Abstract
In all organisms, control of folate homeostasis is of vital importance to sustain the demand for one-carbon (C1) units that are essential in major metabolic pathways. In this study we induced folate deficiency in Arabidopsis (Arabidopsis thaliana) cells by using two antifolate inhibitors. This treatment triggered a rapid and important decrease in the pool of folates with significant modification in the distribution of C1-substituted folate coenzymes, suggesting an adaptive response to favor a preferential shuttling of the flux of C1 units to the synthesis of nucleotides over the synthesis of methionine (Met). Metabolic profiling of folate-deficient cells indicated important perturbation of the activated methyl cycle because of the impairment of Met synthases that are deprived of their substrate 5-methyl-tetrahydrofolate. Intriguingly, S-adenosyl-Met and Met pools declined during the initial period of folate starvation but were further restored to typical levels. Reestablishment of Met and S-adenosyl-Met homeostasis was concomitant with a previously unknown posttranslational modification that consists in the removal of 92 amino acids at the N terminus of cystathionine γ-synthase (CGS), the first specific enzyme for Met synthesis. Rescue experiments and analysis of different stresses indicated that CGS processing is specifically associated with perturbation of the folates pool. Also, CGS processing involves chloroplastic serine-type proteases that are expressed in various plant species subjected to folate starvation. We suggest that a metabolic effector, to date unidentified, can modulate CGS activity in vivo through an interaction with the N-terminal domain of the enzyme and that removal of this domain can suppress this regulation.
Many aspects of plant metabolism and development require the addition or removal of one-carbon (C1) units (C1 metabolism) for biosynthetic or regulatory functions. The role of tetrahydrofolate (THF) derivatives, collectively termed folate(s), is to transport and donate C1 units, which exist under various oxidation states to enable several major anabolic processes (Hanson and Roje, 2001). For example, 10-formyl-THF is involved in purine and 10-formyl-Met-tRNA synthesis, 5,10-methylene-THF is required for thymidylate and pantothenate synthesis and for Gly to Ser conversion, and 5-methyl-THF is the methyl donor for the synthesis of Met. Analyses of the physiological role of folates indicated that a major fate of C1 units in photosynthetic leaves from C3 plants is the conversion of Gly into Ser (Fig. 1), which is a crucial step in photorespiration (Ravanel et al., 2004b). Also, an important anabolic fate of C1 units is the synthesis of Met, which is in part incorporated into protein but mainly converted to S-adenosyl-Met (AdoMet; Fig. 1; Ravanel et al., 2004b). AdoMet is a universal methyl-group donor participating in dozens of methyltransferase reactions and it is involved in the biogenesis of biotin and polyamines. AdoMet also plays unique roles in plants, such as ethylene and nicotianamine syntheses, and regulation of the synthesis of Asp-derived amino acids (Curien et al., 1998, 2007).
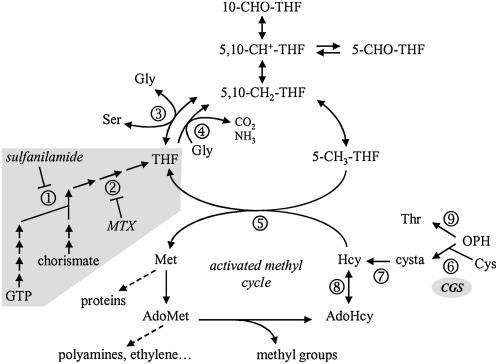
Figure 1.
Synthetic overview of C1 metabolism in plant cells. The synthesis of THF is shown on a gray background. The enzymes that are specifically inhibited by sulfanilamide and MTX are dihydropteroate synthase (1) and dihydrofolate reductase (2), respectively. The enzymes that utilize folate coenzymes are Ser hydroxymethyltransferase (3), Gly decarboxylase (4), and Met synthase (5). CGS (6) and cystathionine β-lyase (7) are involved in de novo synthesis of Met. AdoHcy hydrolase (8) catalyzes the reversible conversion of AdoHcy to Hcy and adenosine. Thr synthase (9) competes with CGS for the common substrate OPH to synthesize Thr. C1 units transported by THF are 10-formyl (10-CHO), 5-formyl (5-CHO), 5,10-methenyl (5,10-CH+), 5,10-methylene (5,10-CH2), and 5-methyl (5-CH3). cysta, Cystathionine.
An attractive challenge is to understand how folate homeostasis is controlled to match the supply of C1 units with demand, and how C1 units are accurately distributed between different anabolic routes. To address this problem, several authors used NMR techniques to measure C1-unit fluxes associated with Ser and Gly metabolism in plants. Thus, Prabhu et al. (1998) exposed Arabidopsis (Arabidopsis thaliana) plants to folate antagonists and determined that a continuous supply of folates was essential to maintain high rates of Ser synthesis. Also, plant cell cultures treated with the folate analog methotrexate (MTX) displayed an important inhibition of their growth (Wu et al., 1993), thus emphasizing the crucial role of folates in cell division and nucleotide synthesis. Apart from these data, the consequences of modification of folates pool size on the other pathways of C1 metabolism, particularly the synthesis of Met, are largely unknown.
The three enzymes involved in de novo synthesis of Met in plants are located in chloroplasts (Ravanel et al., 2004a). The first two enzymes, cystathionine γ-synthase (CGS) and cystathionine β-lyase, convert Cys into homo-Cys (Hcy), which is then methylated to Met by transfer of the methyl group of 5-methyl-THF, a reaction catalyzed by Met synthase (Fig. 1). The principal fate of Met is the synthesis of AdoMet in the cytosol, which is then largely used for methylation reactions in all cell compartments. Utilization of the methyl group of AdoMet by methyltransferases is accompanied by recycling of the homocysteinyl moiety and regeneration of Met, a set of reactions located in the cytosol and designated as the activated methyl cycle (Fig. 1). Until now, four regulatory mechanisms have been implicated in the maintenance of Met and AdoMet homeostasis in plant cells (Goto et al., 2005). The first two processes allow a fine regulation of the metabolic flux to Met and Thr syntheses, the first two enzymes of these pathways, namely CGS and Thr synthase, competing for a common substrate, O-phosphohomo-Ser (OPH). Flux partitioning is controlled by the intracellular level of AdoMet because AdoMet is an allosteric activator of Thr synthase (Curien et al., 1998, 2003) and AdoMet controls the stability of the mRNA coding CGS. Posttranscriptional control of CGS mRNA stability is mediated by a conserved region located in the N-terminal part of the CGS protein, the MTO1 (Met overaccumulation 1) domain (Chiba et al., 1999, 2003; Onouchi et al., 2005). The N-terminal region of the enzyme has also been suggested to contribute to regulate CGS and/or Met metabolism through uncharacterized posttranslational modification(s) (Hacham et al., 2002). The third mechanism accounting for the control of Met and AdoMet pools is the S-methyl-Met (SMM) cycle, which is specific for plants (Ranocha et al., 2001). In this cycle, SMM is synthesized by the AdoMet-dependent methylation of Met and can then donate a methyl group to Hcy, yielding two molecules of Met. Last, Met pool size is controlled by Met γ-lyase, an enzyme that catalyzes the cleavage of Met into methanethiol. Because this enzyme is expressed constitutively and up-regulated in case of Met overflow, it is assumed that Met γ-lyase plays an essential role in maintenance of Met and AdoMet pools (Rébeillé et al., 2006; Goyer et al., 2007).
Altogether, these data indicate a very dynamic regulation of Met homeostasis in which the role of the intracellular status of folates has never been examined. In this work we showed that 5-methyl-THF was the most dramatically reduced folate coenzyme in Arabidopsis cells exposed to antifolate drugs. As a consequence, the homeostasis of Met and derivatives was markedly affected. We showed that in cells starved for folates for a prolonged period the N-terminal domain of CGS was removed by proteolytic cleavage. This original posttranslational modification of CGS was accompanied by restoration of typical Met and AdoMet levels in folate-deficient cells.
RESULTS
Induction of Folate Deficiency in Arabidopsis Cells Using Antifolates
MTX and sulfanilamide are dihydrofolate and p-aminobenzoate analogs, respectively, that specifically inhibit THF synthesis. Sulfanilamide inhibits dihydropteroate synthase whereas the target of MTX is dihydrofolate reductase (Fig. 1). These antifolates have been used to manipulate the availability of folates in plants and plant cell cultures. In particular, Prabhu et al. (1998) showed that the combined action of MTX and sulfanilamide resulted in an important reduction of Ser and Gly metabolism in Arabidopsis plants. In this study, we exposed Arabidopsis cell suspension cultures to both MTX (100 μm) and sulfanilamide (100 μm), and analyzed the consequences of this treatment upon the pool of folates. Folates were extracted and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after enzymatic deconjugation, which converts folylpolyglutamates into the corresponding monoglutamate forms (Zhang et al., 2005). A major weakness of the analytical procedures developed for folates measurements, including the one used in this study, is the pH-dependent conversion of some species (De Brouwer et al., 2007). Thus, 5,10-methylene-THF dissociates to THF and formaldehyde, and 10-CHO-THF and 5-CHO-THF are cyclized to 5,10-methenyl-THF at various rates. Accordingly, the final quantitative LC-MS/MS data we obtained were simplified to three pools of folate: 5-methyl-THF, THF plus 5,10-methylene-THF, and other C1-substituted folates (5-CHO-THF, 10-CHO-THF, and 5,10-methenyl-THF).
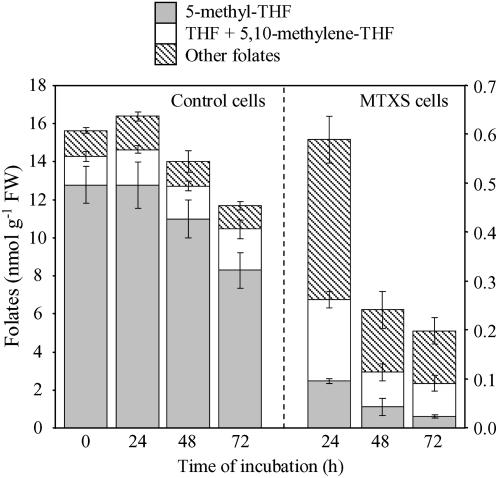
Exponentially growing Arabidopsis cells contained 15.5 ± 2.1 nmol folates g−1 fresh weight (FW), with 5-methyl-THF representing 70% to 80% of the pool, THF/5,10-methylene-THF 10% to 12%, and other C1 derivatives 8% to 11% (Fig. 2). Arabidopsis cells treated with MTX and sulfanilamide (referred to as MTXS cells) displayed a sharp and marked (25-fold) decrease in folates content after 24 h of drugs exposure (Fig. 2). Pools measured in MTXS cells were low but comparable with folate levels recorded in plant tissues with reduced activity in C1 metabolism, namely roots, stems, hypogeal cotyledons, or fruits (0.2–1.2 nmol folates g−1 FW; Jabrin et al., 2003). The decline in folates pool in MTXS cells indicated that THF synthesis was strongly inhibited. This is supported by the important cellular accumulation of MTX all along the examined period (about 5–6 nmol g−1 FW, as measured by LC-MS/MS). Along with the establishment of folate deficiency in MTXS cells, the distribution of C1-substituted pools was also markedly affected. Indeed, the major pool corresponded to 5-CHO-THF, 10-CHO-THF, and 5,10-methenyl-THF (54% ± 4%), THF plus 5,10-methylene-THF accounted for 31% ± 3%, and 5-methyl-THF was reduced to 15% ± 3% of total folates content (Fig. 2). Thus, 5-methyl-THF was the most dramatically reduced folate coenzyme in MTXS cells (more than 100-fold decrease after 24 h of exposure to drugs). This redistribution of C1 pools probably corresponded to a reorientation of C1 units demand in folate-deficient cells, with a probable marked impact on Met synthesis that directly depends on 5-methyl-THF supply (Fig. 1).
Figure 2.
Analysis of folates pools in control and MTXS-treated Arabidopsis cells. Cells were grown in standard conditions or exposed to MTX (100 μm) and sulfanilamide (100 μm) over a 72 h period and folates were analyzed by LC-MS/MS (Zhang et al., 2005). Three pools of folates were considered: 5-methyl-THF, THF and 5,10-methylene-THF, and other derivatives (5-CHO-THF, 10-CHO-THF, and 5,10-methenyl-THF). Data are means of three to six biological replicates and sd. Note that the scale used for folates quantification is different for control and MTXS-treated cells.
The growth of cells supplied with MTX and sulfanilamide was totally stopped (data not shown). This observation could be attributed mainly if not exclusively to MTX, which is known to act as an antiproliferative agent in plant cells (Wu et al., 1993). Indeed, exposure to MTX (100 μm) alone resulted in growth arrest of Arabidopsis cultures whereas cells treated with sulfanilamide (100 μm) alone grew similarly to control cells. Folates measurements in sulfanilamide-treated cells indicated a 2-fold reduction in folates content as compared to controls and no significant change in the distribution of C1 derivatives was observed (data not shown). To further analyze the physiological consequences of folate starvation, we measured respiration rates and determined cell viability in MTXS cultures. During the first 48 h of treatment, the respiration rate of MTXS cells was reduced by approximately 25% as compared to controls (see Supplemental Fig. S1). At 72 h, respiration rates were similar in both conditions; they remained stable in MTXS cells and decreased in untreated cells, probably because of Suc limitation in the medium from control cultures. During the entire period of exposure to antifolates, cell viability measured using the vital dye fluoresceine diacetate was similar in treated and control cultures (85% ± 6% living cells). Taken together, these data indicated that even blocked for division, Arabidopsis cells exposed to MTX and sulfanilamide were viable, metabolically active, and suitable to analyze the effects of folate deficiency on the homeostasis of Met and derivatives.
Modification of the Pool of Ser, Gly, Met, and Derivatives in Folate-Deficient Cells
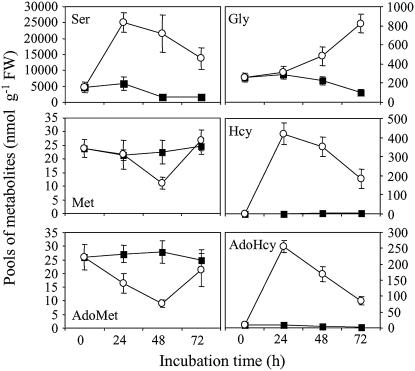
To analyze the consequences of folate starvation on metabolic profiles of Arabidopsis cells we measured free amino acids, thiols, AdoMet, and S-adenosylhomo-Cys (AdoHcy). Our analysis focused on the pools of Ser, Gly, Met, and derivatives that displayed important perturbations in MTXS versus control cells (Fig. 3). Ser, through its folate-dependent conversion to Gly by the reversible enzyme Ser hydroxymethyltransferase (Fig. 1), is the principal donor of C1 units in plants. During the first 24 h of exposure to antifolate drugs, the pool of Ser in Arabidopsis cells was increased by 5-fold (Fig. 3). In the rest of the folate-starvation time course, the level of Ser was about 10-times higher in MTXS than in control cells. The catabolism of Gly by the Gly decarboxylase complex (Fig. 1) located in mitochondria is another source of C1 units through the conversion of THF to 5,10-methylene-THF. In leaves of C3 plants, coupling of Gly decarboxylase and Ser hydroxymethyl transferase activities is the key step of photorespiration, which allows transfer of the α-C of one Gly molecule to a second Gly molecule for Ser synthesis. Photosynthesis and photorespiration in Arabidopsis cell cultures (mixautrophic growth) is not as high as in leaves but the catabolism of Gly through Gly decarboxylase, which is present in all cells (Mouillon et al., 1999), was affected by folate starvation. Indeed, while no significant difference in Gly pool was observed in the first 24 h of exposure to drugs, 48-h and 72-h periods were characterized by 2- and 5-fold increase in Gly content in MTXS versus control cells, respectively (Fig. 3). Taken together, our data indicated that folate deficiency in Arabidopsis cells markedly impaired Ser and Gly metabolism. These results are in accordance with a previous study showing that a continuous supply of THF is essential to maintain elevated rates of Ser hydroxymethyltransferase and Gly decarboxylase activities in Arabidopsis (Prabhu et al., 1998).
Figure 3.
Analysis of key metabolites of C1 metabolism in folate-sufficient and folate-deficient Arabidopsis cells. Free amino acids, thiols, and AdoMet/AdoHcy were extracted from control (▪) and MTX-treated (○) cells collected at different time intervals and analyzed by LC as described in “Materials and Methods.” Data are means ± sd of three biological replicates.
Analysis of the Hcy pool size in MTXS-treated cells indicated that Met synthesis and recycling were markedly reduced by folate deficiency. Indeed, Hcy is low abundant in control cells (about 1.5–2 nmol g−1 FW) but accumulated by 250- to 300-fold in MTXS cells (Fig. 3). The accumulation of Hcy could be attributed to an impairment of the reaction catalyzed by Met synthase (Fig. 1). A similar profile was observed for AdoHcy, the by-product of AdoMet-dependent methyltransferases, which accumulated 30-fold in MTXS versus control cells (Fig. 3). The rise in AdoHcy pool size could be attributed in part to the utilization of AdoMet by methyltransferases but mainly to the activity of AdoHcy hydrolase (Fig. 1). Indeed, this enzyme is reversible and AdoHcy hydrolysis to Hcy and adenosine is only favored by removal of these products (Moffatt and Weretilnyk, 2001). In MTXS cells, if adenosine is not rate limiting, the huge accumulation of Hcy would favor AdoHcy hydrolase in the direction of AdoHcy synthesis. The variations of Met and AdoMet pools during the time course of folate starvation were different from that of Hcy and AdoHcy. In Arabidopsis cells cultured in standard medium, the Met and AdoMet contents were maintained constant (about 25 nmol g−1 FW) all along the exponential growing phase (Fig. 3). In MTXS cells, the pool of AdoMet decreased by 40% and 70% after 24 and 48 h of culture, respectively. The level of Met also decreased in treated cells but after a longer delay period (2-fold reduction after 48 h of starvation). These data suggested that an impairment of the activated methyl cycle at the level of Met synthase imbalanced the rates of Met and AdoMet synthesis and utilization (protein synthesis, methylation reactions, ethylene, and polyamines syntheses). The most striking observation associated with fluctuations in Met and AdoMet was the restoration of pools characteristic of control cells after 72 h of exposure to antifolate drugs (Fig. 3). The recovery of typical levels of Met and AdoMet could not be explained by using only our metabolite profiling data. Indeed, as previously observed in Arabidopsis cell cultures (Rébeillé et al., 2006), the pool of SMM was too low to be determined in our experimental system (below 2 nmol g−1 FW), and thus this amino acid could not act as a reservoir for Met and AdoMet in folate-deficient Arabidopsis cells.
Expression of Met-Synthesizing Enzymes in Folate-Deficient Cells
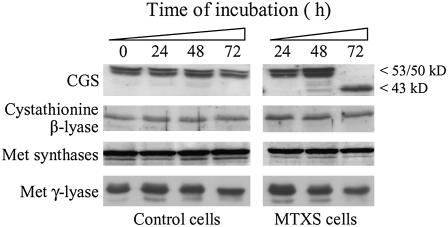
To gain insight into the fluctuations of Met and AdoMet pools in folate-deficient cells we analyzed the expression pattern of the three enzymes involved in the synthesis of Met (Fig. 1). Western-blot analyses performed using soluble protein extracts from MTXS and control Arabidopsis cells indicated that cystathionine β-lyase and Met synthase isoforms were expressed at constant levels in both culture conditions during the whole experimental period (Fig. 4). Thus, the oscillation of Met and AdoMet levels could not be attributed to modifications of the expression of the second (cystathionine β-lyase) and third (plastid Met synthase) enzymes involved in de novo Met synthesis in plastids, neither to the expression of the cytosolic Met synthases involved in the recycling of Met in the activated methyl cycle. The expression pattern of CGS was more fluctuating. It is worth noting that immunodetection or purification of CGS in various plant species resulted in two polypeptides encoded by the same gene (CGS1, At3g01120 in Arabidopsis; Ravanel et al., 1998; Hacham et al., 2006). The first polypeptide migrates with the expected size of the mature CGS (MCGS; 53 kD) while the second has an estimated size of 50 kD. Recently, Hacham et al. (2006) showed that the 50-kD polypeptide resulted from the translation of a CGS transcript bearing a deletion of 90 or 87 nucleotides (about 3 kD located internally in the N terminus of the enzyme). In control Arabidopsis cells, the two polypeptides characteristic for CGS were expressed at constant levels during the first 48 h of culture and then the amount of enzyme was slightly reduced at 72 h (Fig. 4). In folate-deficient cells, CGS expression level was comparable to control cells at 24 h but was increased by approximately 2-fold after 48 h of exposure to antifolates (Fig. 4). After a 72 h period of treatment, the 53- and 50-kD CGS bands were not detected but the CGS antiserum cross-reacted with a polypeptide of 43 ± 1 kD. The accumulation of CGS polypeptides at 48 h could be attributed to a perturbation of the AdoMet-dependent feedback regulation of the expression of the CGS1 gene (Goto et al., 2005). Indeed, we can assume that the decrease in AdoMet level in MTXS cells (9 nmol g−1 FW at 48 h versus 28 nmol g−1 FW in control cells, see Fig. 3) would increase CGS1 mRNA stability, thus allowing mRNA and protein to accumulate. After 72 h of folate starvation, the concomitant disappearance of CGS at 53/50 kD and detection of the 43-kD polypeptide could not be attributed to this regulatory process. A global proteolytic degradation of proteins in MTXS cells could also be rejected because (1) Coomassie Brilliant Blue staining of protein extracts after SDS-PAGE displayed typical patterns (data not shown), and (2) immunodetection of cystathionine β-lyase, Met synthases, and Met γ-lyase in these extracts was not affected (Fig. 4).
Figure 4.
Expression of the enzymes involved in Met synthesis and catabolism in folate-sufficient and folate-deficient cells. Soluble proteins (40 μg per lane) from Arabidopsis cells grown in standard medium (control cells) or exposed to 100 μm MTX and 100 μm sulfanilamide (MTXS cells) were analyzed by western blot using antibodies raised against CGS, cystathionine β-lyase, Met synthases, and Met γ-lyase from Arabidopsis. The 53- and 50-kD polypeptides detected with the CGS serum are characteristic of the mature enzyme, the one at 43 kD being observed only in cells starved for folates for 72 h. Quantitation of CGS polypeptides using chemiluminescence detection reagents and a Typhoon 9400 scanner indicated that the amount of CGS protein was increased by 2-fold between 24 and 48 h of treatment and was maintained constant between 48 and 72 h (titration experiments using recombinant proteins indicated that the signal obtained with the 43-kD polypeptide of CGS was reduced by 30%–40% as compared with the signal measured for the mature protein). The antibodies against Met synthase cross-react with both the cytosolic (top band) and chloroplastic (bottom band) isoforms of the enzyme (Ravanel et al., 2004a).
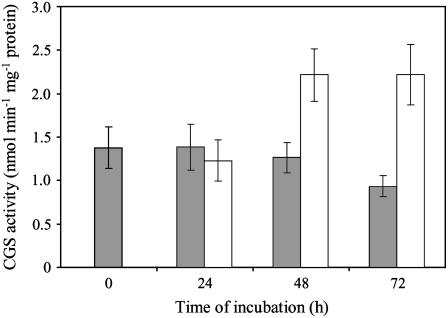
To validate the expression patterns described above, we measured CGS activity using desalted soluble protein extracts prepared from MTXS and control cells. Because CGS activity is low in plant extracts, cystathionine production was monitored by HPLC after derivatization with O-phthaldialdehyde. As shown in Figure 5, CGS activity measured in control cells matched the expression profile of the protein (Fig. 4), with constant cystathionine production during the first 48 h of culture followed by a 30% decrease at 72 h. Also, CGS activity measured in MTXS cells after 24 and 48 h of exposure to drugs fitted well with immunoblots (Fig. 5). More importantly, the 2-fold increase in CGS activity measured at 48 h was maintained at 72 h, thus indicating that the 43-kD polypeptide detected by western blot corresponded to an active CGS enzyme with a modified electrophoretic behavior.
Figure 5.
CGS activity in control and MTXS-treated Arabidopsis cells. Soluble proteins were extracted from control (gray bars) and MTX-treated (white bars) cells collected at different time intervals, desalted through Sephadex G25, and CGS activity was determined by LC after derivatization of cystathionine. Data are means ± sd of three biological replicates.
Expression of Met γ-Lyase in Folate-Deficient Cells
It has been shown recently that Met γ-lyase, which catalyzes Met degradation into methanethiol, α-ketobutyrate, and ammonia, plays an important role in controlling Met homeostasis in plants (Rébeillé et al., 2006; Goyer et al., 2007). To determine whether this enzyme could participate to the oscillation in Met pool size we analyzed its expression in folate-sufficient and folate-deficient cells by western blot. The expression patterns observed in both conditions were similar (Fig. 4), indicating that even if the production of the enzyme is increased in Arabidopsis cells accumulating Met (Rébeillé et al., 2006), Met γ-lyase was not down-regulated in a situation of Met limitation. Thus, restoration of Met and AdoMet pools after 72 h of folate starvation was not a result of a decreased catabolism of Met.
The N-Terminal Domain of CGS Is Cleaved in Folate-Deficient Cells
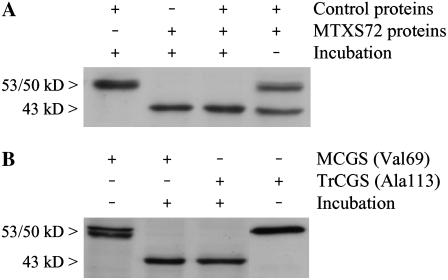
Several hypotheses were envisaged to explain the origin of the 43-kD CGS polypeptide detected in cells starved for folates. First, transcription of the CGS1 gene (At3g01120) could be affected to generate an unusual deleted transcript (e.g. distinct transcription start site and/or alternative splicing). Second, the Arabidopsis genome contains a second gene (At1g33320) coding a putative CGS enzyme, which differs from the CGS1-encoded protein by the absence of 151 residues in the N-terminal region (including the transit peptide and the MTO1 domain). Although the absence of expressed sequence tag for At1g33320 in The Arabidopsis Information Resource database (www.arabidopsis.org) suggests that this gene may not be expressed, the predicted molecular mass of this putative CGS (46 kD, 412 residues) could correspond to the polypeptide detected by western blot in folate-deficient cells. Third, the 43-kD polypeptide could originate from MCGS (53/50 kD) through a proteolytic cleavage. To test this last hypothesis, we used soluble proteins obtained from cells treated with MTX and sulfanilamide for 72 h (MTXS72) as a source of the hypothetical CGS-specific cleavage system. The combination of MTXS72 proteins with proteins extracted from control cells resulted, upon incubation for 2 h at 25°C, in the conversion of the 53/50-kD bands of MCGS into the 43-kD polypeptide (Fig. 6A). These data indicated that CGS was subjected to a posttranslational cleavage in folate-deficient cells. This result was confirmed using recombinant CGS proteins purified from Escherichia coli overproducing cells. Two versions of recombinant CGS were tested as substrates for the CGS-cleavage machinery existing in MTXS72 extracts. The first enzyme corresponded to MCGS (starting with Val-69; Ravanel et al., 1998), the second was a truncated form with a deletion of 44 residues at the N terminus of the protein (starting with Ala-113 and thus lacking the MTO1 domain; G. Curien, unpublished data). Incubation of the two recombinant CGSs with the MTXS72 protein extract resulted in the same cleavage product at 43 ± 1 kD (Fig. 6B). Because the two versions of CGS differed only by their N-terminal regions, this result indicated that the posttranslational cleavage of CGS occurred at the N terminus of the enzyme.
Figure 6.
Evidence for posttranslational cleavage of the N-terminal region of MCGS in folate-deficient cells. A, Soluble proteins prepared from control cells were mixed with an equal amount of proteins from cells treated with MTXS for 72 h (MTXS72) and incubated for 2 h at 25°C. Forty micrograms of proteins were analyzed in each lane. B, Pure recombinant CGS enzymes (25 ng) were incubated for 2 h at 25°C with 2.5 μg soluble proteins from MTXS72 cells. Two versions of the enzyme were analyzed: the MCGS (starting with Val-69) and a truncated CGS (TrCGS, starting with Ala-113) bearing a deletion of 44 residues at the N terminus of the protein. Each combination from sections A and B was analyzed by western blot with polyclonal antibodies raised against CGS. Note that in B only the recombinant CGSs are detected because they are present in excess as compared to the enzyme provided by the MTXS72 extract.
To further characterize the posttranslational modification affecting MCGS, we first tested the effect of several inhibitors acting on the four major classes of proteases, namely metallo proteases (EDTA 20 mm, 1,10-phenanthroline 5 mm), thiol proteases (E64 50 μm), Ser proteases (phenylmethylsulphonyl fluoride [PMSF], 5 mm), and aspartic proteases (pepstatin A 10 μm). With the exception of PMSF that fully abolished MCGS cleavage, none of the inhibitor tested had significant effect on the production of the 43-kD CGS polypeptide in our experimental conditions (see Supplemental Fig. S2). This suggested that at least a Ser protease was involved. In another set of experiments, the different constituents of the reconstituted in vitro cleavage assay were treated at 100°C for 5 min before combining the substrate/protease(s) fractions. Heat denaturation of the MTXS72 extract fully abolished the cleavage of CGS (Supplemental Fig. S2), thus confirming the enzymatic nature of the process. On the other hand, heat denaturation of native or recombinant MCGS did not impair processing of the N-terminal domain of the enzyme by a MTXS72 extract and, moreover, did not lead to a complete degradation of the enzyme (the proteolytic product was the characteristic 43-kD polypeptide; Supplemental Fig. S2). These data suggested that the secondary structure of the N terminus of CGS is not important for recognition by the proteolytic system or that this part of the protein is naturally unfolded or highly thermostable.
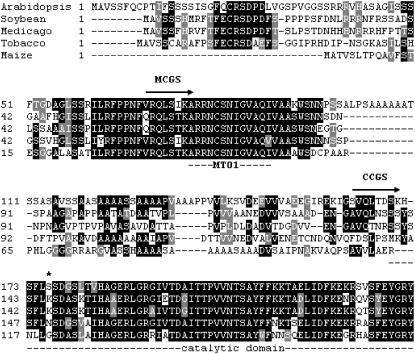
To identify the cleavage site in the N-terminal domain of CGS we used the following strategy. MCGS was overproduced in E. coli as a fusion protein with a C-terminal His tag and purified by affinity chromatography on Ni-agarose column (see “Materials and Methods”). Pure recombinant MCGS-6His was combined with a MTXS72 protein extract, incubated until all the protein has been cleaved, and the mixture was applied onto a Ni-agarose column. Cleaved MCGS-6His was then eluted from the column as a pure protein and subjected to Edman's degradation. The N-terminal sequence obtained (SVQLTDSK) corresponded to residues 161 to 168 of CGS. The truncated enzyme derived from MCGS, now referred to as CCGS for cleaved CGS, is a 403-residue protein with a theoretical Mr of 43,988 D. Thus, processing of MCGS to CCGS in folate-deficient Arabidopsis cells corresponds to the removal of 92 residues at the N terminus of the mature enzyme. An alignment of the amino acid sequences of CGS from different plant species is shown in Figure 7. This comparison revealed interesting features on the N-terminal domain of CGS and the consequences of its elimination. First, the highly conserved MTO1 domain that is implicated in the posttranscriptional regulation of CGS1 transcript stability by AdoMet (Chiba et al., 1999, 2003; Onouchi et al., 2005) is removed in CCGS. Second, the region located between the end of the MTO1 domain and the site of cleavage (Ser-161) is poorly conserved, particularly in plants from different families and genus (e.g. maize [Zea mays] versus Arabidopsis), and consists of Ala repeats typical of low-complexity sequences. Thus, a canonical consensus cleavage site cannot be determined. Third, removal of the N-terminal domain of CGS does not affect the catalytic core domain of the enzyme, as determined using the three-dimensional structure of CGS from tobacco (Nicotiana tabacum; Steegborn et al., 1999), suggesting therefore that CCGS is active. This is supported by the observation that the CCGS enzyme detected in MTXS72 protein extracts was associated with CGS activity (see Figs. 4 and 5).
Figure 7.
Alignment of amino acid sequences of the N-terminal region of CGS from various plant species. The alignment was generated with ClustalW using the amino acid sequences of CGS from Arabidopsis (GenBank accession no. ATU83500), soybean (Glycine max; AAD34548), Medicago truncatula (ABE79443), tobacco (AB035300 and AF097180), and maize (AAB61347). Only the N-terminal region of CGSs is shown in the alignment. The mature (MCGS) and cleaved (CCGS) versions of the Arabidopsis enzyme start with residues Val-69 and Ser-161, respectively. The MTO1 (Goto et al., 2005) and catalytic (Steegborn et al., 1999) domains are underlined. The truncated Arabidopsis CGS enzyme overexpressed in transgenic tobacco plants by Hacham et al. (2002) starts with Ser-173 (asterisk).
Comparison of the Kinetic Properties of MCGS and CCGS
The above-mentioned data indicated that the N-terminal domain of CGS is not essential for catalytic activity. To analyze further the role of this domain and its processing in folate-deficient cells, we compared the biochemical and kinetic properties of MCGS and CCGS. To this aim, we purified both proteins fused with a C-terminal His tag and overproduced in E. coli cells. First, gel filtration experiments indicated that both forms of the enzyme behave as tetramers (data not shown), thus indicating that the oligomerization of CGS was not affected by its N-terminal region. Second, steady-state kinetic analyses were consistent with Michaelis-Menten behavior for both MCGS and CCGS. The kinetic parameters of the two enzymes were similar, with apparent Vmax of 8.5 ± 0.5 μmol min−1 mg−1 protein, and Km values of 2.3 ± 0.4 mm and 500 ± 70 μm for OPH and Cys, respectively. Thus, removal of the N terminus of CGS did not modify the catalytic efficiency of the enzyme.
We have previously shown that several metabolites associated with the metabolism of Met (namely cystathionine, Hcy, Met, AdoMet, AdoHcy, SMM, 5-methylthioadenosine, Thr, and Ile) did not regulate Arabidopsis MCGS activity (Ravanel et al., 1998). These metabolites have been tested separately at elevated (nonphysiological) concentrations in a standard enzyme assay containing saturating substrate levels (1 mm Cys and 10 mm OPH). Because the cleavage of the N terminus of CGS did not change the catalytic properties of the enzyme, we reexamined the possibility of CGS regulation by feedback inhibition or activation using in vitro assay conditions mimicking the metabolite pools that exist in vivo. To set up these assays at near physiological conditions, we fixed the concentration of CGS substrates at 100 μm and used two sets of putative effectors: the one corresponding to folate-sufficient cells (20 μm Met, 25 μm AdoMet, 8 μm AdoHcy, 2 μm Hcy, 10 μm 5-methyl-THFGlu-5 [pentaglutamate form], Ser 3 mm, and Gly 250 μm), the other mimicking folate-deficient cells (10 μm Met, 10 μm AdoMet, 200 μm AdoHcy, 350 μm Hcy, 0.05 μm 5-methyl-THFGlu-5, Ser 25 mm, and Gly 550 μm). Kinetic data indicated that none of the metabolite mixtures used had significant effect on MCGS or CCGS activity (maximal changes were about 10%–15% for both enzymes). To analyze the effect of more complex metabolite mixtures on CGS activity, we collected the salt fractions issued from desalting of protein extracts by gel filtration on Sephadex G-25. Salt fractions significantly inhibited CGS activity in a dose-dependent manner but (1) MCGS and CCGS were similarly affected, and (2) salts obtained from folate-sufficient or folate-deficient cells produced similar effects on both enzymes. It is likely that these inhibitions were due to the previously observed sensitivity of CGS to a variety of salts (Ravanel et al., 1995) rather than to a specific regulation through feedback regulators. To support this assumption, we verified that MCGS and CCGS activities were inhibited by KCl in similar ways (50% inhibition at about 20 mm). Together, these data did not allow us to identify metabolites that could interact in vitro with the N-terminal domain of CGS and modify its kinetic behavior.
Cleavage of the N-Terminal Domain CGS Is Specifically Associated with Perturbation of the Folates Pool and Occurs in Various Plant Species
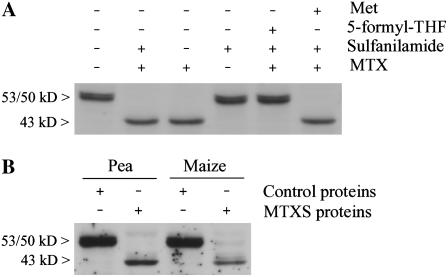
To gain insight into the physiological situations that trigger the cleavage of the N-terminal region of CGS, we analyzed the behavior of the enzyme in Arabidopsis cells cultured in different conditions. First, we showed that processing of the N terminus of MCGS occurred in cells only in conditions of folate deficiency. Indeed, CCGS was produced in cells treated with a combination of MTX and sulfanilamide (each at 100 μm) or MTX alone (at 100 μm), but not in cells in which the folate pool was reduced by 2-fold by using sulfanilamide (100 μm) alone (Fig. 8A). We also performed rescue experiments using 5-CHO-THF to determine whether the cleavage of CGS was attributable to folate deficiency or to another effect of MTX. In a separate control experiment we showed that Arabidopsis cells cultured with 5-CHO-THF, a chemically stable folate derivative, gradually accumulated large amounts of folates (up to 350 nmol g−1 FW after 72 h of incubation), among which 5-methyl-THF accounted for 24% to 36% of the total pool (data not shown). As shown in Figure 8A, the supply of 5-CHO-THF abolished the cleavage of CGS in cells treated with MTX and sulfanilamide. Because the homeostasis of several metabolites linked to Met metabolism was markedly perturbed in folate-deficient cells, we further tested the ability of a Met supply to suppress CGS processing. In a previous study, we showed that Arabidopsis cells fed with Met accumulated large amounts of Met and had an AdoMet pool size increased by 10-fold (Rébeillé et al., 2006). When MTXS-treated cells were supplied with exogenous Met, processing of the N-terminal domain of CGS was not abolished (Fig. 8A). Together, these data indicated that the posttranslational modification of CGS occurred only in cells markedly starved with folates and that replenishment of Met and AdoMet pools through exogenous Met application was ineffective to counter this process.
Figure 8.
Processing of the N-terminal domain of CGS is specifically associated with folates starvation and occurs in several plant species. A, Soluble protein extracts were prepared from Arabidopsis cells grown for 72 h in culture medium supplemented with the following compounds, alone or in combination: 100 μm MTX, 100 μm sulfanilamide, 0.5 mm 5-formyl-THF, and 1 mm Met. Soluble proteins (30 μg) were extracted and analyzed by western blot with polyclonal antibodies against CGS. B, Leaf discs from pea and maize were floated for 8 d in petri dishes containing control medium or supplied with both MTX (100 μm) and sulfanilamide (100 μm). Soluble proteins (5 μg) prepared from control or MTXS-treated leaf discs were combined with an Arabidopsis protein extract (20 μg) containing MCGS and incubated for 1 h at 25°C before performing immunoblot analysis.
To determine whether the production of CCGS occurred in situations different from folate deficiency we exposed Arabidopsis cultures to different stresses. To this aim, cells were supplied with 100 mm NaCl or 44 mm hydrogen peroxide to induce salt and oxidative stresses, respectively (Sweetlove et al., 2002; Kim et al., 2007). Also, sulfur deficiency was induced by growing cells in medium with limiting sulfur (73 μm instead of 1.7 mm in standard conditions, see “Materials and Methods”). Western-blot analyses indicated that salt stress, oxidative stress, and sulfur starvation were not associated with the presence of CCGS (see Supplemental Fig. S3). Therefore, it is likely that none of the important metabolic changes that occur in these situations could mimic the metabolic status of folate-deficient cells and trigger cleavage of the N terminus of CGS.
To examine whether the cleavage of MCGS to CCGS was a conserved regulatory process in several plant species we induced folate deficiency in leaves detached from Arabidopsis, pea (Pisum sativum), or maize plants and floated on a solution of MTX and sulfanilamide (100 μm of each drug). After 6 d of incubation, CCGS was detected in MTXS-treated Arabidopsis leaves whereas MCGS was present in control leaves, thus indicating that the situation observed in cell suspension cultures holds true in leaves. This situation could not be observed in pea and maize leaf extracts because the antibodies against Arabidopsis CGS did not cross-react with CGS from these plants. To overcome this problem, we used the in vitro combination assay described above (Fig. 6) to examine the ability of proteins extracts from pea and maize leaves treated with MTX and sulfanilamide to cleave MCGS from Arabidopsis. As shown in Figure 8B, folate starvation in pea and maize leaves induced a proteolytic machinery that is able to remove the N-terminal domain of the Arabidopsis CGS. These data indicated that this regulatory process is widespread in the plant kingdom and not limited to Arabidopsis.
DISCUSSION
In all living organisms, control of folate homeostasis is of vital importance to sustain the demand for C1 units that are essential in several major metabolic pathways. In this study we induced folate deficiency in Arabidopsis cells by using two inhibitors of THF synthesis. This treatment triggered a rapid and important decrease in the pool of folates with significant modification in the distribution of C1-substituted folate coenzymes. These data indicated that the synthesis of THF was strongly inhibited and that the catabolism of folates was important. Using Arabidopsis plants exposed to sulfanilamide, Orsomando et al. (2006) estimated a total folate breakdown rate of about 10% per day. In our experimental system, Arabidopsis cells exhibited folate catabolism rate of at least 4% per hour. As previously observed by Prabhu et al. (1998), it thus appears that MTX triggers more important perturbations of folate homeostasis (synthesis, recycling, and catabolism) than sulfanilamide, most probably because the later inhibitor blocks only de novo synthesis of THF whereas MTX also affects rereduction of dihydrofolate to THF by the enzyme dihydrofolate reductase. In Arabidopsis cells exposed to MTX and sulfanilamide, depletion was more pronounced for 5-methyl-THF (>100-fold decrease at 24 h) than for THF and 5,10-methylene-THF (10-fold) or 5-CHO-THF, 10-CHO-THF, and 5,10-methenyl-THF (5-fold). This suggested an adaptive response to favor a preferential shuttling of the flux of C1 units to the synthesis of purines and thymidylate over the synthesis and recycling of Met. Despite these changes, folate-deficient cells stopped to divide, suggesting that nucleotides synthesis was limiting. Further investigations, including analysis of the expression of key genes involved in C1 metabolism, will be necessary to decipher accurately the metabolic priorities associated with folate deficiency in Arabidopsis.
Metabolic profiling of folate-deficient cells indicated important perturbation of the activated methyl cycle because of the impairment of Met synthase enzymes that are deprived of their substrate 5-methyl-THF. The immediate consequences of this blockage are the accumulation of Hcy, and then the increase in AdoHcy pool owing to the reversible activity of AdoHcy hydrolase. The pools of AdoMet and particularly Met are more robust to these perturbations, suggesting that the demands for these metabolites in anabolic reactions are also altered by folate deprivation. In regard, MTXS-treated cells ceased to divide and probably reduced their demand for Met to synthesize proteins. Also, it must be stressed that the methyl index (AdoMet to AdoHcy ratio) was 100-fold lower in folate-deficient than in folate-sufficient cells, which suggests an important impairment of AdoMet-dependent methyltransferase reactions. Indeed, AdoHcy strongly inhibits methyltransferases through competition with the substrate AdoMet (Moffatt and Weretilnyk, 2001).
The most intriguing result from our metabolic analysis was the restoration of typical pools of Met and AdoMet after a prolonged period of folate starvation. We propose that the adaptive response of Arabidopsis cells to reestablish Met and AdoMet homeostasis under folate-limiting conditions involves two mechanisms. First, the posttranscriptional regulation of the CGS1 gene coding CGS (Chiba et al., 1999, 2003; Onouchi et al., 2005) is affected by the decrease in AdoMet content, thus leading to a 2-fold increase in CGS protein and activity at 48 h (Figs. 4 and 5). Loosening of the AdoMet-dependent CGS1 mRNA degradation was probably not sufficient however to improve de novo synthesis of Met after long-term folate deficiency. Indeed, this period is characterized by a previously unknown posttranslational regulation of CGS that consists in the removal of 92 amino acids at the N terminus of the enzyme. Kinetic analyses conducted in vitro using pure recombinant enzymes or cellular protein extracts failed to detect any catalytic improvement of CGS as a consequence of N-terminal processing. However, the situation proved to be different in vivo. Indeed, to determine the function of the N-terminal region of CGS, Hacham et al. (2002) have generated transgenic tobacco plants overproducing the mature Arabidopsis CGS or a truncated version of the enzyme. Transgenic plants expressing truncated CGS, which starts 12 residues after the cleavage site we identified (Fig. 7), produced higher amounts of Met (free or in proteins) than plants expressing the mature version of the enzyme and emitted high levels of ethylene and Met catabolic products. Because similar levels of the enzyme were produced in both transgenic lines and since no significant difference was observed regarding CGS transcript or protein stability, it was concluded that the N-terminal region of CGS plays an important regulatory role in Met metabolism in Arabidopsis (Hacham et al., 2002). The concomitant processing of CGS with restoration of Met and AdoMet pools in folate-deficient cells agree with this finding and demonstrate moreover that this regulatory domain of CGS can be processed in vivo to favor Met and then AdoMet production. This suggests also that, in our experimental conditions, the 2-fold increase in CGS protein and the posttranslational processing of the enzyme were essential to allow restoration of Met and AdoMet pools albeit the limitation downstream in the pathway, at the level of Met synthase. To strengthen this hypothesis, it is worth noting that the Km values for 5-methyl-THF of the chloroplastic Met synthase is 4-fold lower than that of the cytosolic isoforms (Ravanel et al., 2004a), thus suggesting that de novo synthesis of Met in plastids is less affected by folate deprivation than recycling of Met in the cytosol. The essential role attributed to CGS processing is also supported by the observation that the key enzyme involved in the catabolism of Met, Met γ-lyase, was not down-regulated in folate-deficient cells and thus could not contribute to restoration of Met and AdoMet levels.
These findings suggest that CGS activity could be regulated in vivo via a feedback inhibition involving the interaction of one or several metabolites with the N-terminal domain of the enzyme (Hacham et al., 2002). We tested this hypothesis in vitro using pure recombinant MCGS and CCGS proteins and several metabolites for which pools were markedly affected in folate-deficient cells. In particular, we analyzed the effect of AdoMet and 5-methyl-THF, alone or in combination, on both versions of the enzyme. These metabolites were of particular interest because Selhub et al. (1971) have shown that, in Neurospora crassa, 5-methyl-THF acts as an essential activator of CGS and antagonizes the feedback inhibition of the enzyme by AdoMet. Also, plant and Neurospora CGSs share a common feature as they possess an N-terminal extension of about 180 to 200 residues that is not found in bacterial CGS. Our kinetic data indicated that CGS from Arabidopsis is not regulated in vitro by AdoMet or 5-methyl-THF. Also, all the other metabolites tested at physiological or elevated concentrations (Met, AdoHcy, Hcy, Ser, Gly, reduced, or oxidized glutathione) did not result to any significant modification of MCGS or CCGS activity. These data, together with previous inhibition studies (Ravanel et al., 1998), failed to identify a metabolic effector of plant CGS. We cannot rule out the possibility, however, that some other metabolites (e.g. ethylene or other anabolic or catabolic products derived from Met) can modulate CGS activity through an interaction with the N terminus of the enzyme and that removal of this domain can suppress this regulation. One example of such a regulation has been described for human cystathionine β-synthase, the first enzyme of the pathway leading to the conversion of Hcy to Cys. It was proposed that the catalytic site of cystathionine β-synthase is partially occluded by the C-terminal autoregulatory domain and that AdoMet binding or proteolytic cleavage displace this domain, thus increasing the enzyme catalytic activity (Miles and Kraus, 2004). Although there are many similarities between the regulatory process described for human cystathionine β-synthase and the processing of Arabidopsis CGS, further studies will be required to improve our knowledge about the regulatory function of the N-terminal domain of the plant enzyme. In particular, we will have to consider the possibility that this regulatory process involves a protein partner or posttranslational modification, which was not present in our in vitro assays that contain only pure recombinant CGS enzymes.
Among the various molecules tested for their ability to inhibit the proteolytic system acting on CGS, only PMSF was able to prevent the cleavage. This result, together with the localization of CGS in the stroma, suggests that at least one chloroplastic Ser protease is involved in the processing of the enzyme. The occurrence of CGS processing outside the chloroplast, i.e. before import of the protein precursor into plastids, is unlikely because (1) its substrate OPH is synthesized only in plastids, (2) the next enzyme involved in de novo Met synthesis is only located in plastids (Ravanel et al., 1996), and (3) the truncated CGS enzyme that is responsible for important perturbation of Met homeostasis in transgenic tobacco plants was expressed in plastids (Hacham et al., 2002). For similar reasons, the possibility that CGS processing may have induced a change in the subcellular location of the enzyme is improbable. As a result of the complete sequencing of the Arabidopsis genome, most (if not all) of the chloroplastic proteases are known (Adam et al., 2006). Among them, several members of the major protease families Clp and DegP, as well as the SppA and Lon proteases, are Ser-type enzymes located in the stroma or attached to the stromal side of the thylakoid membranes. Although the physiological function of some components of the complex proteolytic machinery in plastids has been determined (Adam et al., 2006), data available to date are not sufficient to identify candidate(s) responsible for CGS processing, which is observed only in folate-deprived cells. However, we obtained preliminary data about the mechanism of substrate recognition that will be helpful in the future to identify the enzyme that is involved in CGS processing. First, the three-dimensional structure of CGS from tobacco indicated that the N-terminal region protrudes from the globular body of each monomer of the enzyme (Steegborn et al., 1999), suggesting that this domain is freely accessible to the protease(s) or a regulatory subunit. Second, the N terminus of CGS is probably structurally disordered because (1) it is not visible in the electron density map (Steegborn et al., 1999), and (2) heat denaturation of the enzyme does not abolish CGS recognition and processing by the protease(s). Third, the only part of the N-terminal domain that is highly conserved among plant CGSs, the MTO1 domain (Goto et al., 2005), is not necessary for recognition of the enzyme by the proteolytic system, although it may be requested for the regulatory function of the N-terminal domain of CGS. Last, although the cleavage site is not canonical, the proteases expressed in response to folate starvation in different plant sources (i.e. pea and maize) are able to process correctly the Arabidopsis enzyme.
The intracellular signal that triggers proteolytic removal of the N-terminal regulatory domain of CGS is yet unknown. However, comparison of the metabolic profiles established in various situations with the presence of the mature or cleaved form of the enzyme allowed us to establish a short list of putative candidates. It was shown that sulfur deficiency in Arabidopsis led to important decrease in the levels of sulfur-related metabolites Cys, glutathione, and AdoMet, whereas the pool of AdoHcy remained unchanged (Nikiforova et al., 2005). Also, a time-course metabolic profiling in Arabidopsis cells after salt stress treatment indicated a 40% reduction in the AdoMet pool, a 70% increase in AdoHcy level, and a constant amount of 5-methyl-THF throughout the examined period (Kim et al., 2007). In these two particular situations we did not observe any processing of CGS, suggesting that perturbations were not capable to induce posttranslational regulation of the enzyme. Thus, at this stage of this study, the metabolic signature that is typical to cells expressing a processed form of CGS resides in (1) an important reduction of folates pool, mainly 5-methyl-THF; (2) an elevated level of Hcy; and (3) an increased AdoHcy pool size associated with a marked reduction of the methyl index. One can hypothesize that these metabolic changes are potential intracellular sensors that trigger, alone or in combination, posttranslational regulation of CGS. Identification of the gene(s) coding the proteolytic machinery involved in CGS processing and analysis of its expression pattern should contribute to a better understanding of the physiological conditions in which this original regulatory process controls Met homeostasis.
MATERIALS AND METHODS
Plants, Cells, and Growth Conditions
Arabidopsis (Arabidopsis thaliana; ecotype Columbia) cell suspension cultures were grown under continuous light (40 μE m−2 s−1) at 22°C with rotary agitation at 125 rpm in Gamborg's B5 medium supplemented with 1 μm 2-naphtalene acetic acid and 1.5% (w/v) Suc. Cells were subcultured every 7 d. Arabidopsis (ecotype Columbia) plants were grown for 3 weeks in potting soil irrigated with water (22°C with a 16-h photoperiod and a light intensity of 120 μE m−2 s−1). Pea (Pisum sativum L. var. Douce Provence) and maize (Zea mays ‘Furio’) were grown for 2 weeks in vermiculite irrigated with water under a 12-h photoperiod (140 μE m−2 s−1) at 22°C (day) and 20°C (night).
Chemicals were solubilized in MES-KOH 10 mm, pH 5.6, filter sterilized, and added to cells cultures at the beginning of exponential growing phase (3 d after subcloning). To generate a sulfur-limiting medium, sulfate-containing salts (1.5 mm) were replaced with equimolar amounts of chloride salts and FeSO4/Na-EDTA (0.1 mm) was replaced by Fe-citrate/Na-EDTA. The unique sulfur source left in this modified medium was from sulfur-containing microelements (73 μm). One-week-old Arabidopsis cells were subcultured in sulfur-deficient medium and analyzed after 3 to 7 d of sulfur starvation. Leaves from Arabidopsis and leaf discs from pea and maize were floated in petri dishes containing MES-KOH 10 mm, pH 5.6, supplemented with 0.4 g L−1 Murashige and Skoog Basal medium (Sigma-Aldrich) and incubated at 22°C under continuous light (40 μE m−2 s−1). At each time point cells or leaf materials were collected, extensively washed with distilled water, weighted, and frozen in liquid nitrogen.
Measurements of Respiration and Cell Viability
Measurements of respiration rate were done in 1 mL of Gamborg's B5 medium and oxygen consumption was determined in the darkness using an O2 electrode (Hansatech, Eurosep Instruments) at 25°C. Cell viability assays were performed using protoplasts prepared from Arabidopsis cells. Fluorescein diacetate was added to protoplasts (final concentration 5 μg mL−1), followed by incubation at room temperature for 5 min. Samples were then placed on ice for 5 min and analyzed by epifluorescence microscopy using Green Fluorescent Protein filter sets (Zeiss Axioplan 2, Le Pecq). Cell viability was estimated as the ratio between fluorescing (living) protoplasts versus total protoplasts counted using bright-field illumination.
Protein Extraction and Western-Blot Analysis
Plant material was ground in liquid nitrogen and total soluble proteins were extracted in 20 mm Tricine (pH 7.5), 10 mm 2-mercaptoethanol, 10% (v/v) glycerol, and an EDTA-free complete protease inhibitor cocktail (Roche Applied Science, catalog no. 11873580001). Samples were centrifuged at 16,000g for 20 min at 4°C and the supernatant was used as a source of soluble proteins. Protein concentrations were estimated by the method of Bradford (Bradford, 1976) using the Bio-Rad protein assay reagent with bovine serum albumin as standard. Proteins were resolved by SDS/PAGE and stained with Coomassie Brilliant Blue or electroblotted to nitrocellulose membrane. Membranes were probed with polyclonal antibodies raised against recombinant Arabidopsis CGS (Ravanel et al., 1998), cystathionine β-lyase (Ravanel et al., 1996), Met synthase (Ravanel et al., 2004a), and Met γ-lyase (Rébeillé et al., 2006). Detection was performed by chemiluminescence. For N-terminal protein sequence determination, purified proteins were resolved by SDS/PAGE, electroblotted to polyvinylidene difluoride membrane, stained with Coomassie Brillant Blue, and then subjected to Edman degradation using an Applied Biosystem gas-phase sequencer model 492.
Overexpression and Purification of Recombinant CGS
DNA manipulations were conducted in Escherichia coli DH5α cells and expression of recombinant proteins was performed in the E. coli strain BL21 CodonPlus (DE3) RIL (Stratagene). The pET29-MCGS plasmid coding the mature (without its chloroplastic transit peptide) CGS from Arabidopsis (Ravanel et al., 1998) was used as a template to amplify MCGS and CCGS coding sequences for subcloning into the pET20b(+) expression vector (Novagen, Merck Biosciences). PCR were performed using the high fidelity Pfu DNA polymerase and primers MCGS5′ (5′-ATATACATATGGTCCGTCAG-3′), CCGS5′ (5′-GAGACATATGAGTGTACAGCTGA-CGGATTCC-3′), and CGS3′ (5′-GAGACTCGAGGATGGCTTCGAGAGCTTG-3′). The amplification products obtained with primer pairs MCGS5′/CGS3′ and CCGS5′/CGS3′ encode mature (starting with Val-69) and cleaved (starting with Ser-161) CGS, respectively. Amplicons were digested with NdeI and XhoI and cloned into the pET20b(+) vector digested by the same enzymes. The pET-MCGS-6His and pET-CCGS-6His constructs code for proteins fused with a C-terminal hexa-His tag. The DNA inserts were further sequenced to ensure that no mutation had been introduced during the course of the PCR amplification (Genome Express). E. coli BL21 CodonPlus (DE3) RIL cells transformed with pET-MCGS-6His or pET-CCGS-6His were grown at 37°C in Luria-Bertani medium supplemented with carbenicillin (100 μg mL−1). When A600 reached 0.6, 0.4 mm isopropylthio-β-galactoside was added and growth continued for 16 h at 20°C or 28°C for cells transformed with pET-MCGS-6His or pET-CCGS-6His, respectively. Cells were collected by centrifugation, resuspended in buffer A (KH2PO4-K2HPO4, pH 8.0, 0.3 m NaCl) containing 10 mm imidazole and an EDTA-free complete protease inhibitor cocktail (Roche Applied Science), and disrupted by sonication. The soluble protein extract was applied onto a nickel-nitrilotriacetic acid-agarose column (Qiagen) previously equilibrated with buffer A containing 10 mm imidazole. After successive washes with buffer A supplemented with 10, 30, and 50 mm imidazole, the recombinant protein was eluted with buffer A containing 250 mm imidazole. Fractions containing CGS were pooled, dialyzed against HEPES-K 20 mm, pH 7.5, 10% (v/v) glycerol for 2 h at 4°C, and concentrated by centrifugation (Microsep, 30 kD cutoff, Pall Filtron).
Measurements of Metabolites
Determination of folates was done by LC-MS/MS as described by Zhang et al. (2005). Following extraction, folylpolyglutamates were deconjugated in presence of rat serum to generate the corresponding monoglutamate derivatives. Reverse phase (RP)-HPLC was performed using a Purosphere Star RP-18 column (150 × 4.6 mm, 5 μm particle size, Merck) and elution of folates was done using a gradient of acetonitrile in 0.1% (v/v) formic acid (1 mL min−1, 35°C). Folates and MTX were detected by electrospray ionization on an Applied Biosystems API4000 tandem quadrupole mass spectrometer.
Extraction of soluble amino acids was performed as described by Kreft et al. (2003). Amino acids were derivatized using O-phthaldialdehyde and analyzed by RP-HPLC using an octadecyldimethylsilica column (Hypersil C18; 150 × 4.6 mm; 3 μm; Knauer GmbH) connected to an HPLC system (Agilent Technologies 1100 series). Amino acids were eluted with a linear gradient of methanol in 50 mm sodium acetate, pH 5.7, at 0.8 mL min−1 and 37°C, and detected by fluorescence (excitation 340 nm, emission 455 nm).
Thiols were extracted and derivatized with monobromobimane using the procedure described by Kreft et al. (2003). Derivatized thiols were subjected to RP-HPLC using an Atlantis dC18 column (250 × 4.6 mm; 5 μm; Waters) connected to an HPLC system. Elution was done using an acetonitrile gradient in 57 mm sodium perchlorate, 0.25% (v/v) acetic acid, pH 3.4, at 1 mL min−1 and 25°C. Derivatized thiols were detected by fluorescence (excitation 388 nm, emission 480 nm).
The procedure used for AdoMet and AdoHcy measurements has been adapted from Castro et al. (2002). Metabolites were derivatized with chloroacetaldehyde and separated by RP-HPLC using a Nucleodur C18 Pyramid column (250 × 4 mm, 5 μm, Macherey-Nagel) connected to an HPLC system. Elution was performed at 1 mL min−1 and 25°C with a linear gradient of acetonitrile in 50 mm sodium phosphate, pH 4.5. 1,N6-etheno derivatives of AdoMet and AdoHcy were detected by fluorescence (excitation 270 nm, emission 410 nm).
CGS Activity Measurements
CGS activity in cell extracts was assayed as described by Ravanel et al. (1995) using the O-phthaldialdehyde derivatization procedure to measure the production of cystathionine. Prior to kinetic analysis, cellular soluble proteins (see above for extraction) were desalted on Sephadex G-25 (Nap-5 columns, GE Healthcare) equilibrated in 20 mm Tricine-K, pH 7.5, and 10% (v/v) glycerol. The assay mixture (100 μL) contained 50 mm Tricine-K, pH 7.5, 0.1 mm dithiothreitol, 0.1 to 1 mm Cys, 0.1 to 10 mm OPH, 20 μm pyridoxal 5′-P, and 100 μm aminoethoxyvinyl Gly (an inhibitor of cystathionine β-lyase used to avoid cystathionine consumption by this enzyme). The ability of purified recombinant CGSs to catalyze cystathionine production was assayed either by the O-phthaldialdehyde-derivatization procedure or by coupling CGS, cystathionine β-lyase, and lactate dehydrogenase activities using the procedure described by Curien et al. (2003). Assays were conducted at 30°C.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number ATU83500.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Respiration rates in control and MTXS-treated Arabidopsis cells.
Supplemental Figure S2. Biochemical analysis of CGS processing by a protein extract from folate-deficient Arabidopsis cells.
Supplemental Figure S3. Analysis of CGS in Arabidopsis cells exposed to different stresses.
Supplementary Material
Acknowledgments
We thank Dr. Jean-Pierre Andrieu (laboratoire d'Enzymologie Moléculaire, Institut de Biologie Structurale, Grenoble) for performing amino acid sequence analyses. We are grateful to Drs. Claude Alban, Renaud Dumas, Eric Maréchal, and Michel Matringe (laboratoire de Physiologie Cellulaire Végétale, Grenoble) for helpful discussions and critical reading of the manuscript.
This work was supported by a PhD fellowship from the French Ministry of Research (to K.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stéphane Ravanel (sravanel@cea.fr).
The online version of this article contains Web-only data.
References
- Adam Z, Rudella A, van Wijk KJ (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9 234–240 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Castro R, Struys EA, Jansen EEW, Blom HJ, de Almeida IT, Jakobs C (2002) Quantification of plasma S-adenosylmethionine and S-adenosylhomocysteine as their fluorescent 1,N-6-etheno derivatives: an adaptation of previously described methodology. J Pharm Biomed Anal 29 963–968 [DOI] [PubMed] [Google Scholar]
- Chiba Y, Ishikawa M, Kijima F, Tyson RH, Kim J, Yamamoto A, Nambara E, Leustek T, Wallsgrove RM, Naito S (1999) Evidence for autoregulation of cystathionine gamma-synthase mRNA stability in Arabidopsis. Science 286 1371–1374 [DOI] [PubMed] [Google Scholar]
- Chiba Y, Sakurai R, Yoshino M, Ominato K, Ishikawa M, Onouchi H, Naito S (2003) S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine gamma-synthase gene in Arabidopsis. Proc Natl Acad Sci USA 100 10225–10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G, Job D, Douce R, Dumas R (1998) Allosteric activation of Arabidopsis threonine synthase by S-adenosylmethionine. Biochemistry 37 13212–13221 [DOI] [PubMed] [Google Scholar]
- Curien G, Laurencin M, Robert-Genthon M, Dumas R (2007) Allosteric monofunctional aspartate kinases from Arabidopsis. FEBS J 274 164–176 [DOI] [PubMed] [Google Scholar]
- Curien G, Ravanel S, Dumas R (2003) A kinetic model of the branch-point between the methionine and threonine biosynthesis pathways in Arabidopsis thaliana. Eur J Biochem 270 4615–4627 [DOI] [PubMed] [Google Scholar]
- De Brouwer V, Zhang G-F, Storozhenko S, Van Der Straeten D, Lambert WE (May 29, 2007) pH stability of individual folates during critical sample preparation steps in prevision of the analysis of plant folates. Phytochem Anal 10.1002/pca.1006 [DOI] [PubMed]
- Goto DB, Onouchi H, Naito S (2005) Dynamics of methionine biosynthesis. Plant Biotechnol 22 379–388 [Google Scholar]
- Goyer A, Collakova E, Shachar-Hill Y, Hanson AD (2007) Functional characterization of a methionine gamma-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway. Plant Cell Physiol 48 232–242 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Avraham T, Amir R (2002) The N-terminal region of Arabidopsis cystathionine γ-synthase plays an important regulatory role in methionine metabolism. Plant Physiol 128 454–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Schuster G, Amir R (2006) An in vivo internal deletion in the N-terminus region of Arabidopsis cystathionine gamma-synthase results in CGS expression that is insensitive to methionine. Plant J 45 955–967 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52 119–137 [DOI] [PubMed] [Google Scholar]
- Jabrin S, Ravanel S, Gambonnet B, Douce R, Rébeillé F (2003) One-carbon metabolism in plants: regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol 131 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Bamba T, Harada K, Fukusaki E, Kobayashi A (2007) Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J Exp Bot 58 415–424 [DOI] [PubMed] [Google Scholar]
- Kreft O, Hoefgen R, Hesse H (2003) Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiol 131 1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles EW, Kraus JP (2004) Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem 279 29871–29874 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Weretilnyk EA (2001) Sustaining S-adenosyl-L-methionine-dependent methyltransferase activity in plant cells. Physiol Plant 113 435–442 [Google Scholar]
- Mouillon JM, Aubert S, Bourguignon J, Gout E, Douce R, Rébeillé F (1999) Glycine and serine catabolism in non-photosynthetic higher plant cells: their role in C1 metabolism. Plant J 20 197–205 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H, Nagami Y, Haraguchi Y, Nakamoto M, Nishimura Y, Sakurai R, Nagao N, Kawasaki D, Kadokura Y, Naito S (2005) Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev 19 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsomando G, Bozzo GG, de la Garza RD, Basset GJ, Quinlivan EP, Naponelli V, Rébeillé F, Ravanel S, Gregory JF III, Hanson AD (2006) Evidence for folate-salvage reactions in plants. Plant J 46 426–435 [DOI] [PubMed] [Google Scholar]
- Prabhu V, Chatson KB, Lui H, Abrams GD, King J (1998) Effects of sulfanilamide and methotrexate on 13C fluxes through the glycine decarboxylase/serine hydroxymethyltransferase enzyme system in Arabidopsis. Plant Physiol 116 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P, McNeil SD, Ziemak MJ, Li C, Tarczynski MC, Hanson AD (2001) The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J 25 575–584 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, Douce R (2004. a) Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J Biol Chem 279 22548–22557 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Douce R, Rébeillé F (2004. b) The uniqueness of tetrahydrofolate synthesis and one-carbon metabolism in plants. In DA Day, AH Millar, J Whelan, eds, Advances in Photosynthesis and Respiration. Plant Mitochondria, from Genome to Function, Vol 17. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 277–292
- Ravanel S, Droux M, Douce R (1995) Methionine biosynthesis in higher plants. I. Purification and characterization of cystathionine gamma-synthase from spinach chloroplasts. Arch Biochem Biophys 316 572–584 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R (1998) Cystathionine gamma-synthase from Arabidopsis thaliana: purification and biochemical characterization of the recombinant enzyme overexpressed in Escherichia coli. Biochem J 331 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Job D, Douce R (1996) Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia coli. Biochem J 320 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S (2006) Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selhub J, Savin MA, Sakami W, Flavin M (1971) Synchronization of converging metabolic pathways: activation of the Cystathionine gamma-synthase of Neurospora crassa by methyltetrahydrofolate. Proc Natl Acad Sci USA 68 312–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegborn C, Messerschmidt A, Laber B, Streber W, Huber R, Clausen T (1999) The crystal structure of cystathionine gamma-synthase from Nicotiana tabacum reveals its substrate and reaction specificity. J Mol Biol 290 983–996 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32 891–904 [DOI] [PubMed] [Google Scholar]
- Wu K, Atkinson IJ, Cossins EA, King J (1993) Methotrexate resistance in Datura innoxia (uptake and metabolism of methotrexate in wild-type and resistant cell lines). Plant Physiol 101 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GF, Storozhenko S, Van Der Straeten D, Lambert WE (2005) Investigation of the extraction behavior of the main monoglutamate folates from spinach by liquid chromatography-electrospray ionization tandem mass spectrometry. J Chromatogr A 1078 59–66 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.