl2 (STM buffer) at 30°C for 1 h. The reaction was then stopped and the methylated products precipitated by the addition of one volume of 20% (w/v) TCA and 5 μL of a 10% (w/v) bovine serum albumin solution. The resulting suspension was centrifuged for 10 min at 4,000g. Unincorporated SAM was removed by washing the pellets twice with 200 μL of 2% (w/v) TCA. The washed pellets were boiled for 10 min with 25 μL of 2 m NaOH, neutralized with 30 μL of 2 m HCl, and the radioactivity incorporation measured by liquid scintillation.
To measure glucosidase II activity, aliquots of 1 mL from the gradients were centrifuged with 1 volume of water for 60 min at 100,000g. The pellets obtained were then resuspended in 100 μL of STM buffer. Glucosidase II activity was determined as described by Trombetta et al. (1996) using 50 μL of resuspended pellet with 1 volume of distilled water in the presence of 20 mm HEPES, pH 7.5, 5 mm p-nitrophenyl-α-glucopiranoside, and Triton X-100 0.02% for 30 min at room temperature. The reaction was terminated by adding 300 μL of 2 m Tris base containing 2% SDS and the absorbance was then measured at 405 nm.
To determine cytochrome c oxidase, 40 μL of the gradient fractions were used to measure the activity as described by Briskin et al. (1987).
TOC 33 was determined by western blot using a specific antibody kindly donated by Dr. Danny Schnell.
Proteolysis of Golgi Vesicles
Golgi vesicles (200 μg of protein) were incubated with 5 or 25 μg of trypsin in a solution containing 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5, for 10 min at 30°C in a final volume of 25 μL. These reactions were also incubated in the presence or absence of 0.02% (v/v) Triton X-100 and were stopped by the addition of 1.25 μL of a 10 μg/μL concentration of soybean trypsin inhibitor, followed by an incubation for 10 min at 30°C. As a control, only the trypsin inhibitor was added. The samples were kept in ice until the enzymatic reactions were performed.
Uptake of SAM and GDP-Fuc into Golgi Vesicles
Golgi vesicle preparations (100 μg of protein) were incubated for 5 min at 25°C (or as indicated) with 3 μm [methyl-14C]SAM or [carboxyl-14C]SAM in 100 μL of a buffer containing 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5. The incubation was terminated by dilution with 10 volumes of an ice-cold solution containing 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5, and immediately filtered through 0.45-μm cellulose ester filters using the filtration system described above. The filters were then washed with an additional 10 volumes of the same solution, dried, and subjected to liquid scintillation counting.
The uptake of GDP-Fuc was performed using a filtration assay as described previously by Wulff et al. (2000). Golgi vesicles (100 μg of protein) were incubated with 1 μm GDP-[3H]Fuc in the same incubation buffer used for the SAM uptake assay. The reaction was terminated in the same manner as described above and the radioactivity associated with the filters was determined by liquid scintillation counting.
Enzymatic Treatments
Golgi vesicles (50 μg of protein) or aliquots from the gradients were incubated at 30°C for 1 h in 50 μL of a solution containing 6 μm [methyl-14C]SAM, 0.25 m Suc, 10 mm Tris-HCl, pH 7.5, and 1 mm MgCl2. The reaction was stopped and the methylated products precipitated by the addition of 1 volume of 20% (w/v) TCA and 5 μL of a 10% (w/v) bovine serum albumin solution. The resulting suspension was centrifuged for 10 min at 4,000g. The TCA insoluble fraction was resuspended in 50 μL of 0.1 m NH4Ac, pH 4.0, for pectinase treatment or 0.1 m Tris-Cl, pH 7.5, for PME or trypsin treatment. For these analyses, either 0.3 units of pectinase, 1 unit of PME, or 25 μg of trypsin was added to the samples, followed by an incubation for both 30 min and 4 h at 25°C for pectinase or 30°C for PME and trypsin. The reaction was stopped by adding 1 volume of 20% TCA. The samples were then centrifuged for 15 min at 10,000g, and the radioactivity associated with the pellet and the supernatant was determined by liquid scintillation counting.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of SAH in the incorporation of SAM into Golgi vesicles and HGA-MT activity.
Supplemental Table S1. Incorporation of SAM is sensitive to osmotic changes.
Supplementary Material
Acknowledgments
We are very grateful to Francisca Reyes for her help with the subcellular fractionation experiments.
This work was supported by Fondecyt 1030551 and the Millennium Nucleus in Plant Cell Biology (ICM P 02–009–F).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ariel Orellana (aorellana@unab.cl).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albersheim P, Darvill AG, O'Neill MA, Schols HA, Voragen AGJ (1996) An hypothesis: the same six polysaccharides are components of the primary cell walls of all higher plants. In J Visser, AGJ Voragen, eds, Pectins and Pectinases. Elsevier, Amsterdam, pp 47–55
- Agrimi G, Di Noia MA, Marobbio CMT, Fiermonte G, Lasorsa FM, Palmieri F (2004) Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem J 379 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker H, Routier F, Oelmann S, Jordi W, Lommen A, Gerardy-Schahn R, Bosch D (2005) Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology 15 193–201 [DOI] [PubMed] [Google Scholar]
- Baldwin TC, Handford MG, Yuseff MI, Orellana A, Dupree P (2001) Identification and characterization of GONST1, a Golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13 2283–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard T, Schaumann-Gaudinet A, Bruyant-Vannier MP, Morvan C (1997) Various pectin methyltransferase activities with affinity for low and highly methylated pectins. Plant Cell Physiol 38 259–267 [Google Scholar]
- Bouvier F, Linka N, Isner JC, Mutterer J, Weber APM, Camara B (2006) Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briskin DP, Leonard RT, Hodges TK (1987) Isolation of the plasma membrane: membrane markers and general principles. Methods Enzymol 148 542–558 [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M, Atta M, Mulliez E (2004) S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci 29 243–249 [DOI] [PubMed] [Google Scholar]
- Goubet F, Council LN, Mohnen D (1998) Identification and partial characterization of the pectin methyltransferase “homogalacturonan methyltransferase” from membranes of tobacco cell suspensions. Plant Physiol 116 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Mohnen D (1999. a) Subcellular localization and topology of homogalacturonan methyltransferase in suspension-cultured Nicotiana tabacum cells. Planta 209 112–117 [DOI] [PubMed] [Google Scholar]
- Goubet F, Mohnen D (1999. b) Solubilization and partial characterization of homogalacturonan-methyltransferase from microsomal membranes of suspension-cultured tobacco cells. Plant Physiol 121 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52 119–137 [DOI] [PubMed] [Google Scholar]
- Horne DW, Holloway RS, Wagner C (1997) Transport of S-adenosylmethionine in isolated rat liver mitochondria. Arch Biochem Biophys 343 201–206 [DOI] [PubMed] [Google Scholar]
- Kende H (1993) Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 44 283–307 [Google Scholar]
- Kauss H, Hassid WZ (1967) Enzymatic introduction of the methyl ester groups of pectin. J Biol Chem 242 3449–3453 [Google Scholar]
- Marobbio CM, Agrimi G, Lasorsa FM, Palmieri F (2003) Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J 22 5975–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50 605–614 [DOI] [PubMed] [Google Scholar]
- Muñoz P, Norambuena L, Orellana A (1996) Evidence for a UDP-glucose transporter in Golgi apparatus derived vesicles from pea and its possible role in polysaccharide biosynthesis. Plant Physiol 112 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckelmann G, Orellana A (1998) Metabolism of uridine 5′-diphosphate glucose in Golgi vesicles from pea stems. Plant Physiol 117 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norambuena L, Nilo R, Handford M, Reyes F, Marchant L, Meisel L, Orellana A (2005) AtUTr2 is an Arabidopsis thaliana nucleotide sugar transporter located in the Golgi apparatus capable of transporting UDP-galactose. Planta 222 521–529 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, Albersheim P, Darvill A (1990) The pectic polysaccharides of primary cell walls. In PM Dey, ed, Methods in Plant Biochemistry, Vol 2. Academic Press, London, pp 415–441
- Ordenes VR, Reyes FC, Wolff D, Orellana A (2002) A thapsigargin-sensitive Ca2+ pump is present in the pea Golgi apparatus membrane. Plant Physiol 129 1820–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Arrigoni R, Blanco E, Carrari F, Zanor MI, Studart-Guimareas C, Fernie AR, Palmieri F (2006) Molecular identification of an Arabidopsis S-adenosylmethionine transporter: analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol 142 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284 1976–1979 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, Douce R (2004) Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the citosol. J Biol Chem 279 22548–22557 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollwitz I, Santaella M, Hille D, Flugge UI, Fischer K (2006) Characterization of AtNST-T1, a novel UDP-galactose transporter from Arabidopsis thaliana. FEBS Lett 580 4246–4251 [DOI] [PubMed] [Google Scholar]
- Schröder G, Eichel J, Breinig S, Schröder J (1997) Three differentially expressed S-adenosylmethionine synthetases from Catharanth roseus: molecular and functional characterization. Plant Mol Biol 33 211–222 [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kolli VSK, Quigley HF, Hahn MG, Mohnen D (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Quigley HF, Orellana A, Mohnen D (2001) The catalytic site of the pectin biosynthetic enzyme α-1,4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol 127 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES, Simons JF, Helenius A (1996) Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J Biol Chem 271 27509–27516 [DOI] [PubMed] [Google Scholar]
- Vannier MP, Thoiron B, Morvan C, Demarty M (1992) Localization of methyltransferase activities throughout the endomembrane complex system of flax (Linum usitatissimum L) hypocotyls. Biochem J 286 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallsgrove RM, Lea PJ, Miflin BJ (1983) Intracellular localization of aspartate kinase and the enzymes of threonine and methionine biosynthesis in green leaves. Plant Physiol 71 780–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff C, Norambuena L, Orellana A (2000) GDP-fucose uptake into the Golgi apparatus during xyloglucan biosynthesis requires the activity of a transporter-like protein other than the UDP-glucose transporter. Plant Physiol 122 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GF, Staehelin LA (1992) Functional compartmentalization of the Golgi apparatus of plant cells. An immunocytochemical analysis of high pressure frozen and freeze-substituted sycamore maple suspension culture cells. Plant Physiol 99 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.