Abstract
The acetyl-coenzyme A carboxylase (ACCase)-inhibiting cyclohexanedione herbicide clethodim is used to control grass weeds infesting dicot crops. In Australia clethodim is widely used to control the weed Lolium rigidum. However, clethodim-resistant Lolium populations have appeared over the last 5 years and now are present in many populations across the western Australian wheat (Triticum aestivum) belt. An aspartate-2078-glycine (Gly) mutation in the plastidic ACCase enzyme has been identified as the only known mutation endowing clethodim resistance. Here, with 14 clethodim-resistant Lolium populations we revealed diversity and complexity in the molecular basis of resistance to ACCase-inhibiting herbicides (clethodim in particular). Several known ACCase mutations (isoleucine-1781-leucine [Leu], tryptophan-2027-cysteine [Cys], isoleucine-2041-asparagine, and aspartate-2078-Gly) and in particular, a new mutation of Cys to arginine at position 2088, were identified in plants surviving the Australian clethodim field rate (60 g ha−1). Twelve combination patterns of mutant alleles were revealed in relation to clethodim resistance. Through a molecular, biochemical, and biological approach, we established that the mutation 2078-Gly or 2088-arginine endows sufficient level of resistance to clethodim at the field rate, and in addition, combinations of two mutant 1781-Leu alleles, or two different mutant alleles (i.e. 1781-Leu/2027-Cys, 1781-Leu/2041-asparagine), also confer clethodim resistance. Plants homozygous for the mutant 1781, 2078, or 2088 alleles were found to be clethodim resistant and cross resistant to a number of other ACCase inhibitor herbicides including clodinafop, diclofop, fluazifop, haloxyfop, butroxydim, sethoxydim, tralkoxydim, and pinoxaden. We established that the specific mutation, the homo/heterozygous status of a plant for a specific mutation, and combinations of different resistant alleles plus herbicide rates all are important in contributing to the overall level of herbicide resistance in genetically diverse, cross-pollinated Lolium species.
Acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) is a key enzyme involved in the first step of fatty acid biosynthesis. In plants, ACCase is also the target enzyme for important herbicides used in world agriculture. Three chemically distinct classes of herbicides that are known to inhibit ACCase are aryloxyphenoxypropionates (APP), cyclohexanediones (CHD), and the more recent (Hofer et al., 2006) phenylpyrazolin class herbicide pinoxaden (hereinafter referred to as ACCase herbicides). In plants, there are two isoforms of ACCase: The plastid ACCase is essential in biosynthesis of primary fatty acids and the cytosolic ACCase is involved in biosynthesis of long chain fatty acids. The homomeric ACCase in the cytosol of nearly all plant species and the heteromeric ACCase in the chloroplasts of dicots are insensitive to APP, CHD, and pinoxaden herbicides. In contrast, the plastidic homomeric ACCase in nearly all grass species is herbicide sensitive, and this is the basis for selective control of grass weeds by ACCase herbicides. All ACCase isoforms contain three catalytic domains: the biotin carboxyl carrier, the biotin carboxylase, and the carboxyl transferase (CT) domains (Nikolau et al., 2003). Molecular and biochemical studies have clearly established that the CT domain of the plastidic homomeric ACCase is the primary target site for APP and CHD herbicides, and two regions of the CT domain of the plastidic ACCase are critical for sensitivity to these herbicides (Zhang et al., 2004; for review, see Délye, 2005).
The obligate cross-pollinated grass weed Lolium rigidum has demonstrated in Australia an ability to rapidly evolve resistance to ACCase herbicides and other herbicide groups. Within a few years of initial ACCase herbicide use (1978), the first ACCase herbicide-resistant L. rigidum population was evident (Heap and Knight, 1986). Since then, ACCase herbicide-resistant L. rigidum populations have evolved across huge areas of Australian croplands (Llewellyn and Powles, 2001; Owen et al., 2007). The biochemical basis of ACCase herbicide resistance has been revealed in several populations to involve resistant ACCase (Matthews et al., 1990; Holtum et al., 1991; Tardif et al., 1993, 1996). Many resistant populations can also have a nontarget site-based resistance mechanism of enhanced rates of ACCase herbicide metabolism (Tardif and Powles, 1994; Preston et al., 1996; Preston and Powles, 1998). L. rigidum is an obligate cross-pollinated plant and it is emphasized that individual plants and populations can accumulate resistance mechanisms.
Recently, we have identified molecular mutations in the ACCase gene endowing target site based herbicide resistance in some ACCase herbicide-resistant L. rigidum populations. We have identified mutations causing resistance-endowing amino acid substitutions at amino acids 1781 and 2041 (Zhang and Powles, 2006a, 2006b). Both of these amino acid substitutions have been previously identified to endow ACCase herbicide resistance in studies by others. Thus far, six distinct amino acid substitutions in the CT domain of the plastidic ACCase gene that individually endow resistance to certain ACCase herbicides have been characterized in Alopecurus myosuroides and other grass weed species, as reviewed by Délye et al. (2005) and updated in Table I. Specific ACCase mutations confer specific cross-resistance patterns to ACCase herbicides (for review, see Délye, 2005). The Ile-1781-Leu mutation is associated with resistance to APP and some CHD herbicides (not including clethodim). The Trp-2027-Cys, Ile-2041-Asn, or Gly-2096-Ala mutations confer resistance only to APP herbicides. The Asp-2078-Gly mutation confers resistance to many APP and CHD herbicides including clethodim. The Trp-1999-Cys mutation confers resistance only to the APP herbicide fenoxaprop (Liu et al., 2007).
Table I.
Summary of published resistance-endowing amino acid substitutions identified within the CT domain of the plastidic ACCase gene from resistant grass populations
*, Amino acid positions correspond to the full-length plastidic ACCase in A. myosuroides.
| Amino Acid Substitution* | Grass Species | References |
|---|---|---|
| Ile-1781-Leu | L. rigidum | Zagnitko et al. (2001); Délye et al. (2002b); Brown et al. (2002); Tal and Rubin (2004); Zhang and Powles (2006b) |
| Setaria viridis | Délye et al. (2002c) | |
| A. myosuroides | Délye et al. (2002a, 2002b); Brown et al. (2002) | |
| A. fatua | Christoffers et al. (2002) | |
| L. multiflorum | White et al. (2005) | |
| A. sterilis | Liu et al. (2007) | |
| Ile-2041-Asn | A. myosuroides | Délye et al. (2003) |
| L. rigidum | Délye et al. (2003); Zhang and Powles (2006a) | |
| A. sterilis | Liu et al. (2007) | |
| Ile-2041-Val | L. rigidum | Délye et al. (2003) |
| Trp-2027-Cys | A. myosuroides | Délye et al. (2005) |
| A. sterilis | Liu et al. (2007) | |
| Gly-2096-Ala | A. myosuroides | Délye et al. (2005) |
| Asp-2078-Gly | A. myosuroides | Délye et al. (2005) |
| A. sterilis | Liu et al. (2007) | |
| Trp-1999-Cys | A. sterilis | Liu et al. (2007) |
Despite widespread resistance to certain ACCase herbicides, our 1998 survey across 300 western Australian crop fields confirmed that the CHD herbicide clethodim was still effective on many otherwise ACCase herbicide-resistant L. rigidum populations (Llewellyn and Powles, 2001). Five years later, however, our random survey of 452 ryegrass populations from the same region revealed clethodim resistance to be present in 8% of these populations (Owen et al., 2007). Thus far, the Asp-2078-Gly mutation in the plastidic ACCase enzyme is the only known mutation endowing clethodim resistance (Délye et al., 2005). As L. rigidum is a highly genetically variable species we expect that all possible herbicide resistance-endowing mechanisms can be present and enriched in large populations of this species under herbicide selection (Powles and Matthews, 1992). Thus, we expect that a number of different mutations endowing ACCase herbicide resistance (Table I) could be enriched both within and between different resistant populations. Our hypothesis tested here is that field evolved ACCase herbicide-resistant L. rigidum populations would be comprised of individuals carrying a diverse range of resistance-endowing mutations and that individuals would be heterozygous or homozygous for one or any two possible combinations of different mutations. To examine this in depth we selected 12 clethodim-resistant Australian L. rigidum populations, together with two resistant Italian Lolium populations. We reveal diversity and complexity in ACCase mutations in this weed species. We identify five ACCase mutations and reveal 12 combination patterns of mutant alleles (genotypes) from these 14 clethodim-resistant Lolium populations. We demonstrate that a new mutation, Cys-2088-Arg, and the known mutations, Ile-1781-Leu and Asp-2078-Gly, endow resistance to clethodim and other ACCase herbicides. We establish and emphasize that the specific mutation, the homo/heterozygous status of a plant for a specific mutation, and combinations of different resistant alleles plus herbicide rates are all important in contributing to the overall level of herbicide resistance. In addition, we developed (derived) cleaved amplified polymorphic sequence ([d]CAPS) markers for the 2041, 2078, and 2088 mutations to enable rapid detection of these mutations in the Lolium populations.
RESULTS
ACCase Mutations Revealed in Clethodim-Resistant Lolium Populations
At least six clethodim-resistant individuals from each clethodim-resistant population were initially sequenced. Subsequently, a total of 124 individual plants were sequenced from 12 clethodim-resistant Australian L. rigidum populations and two Italian Lolium populations. Using three overlapping primer pairs (Table II), we were able to amplify three regions containing all known potential ACCase mutation sites (Délye and Michel, 2005) in the CT domain of the plastidic ACCase genes. A contig of 1,513 bp was clearly identified and assembled from sequence results of all individual resistant and susceptible (bulked) plants. When compared with other ACCase gene sequences in GenBank, this assembled contig showed 99% identity with the plastidic ACCase gene from L. rigidum (accession no. AY995232) and Lolium multiflorum (AY710293), 95% identity with Avena fatua (AF231335), 93% identity with A. myosuroides (AJ310767) and Phalaris minor (AY196481), and 91% with wheat (Triticum aestivum; AF029895). However, only 77% and 76% identity was shared with cytosolic ACCase genes of A. myosuroides (AJ632096) and wheat (U39321), respectively. Sequence comparison between individual resistant plants from each population and two susceptible populations revealed four mutations previously established to endow ACCase herbicide resistance (Table III): Ile-1781-Leu (referred to as 1781-Leu), Trp-2027-Cys (2027-Cys), Ile-2041-Asn (2041-Asn), and Asp-2078-Gly (2078-Gly). In addition, a new mutation of Cys-2088-Arg (2088-Arg) was also detected in five populations (sequences have been deposited in GenBank with accession numbers EF538937–EF538943). Sequence alignment of 1,513 bp contigs from susceptible controls and resistant plants containing the 2088 mutation revealed 18 synonymous single nucleotide polymorphisms (SNPs) and 10 nonsynonymous SNPs. Among the 10 nonsynonymous SNPs, only the SNP at 2088 differs between resistant and susceptible sequences (Table IV). In addition, the sequence results containing the Cys to Arg mutation were also validated by restriction analysis (CAPS; see “Materials and Methods”). Therefore, the mutation Cys to Arg at position 2088 is very likely a newly identified mutation endowing resistance to ACCase herbicides (including clethodim).
Table II.
Primers
In the second column, an introduced point mutation in the designed dCAPS primer is in bold. Nucleotides discriminating grass plastid ACCase sequences from cytosolic sequences at the 3′ end of the designed primers are underlined.
| Primer | Sequence 5′-3′ | Usage | References |
|---|---|---|---|
| ACCF5 | AATGGGTCGTGGGGCACTCCTATAATTCC | Gene-specific PCR | Délye et al. (2002b) |
| ACCR5 | GCTGAGCCACCTCAATATATTAGAAACACC | – | – |
| ACCF6 | CATACAGCGTGAAGATCAGC | – | This article |
| ACCR6 | TCCTGGATCAGCTGGGACG | – | – |
| ACCF1 | CACAGACCATGATGCAGCTC | – | – |
| ACCR1 | CTCCCTGGAGTTGTGCTTTC | – | – |
| NsiI1781f | CTGTCTGAAGAAGACTATGGCCG | dCAPS for 1781 | Kaundun and Windass (2006) |
| NsiI1781r | AGAATACGCACTGGCAATAGCAGCACTTCCATGCA | – | – |
| EcoRV2078r | GCACTCAATGCGATCTGGATTTATCTTGATA | dCAPS for 2078 | This article |
Table III.
Combinations of ACCase mutant alleles that were identified in individual plants that survived the field rate of clethodim treatment (60 g ha−1 in Australia) from 14 clethodim-resistant Lolium populations
Twenty-one plants in population R7, 11 plants in R12, 12 plants in R13, 15 plants in R14, and six plants in each of the other populations were analyzed. *, 2078-Asp and 2088-Cys are wild-type alleles.
| Group | Genotype | Population (and No. of Plants) Where Detected |
|---|---|---|
| 1 | 1781-Leu/1781-Leu | R6 (2), R7 (21), R8 (4), R10 (2) |
| 2 | 1781-Leu/2027-Cys | R2 (3) |
| 3 | 1781-Leu/2041-Asn | R8 (1), R9 (1), R10 (4), R11 (1) |
| 4 | 2078-Gly/2078-Gly | R1 (4), R5 (6), R9 (1), R11 (4), R12 (11), R14 (1) |
| 5 | 2078-Gly/2078-Asp* | R3 (2), R4 (1), R9 (2), R13 (2), R14 (2) |
| 6 | 2078-Gly/1781-Leu | R1 (3), R4 (1), R6 (4), R8 (1), R9 (3), R11 (1), R14 (1) |
| 7 | 2078-Gly/2041-Asn | R9 (2) |
| 8 | 2088-Arg/2088-Arg | R2 (1), R3 (2), R4 (3), R13 (10), R14 (8) |
| 9 | 2088-Arg/2088-Cys* | R14 (2) |
| 10 | 2088-Arg/1781-Leu | R2 (2) |
| 11 | 2088-Arg /2041-Asn | R4 (1) |
| 12 | 2088-Arg/2078-Gly | R2 (1), R3 (2), R14 (1) |
Table IV.
Nonsynonymous SNPs with corresponding amino acid substitutions in the CT domain of plastidic ACCase of susceptible (S) populations S1 and S2 and resistant (R) populations containing a Cys to Arg mutation
Amino acid positions correspond to the full-length plastid ACCase in A. myosuroides. Nucleotide position numbers refer to the sequenced region (1,513 bp) of the plastidic ACCase gene (GenBank accessions EF538937–EF538943).
| Nucleotide Position | 221 | 729 | 741 | 752 | 884 | 933 | 957 | 1154 | 1281 | 1382 |
|---|---|---|---|---|---|---|---|---|---|---|
| SNP alleles | A, T | G, C | A, C | A, C | A, G | A, G | A, G | T, A | C, T | T, C |
| Amino acid position | 1701 | 1870 | 1874 | 1878 | 1922 | 1938 | 1946 | 2012 | 2054 | 2088 |
| S1-bulk | Leu/Met | Arg/Pro | Glu/Ala | Asn/His | Ser | Lys | Glu/Asp | Met/Leu | Thr/Ile | Cys |
| S2-bulk | Leu/Met | Arg/Pro | Glu/Ala | Asn/His | Ser/Gly | Lys/Arg | Glu/Asp | Met/Leu | Thr/Ile | Cys |
| R2-3 | Leu/Met | Arg/Pro | Glu/Ala | Asn/His | Ser | Lys | Glu/Asp | Met/Leu | Thr/Ile | Arg |
| R3-6 | Leu/Met | Arg/Pro | Glu/Ala | Asn/His | Ser | Lys | Glu/Asp | Met/Leu | Thr/Ile | Arg |
| R4-5 | Leu | Pro | Ala | His | Ser | Lys | Glu | Leu | Ile | Arg |
| R13-2 | Met | Arg | Glu | Asn | Ser | Lys | Asp | Met | Thr | Arg |
| R14-6 | Leu | Pro | Ala | His | Ser | Lys | Glu | Leu | Ile | Arg |
Combination Patterns of ACCase Mutant Alleles Endowing Clethodim Resistance
To facilitate quick and accurate identification of mutant ACCase alleles, we used a published CAPS/dCAPS marker for the 1781 allele and developed dCAPS markers for 2041, 2078, and 2088 alleles (see “Materials and Methods”). Marker analysis for the 2027 allele was not developed in this study due to limited numbers of clethodim survivors carrying the mutation. Among 14 clethodim-resistant populations tested, the mutant 2078-Gly allele(s) was found in clethodim survivors from 12 populations and 1781-Leu from 10 populations, while mutant 2088-Arg or 2041-Asn alleles were identified in five populations, and 2027-Cys only in one population (Table III). Clearly, clethodim resistance can be related to one or more of several specific mutant alleles.
We found that at least two types of mutant ACCase alleles were present in most populations except for some populations (R5, R7, and R12) in which only one type of mutant allele was detected (Table III). Moreover, different mutant alleles can be present in the same Lolium individual, as has already been observed in cross-pollinated A. myosuroides (Délye et al., 2005). For example, individual resistant Lolium plants could possess one 2078-Gly allele together with a 2088-Arg, 1781-Leu, or Asn-2041 allele. It is emphasized that diploid L. rigidum is obligate cross-pollinated, and therefore resistant individuals easily hybridize in the field and therefore there is enrichment of all possible resistance alleles in the progeny. A given individual L. rigidum plant can contain, at most, two distinct mutant ACCase alleles. As summarized in Table III, 12 combinations of mutant alleles (genotypes) in individual plants were found in 14 resistant populations. Most individuals surviving clethodim treatment usually had two mutant ACCase alleles (either a single type or two types), although a few surviving plants were heterozygous for the mutant 2078-Gly or 2088-Arg allele (Table III, groups 5 and 9). Additionally and unexpectedly, genotype groups 1 (1781-Leu/1781-Leu), 2 (1781-Leu/2027-Cys), and 3 (1781-Leu/2041-Asn) were also found to be associated with clethodim resistance. This is intriguing and worth further attention (see “Discussion”).
In Vitro Inhibition of ACCase Activity by ACCase Herbicides
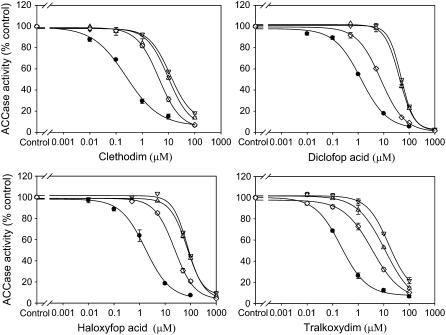
ACCase assays were conducted to confirm that different mutations/combinations of mutant alleles displayed resistant ACCase. Thus, ACCase was partially purified from plants homozygous for the 1781-Leu, 2078-Gly, or 2088-Arg alleles and from plants with two different mutant alleles (1781-Leu/2027-Cys or 1781-Leu/2041-Asn alleles). ACCase activity was evaluated in the presence of clethodim or other ACCase herbicides. Figure 1 shows that, as expected, ACCase from plants with mutant alleles was significantly less inhibited by APP herbicides (diclofop and haloxyfop acid) or CHD herbicides (clethodim or tralkoxydim). The herbicide concentration causing 50% inhibition of ACCase activity (I50) was determined for each herbicide and each genotype, to give a resistant/susceptible (R/S) ratio (Table V). High level resistance to ACCase herbicides was found for ACCase from homozygous 2078 or 2088 mutants (with the R/S ratio ranging from 32–75). Clear but lower level resistance was found for ACCase from homozygous 1781 mutants (R/S ratio from 6–17). A 7- to 13-fold resistance to clethodim was also observed for ACCase from mutant genotypes of 1781-Leu/2027-Cys and 1781-Leu/2041-Asn (Fig. 1; Table V). Clearly, different ACCase mutations/combinations can endow different levels and patterns of ACCase herbicide resistance.
Figure 1.
In vitro inhibition of ACCase activity by ACCase herbicides for susceptible plants (S1, •), resistant plants homozygous for 1781-Leu (⋄), 2078-Gly (▵), and 2088-Arg (▿) from population R7, R12, and R14, respectively. Data are pooled from two extractions per population per herbicide and each was assayed in duplicate.
Table V.
Parameter estimates for logistic analysis of in vitro inhibition of ACCase enzyme activity by various ACCase inhibiting herbicides for the susceptible population S1 and resistant (R) populations
ses are in parentheses. Data are pooled from two extractions per population per herbicide and each assayed in duplicate.
| Herbicide | Genotype (Population) | C | D | b | I50 | P Value for I50 | R/S Ratio for I50 |
|---|---|---|---|---|---|---|---|
| μm | |||||||
| Diclofop acid | 1781/1781a (R7) | 0.40 (1.06) | 100 (0.85) | 0.94 (0.04) | 7.67 (0.41) | <0.01 | 6 |
| 2078/2078a (R12) | 1.57 (1.91) | 102 (1.17) | 1.52 (0.16) | 40.9 (2.52) | <0.01 | 32 | |
| 2088/2088a (R14) | 2.56 (1.62) | 101 (0.93) | 1.75 (0.16) | 48.2 (2.02) | <0.01 | 38 | |
| Wild type (S1) | 3.22 (3.87) | 98 (2.26) | 0.84 (0.13) | 1.27 (0.24) | <0.05 | ||
| Haloxyfop acid | 1781/1781 (R7) | 3.37 (2.36) | 99 (1.59) | 1.11 (0.09) | 23.4 (2.53) | 0.01 | 14 |
| 2078/2078 (R12) | 3.57 (1.63) | 99 (0.72) | 1.30 (0.10) | 75 (3.06) | <0.01 | 44 | |
| 2088/2088 (R14) | 7.63 (3.86) | 102 (1.92) | 1.51 (0.31) | 73.5 (6.92) | <0.01 | 43 | |
| Wild type (S1) | 3.96 (3.80) | 98 (2.01) | 0.89 (0.13) | 1.70 (0.31) | <0.05 | ||
| Tralkoxydim | 1781/1781 (R7) | 3.56 (4.60) | 98 (1.71) | 0.77 (0.10) | 3.53 (0.72) | <0.05 | 17 |
| 2078/2078 (R12) | 0.53 (6.22) | 102 (1.16) | 0.80 (0.10) | 11.2 (2.31) | <0.05 | 53 | |
| 2088/2088 (R14) | 9.48 (7.45) | 102 (1.03) | 1.02 (0.19) | 15.8 (3.65) | <0.05 | 75 | |
| Wild type (S1) | 7.41 (1.33) | 100 (1.35) | 0.86 (0.06) | 0.21 (0.019) | <0.01 | ||
| Clethodim | 1781/2027b (R2) | 6.4 (1.36) | 100 (0.70) | 0.79 (0.04) | 1.62 (0.11) | <0.01 | 7 |
| 1781/2041c (R10) | 2.42 (3.92) | 100 (1.21) | 0.58 (0.05) | 3.20 (0.56) | <0.05 | 13 | |
| 1781/1781 (R7) | 3.56 (1.4) | 99 (0.64) | 1.04 (0.04) | 4.26 (0.26) | <0.01 | 18 | |
| 2078/2078 (R12) | 5.20 (3.71) | 98 (1.04) | 1.07 (0.14) | 9.76 (1.11) | <0.05 | 41 | |
| 2088/2088 (R14) | 10.46 (4.64) | 98 (1.05) | 1.07 (0.17) | 11.57 (1.70) | <0.05 | 48 | |
| 2088/2088 (R3) | 4.89 (4.87) | 99 (0.98) | 0.93 (0.11) | 11.75 (1.85) | <0.05 | 49 | |
| Wild type (S1) | 5.91 (3.51) | 99 (3.26) | 0.68 (0.10) | 0.24 (0.06) | 0.05 |
Plants homozygous for the mutant alleles of 1781-Leu, 2078-Gly, or 2088-Arg.
Plants containing two types of mutant alleles (1781-Leu/2027-Cys).
Plants containing two types of mutant alleles (1781-Leu/2041-Asn).
It is notable that we consistently observed that specific ACCase activity (in the absence of herbicides) was lower in extracts from plants homozygous for the 2078 or 2088 mutant allele (three resistant populations) compared to that from plants homozygous for the wild-type allele (Table VI). Conversely, ACCase activity in extracts from plants homozygous for the 1781 mutant allele or plants of other genotypes (1781/2027 or 1781/2041) was not significantly different from that of susceptible controls (Table VI). These results were obtained by carefully conducted experiments in which protein concentration in the assay mixture was normalized for each genotype, and by using two susceptible controls.
Table VI.
ACCase activities in the absence of ACCase herbicides in the shoots of susceptible (S) and resistant (R) populations with known ACCase mutations
Data are means ± se of two to six enzyme extractions per population and each assayed in duplicate. Means followed by different letters are significantly different at the 5% level by the lsd test. *, ACCase activities from two S populations (S1 and S2) were averaged for calculation of R/S ratios.
| Genotype Used | Population | ACCase Activity | R/S* Ratio |
|---|---|---|---|
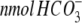 mg protein−1 min−1 mg protein−1 min−1
|
|||
| 1781-Leu/1781-Leu | R7 | 9.10 ± 0.26 a | 1.12 |
| 1781-Leu/2027-Cys | R2 | 9.97 ± 0.25 a | 1.23 |
| 1781-Leu/2041-Asn | R10 | 7.88 ± 2.27 ab | 0.97 |
| 2078-Gly/2078-Gly | R12 | 5.86 ± 0.32 bc | 0.72 |
| 2088-Arg/2088-Arg | R3 | 4.38 ± 0.55 c | 0.54 |
| 2088-Arg/2088-Arg | R13 | 4.46 ± 0.97 c | 0.55 |
| 2088-Arg/2088-Arg | R14 | 4.45 ± 0.42 c | 0.55 |
| Wild type | S1 | 7.38 ± 0.06 ab | |
| Wild type | S2 | 8.85 ± 0.22 a |
Genotyping Resistant Populations for the 1781 or 2078 Mutant Alleles Using dCAPS Markers
A published dCAPS marker (Kaundun and Windass, 2006) was tested for suitability for genotyping Lolium populations under our modified PCR conditions. For a total of 84 samples of known genotypes tested, the accuracy was >97%. This dCAPS marker was, therefore, employed to genotype the clethodim-resistant population R7 (n = 40). Remarkably, all plants in this population were homozygous for the resistant 1781-Leu allele. When these genotyped homozygous plants were tested with the field rate of clethodim (at 3–4 leaf stage), they all survived, while all susceptible plants (S1) died. Clearly, homozygous mutants for the 1781-Leu allele can withstand the Australian field rate of clethodim, whereas heterozygous mutants cannot. Therefore, the specific mutation, homozygosity versus heterozygosity, and the rate of herbicide used in testing for resistance are all important in determining the level of resistance in an individual or population.
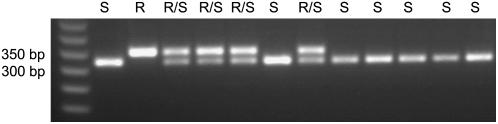
We developed a dCAPS marker for detection of the mutant 2078-Gly allele (Fig. 2; see “Materials and Methods”) in Lolium populations. The robustness and accuracy of this marker was tested with a total of 120 samples of known genotypes from across 14 resistant Lolium populations and the results obtained matched sequencing results by >98%. This dCAPS marker was therefore used to genotype the clethodim-resistant population R12 (n = 45). Genotype frequencies were found to be 0.60 for homozygous-resistant 2078-Gly individuals, 0.02 for homozygous susceptible 2078-Asp individuals, and 0.38 for heterozygous individuals. The heterozygous individuals were further analyzed by the 1781 dCAPS marker and it was found that all heterozygous individuals for the resistant 2078 allele also contained one resistant 1781 allele. Therefore, at the commercial herbicide use rate, 98% of the population was found to be clethodim resistant.
Figure 2.
dCAPS analysis of individual L. rigidum plants homozygous susceptible 2078-Asp (S), homozygous-resistant 2078-Gly (R), or heterozygous 2078-Gly/Asp (R/S). The sizes of restriction enzyme (EcoRV) digested fragments are 353 and 323 bp, respectively.
A CAPS marker for the 2088 mutation was designed and tested against known genotypes. However, this marker is not ideal for large-scale genotyping due to the cost limitation of an expensive restriction enzyme.
Dose Response to Clethodim and Cross Resistance to Other ACCase Herbicides at the Whole Plant Level
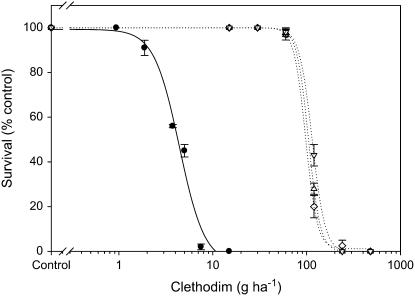
Purified populations with plants homozygous for the mutant resistant 1781, 2078, or 2088 alleles were used to determine their clethodim resistance levels. As shown in Figure 3, the susceptible population (S1) was killed at 7.5 g clethodim ha−1 or higher. In contrast, homozygous-resistant plants were markedly less affected by clethodim, requiring a high rate (240 g ha−1) for substantial mortality. The clethodim rate causing 50% mortality (LD50) for the susceptible population S1 was 4.4 ± 0.43 g ha−1 versus 98 ± 1.68, 105 ± 0.23, and 115 ± 0.45 for the homozygous-resistant populations containing the mutant resistant 1781, 2078, or 2088 alleles, respectively. Therefore, based on the R/S LD50 ratio, the homozygous-resistant populations are more than 20-fold resistant to clethodim at the whole plant level. Clearly, plants homozygous for the mutant resistant 1781, 2078, or 2088 alleles are all resistant to clethodim at the commercial Australian field rate. Cross resistance pattern to other ACCase herbicides was also determined. As shown in Table VII, at field or higher rates, plants homozygous for the mutant resistant 1781, 2078, or 2088 alleles were resistant to APP herbicides clodinafop, diclofop, fluazifop, and haloxyfop, CHD herbicides sethoxydim and tralkoxydim, and the phenylpyrazolin herbicide pinoxaden. About 50%, 70%, and 87% of plants homozygous for the mutant resistant 1781, 2088, or 2078 alleles, respectively, were cross resistant to the CHD herbicide butroxydim (Table VII).
Figure 3.
Clethodim dose response of the known susceptible population S1 (wild type, •) and purified homozygous-resistant populations R7P (1781-Leu, ⋄), R12P (2078-Gly, ▵), and R14P (2088-Arg, ▿). Data for the susceptible population S1 were pooled from two experiments and data for the purified populations was each from a single experiment.
Table VII.
Percentage survival of plants from purified homozygous-resistant populations for the three ACCase mutations at herbicide application rates known to control the susceptible population S1
Forty to 50 plants per population were treated with the respective herbicide.
| Herbicides | Application Rate | Genotype (Population), % Survival
|
|||
|---|---|---|---|---|---|
| Wild Type (S1) | 1781-Leu (R7P) | 2078-Gly (R12P) | 2088-Arg (R14P) | ||
| g ha−1 | |||||
| APP | |||||
| Diclofop | 1,000 | 1 | 100 | 100 | 100 |
| Clodinafop | 50 | 3 | 100 | 100 | 100 |
| Fluazifop | 100 | 0 | 100 | 100 | 100 |
| Haloxyfop | 52 | 0 | 100 | 100 | 100 |
| CHD | |||||
| Sethoxydim | 186 | 0 | 100 | 100 | 100 |
| Tralkoxydim | 800 | 4 | 100 | 100 | 100 |
| Butroxydim | 45 | 2 | 50 | 87 | 70 |
| Phenylpyrazolin | |||||
| Pinoxaden | 30 | 0 | 100 | 100 | 100 |
DISCUSSION
Substitution of Amino Acid Asp-2078-Gly Endowing Resistance to Clethodim and Other ACCase Herbicides in Lolium Populations
Until now, the Asp-2078-Gly substitution has been the only ACCase mutation known to endow clethodim resistance and only reported in A. myosuroides (Délye et al., 2005). Here, we identified this Asp-2078-Gly mutation present in individuals within 12 of 14 (86%) clethodim-resistant Lolium populations examined (Table III), indicating that this mutation is relatively commonly associated with clethodim resistance. We confirmed that this Asp-2078-Gly substitution results in an ACCase enzyme resistant to clethodim and the other ACCase herbicides tested (Fig. 1; Table V). The level of resistance conferred by the Asp-2078-Gly mutation at the enzyme level in Lolium (Table V) was found to be similar to the level of resistance confirmed by this mutation in A. myosuroides (Délye et al., 2005). The purified population (R12P) consisting of individuals homozygous for the mutant 2078-Gly allele was 24-fold more resistant to clethodim than the susceptible population (Fig. 3), and found to be cross resistant to all the APP and CHD herbicides tested, as well as the phenylpyrazolin herbicide pinoxaden (Table VII). Therefore, we conclude that the Asp-2078-Gly substitution endows resistance in Lolium to clethodim and the other ACCase herbicides tested.
Substitution of Amino Acid Cys-2088-Arg Endowing Resistance to Clethodim and Other ACCase Herbicides in Lolium Populations
Notably, in this study we have identified and characterized a new ACCase mutation, a Cys to Arg substitution at position 2088, in five resistant Lolium populations (Tables III and IV). This mutation confers an ACCase herbicide resistance profile (determined at the enzyme and whole plant level) similar to the Asp-2078-Gly mutation (Tables V and VII; Figs. 1 and 3). We, therefore, conclude that this Cys-2088-Arg mutation confers resistance to clethodim and other ACCase herbicides. In fact, the amino acid residue at position 2088 was largely conserved as Cys among 28 grass species putatively susceptible to ACCase herbicides, with only a few species displaying Phe at this position (Fig. 4). Using ACCase three-dimensional models derived from the structure of the yeast (Saccharomyces cerevisiae) CT-APP complex, Délye et al. (2005) assessed the consequences of various amino acid substitutions identified in A. myosuroides, and predicted that a region including amino acids 2027 to 2096 may contain more unknown amino acid residues involved in sensitivity to ACCase herbicides. Our finding supports this hypothesis. Plants homozygous for the mutant 2088 allele survived the field or higher rate of clethodim (60–120 g ha−1; Fig. 3). However, only two individuals (from population R14) heterozygous for this mutation survived the commercial field rate (60 g ha−1) of clethodim (Table III, group 9). This indicates the strong interaction between the specific resistance-endowing mutation, homozygosity versus heterozygosity of this mutation, and the rate of herbicide use. These results show that homozygous and heterozygous plants have different levels of resistance and that the resistant 2088 allele is incompletely dominant above the field rate of clethodim. The same 2078-Gly and 2088-Arg mutations were reported in A. fatua by Christoffers et al. (2000) in their preliminary studies, and proposed as being responsible for low level clethodim resistance (below the field rate of 140 g ha−1 in U.S.; Christoffers et al., 2005).
Figure 4.
Alignment of partial amino acid sequences of chloroplastic homomeric ACCases from 28 grass species that are putatively susceptible to ACCase herbicides. Numbers above the sequences indicate amino acid positions within the A. myosuroides full ACCase sequence (GenBank accession AJ310767). Amino acid residues 2078, 2088, and 2096 are in bold and conserved amino acids are indicated by dots. The boxed 2088 residue was conserved as a Cys (C) among most grass species except for a few species as a Phe (F).
Combination of Two Mutant 1781 Alleles Endowing Resistance to Clethodim and Other ACCase Herbicides in Lolium Populations
In addition to mutant 2078 and 2088 alleles, the mutant 1781-Leu allele was found in many individuals within most (71%) of the clethodim-resistant populations, usually in combination with another mutant allele of the same or different type (Table III). Plants homozygous for the mutant 1781-Leu allele were able to survive clethodim at the field rate, whereas heterozygous plants could not survive this rate. The homozygous-resistant genotype (1781-Leu/1781-Leu) was detected in four populations (Table III) and its resistance to clethodim was confirmed by an ACCase in vitro assay in which a moderate level of resistance (17-fold) was observed (Table VI). This genotype was found to be equally resistant to clethodim at the whole plant level, as compared to plants homozygous for the mutant 2078 or 2088 alleles (Fig. 3). In addition, this genotype was found to be cross resistant to APP herbicides clodinafop, diclofop, fluazifop, and haloxyfop, CHD herbicides sethoxydim and tralkoxydim, and the phenylpyrazolin herbicide pinoxaden (Table VIII). Therefore, resistance at field clethodim rates requires homozygosity of plants for the mutant 1781 alleles. Remarkably, one field evolved clethodim-resistant population (R7) was found to be 100% homozygous for the 1781 mutant alleles.
Table VIII.
Restriction enzymes used in dCAPS/CAPS analysis
| Enzyme | Commercial Isoschizomers | Restriction Site | Experiment | Reference |
|---|---|---|---|---|
| NsiI | AvaIII, EcoT22I, Mph11031, Zsp2I | 5′-ATGCA^T-3′ | dCAPS (1781) | Kaundun and Windass (2006) |
| 3′-T^ACGTA-5′ | ||||
| EcoRI | FunII | 5′-G^AATTC-3′ | CAPS (2041) | This article |
| 3′-CTTAA^G-5′ | ||||
| EcoRV | Eco32I | 5′-GAT^ATC-3′ | dCAPS (2078) | This article |
| 3′-CTA^TAG-5′ | ||||
| Eco47III | AfeI, Aor51HI, FunI | 5′-AGC^GCT-3′ | CAPS (2088) | This article |
| 3′-TCG^CGA-5′ |
Combination of Mutant 1781/2027 or 1781/2041 Alleles Endowing Clethodim Resistance in Lolium Populations
In this study with field evolved resistant Lolium populations, we reveal 12 patterns of mutant ACCase allele combinations endowing ACCase herbicide resistance (Table III). This is to be expected in this highly genetically diverse, obligate cross-pollinated Lolium. Within a large herbicide-treated field, Lolium individuals homo/heterozygous for different specific mutations of ACCase survive herbicide treatment, and in the absence of (killed) susceptible individuals, cross-pollination occurs among resistant survivors. What emerges are resistant populations comprised of individuals containing diverse ACCase mutations (a maximum of two). As expected, the genotype groups 4 to 12 would confer clethodim resistance (Table III). What is interesting is that the 2027-Cys or 2041-Asn allele is known to be mainly associated with APP herbicide resistance (Délye et al., 2005); however, combinations of 1781-Leu/2027-Cys alleles or 1781-Leu/2041-Asn alleles were found to confer clethodim resistance in Lolium at the field rate (Table III). This was also confirmed at the ACCase enzyme level with an I50 R/S ratio of 7 and 13 (Fig. 1; Table V) for the mutant allele combinations of 1781/2027 and 1781/2041, respectively.
In studies with resistant A. myosuroides and L. rigidum populations from France, neither heterozygous nor homozygous mutants of 1781-Leu were found to be resistant to clethodim, haloxyfop, or clodinafop, and the genotype 1781-Leu/2041-Asn was not found to be resistant to clethodim in a seed germination assay (Délye et al., 2002b, 2005). Conversely, in a recent study in Avena sterilis, 2027-Cys and 2041-Asn mutations were found to be associated with resistance to the CHD herbicides tralkoxydim and sethoxydim, respectively (Liu et al., 2007). Similarly, the 2041-Asn mutation in Phalaris paradoxa was found to confer resistance to most CHD herbicides with a lower level resistance to clethodim (Hochberg et al., 2007). These discrepancies in the cross-resistance pattern endowed by a specific ACCase mutation are likely due to difference in plant species, methods of testing herbicide sensitivity, and/or especially herbicide rates used to discriminate between resistant and susceptible individuals. For example, in seed germination assay, germinating seedlings are exposed continuously to the herbicide, which is quite different from the field-simulating herbicide spray treatment used in our research. Therefore, it is possible that A. myosuroides plants containing 2027 or 2041 mutant alleles could survive field rates of CHD herbicides (Délye, 2005).
In assessing herbicide resistance there is often insufficient attention paid to the importance of the rate of herbicide used. Herbicide rate is a potent factor in resistance evolution. Where selection occurs at a high herbicide rate, only individuals endowed with relatively strong resistance mechanisms survive. Conversely, selection at a lower herbicide rate enables survival of both individuals with strong resistance mechanisms and individuals with weaker resistance mechanisms (Neve and Powles, 2005). At the relatively low field rate of clethodim (60 g ha−1 used in Australia, compared with >140 g/ha in North America and Europe), Lolium plants with certain ACCase mutations can survive. The survival of individuals carrying weaker ACCase mutations is dependent on the rate of herbicide used and whether the individuals are homozygous or heterozygous for the particular resistance mutation. A particular herbicide dose can be lethal to heterozygous individuals, whereas homozygous individuals survive. This is evident for 1781-Leu alleles in relation to clethodim resistance in Lolium species. In this study, the resistant 1781-Leu allele was detected in many of the clethodim-resistant populations tested. However, heterozygous individuals do not survive clethodim resistance at the field rate, whereas homozygous mutant individuals do (Table III). Indeed, different levels of resistance conferred by one or two resistant 1781 alleles have been demonstrated in a L. rigidum population in relation to resistance to the ACCase herbicide fenoxaprop (Tal and Rubin, 2004).
Taken together, we have revealed that 12 field evolved clethodim-resistant Lolium populations have the resistant 2078 mutation and five populations have the resistant 2088 mutation, and these two mutations endow a sufficient level of resistance to clethodim and other ACCase herbicides. We found 12 combination patterns of mutant alleles are present in Lolium populations in relation to clethodim resistance. Additionally, we have established that resistance mutation at 1781, 2027, or 2041 can also confer clethodim resistance under certain conditions. Therefore, clethodim resistance was found to be determined by the specific mutation, homo/heterozygous status of plants for a specific mutation, and combinations of different resistant alleles. This diversity and complexity should be recognized.
In this research, we have not examined for any nontarget site-based clethodim resistance mechanism in these populations. However, we know that multiple resistance mechanisms (target site and nontarget site based) can be simultaneously expressed in individual plants of genetically diverse, cross-pollinated L. rigidum (Tardif and Powles, 1994; Yu et al., 2007).
ACCase Activity Associated with Specific ACCase Mutations/Genotypes
It was found in this study that plants homozygous for the resistant allele 2088 or 2078 showed lower ACCase specific activity (Table VI). Low ACCase activity was also observed in A. myosuroides for resistant alleles 2027-Cys and 2078-Gly (Délye et al., 2005). Low ACCase activity indicates that these residues are important for CT catalytic activity and these amino acid substitutions, although conferring herbicide resistance, may reduce enzyme catalytic activity and impose a fitness penalty at the whole plant level. From the three-dimensional models of A. myosuroides CT-herbicide complexes reconstructed from yeast, it was indicated that 2027 and 2078 mutations did not directly interfere with herbicide binding; instead, they may change the three-dimensional shape of the cavity of the CT active site by inducing a number of small allosteric changes (Délye et al., 2005). We intend to examine whether this low enzyme activity associated with the 2078 or 2088 mutation (Table VI) has consequences for ecological fitness of resistant plants. For resistance to acetolactate synthase-inhibiting herbicides certain acetolactate synthase gene mutations cause fitness penalties (Roux et al., 2004; Tardif et al., 2006). Table VI indicates this may also be the case for particular resistant mutant ACCase alleles. We have demonstrated that the 1781 mutation does not result in lower ACCase activity (Table VI) and our fitness studies in one Lolium population containing the 1781 mutation showed no, or negligible, resistance fitness cost (Vila-Aiub et al., 2005a, 2005b). However, a fitness penalty was detected in A. myosuroides in association with the 2078 mutation (Délye et al., 2007). On the basis of the lower ACCase activity in Lolium plants with the 2088 or 2078 mutation (Table VI) we speculate that plants carrying these mutations may suffer a fitness penalty. Since clethodim resistance in our Lolium populations largely involved mutations at 1781, 2041, 2078, and 2088 (Table III), we have developed CAPS/dCAPS markers for each of the mutations, and these markers will be of use in fitness studies.
In summary, we have identified five ACCase mutations (1781-Leu, 2027-Cys, 2041-Asn, 2078-Gly, and 2088-Arg) and revealed 12 genotypes in 14 clethodim-resistant Lolium populations. We established that the mutation 2078-Gly or 2088-Arg confers sufficient level of resistance to clethodim and other ACCase herbicides. In addition, other mutations (especially 1781-Leu) also confer clethodim resistance if plants are homozygous for this mutation or in combination with 2027-Cys or 2041-Asn. This is very important because the 1781 mutation is a relatively common mutation in Lolium populations. Thus, we have established that resistance to ACCase herbicides depends on the specific resistant allele(s), the homo/heterozygous status of plants for the specific resistant allele(s), and combinations of different resistant alleles plus herbicide rates are all important. To fully understand resistance, knowledge of all these factors is essential.
MATERIALS AND METHODS
Plant Materials
Several Lolium rigidum populations resistant to clethodim were identified during herbicide screening in a large random survey across the western Australian wheat (Triticum aestivum) belt (Owen et al., 2007). Seedlings of these field populations were sprayed with clethodim at the commercial rate of 60 g ha−1 using a cabinet sprayer delivering 113 L ha−1 water at a pressure of 200 kPa. The survivors of each population were grown to maturity and allowed to cross-pollinate only within the population. Seeds of 12 clethodim-resistant populations of L. rigidum from Australia (H1/2, H1/10, H1/19, H1/25, H2/2, M1/23, M1/25, M2/3, M2/15, M2/19, M2/23, and M3/4, hereinafter referred to as R1 to R12, respectively) and two Lolium spp. populations from Italy (259 and 281, referred to as R13 and R14) were used in this research. A known herbicide susceptible L. rigidum population (VLR1, referred to as S1) from Australia and a susceptible Lolium population (204, referred to as S2) from Italy were used as controls. Seeds of resistant and susceptible populations were germinated in plastic trays containing potting soil and seedlings grown in a glasshouse at 20°C/15°C day/night temperature under natural sunlight. At the two to three leaf stage, these seedlings were treated with 60 g ha−1 of clethodim. This rate killed all the plants in susceptible populations. Individual survivors from resistant populations were used for subsequent molecular and biochemical analysis.
Sequencing of the Plastidic ACCase Gene CT Domain
Shoot material of individual survivors from resistant populations was used for DNA extraction. Bulked shoot material from two susceptible populations without herbicide treatment was used as a control. Genomic DNA was extracted from shoot tissues using a Nucleon Phytopure DNA extraction kit (Amersham Biosciences). Primers were used or designed to amplify regions in the CT domain known to be involved in sensitivity to ACCase herbicides (Délye and Michel, 2005). Plastidic ACCase sequences used for the primer design were from L. rigidium (GenBank accession numbers are AF359516, AY995225, AY995232, AY995233, DQ184633, DQ184640, and DQ184646), Lolium multiflorum (AY710293), and Alopecurus myosuroides (AJ310767). Cytosolic ACCase sequences were from A. myosuroides (AJ632096) and wheat (U39321). Because of the high level of similarity between plastidic and cytosolic ACCase DNA sequences (about 74%), when designing primers, particular attention was given to consensus sequences of plastidic and cytosolic ACCase sequences, and each primer contained at least one specific nucleotide at the 3′ end to discriminate plastidic and cytosolic sequences. The primer pair ACCF5/ACCR5 from Délye et al. (2002b) was used to amplify a 785-bp region of the plastidic ACCase gene containing codon 1781 (Table II). The primer pair ACCF1/ACCR1 was designed to amplify a 492-bp region containing codons 2027, 2041, 2078, 2088, and 2096 (Table II). The primer pair ACCF6/ACCR6 was designed to amplify a 484-bp region bridging the above mentioned two regions. The PCR was conducted in a 25 μL volume that consisted of about 300 ng of genomic DNA, 0.5 μm of each primer, and 12.5 μL of 2× GoTaq Green Master mix (Promega). The PCR was run in a Mastercycler (Eppendorf) with the following profile: 94°C 4 min, 35 cycles of 94°C 30 s, 62°C 30 s, and 72°C 30 s, followed by a final extension step of 5 min at 72°C. The PCR product was directly purified or purified from agarose gel with Wizard SV gel and PCR Clean-up system (Promega), and sequenced from both ends with the AB-Big Dye Terminator system using a commercial sequencing service. At lease six survivors from each clethodim-resistant population were sequenced. All sequences were visually checked with chromatogram files and assembled and aligned using the DNAMAN software. Heterozygous individuals were recognized by double peaks at the same position in nucleotide chromatograms of both forward and reverse sequencing. Heterozygosity at position 2041, 2078, or 2088 was also further verified by using CAPS or dCAPS analysis (see below).
CAPS Analysis
The nucleotide T to A mutation at codon 2041 in the plastidic ACCase gene, causing an amino acid Ile to Asn change, removes an EcoRI restriction site (Table VIII). Sequence results showed no other SNPs around the restriction site. Thus, we used the primer pair ACCF1/ACCR1 (Table II) to amplify a 492-bp fragment followed by EcoRI digestion at 37°C for 3 h (all restriction enzymes were obtained from Fermentas Life Science). Homozygous-resistant plants with two mutant 2041-Asn alleles would display a single undigested band of 492 bp. In contrast, homozygous susceptible plants with two 2041-Ile alleles would have two resolvable bands of 208 and 282 bp. Heterozygous plants with both 2041-Asn and 2041-Ile alleles would have all three bands.
The nucleotide T to C mutation at codon 2088, causing an amino acid Cys to Arg substitution, creates an Eco47III restriction site (Table VIII). Sequence results revealed no other SNPs around this restriction site. Therefore, the same primer pair ACCF1/ACCR1 (Table II) was used to amplify a 492-bp fragment followed by Eco47III digestion. Homozygous susceptible plants with two 2088-Cys alleles would display a single undigested band of 492 bp. Homozygous-resistant plants with two mutant 2088-Arg alleles would have two resolvable bands of 141 and 351 bp, and heterozygous plants with both wild-type and mutant alleles would have all three bands.
dCAPS Analysis
A dCAPS marker for the 2078 mutation (Asp to Gly) was developed in this research to facilitate rapid and accurate identification of mutant 2078-Gly alleles. A 31-bp reversed dCAPS primer EcoRV2078r was designed (Table II) using the dCAPS Finder 2.0 software (Neff et al., 1998) based on highly conserved sequences around and especially toward the 3′ end of the 2078 codon of all sequenced plants. An A:G mismatch was introduced in the reverse primer to create a restriction site for EcoRV in the susceptible sequence (Table II). The primer pair ACCF1/EcoRV2078r (Table II) amplifies a 353-bp fragment using the same PCR conditions as for sequencing. Following EcoRV digestion, individuals with homozygous-resistant 2078-Gly alleles would have an uncut band of 353 bp, while individuals with homozygous susceptible 2078-Asp alleles would have a digested band of 323 bp (Fig. 2). Individuals with both susceptible and resistant alleles would have a combination of two resolvable bands (Fig. 2).
The published dCAPS marker for the 1781 mutation (Ile to Leu; Kaundun and Windass, 2006) was used with primer pair NsiII1781f/NsiI1781r (Table II) with modified PCR conditions as described for sequencing.
In Vitro Inhibition of ACCase Activity by ACCase Herbicides
Individual clethodim survivors containing two mutant 1781, 2078, or 2088 alleles, and individual survivors containing two types of mutant alleles (1781-Leu/2027-Cys or 1781-Leu/2041-Asn) were identified by marker analyses and sequencing. These plants were transplanted, fertilized, and maintained in a glasshouse at 20°C/15°C day/night temperature. Shoot tissue of each genotype was harvested, snap frozen in liquid nitrogen, and immediately used for the enzyme assay. Herbicide susceptible plants from population S1 or S2 at the same developmental stage were used as controls. ACCase extraction and partial purification and enzyme inhibition by ACCase herbicides were performed as described (Yu et al., 2004). Two subsamples from each extraction were assayed and there were at least two extractions per population per herbicide treatment.
Response of Purified Resistant Populations to ACCase Herbicides at the Whole Plant Level
Three purified populations were obtained by bulk cross-pollinating at least six plants homozygous for the mutant 1781, 2078, or 2088 alleles. Mutant 1781, 2078, or 2088 alleles were therefore purified and fixed in three subpopulations R7P, M12P, and R14P, respectively. Seeds of purified populations were germinated on 0.6% agar-solidified water for 7 d. Germinated seedlings were transplanted to plastic pots (20–25 seedlings per pot) or trays (40–50 seedlings per tray) containing potting mix and were kept in naturally illuminated glasshouses at 25°C/15°C day/night temperature. Seedlings in pots were treated at the two to three leaf stage with rates of clethodim (0, 0.94, 1.88, 3.75, 7.5, and 15 g ha−1 for the susceptible population S1; 0, 15, 30, 60, 120, 240, and 480 g ha−1 for purified resistant populations) using a cabinet sprayer and each treatment contained three replicates. Seedlings in trays were sprayed with a number of other ACCase herbicides at a rate known to control susceptible plants (see Table VII). Herbicides were applied as commercial formulations plus adjuvant as required, using a cabinet sprayer. Plants were returned to the glasshouse after treatment and the mortality was recorded 21 d after herbicide application. Plants were recorded as alive if they had strongly tillered since herbicide application.
Statistical Analysis
The herbicide concentration causing 50% inhibition of enzyme activity (I50), or the herbicide rate causing 50% mortality (LD50), was estimated by nonlinear regression using the logistic model (Seefeldt et al., 1995):
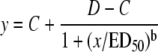 |
where C = lower limit, D = upper limit, ED50 = dose giving 50% response, and b = slope around ED50. Estimates were obtained using the Sigmaplot software (version 8.02, SPSS). A t test (P = 0.05) was used to test significance of the regression parameters. Analysis of variance was performed by ANOVA and significant differences in ACCase specific activities among genotypes in the absence of inhibitor herbicides were determined by the lsd test.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF538937 to EF538943.
This work was supported by the Grains Research and Development Corporation of Australia (to the Western Australian Herbicide Research Initiative) and by a research exchange scholarship from the Western Australia Association for Research between Italy and Australia (to A.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stephen B. Powles (spowles@plants.uwa.edu.au).
Open Access articles can be viewed online without a subscription.
References
- Brown AC, Moss SR, Wilson ZA, Field LM (2002) An isoleucine to leucine substitution in the ACCase of Alopecurus myosuroides (black grass) is associated with resistance to the herbicide sethoxydim. Pestic Biochem Physiol 72 160–168 [Google Scholar]
- Christoffers M, Berg ML, Messersmith CG (2002) An isoleucine to leucine mutation in acetyl-CoA carboxylase confers herbicide resistance in wild oat. Genome 45 1049–1056 [DOI] [PubMed] [Google Scholar]
- Christoffers MJ, Berg ML, Messersmith CG (2000) Analysis of acetyl-CoA carboxylase gene sequences from fenoxaprop-p-resistant wild oat biotypes. NCWSS Proceedings 55 67 [Google Scholar]
- Christoffers MJ, Pederson SN, Kandikonda AV (2005) Herbicide dose-response of wild oat with altered acetyl-CoA carboxylase genes. NCWSS Proceedings 60 35 [Google Scholar]
- Délye C (2005) Weed resistance to acetyl coenzyme A carboxylase inhibitors: an update. Weed Sci 53 728–746 [Google Scholar]
- Délye C, Calmes E, Matejicek A (2002. a) SNP markers for black-grass (Alopecurus myosuroides Huds.) genotypes resistant to acetyl CoA-carboxylase inhibiting herbicides. Theor Appl Genet 104 1114–1120 [DOI] [PubMed] [Google Scholar]
- Délye C, Matejicek A, Gasquez J (2002. b) PCR-based detection of resistance to acetyl-CoA carboxylase-inhibiting herbicides in black-grass (Alopecurus myosuroides Huds.) and ryegrass (Lolium rigidum Gaud). Pest Manag Sci 58 474–478 [DOI] [PubMed] [Google Scholar]
- Délye C, Menchari Y, Cadet E, Chauvel B, Darmency H (2007) Fitness variation associated with herbicide-resistant acetyl-CoA carboxylase alleles in black-grass (Alopecurus myosuroides Huds.). In E Fløistad, ed, 14th EWRS Symposium. Hamar, Norway, p 144
- Délye C, Michel S (2005) “Universal” primers for PCR-sequencing of grass chloroplastic acetyl-CoA carboxylase domains involved in resistance to herbicides. Weed Res 45 323–330 [Google Scholar]
- Délye C, Wang TY, Darmency H (2002. c) An isoleucine-leucine substitution in chloroplastic acetyl-CoA carboxylase from green foxtail (Setaria viridis L. Beauv.) is responsible for resistance to the cyclohexanedione herbicide sethoxydim. Planta 214 421–427 [DOI] [PubMed] [Google Scholar]
- Délye C, Zhang XQ, Chalopin C, Michel S, Powles SB (2003) An isoleucine residue within the carboxyl-transferase domain of multidomain acetyl-coenzyme A carboxylase is a major determinant of sensitivity to aryloxyphenoxypropionate but not to cyclohexanedione inhibitors. Plant Physiol 132 1716–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Délye C, Zhang XQ, Michel S, Matejicek A, Powles SB (2005) Molecular bases for sensitivity to acetyl-coenzyme A carboxylase inhibitors in black-grass. Plant Physiol 137 794–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap IM, Knight R (1986) The occurrence of herbicide cross-resistance in a population of annual ryegrass, Lolium rigidum, resistant to diclofop-methyl. Aust J Agric Res 41 121–128 [Google Scholar]
- Hochberg O, Sibony M, Tal A, Rubin B (2007) Molecular bases for the resistance to ACCase inhibiting herbicides in Phalaris paradoxa. In E Fløistad, ed, 14th EWRS Symposium, Hamar, Norway, p 150
- Hofer U, Muehlebach M, Hole S, Zoschke A (2006) Pinoxaden-for broad spectrum grass weed management in cereal crops. J Plant Dis Prot 20 989–995 [Google Scholar]
- Holtum JAM, Matthews JM, Liljegren DR, Powles SB (1991) Cross-resistance to herbicides in annual ryegrass (Lolium rigidum). III. On the mechanism of resistance to diclofop-methyl. Plant Physiol 97 1026–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundun SS, Windass JD (2006) Derived cleaved amplified polymorphic sequence, a simple method to detect a key point mutation conferring acetyl CoA carboxylase inhibitor herbicide resistance in grass weeds. Weed Res 45 34–39 [Google Scholar]
- Liu WJ, Harrison DK, Chalupska D, Gornicki P, O'Donnell CC, Adkins SW, Haselkorn R, Williams RR (2007) Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Natl Acad Sci USA 104 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn R, Powles SB (2001) High levels of herbicide resistance in rigid ryegrass (Lolium rigidum) across the Western Australian wheatbelt. Weed Technol 15 242–248 [Google Scholar]
- Matthews JM, Holtum JAM, Liljegren DR, Furness B, Powles SB (1990) Cross-resistance to herbicides in annual ryegrass (Lolium rigidum). 1. Properties of the herbicide target enzymes acetyl-CoA carboxylase (ACC) and acetolactate synthase (ALS). Plant Physiol 94 1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14 387–392 [DOI] [PubMed] [Google Scholar]
- Neve P, Powles SB (2005) Recurrent selection with reduced herbicide rates results in the rapid evolution of herbicide resistance in Lolium rigidum. Theor Appl Genet 110 1154–1166 [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Ohlrogge JB, Wurtele ES (2003) Plant biotin-containing carboxylases. Arch Biochem Biophys 414 211–222 [DOI] [PubMed] [Google Scholar]
- Owen M, Walsh M, Llewellyn R, Powles SB (2007) Widespread occurrence of multiple herbicide resistance in Western Australian annual ryegrass (Lolium rigidum) populations. Aust J Agric Res 58 711–718 [Google Scholar]
- Powles SB, Matthews JM (1992) Multiple herbicide resistance in annual ryegrass (Lolium rigidum): a driving force for the adoption of integrated weed management strategies. In I Denholm, AL Devonshire, DW Hollomon, eds, Resistance 91: Achievements and Developments in Combating Pesticide Resistance. Elsevier, New York, pp 75–87
- Preston C, Powles SB (1998) Amitrole inhibits diclofop metabolism and synergises diclofop-methyl in a diclofop-methyl-resistant biotype of Lolium rigidum. Pestic Biochem Physiol 62 179–189 [Google Scholar]
- Preston C, Tardif FJ, Christopher JT, Powles SB (1996) Multiple resistance to dissimilar herbicide chemistries in a biotype of Lolium rigidum due to enhanced activity of several herbicide degrading enzymes. Pestic Biochem Physiol 54 123–134 [Google Scholar]
- Roux F, Gasquez J, Rebound X (2004) The dominance of the herbicide resistance cost in several Arabidopsis thaliana mutant lines. Genetics 166 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeldt S, Jensen JE, Fuerst EP (1995) Log-logist analysis of herbicide dose-response relationships. Weed Technol 9 218–227 [Google Scholar]
- Tal A, Rubin B (2004) Molecular characterization and inheritance of resistance to ACCase herbicides in Lolium rigidum. Pest Manag Sci 60 1013–1018 [DOI] [PubMed] [Google Scholar]
- Tardif FJ, Holtum JAM, Powles SB (1993) Occurrence of a herbicide-resistant acetyl-coenzyme A carboxylase mutant in annual ryegrass (Lolium rigidum) selected by sethoxydim. Planta 190 176–181 [Google Scholar]
- Tardif FJ, Powles SB (1994) Herbicide multiple-resistance in a Lolium rigidum biotype is endowed by multiple mechanisms: isolation of a subset with resistant acetyl-CoA carboxylase. Physiol Plant 91 488–494 [Google Scholar]
- Tardif FJ, Preston C, Holtum JAM, Powles SB (1996) Resistance to acetyl-coenyme A carboxylase-inhibiting herbicides endowed by a single major gene encoding a resistant target site in a biotype of Lolium rigidum. Aust J Plant Physiol 23 15–23 [Google Scholar]
- Tardif FJ, Rajcan I, Costea M (2006) A mutation in the herbicide target site acetohydroxyacid synthase produce morphological and structural alterations and reduces fitness in Amaranths powellii. New Phytol 169 251–264 [DOI] [PubMed] [Google Scholar]
- Vila-Aiub MM, Neve P, Powles SB (2005. a) Resistance cost of a cytochrome P450 herbicide metabolism mechanism but not an ACCase target site mutation in a multiple resistant Lolium rigidum population. New Phytol 167 787–796 [DOI] [PubMed] [Google Scholar]
- Vila-Aiub MM, Neve P, Steadman KJ, Powles SB (2005. b) Ecological fitness of a multiple herbicide-resistant Lolium rigidum population: dynamics of seed germination and seedling emergence of resistant and susceptible phenotypes. J Appl Ecol 42 288–298 [Google Scholar]
- White GM, Moss SR, Karp A (2005) Differences in the molecular basis of resistance to the cyclohexanedione herbicide sethoxydim in Lolium multiflorum. Weed Res 45 440–448 [Google Scholar]
- Yu Q, Cairn A, Powles SB (2007) Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta 225 499–513 [DOI] [PubMed] [Google Scholar]
- Yu Q, Friesen LJS, Zhang XQ, Powles SB (2004) Tolerance to acetolactate synthase and acetyl-coenzyme A carboxylase inhibiting herbicides in Vulpia bromoides is conferred by two co-existing resistance mechanisms. Pestic Biochem Physiol 78 21–30 [Google Scholar]
- Zagnitko O, Jelenska J, Tevzadze G, Haselkorn R, Gornicki P (2001) An isoleucine/leucine residue in the carboxyltransferase domain of acetyl-CoA carboxylase is critical for interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors. Proc Natl Acad Sci USA 98 6617–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tweel B, Tong L (2004) Molecular basis for the inhibition of the carboxytransferase domain of acetyl-coenzyme-A carboxylase by haloxyfop and diclofop. Proc Natl Acad Sci USA 101 5910–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Powles SB (2006. a) Six amino acid substitutions in the carboxyl-transferase domain of the plastidic acetyl-CoA carboxylase gene are linked with resistance to herbicides in a Lolium rigidum population. New Phytol 172 636–645 [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Powles SB (2006. b) The molecular bases for resistance to acetyl co-enzyme A carboxylase (ACCase) inhibiting herbicides in two target-based resistant biotypes of annual ryegrass (Lolium rigidum). Planta 223 550–557 [DOI] [PubMed] [Google Scholar]