Abstract
Plant endosperm cells have a nuclear ratio of two maternal genomes to one paternal genome. This 2 to 1 dosage relationship provides a unique system for studying the additivity of gene expression levels in reciprocal hybrids. A combination of microarray profiling and allele-specific expression analysis was performed using RNA isolated from endosperm tissues of maize (Zea mays) inbred lines B73 and Mo17 and their reciprocal hybrids at two developmental stages, 13 and 19 d after pollination. The majority of genes exhibited additive expression in reciprocal hybrids based on microarray analyses. However, a substantial number of genes exhibited nonadditive expression patterns, including maternal like, paternal like, high parent like, low parent like, and expression patterns outside the range of the parental inbreds. The frequency of hybrid expression patterns outside of the parental range in maize endosperm tissue is much higher than that observed for vegetative tissues. For a set of 90 genes, allele-specific expression assays were employed to monitor allelic bias and regulatory variation. Eight of these genes exhibited evidence for maternally or paternally biased expression at multiple stages of endosperm development and are potential examples of differential imprinting. Our data indicate that parental effects on gene expression are much stronger in endosperm than in vegetative tissues.
Endosperm tissue, a terminally differentiated tissue that functions as the storage organ of the seed and provides a large proportion of the world's food supply, provides a unique system for studying the effect of gene dosage on expression levels in reciprocal hybrids. The endosperm is a triploid tissue produced from the double-fertilization event in plants and consists of two maternal genomes and one paternal genome (denoted 2m:1p). This unbalanced dosage of the maternal and paternal genomes provides a controlled biological system for studying the effects of allelic dosage and parental effects on gene expression.
Studies of F1 hybrid vegetative tissues from maize (Zea mays) have revealed high levels of additive (equal to the average of the two parents) expression with low levels of nonadditive (different from the average of the two parents) expression levels (Guo et al., 2006; Stupar and Springer, 2006; Swanson-Wagner et al., 2006; Springer and Stupar, 2007a). However, several other studies have found higher levels of nonadditive expression (Auger et al., 2005; Uzarowska et al., 2007). There is relatively little information on the prevalence of additive and nonadditive expression in the triploid endosperm tissue. Guo et al. (2003) examined additivity in endosperm gene expression using a cDNA differential display approach. The majority of genes exhibited additive expression in the hybrid maize endosperm, meaning that hybrid gene expression was approximately equal to the average level of the inbred parents when factoring for 2m:1p transcript contributions (Guo et al., 2003). However, a portion of genes exhibited nonadditive expression patterns in the hybrids. Specifically, Guo et al. (2003) reported that approximately 8% of the genes that were differentially expressed between inbred parents displayed either maternal-like expression (MLE; pattern in which the reciprocal hybrids display expression levels similar to the maternal inbred level) or paternal-like expression (PLE; pattern in which the reciprocal hybrids display expression levels similar to the paternal inbred level) patterns in the hybrids.
A growing body of evidence suggests that the endosperm is subject to genetic mechanisms that differ from vegetative tissues. The most abundantly expressed mRNA gene families in the maize endosperm, the zein protein storage genes, exhibit variable expression patterns among gene family members and nonadditive expression patterns in F1 hybrid endosperm tissue (Woo et al., 2001; Song and Messing, 2003). Analysis of DNA methylation patterns in endosperm reveal programmed alterations to the maternal or paternal alleles (Lauria et al., 2004; Gutierrez-Marcos et al., 2006; Haun et al., 2007). There is also extensive evidence that the maize endosperm is highly sensitive to parental genome imbalances. Disruptions of the 2m:1p genome ratio, as a consequence of interploidy crosses or crosses involving aneuploid parents, can severely affect endosperm phenotype (for review, see Gehring et al., 2004). Perhaps the most distinctive transcriptional feature observed in the endosperm is that it is the only plant tissue in which gene-specific imprinting has been observed (for review, see Baroux et al., 2002; Gehring et al., 2004; Feil and Berger, 2007). Imprinting occurs when the expression of a gene is influenced by the maternal or paternal transmission such that one allele is preferentially expressed. A small number of imprinted plant genes and alleles have been identified, exhibiting a wide range of putative functions (Feil and Berger, 2007). Gene-specific imprinting can be broadly divided into two categories, binary imprinting and differential imprinting (Dilkes and Comai, 2004). Binary imprinting is the more extreme case in which one allele is active and the homologous allele is completely silent based upon the parent of origin. The majority of imprinted genes characterized to date are putative examples of binary imprinting in which only the maternal allele is expressed. Differential imprinting refers to biased allelic expression dependent upon the parent of origin (Dilkes and Comai, 2004). For example, a differentially imprinted gene may exhibit preferential expression of the maternal allele in all F1 hybrids, but the paternal allele will still exhibit some degree of transcription. There is evidence for differential imprinting at the Nrp1 locus of maize (Guo et al., 2003). However, the techniques used to characterize imprinted genes are often insufficient to detect or distinguish between binary and differential imprinting.
In this study, we investigated the gene expression of inbred and hybrid maize endosperm tissues at two developmental time points using the Affymetrix 18 K maize GeneChip microarray. We have found that endosperm tissues at both developmental stages exhibit a greater range of nonadditive expression in hybrids than was previously observed using the same experimental approaches in three vegetative tissues (Stupar and Springer, 2006). Furthermore, hybrid endosperm expression patterns commonly exhibit parental effects that were not observed in vegetative tissues. We also investigated the relative allelic expression of genes in hybrid endosperm tissue at several developmental stages. Through the use of quantitative allele-specific expression assays, we could assess the contribution of cis- and trans-acting regulatory variation as well as the prevalence of parental effects and imprinting on gene expression.
RESULTS
Affymetrix 18 K maize GeneChips were used to profile gene expression in three biological replicates of maize endosperm tissues isolated at 13 and 19 d after pollination (DAP) from B73, Mo17, and the reciprocal F1 crosses. The two developmental time points of 13 and 19 DAP were chosen based on the developmental program of the endosperm in maize (Kowles and Phillips, 1985; Schweizer et al., 1995). Mitotic activity often peaks around 12 to 14 DAP, but thereafter diminishes and is essentially absent by 18 to 20 DAP. However, nuclear endoreduplication occurs between 14 to 20 DAP and the peak of DNA content per nucleus occurs sometime between 18 and 22 DAP.
The four different endosperm genotypes can be represented as follows: BBB, MMM, BBM, and MMB (the first two letters represent the maternal contribution and the final letter represents the paternal contribution). The presence or absence of a given transcript was determined using MAS 5.0 data processing, as described previously (Stupar and Springer, 2006). Expression was consistently detected for 60% of genes in 13-DAP and 55% of genes in 19-DAP endosperm tissue (Supplemental Table S1), indicating that many of the genes represented on this array platform are transcribed in maize endosperm tissues. Comparisons of the microarray data from different tissue types or developmental stages can be useful to identify developmentally regulated transcripts. Analysis of the presence/absence for transcripts in these endosperm tissues as compared to three vegetative tissues (Stupar and Springer, 2006) allowed for the identification of a set of endosperm-specific transcripts (Supplemental Data; validations for a subset of endosperm-specific transcripts are shown in Supplemental Fig. S1 and Supplemental Table S2). We also identified numerous genes with evidence for differential expression in 13-DAP endosperm relative to 19-DAP endosperm (Supplemental Fig. S2; Supplemental Table S3).
Identification of Genes That Are Differentially Expressed in B73, Mo17, and Hybrid Endosperm
Correlation plots indicate high levels of differential expression between the genotypes in both 13- and 19-DAP endosperm tissues (Supplemental Fig. S3). We performed statistical tests (ANOVA across the BBB, MMM, BBM, and MMB genotypes; see “Materials and Methods” for details) to identify genes that are differentially expressed among genotypes in 13- or 19-DAP tissues (Table I). Following the statistical tests, the lists of differentially expressed genes were further filtered by imposing minimal expression (genotype with highest expression must have a microarray signal >50) and fold change (must be >2-fold change between any highest and lowest genotypes) criteria. These filtering criteria were employed to limit our set of differentially expressed genes to those with robust evidence for differential expression and to allow for classification of hybrid expression patterns as additive or nonadditive. Whereas it is likely that some examples of differential expression with less than 2-fold change are biologically significant, it is quite difficult to classify the specific mode of additive or nonadditive expression of such patterns using the tests described below.
Table I.
Differentially expressed genes between BBB, MMM, BBM, and MMB endosperm genotypes
DE, Differentially expressed.
| 13 DAP | 19 DAP | Both 13 and 19 DAP | |
|---|---|---|---|
| No. DE genes | 2,746 | 3,140 | 1,387 |
| No. DE genes that pass filtersa | 1,725 | 1,557 | 939 |
| Percent DE genes in BBB versus MMMb | 97.4% | 98.2% | 98.0% |
| Percent genes with >8/15 probes significant | 76.5% | 88.1% | 84.2% |
| Percent genes with BBB > MMM | 46.8% | 49.1% | 51.1% |
Number of genes that exhibit minimal signal (>50) and minimal fold-change (>2-fold) requirements.
Percentage of filtered DE genes that exhibit statistically significant differences in BBB relative to MMM.
A list of 1,725 genes meeting the statistical and filtering criteria were identified in 13-DAP endosperm (Supplemental Table S4) and 1,557 such genes were identified at 19-DAP endosperm (Supplemental Table S5). These 13- and 19-DAP endosperm gene lists were compared to determine their overlap (Table I). Approximately 60% of these genes (939) were identified in both 13- and 19-DAP endosperm tissue, indicating that many genes exhibit differential expression among genotypes at multiple stages of endosperm development. The vast majority of genes that were identified as differentially expressed in comparisons of these four genotypes were differentially expressed in B73 relative to Mo17 in both 13-DAP (97.4%) and 19-DAP (98.2%) endosperm (Table I). This indicates that there are relatively few examples of genes with similar expression in the inbred parents and novel expression states in one or both of the hybrids.
Cross-Platform Validation of Microarray Data and Tests for Sequence Polymorphism Effects
The use of microarrays to compare gene expression levels in different genotypes can be complicated by sequence polymorphisms in the regions detected by array features (Borevitz et al., 2003; Kirst et al., 2006). Whereas it is true that a subset of the features on the Affymetrix microarray will exhibit polymorphisms in the B73 and Mo17 targets, it is unclear to what extent this will affect the probe-level comparisons in this study. We have employed an analysis of the individual probe signals for genes that were identified as differentially expressed to assess the prevalence of polymorphisms that affected a small number of probes for a gene and resulted in a false call of differential expression. For each differentially expressed gene, we compared the signal of the perfect match in B73 relative to Mo17 using a t test (P < 0.05). The majority of genes exhibit at least 8/15 probes with a significant difference between the two genotypes (Supplemental Fig. S4A). This finding suggests that sequence polymorphisms were not a major contributor to false positives for differential expression because it is unlikely that sequence polymorphisms will occur in the majority of the probe features for a gene. We also performed this analysis to compare the perfect match-mismatch values for each probe and found similar results.
An allele-specific detection platform (Jurinke et al., 2005) was also used to validate the microarray data. Total RNA isolated from three biological replicates of B73 and Mo17 endosperms were mixed in equal concentrations to provide three different biological replicates of a 1:1 mix of the inbred RNA. Allele-specific expression analysis was used to detect the proportion of the B73 allele in the 1:1 mixes. This value was then compared to the proportion of the B73 allele that should be present based on the microarray data [B73 signal/(B73 signal + Mo17 signal)]. The allele-specific expression data and microarray data were compared for a set of 81 genes, 15 of which exhibited differential expression with at least 2-fold change based on the microarray data. This gene set included nine genes that were differentially expressed among the inbreds in either the 13- or 19-DAP tissues and six genes that were differentially expressed in both developmental stages, resulting in a total of 21 genes by tissue combinations. A scatter plot of the allele-specific expression data versus the microarray data provides evidence for strong correlation between the two platforms (Supplemental Fig. S4B). The allelic proportions for all 21 assays supported the microarray determination of differential expression. There were another 24 genes for which allele-specific expression data were available that were differentially expressed in the microarray analysis at levels below 2-fold change. The differential expression was validated for 20 of these 24 genes using allele-specific expression data.
We proceeded to compare allele-specific expression data and microarray data for all 81 genes analyzed by both methods. The allele-specific expression data and the microarray data exhibited strong correlation between the two platforms (Supplemental Fig. S4C). However, a few genes displayed significant differences in the microarray data relative to allele-specific detection assays. In some cases, such as AI600480 and BM381185, the microarray data show much more severe differences in B73 relative to Mo17 than is detected by allele-specific assays. This may be the result of polymorphisms that affect the hybridization of one of the alleles to the array. In other instances, such as CA403984 and CF009268, analysis of both 13- and 19-DAP tissues finds evidence for strong allelic bias with relatively little differential signal between B73 and Mo17 on the microarrays. The general finding from these cross-platform comparisons was that both platforms found similar relative expression levels in B73 and Mo17 for the majority of genes with a few instances in which the two platforms do not agree.
Observations of Specific Nonadditive Expression Patterns in Hybrid Endosperm
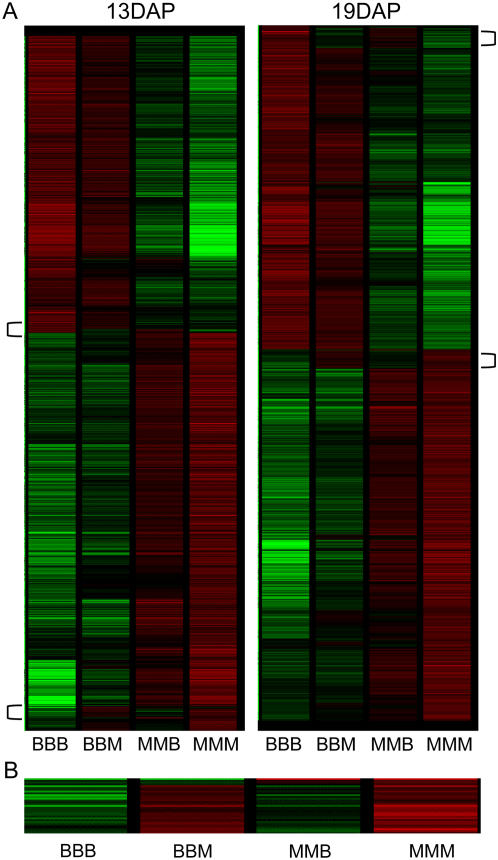
Due to the 2m:1p genome dosage in the endosperm, we expected that the majority of genes would exhibit additive expression patterns in hybrid endosperm such that the hybrids are more similar to the maternal parent than to the paternal parent. Clustering analysis of the gene expression levels for all differentially expressed genes provided evidence that many genes exhibit this type of additive expression pattern reciprocal hybrid (Fig. 1). However, this clustering analysis reveals evidence for some groups of genes with specific types of nonadditive expression. For example, the gene expression profiles indicated by brackets in Figure 1A (and magnified in Fig. 1B) exhibited hybrid expression levels more similar to the paternal parent than the maternal parent. Statistical analyses of inbred and hybrid expression levels were used to identify the proportion of additive and nonadditive expression profiles. Each gene was tested by comparing the hybrid expression value against the expected additive expression value based on the parental expression, while factoring for the 2m:1p dosage (see “Materials and Methods”). Most genes (58.8%) in the 13-DAP endosperm tissues did not exhibit nonadditive patterns in either reciprocal hybrid; however, many genes (30.8%) exhibited nonadditive expression in one reciprocal hybrid and fewer (10.3%) exhibited nonadditive expression in both reciprocal hybrids. Similar proportions were identified in the 19-DAP endosperm tissues (61.1% were nonadditive in neither, 29.7% were nonadditive in one, and 9.1% were nonadditive in both reciprocal hybrids).
Figure 1.
Gene expression and sources of variation among endosperm genotypes. A, Hierarchical clustering of the genes that are differentially expressed among genotypes in 13- and 19-DAP endosperm. Green indicates low relative expression, whereas red indicates high relative expression and black indicates no difference relative to the profile average. A subset of exceptional genes with hybrid expression profiles similar to the male parent are indicated by brackets. B, The bottom bracketed region from the 19-DAP clustering is enlarged to show that the hybrid expression level is more similar to the male parent than to the female parent.
We defined 14 specific types of nonadditive expression that could occur in endosperm tissue (see “Materials and Methods” for definitions and descriptions of each class; sample visualizations are provided in Supplemental Fig. S5). A set of fold-change and statistical significance criteria (as described in Supplemental Table S6) were applied to all 13- and 19-DAP differentially expressed genes to identify genes with strong evidence for any of the 14 different classes of nonadditive expression (Table II). Note that many of the genes that exhibit nonadditive expression (based on a per-gene test for additivity) do not classify into these 14 specific types of nonadditive expression.
Table II.
Identification of specific nonadditive hybrid expression classes among endosperm genotypes
| 13 DAP | 19 DAP | Combineda | |
|---|---|---|---|
| PLE | 5 | 7 | 12 |
| MLE | 79 | 46 | 120 |
| LP | |||
| LP (B73) | 3 | 2 | 5 |
| LP (Mo17) | 1 | 6 | 7 |
| HP | |||
| HP (B73) | 3 | 5 | 8 |
| HP (Mo17) | 20 | 4 | 24 |
| Partial-to-complete expression outside of the parental range | |||
| One hybrid outside of parental range | |||
| BBM expressed AHP | 15 | 7 | 21 |
| BBM expressed BLP | 33 | 21 | 51 |
| MMB expressed AHP | 13 | 16 | 28 |
| MMB expressed BLP | 8 | 9 | 15 |
| Both hybrids outside of parental range | |||
| Both hybrids expressed AHP | 0 | 0 | 0 |
| Both hybrids expressed BLP | 0 | 0 | 0 |
| Hybrid bracket parents | |||
| BBM expressed AHP and MMB expressed BLP | 3 | 3 | 6 |
| MMB expressed AHP and BBM expressed BLP | 6 | 3 | 9 |
| Total number of differentially expressed genes assigned to these expression classes | 189 | 129 | 306 |
| Number of differentially expressed genes not assigned to specific expression class | 1,536 | 1,428 | 2,037 |
The combined list is the nonredundant genes identified at either time point.
We identified a total of 306 genes (out of 2,343 differentially expressed genes tested) belonging to one of the 14 specific types of nonadditive expression in at least one endosperm developmental stage (Supplemental Table S7). Despite the use of relatively stringent filters, the MLE patterns were the most prevalent type of nonadditive expression observed (Table II). Expression patterns with one hybrid outside the range of the parents, either above the high parent (AHP) or below the low parent (BLP), were also relatively common. PLE and hybrid expression outside the range of the parents in opposite directions (AHP and BLP) were rarely detected. We did not find any examples with consistent AHP or BLP expression patterns in both hybrids (Table II). To further assess and confirm whether the same patterns of nonadditive expression were consistently observed for a gene at both 13 and 19 DAP, we visually inspected the relative transcript levels for the four genotypes at both 13 and 19 DAP to determine whether the same type of nonadditive expression patterns were consistently observed for a gene at both developmental stages (Supplemental Figs. S5–S12). We observed consistent evidence for a similar pattern of nonadditive expression at both stages of development for many of these genes.
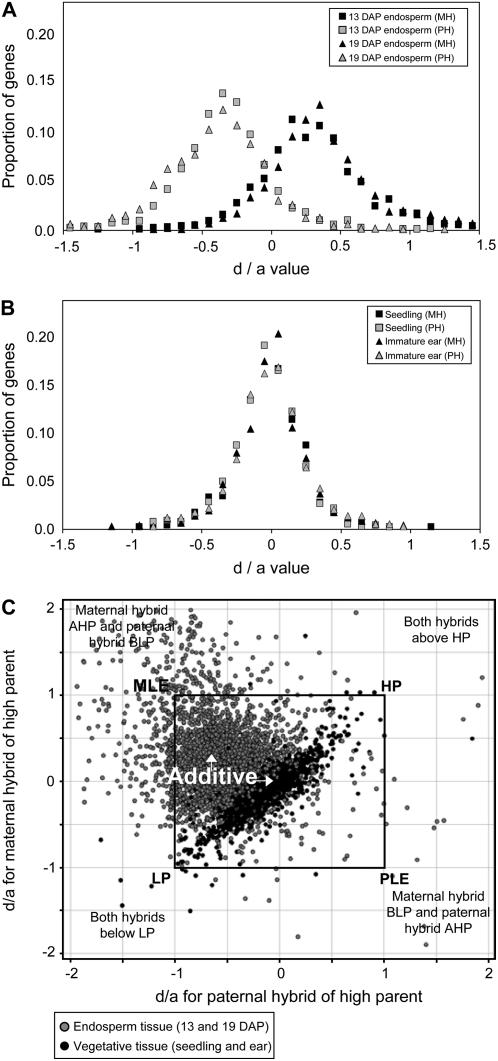
Previous studies have identified unique expression phenomena in endosperm tissues (see introduction). We utilized previously published microarray data on 11-d seedling and immature ear tissues (Stupar and Springer, 2006) to compare the inbred-hybrid gene expression profiles of endosperm versus these vegetative tissues. We used the d/a ratio as a means to compare the global expression patterns of hybrid endosperm and vegetative tissues. The d/a ratio is a measure that is frequently used to compare the expression value of the hybrid relative to the parents. As applied to gene expression studies, the d/a value often represents the hybrid expression value relative to parent 1 and parent 2. In this study, we have slightly modified the calculation such that the d/a value represents the hybrid expression value relative to the high parent (HP) and low parent (LP) for each gene. The d value is the difference between the hybrid and the midparent expression levels, whereas the a value is the difference between the HP and the midparent expression levels. Genes with d/a values equal to 1.0 or −1.0 exhibit hybrid expression levels equal to the HP or LP, respectively. Values above 1.0 or below −1.0 indicate genes with hybrid expression levels outside the range of the parents. In vegetative tissues, additive expression will result in a d/a value of 0. However, due to the dosage imbalance in endosperm tissue, additive expression would result in a maternal hybrid (relative to the higher expressing parent) d/a value of 0.33 and a paternal hybrid (relative to the higher expressing parent) d/a value of −0.33.
Figure 2, A and B, shows the distribution of d/a values for genes exhibiting differential expression in endosperm and vegetative tissues (11-d seedling and immature ear), respectively. As expected, the maternal and paternal hybrid endosperm values centered on 0.33 and −0.33, respectively. The maternal and paternal hybrids in vegetative tissues exhibit a distribution centered on 0. The endosperm tissues exhibit a larger spread around the peak and display more values that are outside the parental range (above 1.0 or below −1.0). In Figure 2C, we have compared the d/a values for the reciprocal hybrids in endosperm and vegetative tissues; the d/a values for the maternal hybrid (relative to the higher expressing parent) are on the y axis and the d/a values for the paternal hybrid are on the x axis. Genes that plot within the black square represent genes that exhibit hybrid expression values within the range of the parents. Deviation toward the top left (MLE) and bottom right (PLE) indicates parental effects on hybrid expression levels. Deviation toward the top right (HP) and bottom left (LP) indicates hybrid expression patterns that are consistently toward the high or low expressing parent. Whereas the majority of data points are near the expected additive values for both tissue types, there are numerous points that exhibit deviation from this center. Interestingly, there is much greater evidence for parental effects in endosperm than in vegetative tissues (the distribution of nonadditive endosperm genes tend to plot from PLE to MLE, whereas the distribution of nonadditive vegetative genes tend to plot from HP to LP). To further compare nonadditive expression patterns in endosperm and vegetative tissues, we determined the number of genes in the 14 specific nonadditive expression classes (described above) in seedling and immature ear tissues. The total percentage of genes with nonadditive expression in both hybrids was similar in endosperm and vegetative tissues (Table III). However, as was observed in the d/a plot (Fig. 2B), vegetative tissues display a markedly different distribution of nonadditive expression patterns with essentially no examples of nonadditive expression influenced by the parent of origin (Table III). Many of the nonadditive expression patterns in hybrid vegetative tissues are examples of HP or LP (Table III).
Figure 2.
Comparison of additive and nonadditive hybrid expression patterns in endosperm and vegetative tissues. A, The d/a values are plotted for all differentially expressed genes in endosperm tissue. For each differentially expressed gene, the d/a value was calculated for the maternal hybrid relative to the higher expressing parent (e.g. BBM if BBB > MMM) and for the paternal hybrid relative to the higher expressing parent (e.g. MMB if BBB > MMM). The expected values for additive expression are 0.33 for the maternal hybrid and −0.33 for the paternal hybrid. Black symbols indicate the values for the maternal hybrid and gray symbols indicate the values for the paternal hybrid. Symbol shape indicates the tissue stage. B, A similar plot for d/a values is presented for seedling and immature ear vegetative tissues. Due to the equal dosage of the two parental genomes, the expected values for additive expression are 0.0 for both the maternal and paternal hybrids. C, A scatter plot of d/a expression values from reciprocal hybrids in endosperm (gray spots) and vegetative (black spots) tissues shows the density and range of nonadditive expression among tissue types. Each spot represents the d/a value of a gene for the two reciprocal hybrids. The y axis represents d/a values of the maternal hybrid of the HP and the x axis represents d/a values of the paternal hybrid of the HP. The approximate position of the additive and nonadditive classes in this display is indicated by the text. Values that plot within the black square represent genes in which both hybrids exhibit expression levels within the parental range. Genes for which both hybrids display AHP or BLP expression would plot in the top right and bottom left quadrants. Genes exhibiting parent-of-origin effects on hybrid expression will plot in the top left and bottom right quadrants. A small portion of the data plotted outside the range of the graph and is therefore not shown in this display.
Table III.
Classification of nonadditive expression for different maize tissues
| Endosperm (13 DAP) | Endosperm (19 DAP) | Seedling | Immature Ear | |
|---|---|---|---|---|
| No. differentially expressed genesa | 1,725 | 1,557 | 860 | 693 |
| No. genes nonadditive in both hybridsb | 178 (10%) | 142 (9%) | 118 (14%) | 115 (17%) |
| No. genes with MLE patternc | 79 | 46 | 0 | 0 |
| No. genes with PLE patternc | 5 | 7 | 1 | 0 |
| No. genes AHP and BLP in both hybridscd | 9 | 6 | 1 | 0 |
| No. genes AHP or BLP in one hybridce | 59 | 44 | 2 | 3 |
| No. HP or LPc | 27 | 17 | 49 | 58 |
This is the number of genes that are differentially expressed and pass the minimal signal and fold-change filtering requirements (as described in “Results”).
The number of differentially expressed genes that exhibit nonadditive expression was calculated based on a per-gene test for additive expression. The number in parentheses indicates the percentage of differentially expressed genes that exhibit nonadditive expression in both the BBM and MMB hybrids.
The number of genes that exhibit specific types of nonadditive expression patterns are reported. Only genes with nonadditive expression in both reciprocal hybrids are included. Values in bold indicate types of nonadditive expression that exhibit different prevalence in endosperm and vegetative tissues.
All genes in this category exhibit AHP expression in one hybrid and BLP expression in the other hybrid. No genes with AHP or BLP expression in both hybrids were observed.
The number reported here are the number of genes in which both hybrids exhibit nonadditive expression, but only one of the hybrids exhibits AHP or BLP expression. Note that this is smaller than the sum of values in Table II because some values in Table II represent genes that exhibit nonadditive expression in only one of the two hybrids.
Allelic Variation Affecting Expression Levels of the B73 and Mo17 Alleles
We were interested in further studying the maternal and paternal contributions to gene expression in developing endosperm tissue. Whereas the microarray hybridizations were quite useful for assessing the steady-state transcript levels, they did not provide information on the relative expression of the maternal and paternal alleles. Allele-specific expression assays can be used to determine the ratio of maternal and paternal transcripts. A set of 90 quantitative allele-specific expression assays were derived from 72 randomly selected endosperm-expressed genes, 15 genes with differential expression in B73 relative to Mo17 (according to statistical analysis of the Affymetrix data), and three genes with observed nonadditive expression in the microarray data, including two MLE and one PLE gene (see “Materials and Methods” for further details on assay development). Many (81/90) of these allele-specific expression assays represented genes that are also on the Affymetrix maize GeneChip; data from these genes were initially used to assess the prevalence of differential expression of the B73 and Mo17 genes in 13- and 19-DAP endosperm of inbred plants (see section above on cross-platform validation of microarray data and tests for sequence polymorphism effects).
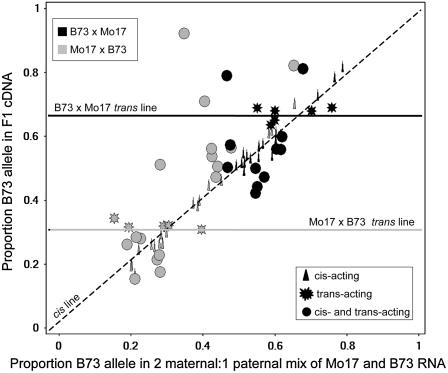
For differentially expressed genes, it is possible to evaluate the relative contribution of cis- and trans-acting regulatory variation by comparing the allele-specific expression ratios in F1 hybrids relative to mixes of the parental RNA (Wittkopp et al., 2004; Stupar and Springer, 2006). In hybrid diploid tissues, cis-acting regulatory variation will result in equally biased expression of the two alleles in a mix of the parental RNA and in the F1 tissue. However, trans-acting variation will result in biased expression of the two alleles in a mix of the parental RNA, but unbiased expression of the two alleles in the F1 because both alleles have access to the same set of trans-acting factors in the F1. The 2m:1p dosage imbalance in endosperm tissue slightly changes the expected results for a cis-/trans-test by allele-specific expression. If a gene is equally expressed in the inbred endosperm of B73 and Mo17, then 1:2 and 2:1 mixes of parental RNA will result in a B73 allelic proportion of 0.33 and 0.66, respectively; deviation from these values indicates that the gene is differentially expressed in the two inbred lines. The cis- and trans-acting variation of differentially expressed genes can subsequently be assessed by comparing the allele-specific expression of the parental mixes versus the hybrids. If the differential expression is caused by trans-acting regulatory variation, then the proportion of the B73 allele in the Mo17 × B73 F1 endosperm tissue will be 0.33 and the proportion of the B73 allele in the B73 × Mo17 F1 will be 0.66. These values will occur because the alleles will all be expressed at the same level because they have access to an identical set of trans-acting factors. However, if differential expression is the result of cis-acting regulatory variation, then the F1 endosperm tissue will exhibit allelic biases similar to those observed in the parental mix samples and will be different from 0.33 (for Mo17 × B73) or 0.66 (for B73 × Mo17).
We assessed regulatory variation for a set of 46 genes that exhibit evidence for differential expression in both microarray and allele-specific expression analyses (including 21 genes with >2-fold change). This included 19 genes with differential expression in both 13- and 19-DAP tissue, 11 genes with differential expression only in 13-DAP tissue, and 16 genes with differential expression only in 19-DAP tissue. A series of statistical tests (as described in Stupar and Springer, 2006; Springer and Stupar, 2007b) were employed to determine the type of regulatory variation causing the differential expression for each gene. For this analysis, we performed separate tests on the BBM and MMB hybrids. Graphic analysis of the endosperm data shows the distribution of regulatory variation observed for these genes (Fig. 3). The symbol shape on this graph indicates the type of regulatory variation as determined by statistical analyses. For 16 genes, there was evidence that cis-acting regulatory variation caused the differential expression and for another five genes the differential expression was due to trans-acting variation. The remaining 25 genes exhibited evidence for both cis- and trans-acting variation contributing to differential expression. Similar to the findings of West et al. (2007), we noted that genes with higher levels of differential expression are frequently the result of cis-acting regulatory variation.
Figure 3.
cis- and trans-acting regulatory variation affecting gene expression levels in endosperm. A comparison of the proportion of the transcript derived from the B73 allele in the F1 RNA (y axis) and in a mix of RNA from the parents (x axis) reveals the type of regulatory variation. Different colors are used to signify whether a data point is derived from the BBM hybrid (black) or the MMB hybrid (gray). Symbol shape indicates the type of regulatory mode ascribed to each gene using a series of statistical tests (as described in Stupar and Springer, 2006). The diagonal line indicates the expected positions for cis-acting regulatory variation, whereas horizontal lines indicate the expected positions for trans-acting variation.
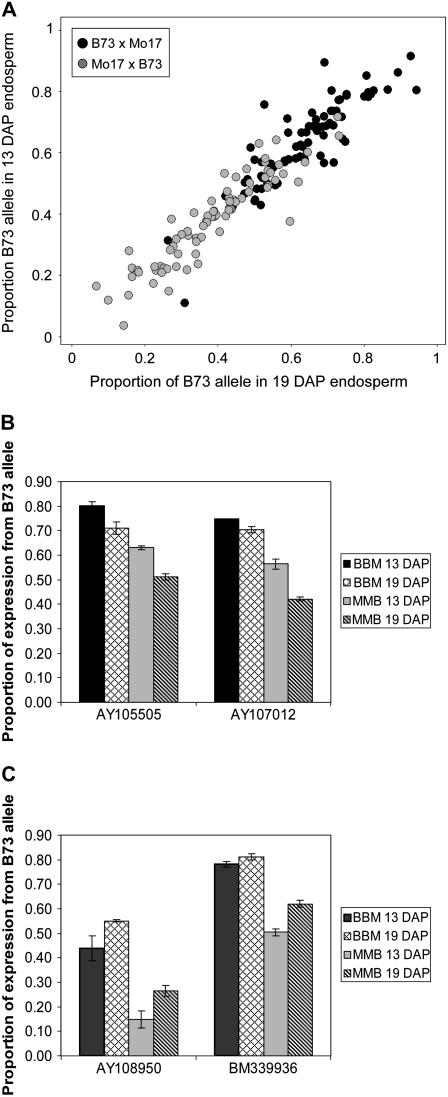
We investigated developmental effects on allelic expression ratios by comparing the transcribed allelic ratios in 13- and 19-DAP hybrid endosperm. Previous studies have demonstrated variation in allelic bias in different tissues or environmental conditions (Guo et al., 2004; Adams and Wendel, 2005; Adams, 2007; Springer and Stupar, 2007b). We compared the proportion of the B73 allele transcribed in hybrid RNA isolated from 13- and 19-DAP endosperm (Fig. 4A). Whereas most genes display equivalent allelic proportions at 13 and 19 DAP, there was evidence for a subset of genes with differential allelic proportions at these two developmental stages. A t test (P < 0.05) was used to compare the relative proportion of transcript derived from the B73 allele at 13 DAP relative to 19 DAP. For 15 genes, we found evidence for a significant difference in the allelic proportion between 13 and 19 DAP for both B73 × Mo17 and Mo17 × B73 F1 hybrids (another 10 genes exhibited a significant difference in one of the two reciprocal hybrids). These 15 genes include seven genes with a decrease in the expression of the B73 allele from 13- to 19-DAP endosperm (two examples are shown in Fig. 4B). The other eight genes displayed evidence for an increase in the relative proportion of the B73 allele from 13- to 19-DAP endosperm (two examples in Fig. 4C).
Figure 4.
Differential expression across endosperm developmental stages. A, The proportion of B73 transcript detected in cDNA isolated from B73 × Mo17 (black) or Mo17 × B73 (gray) is plotted for 13-DAP (y axis) and 19-DAP (x axis) endosperm. Data for all informative genes (86 genes with data in both 13- and 19-DAP endosperm) are presented. B, The proportion of the B73 transcript in B73 × Mo17 and Mo17 × B73 cDNAs is displayed for two genes that show evidence of higher relative expression of the B73 allele in 13- compared to 19-DAP endosperm. C, The proportion of the B73 transcript is displayed for two genes that show evidence of lower relative expression of the B73 allele in 13- compared to 19-DAP endosperm.
Differential Imprinting in Hybrid Endosperms
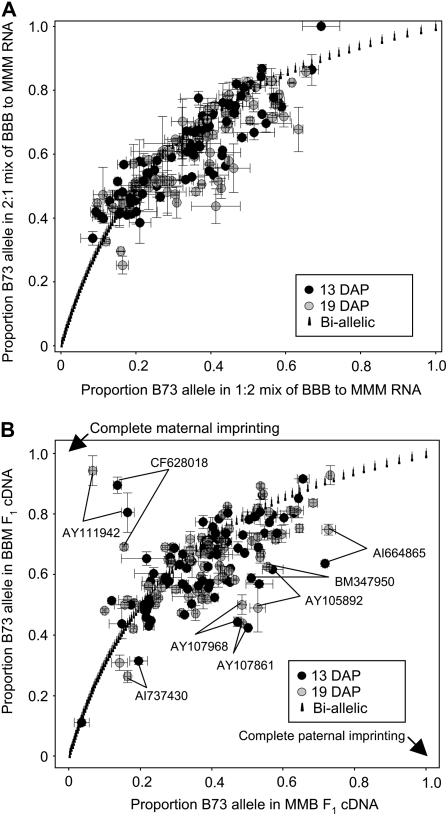
The availability of allele-specific expression data from reciprocal hybrid endosperm tissue allowed us to address the prevalence of parental effects on the relative expression of the two alleles, or imprinting. As a control to determine the noise in a comparison of reciprocal hybrids using this technique, we first compared the B73:Mo17 allelic proportions in a 2:1 mix relative to a 1:2 mix of B73 to Mo17 RNA (Fig. 5A). This comparison between 2:1 and 1:2 mixes of inbred RNA simulates a situation in which hybrid endosperms will exhibit biallelic, nonimprinted expression of the two alleles. For any given gene, the proportions of the B73 allele present in the 2:1 and 1:2 inbred mixes are necessarily dependent upon one another and will theoretically plot along the curve displayed in Figure 5A (labeled as the biallelic curve). The control comparison plotted in Figure 5A represents an estimate of the degree of variance from the biallelic curve that can be attributed to the experimental approach. Whereas we observed some instances of assays deviating from the curve (toward the underside, in particular), the majority of the experimental data exhibit a trend similar to the expected biallelic curve.
Figure 5.
Allele-specific expression to identify transcriptional parent-of-origin effects in hybrid endosperm. A, A control plot was performed by comparing the allele-specific detection of 90 genes in a 1:2 mix versus a 2:1 mix of B73 to Mo17 endosperm RNA. Values are plotted as the proportion of the B73 allele detected in each RNA mix. All genes should plot along the biallelic curve. Values plotting in the top right corner indicate higher expression of the B73 allele, whereas values toward the bottom left corner indicate higher expression of the Mo17 allele. Spots that deviate from the curve indicate the degree of error associated with the experimental procedure. B, Comparison of the allelic expression of reciprocal hybrid endosperm. The majority of the 90 genes analyzed exhibited B73 allelic proportions that plot around the biallelic curve, with deviation similar to the control inbred mixes in A. However, eight genes exhibited B73 allelic proportions that deviate from the curve in both 13- and 19-DAP RNA, suggesting parent-of-origin effects. Accession numbers are shown for these eight genes. The allelic proportions expected for complete maternal and paternal imprinting are also indicated on the plot.
Allele-specific expression analysis was performed on the reciprocal hybrid RNA and a similar graph was produced (Fig. 5B). In this graph, genes that plot along the curve displayed biallelic patterns, genes that plot above the curve toward the top left displayed biased expression of the maternal alleles in both hybrids, and genes that plot below the curve toward the bottom right displayed biased expression of the paternal alleles in both hybrids. Our data indicate that 82 of the 90 genes displayed biallelic expression patterns along or near the curve at both 13- and 19-DAP time points. However, eight of the 90 genes consistently displayed either maternally or paternally biased allelic expression in both the 13- and 19-DAP endosperms (Fig. 5B). Interestingly, only one of these eight genes, BM347950, was a gene that was selected based on displaying nonadditive expression. The other two nonadditive genes for which we performed allele-specific expression assays each displayed allelic expression patterns bias along the biallelic curve.
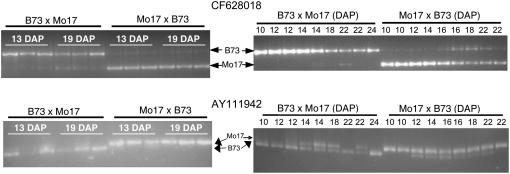
Two genes, AY111942 and CF628018, displayed maternally biased expression in reciprocal hybrids at both 13 and 19 DAP. In both cases, there was also evidence for low levels of expression from the paternal allele, suggesting that these are cases of differential imprinting. We proceeded to confirm the maternally biased expression for both of these genes using cleaved amplified polymorphic sequence (CAPS) assays on the same RNA samples used for the single-nucleotide polymorphism-based analyses (Fig. 6). For each of the 12 samples, the maternal allele is highly enriched, but there are low levels of paternal allele present. We proceeded to determine whether this differential imprinting could be consistently observed throughout endosperm development. RNA samples isolated from a series of endosperm tissue samples collected between 10 and 24 DAP were analyzed using the CAPS assays (Fig. 6). The gene CF628019, a putative polygalacturonase-inhibiting protein, exhibits evidence for differential imprinting in most of the samples analyzed. However, there is evidence for an increase in the expression of the paternal allele in some of the samples from later developmental time points (22 DAP in BBM; 16, 18, and 22 DAP in MMB). Similar results are obtained for the putative sugar transporter gene AY111942; differential imprinting (low levels of paternal allele observed) in some samples and potentially biallelic expression in other samples. There was not a strict developmental relationship that was observed such that imprinting was becoming more or less pronounced in later time points for this gene. Instead, there is evidence for variable parent-of-origin effects on the expression level of the maternal and paternal alleles of this gene.
Figure 6.
Validation of differential imprinting of CF628018 and AY111942. CAPS assays were used to validate the differential imprinting in the same RNA samples used for the allele-specific expression assays shown in Figure 5. The top two images display the results obtained for gene CF628018, whereas the bottom two images display results obtained for gene AY111942. On the left, amplicons were produced from the three biological replicates of 13- and 19-DAP endosperm from either BBM or MMB and were subsequently digested with an appropriate restriction enzyme (Bme1580I for CF628018 and PleI for AY111942) to distinguish the two alleles. The enrichment for the maternal allele in each sample and the presence of low levels of the paternal allele supports the differential maternal imprinting observed by allele-specific expression assays. The two images on the right display the results obtained using the same CAPS assays to assess the differential imprinting of these two genes in a series of developmental stages of endosperm tissue. RNA from endosperm tissue at various developmental stages was isolated from B73 × Mo17 and Mo17 × B73 reciprocal hybrids. The number of DAP for each endosperm tissue sample is indicated above each lane of the gel. The location of the B73 and the Mo17 allelic bands is indicated on the left-hand side of the gel image.
Six genes exhibited consistent evidence for biased expression of the paternal allele in reciprocal hybrid endosperm tissue at 13 and 19 DAP. For each of these genes, we were able to detect expression of both the maternal and paternal alleles. However, instead of being expressed in a manner consistent with the 2:1 allelic dosage, we observed similar allelic ratios between the reciprocal hybrids. Allele-specific assays performed on endosperm DNA controls confirm that the 2:1 allelic dosage is maintained in genomic DNA. These genes are potential examples of differential imprinting favoring the paternal allele or may be genes that are subject to dosage compensation. These genes include a putative zinc-dependent protease, a putative casein kinase, a putative lipoxygenase, a ubiquitin-associated domain protein, and two hypothetical proteins.
DISCUSSION
The endosperm tissue plays an important role in seed development in angiosperms and is a critical part of the world's food supply. Despite this importance, we still have a limited understanding of the basic processes that affect gene expression patterns in the endosperm. The endosperm expression profiles in this study were compared with previous profiling experiments using maize vegetative tissues (Stupar and Springer, 2006). This comparison provided evidence for transcripts that are specific to the endosperm. This finding is consistent with previous cDNA and EST-based studies that reported a large subset of endosperm-specific and endosperm-preferred transcripts in maize (Woo et al., 2001; Lai et al., 2004; Verza et al., 2005). We also noted many differences in gene expression between 13- and 19-DAP endosperm tissue and a substantial level of variation for gene expression between two maize inbred lines, B73 and Mo17. Our findings have allowed us to address important topics, including the prevalence of specific classes of nonadditive expression and the prevalence of differential imprinting in endosperm tissue.
Potential Mechanisms for Nonadditive Expression Patterns
Most of the differentially expressed genes in our study exhibited additive levels of gene expression in hybrid endosperm tissues. However, approximately 10% of the differentially expressed genes exhibit nonadditive expression in both reciprocal hybrids (Table III). Some of these genes with nonadditive expression could be classified into nonadditive expression patterns that can be divided into four main categories: MLE, PLE, dominant patterns (HP and/or LP), and hybrid outside the range of the parents (AHP and/or BLP). For each category, we will provide a short description and discuss potential mechanisms that could be involved.
MLE patterns, defined here as reciprocal hybrid expression levels that are each statistically indistinguishable from the maternal parent, were the most common type of nonadditive expression in endosperm tissue. There are several potential mechanisms that might account for MLE patterns. Binary or differential imprinting of a gene that is differentially expressed in B73 relative to Mo17 may produce a MLE pattern (Dilkes and Comai, 2004). However, allele-specific expression analyses for two of the MLE genes in our study did not find any evidence for maternal imprinting; thus, other mechanisms must also be generating the MLE patterns. For example, maternal influences on endosperm development may also result in MLE patterns. It is known that many seed phenotype characteristics are heavily influenced by the maternal parent with very little contribution of the paternal parent.
PLE patterns, classified here as reciprocal hybrid expression levels that are more similar to the paternal parent than to the maternal parent, were much more rare than MLE patterns, occurring in only 12 genes (<0.5%) of the differentially expressed gene subset. PLE patterns may be the result of paternal imprinting or may result from complex interactions between the two parental genomes. Allele-specific expression for one of the PLE genes identified in this study, BM347950, indicated an expression bias for the paternal allele in hybrid maize endosperm, suggesting differential imprinting for this gene.
It is worthwhile to note the limitations in using microarray expression profiling for the discovery of imprinted expression. MLE and PLE patterns do not necessarily imply imprinting and many genes with imprinted expression will not be classified as MLE or PLE. Microarray expression profiling can only classify MLE or PLE patterns when the gene is differentially expressed in the two parental genotypes. Therefore, examples of imprinting in which the gene is expressed at similar levels in the two parents will not be classified as MLE. Indeed, the two genes in our study with differential maternal imprinting displayed roughly equivalent microarray expression levels in both parental inbreds and therefore could not be assessed for MLE or PLE patterns. Current hypotheses suggest that imprinting may function as a mechanism for controlling the expression level for the imprinted genes (Dilkes and Comai, 2004; Wood and Oakey, 2006). Therefore, imprinted genes may be less likely to exhibit total expression level differences between maize inbred lines in a microarray expression-profiling experiment.
Dominant HP and LP expression patterns were observed for a set of 44 genes in 13- and/or 19-DAP endosperm. Genes were classified as dominant HP or LP expression patterns if both reciprocal hybrids display expression levels that were not statistically different from the HP or LP level, respectively. These genes are putative examples in which the expression level might be controlled by a dominant trans-acting regulator. Cases in which both hybrids are expressed at the level of the HP may be examples of variation in which one of the two inbreds possesses a dominant trans-acting activator. Conversely, expression of both hybrids at the level of the LP may stem from variation in which one of the two inbreds possesses a dominant trans-acting repressor.
Some genes exhibited hybrid endosperm expression levels that were outside the parental range, either AHP or BLP. In some studies, these genes have been referred to as overdominant or underdominant expression patterns. We have avoided the use of these terms because they are often used to refer to the genetic action of a locus; thus, the application of these terms to both expression patterns and gene action may be nonsynonymous (Springer and Stupar, 2007a). In a previous study, we had found very few examples of this type of expression pattern in 11-d seedlings, immature ears, or 19-DAP embryo tissues. Therefore, we were surprised to note a substantial number of genes (78 genes in 13 DAP and 59 genes in 19 DAP) that display hybrid expression patterns that were significantly outside the range of the parents. However, it is worth noting that there are no examples in which both reciprocal hybrids exhibit expression levels significantly AHP or BHP. Instead, the examples of hybrid endosperm expression outside the range of the two parents include cases in which only one hybrid is outside the range of the parents or in which one hybrid is above the higher parent and the other hybrid is below the lower parent. This finding can be seen in Figure 2B. Genes for which both hybrids display AHP or BLP expression would plot in the top right and bottom left quadrants. However, there are essentially no genes in these areas (one endosperm gene does plot AHP for both hybrids, but it was not statistically significant for AHP in both hybrids). Instead, the genes with hybrid endosperm expression outside the range of the parents tend to cluster in the top left and bottom right quadrants.
Differential Imprinting in Maize Endosperms
Dilkes and Comai (2004) proposed classifying examples of imprinting as binary or differential. In F1 hybrids, binary imprinted alleles exhibit monoallelic expression of the maternal or paternal allele, whereas differentially imprinted alleles exhibit maternally or paternally biased biallelic expression. Most previous examples of imprinting have described binary imprinting. However, Guo et al. (2003) reported on the differential imprinting of the nrp1 gene in maize endosperm. The nrp1 gene displayed strong maternal expression bias in the reciprocal hybrids of B73 with Mo17 and the reciprocal hybrids made between inbred lines N1 and S1. However, some paternal allelic expression was observed at relatively low levels throughout development (Guo et al., 2003). The Mo17 × B73 cross showed a stronger maternal expression bias than the B73 × Mo17 cross (Guo et al., 2003).
It is likely that there are relatively few characterized examples of differential imprinting because such discoveries require reliable quantitative allele-specific expression analyses. The use of mass spectrometry-based allele-specific analysis provides a highly quantitative assay for studying the relative expression of two alleles in the same individual. Using this technique, we studied the relative expression of 90 genes for which quantitative assays were available. Interestingly, we found evidence to suggest differential imprinting for eight of these genes. Six cases reflect examples of paternal bias and two cases are examples of maternal bias. Further characterization of the two maternally biased genes provided additional evidence for differential imprinting. However, there is also evidence for some variability in the strength of the parent-of-origin effects for both of these genes. We did not perform further characterization on the examples of putative paternal differential imprinting. Each of these six cases resulted in a change from the expected 2m:1p ratio to an observed approximately 1m:1p ratio (Fig. 5). CAPS markers are not sufficiently quantitative to accurately discriminate between allelic proportions of 0.66 and 0.50. However, for each of these six genes, we observed a consistent paternal bias in both 13 and 19 DAP in all biological replicates studied.
Our data indicate that differential imprinting may be more common than binary imprinting in maize endosperm, raising new questions about the molecular mechanisms that may generate high rates of differential imprinting. There is evidence that imprinting in maize may be caused by allele-specific modifications within relatively small regions (Gutierrez-Marcos et al., 2006; Haun et al., 2007). However, for many genes, there is evidence for the existence of multiple enhancers of expression and, in some cases, these enhancers may be many kilobases upstream or downstream of the transcription start site. Therefore, the multiple enhancers for a given gene may be subject to different types of regulation. For example, one enhancer may be subject to imprinting, whereas the other enhancers may not be subject to imprinting. In this case, allelic expression levels would be influenced by the parent of origin, but the down-regulated allele would exhibit some level of expression because the nonimprinted enhancer remains functional in both alleles. This would result in differential imprinting. Because quantitative allele-specific expression studies are applied in a high-throughput manner to further define parent-of-origin effects in endosperm tissue, we would predict the identification of a substantial number of genes exhibiting differential imprinting.
Differences in Hybrid Expression Patterns of Endosperm and Vegetative Tissues
Several studies have documented prevalent additive expression in maize hybrids (Guo et al., 2006; Stupar and Springer, 2006; Swanson-Wagner et al., 2006). However, several other groups have reported higher frequencies of nonadditive expression, including frequent observations of hybrid expression outside of the parental range (Auger et al., 2005; Meyer et al., 2007; Uzarowska et al., 2007). The differences observed in these studies may be attributed to differences in expression platforms, statistical methods, or tissues sampled. In this study, we have used common expression platforms and methods to assess additive and nonadditive expression in several vegetative and endosperm tissues. Whereas similar frequencies of nonadditive expression are observed in endosperm and vegetative tissues, there are striking differences in the type of nonadditive expression that is observed. Hybrid expression outside the parental range was relatively common in endosperm tissues, but was very rarely observed in vegetative tissues. Gene expression in hybrid endosperms rarely exhibited HP and LP patterns, but relatively frequently displayed novel expression patterns with parental effects between reciprocal hybrids. In contrast, HP and LP expression patterns were the most common form of nonadditive expression observed in hybrid vegetative tissues, and expression patterns with parental effects (MLE and PLE) were not observed.
The dosage imbalance between the maternal and paternal genomes may contribute to the novel types of expression observed in endosperm tissue. The types of nonadditive expression observed in endosperm suggest that dosage compensation mechanisms in endosperm tissues differ from those in vegetative tissues. Studies of dosage dependence and gene expression in aneuploids have noted differences in the response of endosperm and embryo tissues (Guo and Birchler, 1994; Birchler and Veitia, 2007). Analysis of dosage dependence in vegetative tissues of triploid maize provided evidence for dosage-dependent expression (Auger et al., 2005). It is possible that specialized mechanisms have evolved to compensate for the triploid nature of endosperm tissue. A recent study by Baroux et al. (2007) found that endosperm nuclei exhibit chromatin organization that is distinct from those observed in vegetative tissues. This endosperm-specific chromatin organization may lead to higher frequencies of nonadditive expression.
In this study, we have found that additive patterns are the most common form of gene expression in hybrid endosperm; however, these additive patterns inherently generate expression differences between the reciprocal hybrids due to the unequal distribution of parental alleles in the triploid tissues. We have also observed numerous genes with novel hybrid expression patterns, with a far greater frequency of hybrid gene expression outside the parental range than had been previously observed in vegetative tissues. These observations highlight the exceptional diversity of gene expression in maize endosperm. Previous studies have documented unique genetic and epigenetic alterations that occur in the endosperm, including imprinting and altered DNA methylation patterns (Baroux et al., 2002, 2007; Gehring et al., 2004; Lauria et al., 2004; Gutierrez-Marcos et al., 2006; Haun et al., 2007; Zhang et al., 2007). The unique transcriptional and epigenetic processes of the endosperm raise intriguing questions about whether these novel expression patterns are simply a consequence of relaxed selection constraints on a terminal tissue or whether these unique expression patterns provide functional and/or advantageous qualities.
MATERIALS AND METHODS
Microarray Platform
18 K oligonucleotide microarrays designed for detection of maize (Zea mays) genes were purchased from Affymetrix. This microarray was designed using EST contig sequences. There are 15 probes for each represented gene on the microarray and, in most cases, these probes are contiguous adjacent sequences of a transcript. The EST sequences are derived from multiple inbred lines and contain sequence polymorphisms. All polymorphic positions were masked such that, when possible, the probe sets will be robust for multiple genotypes. The maize Affymetrix array contains 17,622 probe sets that are designed to detect the expression of 13,495 genes. Some genes are represented by multiple probes sets designed to detect sense and antisense expression or the expression of alternative transcripts.
Plant Growth and Tissue Collection
Endosperm tissues were isolated from kernels of inbreds MMM and BBB and hybrids MMB and BBM for gene expression analysis. Samples were collected during August 2005 from field-grown maize plants grown on the St. Paul campus Agricultural Experiment Station. Inbred and hybrid crosses were performed at three different dates to produce biological replicates. The crosses for all genotypes within a biological replicate were performed within 30 min of each other. Six ears of each genotype were harvested at 13 and 19 DAP per biological replicate. Six endosperms were collected from each of the six ears and pooled for a total of 36 endosperms from each genotype by biological replicate combination. All tissues were flash frozen in a dry-ice-cooled ethanol bath and subsequently stored at −80°C.
Information on the plant growth and tissue collection of 11-d seedling and immature ear tissues is described in Stupar and Springer (2006).
RNA Isolation
RNA for 13-DAP endosperm tissues was isolated using the plant RNeasy kit, according to the manufacturer's instructions (Qiagen). Initial isolation of 19-DAP endosperm RNA involved a combination of phenol-chloroform extraction and the TRIzol procedure, and essentially followed published methods (Leiva-Neto et al., 2004). Briefly, powders for 19-DAP endosperm were homogenized in extraction buffer (50 mm Tris, pH 8.0, 150 mm LiCl, 5 mm EDTA, pH 8.0, 1% SDS), extracted twice with an equal volume of 1:1 phenol chloroform, extracted with an equal volume of chloroform, and then extracted with TRIzol according to the manufacturer's instructions (Invitrogen). The 19-DAP endosperm RNA was further purified using the RNeasy protocol (Qiagen). All purified RNA samples were quantified and qualified using the Nanodrop spectrophotometer (Nanodrop Technologies) and agarose gel electrophoresis.
The methods used to isolate RNA from 11-d seedling and immature ear tissues are described in Stupar and Springer (2006).
Microarray Hybridizations and Statistical Analysis
Microarray hybridizations were performed to assess the steady-state level of transcripts in endosperm tissue derived from two maize inbred lines and their reciprocal hybrids. Hybridizations for three biological replicates per tissue type were performed for each of four genotypes: inbreds MMM and BBB and hybrids MMB and BBM. Eight micrograms of total RNA were labeled for each hybridization according to the manufacturer's instructions (Affymetrix). Hybridization chemistries were performed at the University of Minnesota Microarray Facility. Previously published microarray data from 11-d seedling and immature ear tissues were obtained using the same methodologies (Stupar and Springer, 2006) and were included in downstream analyses for purposes of comparing the expression profiles of inbred-hybrid vegetative and endosperm tissues.
The signal data from the microarrays was processed using two different methods, MAS 5.0 and GC-robust multiarray average (RMA). The GCOS software package (Affymetrix) was used to produce a MAS 5.0 signal and produce presence-absence calls. The signal from each array was normalized to a value of 1 with a scaling target factor of 500 and the default parameters. After importing the MAS 5.0 values into GeneSpring (Agilent Technologies), per-chip normalization to the fiftieth percentile and per-gene normalization to median were performed. GC-RMA processing of the signal was performed using GeneSpring software with per-gene normalization.
Differentially expressed genes were identified by performing a one-way ANOVA on the GC-RMA or MAS 5.0 values using a parametric test with no assumption of equal variance. A Benjamini and Hochberg multiple testing correction was applied with a false-discovery rate of 0.05 (unless otherwise noted) such that 5% of the genes identified in a test are likely to be falsely identified. Separate ANOVAs were performed to identify genes that were differentially expressed among developmental stages and among genotypes. The lists of differentially expressed genes among genotypes were further filtered using minimal expression (genotype with highest expression must have signal >50) and fold change (must be >2-fold change between any two genotypes) criteria. Clustering analyses were performed using GeneSpring and were based on specified gene lists, conditions, and genotypes.
Statistical analyses were performed on individual microarray probes to test for the possibility of probe-specific polymorphisms resulting in false discovery of differential expression. The individual probe signals were extracted and per-chip normalization was applied. For each differentially expressed gene, the difference between each of the 15 perfect match-mismatch probe signals was determined for each of the three biological replicates and used to perform an independent sample comparison of means assuming normality with a cutoff of P = 0.05. The number of probe pairs that pass this test for each gene was determined.
Identification and Validation of Endosperm-Specific Expression Patterns
Genes that were present in the endosperm transcriptomes and absent in other tissue transcriptomes were identified based on the MAS 5.0 presence-absence calls. BLAST analyses were performed using these sequences to query the National Center for Biotechnology Information genome survey sequences derived from maize. Primers were designed using primer 3 software (Rozen and Skaletsky, 2000). PCR reactions were performed in a 15-μL total volume containing approximately 25 ng of DNA, 2 pmol of each primer, 0.4 units of HotStar Taq polymerase (Qiagen), 1.56 μL of 10× reaction buffer, and 0.2 μL of 25 mm dNTPs. Conditions of PCR were as follows: 94° for 15 min, 35 cycles of 94° for 30 s, 60° for 30 s, 72° for 2 min, followed by 72° for 7 min. Amplified products were separated in a 1% agarose Tris-borate/EDTA gel and visualized by ethidium bromide staining.
Hybrid-Inbred Expression Pattern Analysis
All genes that were determined to be differentially expressed among genotypes based on ANOVA (analysis described above) were investigated for nonadditive expression patterns. The inbred expression values were used to calculate the expected additive expression level of each reciprocal hybrid. To account for the 2m:1p endosperm dosage, the expected additive values were calculated as: additive expression = [(2 × maternal expression level) + paternal expression level]/3. The expected additive values were compared with the appropriate reciprocal hybrid values for all three biological replications. A two-tailed homoscedastic t test was performed and all genes with P < 0.05 were considered to be nonadditively expressed. Both reciprocal hybrids were independently tested for each gene.
The nonadditive patterns were divided into six general classes. (1) PLE, genes that exhibit hybrid transcription levels similar to the male parent; (2) MLE, genes that exhibit hybrid transcription levels that are more similar to the female parent than intermediate levels; (3) HP, genes that exhibit expression patterns in which both hybrids are similar to the HP; (4) LP, genes that exhibit expression patterns in which both hybrids are similar to the LP; (5) AHP, genes in which at least one hybrid is expressed AHP; (6) BLP, genes in which at least one hybrid is expressed BLP. We defined 14 nonadditive expression patterns that a given gene may exhibit among the four genotypes in this study (see Supplemental Fig. S5). There is one pattern that fits the PLE class, one pattern that fits the MLE classes, two different patterns that fit the HP class, two patterns that fit the LP class, and eight patterns that fit the AHP and/or BLP classes. A stringent procedure (see Supplemental Table S6) was applied to assign the nonadditive genes into the 14 classes based on the microarray data.
The d/a ratios were calculated for all genes differentially expressed among genotypes, such that the d/a value would indicate the hybrid expression relative to the HP and LP values for each gene. The d value was calculated as the hybrid signal minus the average signal of the two parents. The a value was calculated as the HP signal minus the average signal of the two parents (thus the a value is equivalent to the absolute value of the difference between either parent and the average of the two parents). Genes with d/a values >1.0 or <−1.0 indicated genes with hybrid expression levels AHP or BLP, respectively. For each gene, a d/a value was calculated for the maternal hybrid, which is the hybrid genotype derived by crossing the genotype with the higher expression as the female parent to the genotype with the lower expression as the male parent. Similarly, a d/a value was calculated for the paternal hybrid, which is the reciprocal hybrid that is generated by crossing the lower expression genotype (as a female) by the higher expression genotype (as a male).
Development of Allele-Specific Expression Assays
A series of allele-specific expression assays were designed to further investigate expression patterns in endosperm tissue. Allele-specific expression assays are quite useful for determining the relative contribution of the maternal and paternal allele to the total level of transcript for a particular gene, whereas microarray hybridizations assess total transcript levels. Allele-specific expression assays were designed for a set of 208 genes that were selected either at random (176 genes), due to differential expression in B73 relative to Mo17 (25 genes), or because they exhibited nonadditive expression levels in reciprocal hybrids (seven genes). The evidence for differential or nonadditive expression was derived from the Affymetrix microarray experiments described above. The single-nucleotide polymorphisms used to design these assays were derived from Panzea (Zhao et al., 2006) and 454-based cDNA sequencing (Emrich et al., 2007). These allele-specific expression assays are performed by single-base extension of a PCR product; the extension products are then separated and quantified using matrix-assisted laser-desorption ionization-time-of-flight mass spectrometry (Jurinke et al., 2005). Each of the assays were then performed on a series of genomic DNA standards that include hybrid DNA from seedlings (1 B73:1 Mo17 allele), DNA from reciprocal hybrid endosperm (1:2 and 2:1 mixes of the B73 and Mo17 alleles), and genomic DNA from inbred B73 and Mo17 seedlings mixed at 4:1 and 1:4 concentrations (Supplemental Fig. S13). Analysis of the data from the genomic DNA standards showed that 88 of the assays were either not polymorphic or not quantitative. For another 30 assays, we were not able to consistently detect expression in endosperm tissues. The remaining 90 assays displayed high R2 correlation (>0.9) between the known proportion of the B73 allele in the genomic DNA standard series and the observed proportion of the B73 allele detected by the assay (full details for these 90 assays are available in Supplemental Table S8). The 90 genes with successful allele-specific expression assays include 72 randomly selected genes, 15 genes with differential expression in B73 relative to Mo17 seedlings (according to statistical analyses of Affymetrix microarray data), and three genes with nonadditive expression levels in the endosperm (according to Affymetrix microarray data).
Allele-Specific Expression
RNA from all tissues was treated with DNAse prior to allele-specific expression analyses. cDNA was synthesized from all three biological replicates of Mo17 × B73 and B73 × Mo17 hybrid RNA from both 13- and 19-DAP endosperms. cDNA was also synthesized from 2:1 and 1:2 mixes of Mo17 and B73 inbred RNA for both 13- and 19-DAP endosperms; these mixed cDNAs were made from all three biological replicates. The cDNA was reverse transcribed using SuperScript III reverse transcriptase, according to the manufacturer's instructions (Invitrogen).
PCR-based assays for allele-specific expression analyses were standardized and performed as described (Stupar and Springer, 2006), with the following modifications. Three measurements of the ratio of the two alleles were performed for each of the three biological replicates of 2:1 and 1:2 mixed RNAs and the six F1 RNAs. Data were graphed using Spotfire version 7.1 software.
CAPS Assays
CAPS assays were designed to validate biased maternal allele expression for two genes. Reverse transcription (RT)-PCR was performed on the same cDNA templates from 13- and 19-DAP hybrid endosperm samples as were used in the allele-specific expression analyses (described above). Additionally, RT-PCR was performed on cDNA samples synthesized from individual hybrid endosperm tissues from six different developmental stages between 10 and 24 DAP.
The primers used for gene CF628018 (a putative polygalacturonase-inhibiting protein): CGTGGAACTACCTCAACTTCGAC (forward) and ACACGTACACAGCTGTCATTTCG (reverse). RT-PCR products from gene CF628018 were digested with enzyme Bme1580I (New England Biolabs), which has a restriction site in the amplicon of the Mo17 allele but does not recognize the B73 allele. The CAPS products were evaluated using agarose gel electrophoresis.
The primers used for gene AY111942 (a putative sugar transporter): AAGGAGAAGGTGGTGGAAATGTC (forward) and TTCGTTGAGAGGGAATTTTCTTG (reverse). RT-PCR products from gene AY111942 were digested with enzyme PleI (New England Biolabs), which has a differential restriction sites for the B73 and Mo17 allele amplicons. The CAPS products were evaluated on high-resolution Metaphor gels.
The microarray data generated for this study are available at Gene Expression Omnibus (GEO). The endosperm microarray data can be accessed under series accession number GSE8308 and the embryo, seedling, and immature ear microarray data can be accessed under series accession number GSE8194.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of endosperm-specific gene expression.
Supplemental Figure S2. Differential expression in 13 and 19 DAP.
Supplemental Figure S3. Genotype effects on gene expression.
Supplemental Figure S4. Microarray validation.
Supplemental Figure S5. Classification of nonadditive expression types.
Supplemental Figure S6. MLE patterns in 13 and 19 DAP.
Supplemental Figure S7. PLE patterns in 13 and 19 DAP.
Supplemental Figure S8. HP expression patterns in 13 and 19 DAP.
Supplemental Figure S9. LP expression patterns in 13 and 19 DAP.
Supplemental Figure S10. Hybrid “bracket” expression patterns in 13 and 19 DAP.
Supplemental Figure S11. BBM outside range of parents in 13 and 19 DAP.
Supplemental Figure S12. MMB outside range of parents in 13 and 19 DAP.
Supplemental Figure S13. Linearity of allele-specific expression assays.
Supplemental Table S1. Transcript detection in 13- and 19-DAP endosperm.
Supplemental Table S2. Validation of endosperm-specific gene expression.
Supplemental Table S3. Genes that are differentially expressed between 13 and 19 DAP.
Supplemental Table S4. Differential expression between genotypes at 13 DAP.
Supplemental Table S5. Differential expression between genotypes at 19 DAP.
Supplemental Table S6. Classification of nonadditive expression patterns.
Supplemental Table S7. Genes with specific nonadditive expression patterns.
Supplemental Table S8. Allele-specific expression assays.
Supplementary Material
Acknowledgments
We would like to thank Irina Makarevitch, Elizabeth Chrans, and Dinesha Walek for providing technical help with sample preparation and/or data collection. The Minnesota Supercomputing Institute provided access to software packages used for data analysis. We are very grateful to Shawn Kaeppler and Ronald Phillips for providing invaluable discussions and feedback. We also appreciate the comments and suggestions of four anonymous reviewers and the journal editor.
This work was supported by the National Science Foundation (grant no. DBI–0227310) and the Minnesota Agriculture Experiment Station (grant to N.M.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nathan M. Springer (springer@umn.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams KL (2007) Evolution of duplicate gene expression in polyploid and hybrid plants. J Hered 98 136–141 [DOI] [PubMed] [Google Scholar]
- Adams KL, Wendel JF (2005) Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics 171 2139–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger DL, Gray AD, Ream TS, Kato A, Coe EH Jr, Birchler JA (2005) Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Pecinka A, Fuchs J, Schubert I, Grossniklaus U (2007) The triploid endosperm genome of Arabidopsis adopts a peculiar, parental, dosage-dependent chromatin organization. Plant 19 1782–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Spillane C, Grossniklaus U (2002) Genomic imprinting during seed development. Adv Genet 46 165–214 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J (2003) Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res 13 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes BP, Comai L (2004) A differential dosage hypothesis for parental effects in seed development. Plant Cell 16 3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich SJ, Barbazuk WB, Li L, Schnable PS (2007) Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res 17 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23 192–199 [DOI] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fischer RL (2004) Imprinting and seed development. Plant Cell 16 S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Birchler JA (1994) Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266 1999–2002 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Danilevskaya ON, Yan X, Hu Z (2003) Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J 36 30–44 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Yang X, Crasta O, Zinselmeier C, Smith OS, Bowen B (2006) Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet 113 831–845 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS (2004) Allelic variation of gene expression in maize hybrids. Plant Cell 16 1707–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Dal Pra M, Scholten S, Kranz E, Perez P, Dickinson HG (2006) Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38 876–878 [DOI] [PubMed] [Google Scholar]
- Haun WJ, Laoueille-Duprat S, O'Connell MJ, Spillane C, Grossniklaus U, Phillips AR, Kaeppler SM, Springer NM (2007) Genomic imprinting, methylation and molecular evolution of maize enhancer of zeste (Mez) homologs. Plant J 49 325–337 [DOI] [PubMed] [Google Scholar]
- Jurinke C, Denissenko MF, Oeth P, Ehrich M, van den Boom D, Cantor CR (2005) A single nucleotide polymorphism based approach for the identification and characterization of gene expression modulation using MassARRAY. Mutat Res 573 83–95 [DOI] [PubMed] [Google Scholar]
- Kirst M, Caldo R, Casati P, Tanimoto G, Walbot V, Wise RP, Buckler ES (2006) Genetic diversity contribution to errors in short oligonucleotide microarray analysis. Plant Biotechnol J 4 489–498 [DOI] [PubMed] [Google Scholar]
- Kowles RV, Phillips RL (1985) DNA amplification patterns in maize endosperm nuclei during kernel development. Proc Natl Acad Sci USA 82 7010–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Dey N, Kim CS, Bharti AK, Rudd S, Mayer KF, Larkins BA, Becraft P, Messing J (2004) Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Res 14 1932–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria M, Rupe M, Guo M, Kranz E, Pirona R, Viotti A, Lund G (2004) Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell 16 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva-Neto JT, Grafi G, Sabelli PA, Dante RA, Woo YM, Maddock S, Gordon-Kamm WJ, Larkins BA (2004) A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell 16 1854–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Pospisil H, Scholten S (2007) Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol Biol 63 381–391 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132 365–386 [DOI] [PubMed] [Google Scholar]
- Schweizer L, Yerk-Davis GL, Phillips RL, Srienc F, Jones RJ (1995) Dynamics of maize endosperm development and DNA endoreduplication. Proc Natl Acad Sci USA 92 7070–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Messing J (2003) Gene expression of a gene family in maize based on non-collinear haplotypes. Proc Natl Acad Sci USA 100 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM (2007. a) Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res 17 264–275 [DOI] [PubMed] [Google Scholar]
- Springer NM, Stupar RM (2007. b) Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell http://dx.doi.org/10.1105/tpc.107.052258 [DOI] [PMC free article] [PubMed]
- Stupar RM, Springer NM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzarowska A, Keller B, Piepho HP, Schwarz G, Ingvardsen C, Wenzel G, Lubberstedt T (2007) Comparative expression profiling in meristems of inbred-hybrid triplets of maize based on morphological investigations of heterosis for plant height. Plant Mol Biol 63 21–34 [DOI] [PubMed] [Google Scholar]
- Verza NC, Silva E, Neto TR, Nogueira GC, Fisch FT, de Rosa PH, Jr VE, Rebello MM, Vettore AL, da Silva FR, et al (2005) Endosperm-preferred expression of maize genes as revealed by transcriptome-wide analysis of expressed sequence tags. Plant Mol Biol 59 363–374 [DOI] [PubMed] [Google Scholar]
- West MA, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St Clair DA (2007) Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430 85–88 [DOI] [PubMed] [Google Scholar]
- Woo YM, Hu DW, Larkins BA, Jung R (2001) Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13 2297–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Oakey RJ (2006) Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet 2 e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MS, Yan HY, Zhao N, Lin XY, Pang JS, Xu KZ, Liu LX, Liu B (2007) Endosperm-specific hypomethylation, and meiotic inheritance and variation of DNA methylation level and pattern in sorghum (Sorghum bicolor L.) inter-strain hybrids. Theor Appl Genet 115 195–207 [DOI] [PubMed] [Google Scholar]
- Zhao W, Canaran P, Jurkuta R, Fulton T, Glaubitz J, Buckler E, Doebley J, Gaut B, Goodman M, Holland J, et al (2006) Panzea: a database and resource for molecular and functional diversity in the maize genome. Nucleic Acids Res 34 D752–D757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.