Abstract
Salicylic acid (SA) plays a central role in plant disease resistance, and emerging evidence indicates that auxin, an essential plant hormone in regulating plant growth and development, is involved in plant disease susceptibility. GH3.5, a member of the GH3 family of early auxin-responsive genes in Arabidopsis (Arabidopsis thaliana), encodes a protein possessing in vitro adenylation activity on both indole-3-acetic acid (IAA) and SA. Here, we show that GH3.5 acts as a bifunctional modulator in both SA and auxin signaling during pathogen infection. Overexpression of the GH3.5 gene in an activation-tagged mutant gh3.5-1D led to elevated accumulation of SA and increased expression of PR-1 in local and systemic tissues in response to avirulent pathogens. In contrast, two T-DNA insertional mutations of GH3.5 partially compromised the systemic acquired resistance associated with diminished PR-1 expression in systemic tissues. The gh3.5-1D mutant also accumulated high levels of free IAA after pathogen infection and impaired different resistance-gene-mediated resistance, which was also observed in the GH3.6 activation-tagged mutant dfl1-D that impacted the auxin pathway, indicating an important role of GH3.5/GH3.6 in disease susceptibility. Furthermore, microarray analysis showed that the SA and auxin pathways were simultaneously augmented in gh3.5-1D after infection with an avirulent pathogen. The SA pathway was amplified by GH3.5 through inducing SA-responsive genes and basal defense components, whereas the auxin pathway was derepressed through up-regulating IAA biosynthesis and down-regulating auxin repressor genes. Taken together, our data reveal novel regulatory functions of GH3.5 in the plant-pathogen interaction.
During the coevolution of plant-microbe interactions, plants have developed a complicated system against microbial pathogens (Chisholm et al., 2006). Plant-pathogen interactions are subjected to genetic control that leads to different outcomes. The presence of a pathogen avirulence (avr) gene and a cognate plant disease resistance (R) gene triggers an incompatible interaction characterized by rapid programmed cell death at sites of infection (called the hypersensitive response) and other defense-related responses (Dangl and Jones, 2001). Following the hypersensitive response, secondary defense responses can be activated in distal tissues, termed systemic acquired resistance (SAR). SAR is a nonspecific, systemic, and long-lasting defense mechanism that enhances resistance to subsequent infections by a variety of pathogens (Ryals et al., 1996; Sticher et al., 1997; Durrant and Dong, 2004). In contrast, compatible interaction occurs if either the host or the pathogen lacks the corresponding R or avr gene, respectively. Under these conditions, plants can still activate basal defense to restrict pathogen growth.
Extensive studies have shown that salicylic acid (SA) plays a central role in plant defense against pathogens (Dempsey et al., 1999). SA levels increase in both infected and distal leaves in response to pathogen attack (Summermatter et al., 1995), and exogenous application of SA can induce a set of pathogenesis-related (PR) genes and establish SAR (Uknes et al., 1992). Transgenic plants expressing the nahG gene, encoding a salicylate hydroxylase that converts SA to inactive catechol, are compromised in SAR, basal resistance, and some R-gene-mediated resistance (Gaffney et al., 1993; Delaney et al., 1994). Furthermore, the Arabidopsis (Arabidopsis thaliana) sid1/eds5 and sid2/eds16 mutants deficient in pathogen-induced SA production are also impaired in SAR, certain R-gene-mediated resistance, and basal resistance (Rogers and Ausubel, 1997; Nawrath and Métraux, 1999).
Despite the presence of effective defense systems, many bacterial pathogens are able to evade or suppress the host defense or modulate the metabolism of the host to obtain nutrients for their colonization through virulence strategies (Nomura et al., 2005; Abramovitch et al., 2006). For example, gram-negative bacteria, such as Pseudomonas syringae, secrete virulence proteins, called effectors, directly into the host cell via a type III secretion system to promote pathogenicity (Mudgett, 2005). In addition, P. syringae also produces the phytohormone auxin that may facilitate its colonization (Glickmann et al., 1998; Katagiri et al., 2002; Abramovitch et al., 2006; Spaepen et al., 2007).
Previous studies have shown that the levels of indole-3-acetic acid (IAA), the primary plant auxin, increased in plant tissues infected by P. syringae pv tomato (Pst) DC3000 (O'Donnell et al., 2003). Moreover, the type III effector AvrRpt2 modulates host IAA levels to promote pathogen virulence and disease development in Arabidopsis (Chen et al., 2004; Kunkel et al., 2005). Recent microarray analysis has shown that infection with Pst DC3000 activates genes related to IAA biosynthesis and represses Aux/IAA family and auxin transporter genes, suggesting that Pst DC3000 impacts auxin signaling probably through activating IAA production, altering IAA movement, and derepressing the auxin pathway (Thilmony et al., 2006). Conversely, the Arabidopsis mRNA miR393a is induced by the flagellin-derived peptide flg22 and represses auxin signaling through down-regulating the auxin receptor genes, resulting in increased resistance to P. syringae (Navarro et al., 2006). Together, these results suggest that auxin plays an important role in disease susceptibility pathways. However, little is known about genes that regulate auxin signaling in response to pathogen infection and the downstream targets of altered auxin signaling responsible for enhanced disease susceptibility.
Auxin rapidly induces numerous genes called early auxin response genes. The best-characterized early auxin response genes include three major classes: Aux/IAAs, SAURs, and GH3s (Hagen and Guilfoyle, 2002). The Arabidopsis GH3 family consists of 19 members, six of which are known to adenylate IAA in vitro (Staswick et al., 2002). Further studies have shown that GH3 genes encode IAA-amido synthetases that are involved in auxin homeostasis through conjugating amino acids to IAA (Staswick et al., 2005). However, the physiological function and regulatory target genes of these GH3 members are still largely unknown. It was also shown that GH3.5 (At4g27260) adenylated both IAA and SA in vitro (Staswick et al., 2002), suggesting that GH3.5 could function in modulating and integrating both auxin and SA signaling in the plant-pathogen interaction.
In this work, we investigate the functions of GH3.5 in response to P. syringae infection. We show that GH3.5 positively regulates the SA signaling pathway in plant defense and modulates the auxin pathway to enhance host susceptibility. Our findings demonstrate that GH3.5 is a key modulator that both positively and negatively affects various aspects of plant defense, revealing another dimension to the complex and dynamic plant-pathogen interaction.
RESULTS
Characterization of Semidominant Gain-of-Function Mutant gh3.5-1D
We have previously generated a large population of transgenic Arabidopsis plants using the plasmid pSKI015 (Weigel et al., 2000; Li et al., 2005). From this population, we identified a dwarf mutant that contains a single T-DNA insertion revealed by cosegregation of the dwarf phenotype with the herbicide resistance marker. Using the plasmid rescue approach, we found that the T-DNA was inserted 826 bp upstream of the GH3.5 gene (At4g27260; Fig. 1A). The mutant was therefore named gh3.5-1D. Whereas the heterozygous mutant exhibits a mild morphological defect, the homozygous gh3.5-1D mutant displays a severe phenotype with smaller curly rosette leaves (Fig. 1B) and shortened primary roots and reduced lateral roots that were more resistant to exogenous auxins (Supplemental Fig. S1), resembling a previously reported activation-tagged mutant, dfl1-D, caused by overexpression of DFL1/GH3.6 known to be involved in auxin signaling/homeostasis (Nakazawa et al., 2001; Staswick et al., 2005).
Figure 1.
Phenotypic and molecular characterization of gh3.5-1D. A, T-DNA insertion in gh3.5-1D. The large arrow indicates the GH3.5 gene. The four cauliflower mosaic virus 35S enhancer elements are indicated with small arrowheads. B, Four-week-old Col-0, heterozygous (gh3.5-1D+/−), homozygous (gh3.5-1D−/−), and representative p35S∷GH3.5 transgenic plant. C, Northern-blot analysis (top) of GH3.5 expression and western-blot detection (bottom) of the GH3.5 protein in Col-0, gh3.5-1D+/−, gh3.5-1D−/−, and p35S∷GH3.5 plants. Rubisco staining was used as a loading control in western blots. The experiments were repeated at least once with similar results.
Northern- and western-blot analysis showed that the gh3.5-1D mutant accumulated higher levels of the GH3.5 transcript and protein, respectively (Fig. 1C). To further confirm that the gh3.5-1D phenotype was caused by GH3.5 overexpression, we generated transgenic plants that overexpress GH3.5 driven by the constitutively active cauliflower mosaic virus 35S promoter (p35S∷GH3.5). Most of the homozygous transgenic lines displayed a phenotype similar to that of gh3.5-1D heterozygous plants (Fig. 1, B and C). We thus concluded that increased expression of GH3.5 gives rise to the phenotypes of gh3.5-1D.
R-Mediated Local Resistance Is Compromised while SAR and Basal Defense Are Normal in gh3.5-1D
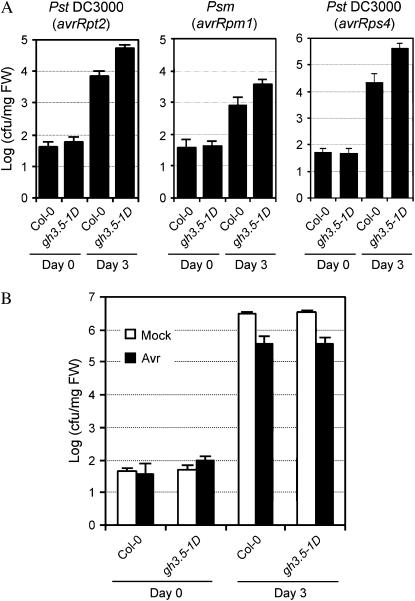
SA plays important roles in R-mediated local resistance and SAR (Delaney et al., 1994). The fact that GH3.5 adenylates SA in vitro serves as a reminder for a role of GH3.5 in defense responses (Staswick et al., 2002). To explore this idea, we first tested the R-mediated resistance in gh3.5-1D plants challenged with avirulent strains Pst DC3000(avrRpt2), P. syringae pv maculicola (Psm)(avrRpm1), and Pst DC3000(avrRps4), which trigger the corresponding RPS2- (Bent et al., 1994; Mindrinos et al., 1994), RPM1- (Grant et al., 1995), and RPS4-mediated resistance in ecotype Columbia-0 (Col-0; Gassmann et al., 1999). As shown in Figure 2A, bacterial growth increased in the gh3.5-1D mutant as compared to wild-type Col-0 at 3 d postinfection (dpi), indicating that overexpression of GH3.5 impaired different R-gene-mediated disease resistance. Consistently, disease symptoms that were typically undetectable on Col-0 plants were visible on gh3.5-1D at 3 dpi with a relatively low inoculum of 105 colony-forming units (cfu)/mL (Supplemental Fig. S2).
Figure 2.
Pathogen growth in leaves of Col-0 and gh3.5-1D plants. A, Growth of avirulent strains Pst DC3000(avrRpt2), Psm(avrRpm1), and Pst DC3000(avrRps4) in Col-0 and gh3.5-1D plants. Plants were infected at a density of 105 cfu/mL. B, Growth of Pst DC3000 in SAR-induced leaves of Col-0 and gh3.5-1D plants. Lower leaves were preinoculated with Psm carrying avrRpm1 at 107 cfu/mL or buffer (mock) 3 d prior to infection with Pst DC3000 at 105 cfu/mL in three systemic leaves. Bacterial titers (A and B) were measured at 0 and 3 dpi. All values are means ± se (n = 6). Similar results were observed in three independent experiments.
To test whether the impaired R-mediated local resistance in gh3.5-1D affected R-induced SAR, both wild-type and gh3.5-1D plants were challenged with Psm(avrRpm1) or with 10 mm MgCl2. Three days later, fully expanded intact leaves were inoculated with Pst DC3000. As shown in Figure 2B, R-mediated local resistance triggered the usual SAR in wild-type plants. There was no detectable difference in the growth of Pst DC3000 on avirulent pathogen-preinfected gh3.5-1D plants in comparison with the corresponding wild-type plants, indicating that SAR developed normally in gh3.5-1D. The same bacterial growth on the MgCl2-treated wild-type and gh3.5-1D plants also indicated intact basal resistance in gh3.5-1D plants (Fig. 2B). Therefore, neither the SAR nor basal resistance was altered in gh3.5-1D plants.
Induction of Defense Genes and SA Levels in gh3.5-1D
Activation of the SA pathway results in the expression of certain PR genes such as PR-1 (Durrant and Dong, 2004). To determine whether impaired R-mediated resistance in gh3.5-1D is associated with decreased PR gene induction, we analyzed the expression of PR-1 after infection with Psm(avrRpm1) in wild-type and gh3.5-1D plants. Surprisingly, expression of PR-1 in response to Psm(avrRpm1) was strongly up-regulated locally and systemically in gh3.5-1D as compared to the wild type (Fig. 3A), indicating that GH3.5 positively regulates expression of the PR-1 gene. In addition, expression of PDF1.2, a known marker gene of the jasmonic acid/ethylene-mediated defense pathway, was obviously decreased both locally and systemically in gh3.5-1D, consistent with the antagonistic interaction between the SA and jasmonic acid pathways as previously described (Kunkel and Brooks, 2002). SA accumulation is necessary for the activation of PR-1 gene expression (Durrant and Dong, 2004). We further analyzed SA levels in wild-type and gh3.5-1D plants after infection with Psm(avrRpm1). As shown in Figure 3B, gh3.5-1D plants accumulated much more both free and total SA in local and systemic tissues than wild-type plants at 48 h postinfection (hpi), indicating that GH3.5 overexpression positively regulates SA accumulation in both local and systemic tissues. A similar result was also observed in plants infected with Pst DC3000(avrRpt2) (Supplemental Fig. S3). Taken together, our results demonstrate that the PR-1 transcript and SA levels were increased rather than decreased in the gh3.5-1D mutant, suggesting that GH3.5 overexpression might also enhance a susceptibility-related pathway that contributes to the compromised R-mediated local resistance.
Figure 3.
Induction of defense genes and SA accumulation in Col-0, gh3.5-1D, and gh3.5-2 plants. A, Local and systemic induction of PR-1 and PDF1.2 in Col-0, gh3.5-1D, and gh3.5-2 plants. Samples were collected at 0, 1, 2, 3, and 4 dpi with Psm(avrRpm1) at 107 cfu/mL. Experiments were repeated twice with similar results. B, Levels of free and total SA in local and systemic leaves of Col-0, gh3.5-1D, and gh3.5-2 plants after infection with Psm(avrRpm1) at 107 cfu/mL. Leaves were harvested at 0 (control) and 48 hpi. Data are means ± se (n = 3). Asterisks indicate significant difference to Col-0 (P < 0.05). Experiments were repeated once with similar results.
Previous in vitro analysis showed that GH3.5 exhibited adenylation function on SA (Staswick et al., 2002). We indeed observed that salicyloyl-aspartate (SA-Asp), the only identified endogenous SA-amido conjugate in planta (Bourne et al., 1991), accumulated to higher levels in the gh3.5-1D leaves infected with Psm(avrRpm1), as compared to Col-0 (Supplemental Fig. S4). Because the only identified active SA form in vivo is free SA (Sticher et al., 1997), there may be a possibility that accumulated SA-Asp might serve as a pool for the increased SA generation observed in gh3.5-1D.
SAR Response Is Impaired in Loss-of-Function Mutants of GH3.5
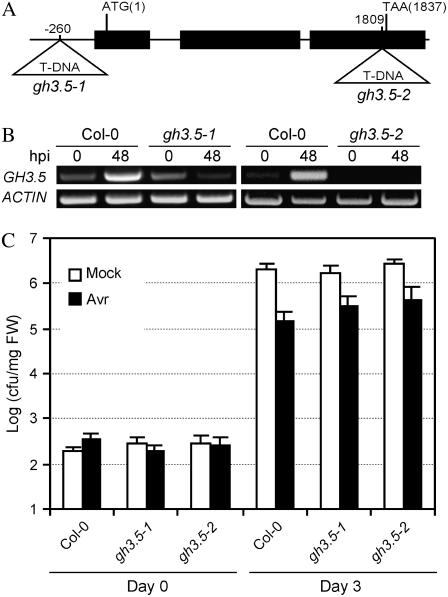
To further investigate the role of GH3.5 in response to pathogens, we obtained two T-DNA insertion lines of GH3.5 from the Salk T-DNA collection (Alonso et al., 2003). The SALK_014376 T-DNA line contains the T-DNA insertion 260 bp upstream of the GH3.5 start codon and was previously named gh3.5-1 (Staswick et al., 2005), whereas SALK_151766 contains the T-DNA insertion 1,809 bp downstream of the GH3.5 start codon and was designated gh3.5-2 (Fig. 4A). Reverse transcription (RT)-PCR analysis showed that the GH3.5 transcript was induced in wild-type plants upon infection with Psm(avrRpm1) (Fig. 4B; see below). Although no difference in the GH3.5 transcript level was detected between the wild-type and the gh3.5-1 plant at 0 hpi, Psm(avrRpm1) infection led to no induction of the GH3.5 gene, indicating that gh3.5-1 is a partial loss-of-function mutant. In contrast, no GH3.5 transcript was detected in gh3.5-2 regardless of pathogen infection, indicating that gh3.5-2 is a loss-of-function mutant. Consistent with the predicted functional redundancy among the members of the GH3 family (Staswick et al., 2005), both mutants display no obvious morphological phenotypes compared with the wild type, although they showed a slight increase in sensitivity to exogenous auxins (Supplemental Fig. S1).
Figure 4.
Characterization of GH3.5 loss-of-function mutants gh3.5-1 and gh3.5-2. A, Insertion sites of T-DNA in gh3.5-1 and gh3.5-2. B, RT-PCR detection of the GH3.5 transcript in gh3.5-1 and gh3.5-2 at 0 and 48 hpi with Psm(avrRpm1) at 107 cfu/mL. ACTIN, Loading control for RT-PCR. The experiment was repeated once with similar results. C, Compromised SAR in gh3.5-1 and gh3.5-2 mutants. Growth of Pst DC3000 was measured in plants either mock treated or pretreated with Psm (avrRpm1). All values are mean ± se (n = 6). Similar results were obtained in three independent experiments.
We next examined disease resistance in gh3.5-1 and gh3.5-2 plants inoculated with Pst DC3000(avrRpt2). Both mutants showed normal resistance similar to wild-type plants (Supplemental Fig. S5). We then tested whether the gh3.5-1 and gh3.5-2 mutants altered R-induced SAR. As shown in Figure 4C, basal resistance to Pst DC3000 in gh3.5-1 and gh3.5-2 plants was not different from that of Col-0 plants at 3 dpi. However, SAR was partially, yet significantly, compromised in both gh3.5-1 and gh3.5-2 compared to the wild type at 3 dpi (0.36 and 0.47 log, t test P < 0.05, respectively), suggesting that GH3.5 is partially required for SAR. Consistent with the compromised SAR in the gh3.5 knockout mutant, systemic induction of PR-1 was decreased, whereas its local induction was almost unaffected in gh3.5-2 plants after infection with Psm(avrRpm1) (Fig. 3A). Moreover, systemic induction of PDF1.2 was increased in the gh3.5-2 mutant (Fig. 3A). However, the SA levels both in local and systemic tissues were not altered in gh3.5-2 compared to the wild type infected by Psm(avrRpm1) and Pst DC3000(avrRpt2) (Fig. 3B; Supplemental Fig. S3).
Overexpression of DFL1 Impairs R-Mediated and Basal Resistance
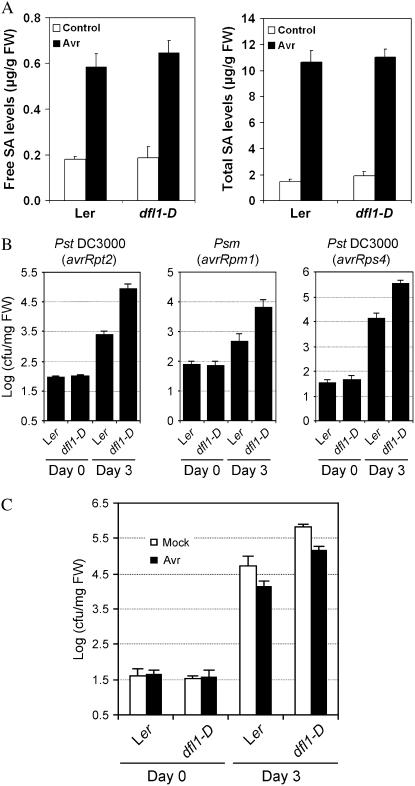
Despite accumulation of SA and the PR-1 transcript in the gh3.5-1D mutant, it still exhibited compromised R-mediated local resistance and had a normal, rather than an increased, SAR and basal resistance. The data suggest that GH3.5 overexpression might enhance a susceptibility-related pathway that counteracts SA-mediated disease resistance. Given the facts that the protein product of the GH3.5 gene can conjugate IAA to amino acids in vitro and a recent revelation of the auxin involvement in disease susceptibility (Staswick et al., 2002; Navarro et al., 2006), we hypothesized that an altered auxin pathway is likely responsible for the enhanced local susceptibility phenotype of the gh3.5-1D mutant. To test our hypothesis, we analyzed the disease resistance of a similar activation-tagged dwarf mutant dfl1-D (Nakazawa et al., 2001). DFL1 (GH3.6) is the closest family member of GH3.5, and a previous study showed that DFL1 exhibited IAA-amido synthetase activity similar to GH3.5 in vitro (Staswick et al., 2005), suggesting that both proteins regulate IAA homeostasis/signaling through a similar mechanism. Unlike GH3.5, GH3.6 did not show activity on SA in vitro (Staswick et al., 2002), so dfl1-D should be suitable to study the effect of the auxin pathway on disease susceptibility. Consistent with the in vitro result, we did not observe changes in SA levels in dfl1-D compared to the corresponding wild type (Fig. 5A).
Figure 5.
SA levels and pathogen growth in dfl1-D plants. A, Levels of free and total SA in local and systemic leaves of Ler and dfl1-D plants after infection with Psm(avrRpm1) at 107 cfu/mL. Leaves were harvested at 0 (control) and 48 hpi. Data are means ± se (n = 3). Experiments were repeated once with similar results. B, Growth of Pst DC3000(avrRpt2), Psm(avrRpm1), and Pst DC3000(avrRps4) in Ler and dfl1-D plants. Leaves were infected at a density of 105 cfu/mL. C, Growth of Pst DC3000 in Ler and dfl1-D plants after preinoculation with Psm(avrRpm1). Data are mean ± se (n = 6). Similar results were observed in three independent experiments (B and C).
Bacterial growth assay showed that dfl1-D was indeed more susceptible to Pst DC3000(avrRpt2), Psm(avrRpm1), and Pst DC3000(avrRps4), resulting in more bacterial growth than in the corresponding wild-type controls at 3 dpi (Fig. 5B). This indicates that dfl1-D also impairs the same set of R-gene-mediated resistance as gh3.5-1D. In contrast to gh3.5-1D, basal resistance to Pst DC3000 (mock) was decreased in dfl1-D plants (Fig. 5C). Although dfl1-D was capable of mounting SAR, enhanced growth of Pst DC3000 was still apparent in the secondary infected leaves of dfl1-D (Fig. 5C), showing a phenotype similar to that of the previously reported eds (enhanced disease susceptibility) mutants (Rogers and Ausubel, 1997).
IAA Levels Are Increased in gh3.5-1D during Pathogen Infection
Considering GH3.5 has IAA-amido synthase activity (Staswick et al., 2002, 2005), we measured levels of free IAA and IAA-amido conjugates in gh3.5-1D in response to virulent and avirulent pathogen attack. As shown in Table I, IAA levels were not different between gh3.5-1D and Col-0 with mock treatment, similar to the observation with dfl-1D (Staswick et al., 2005). Infection with Pst DC3000 but not Pst DC3000(avrRpt2) greatly increased the IAA levels in wild-type plants, suggesting an important role for IAA in susceptibility during virulent pathogen infection. Surprisingly, the IAA levels were significantly increased in gh3.5-1D plants infected with Pst DC3000(avrRpt2) as compared to that of wild-type plants, and were further increased when the mutant was infected with Pst DC3000, indicating that GH3.5 overexpression positively regulates IAA accumulation during pathogen infection. The increased IAA level might contribute to the enhanced susceptibility to avirulent pathogen. We did not observe significant elevation of the three known IAA conjugates in gh3.5-1D compared with the wild-type controls, indicating GH3.5 does not regulate accumulation of those amido-conjugated IAAs under the conditions of our study or they were immediately hydrolyzed after formation. We further measured IAA levels in the systemic tissues of the gh3.5-1D and wild-type plants that were either mock treated or preinoculated with Pst DC3000(avrRpt2). There was no difference in IAA levels between the gh3.5-1D and wild type (data not shown), indicating that gh3.5-1D did not affect systemic IAA levels during SAR establishment.
Table I.
Analysis of IAA and IAA-amido levels
Values are means ± se (four or five biological replicates). All values are pmol/g fresh weight. Means with different letters are significantly different at the 0.05 level.
| Genotype | Treatment | IAA | IAA-Ala | AA-Leu | IAA-Asp |
|---|---|---|---|---|---|
| Col-0 | Mock | 67.9a ± 5.5 | 13.4 ± 4.0 | 5.0 ± 1.2 | 7.8 ± 2.2 |
| gh3.5-1D | Mock | 79.8a ± 8.9 | 15.4 ± 4.7 | 4.9 ± 1.3 | 7.5 ± 1.4 |
| Col-0 | Pst DC3000(avrRpt2) | 70.2a ± 7.6 | 15.2 ± 1.8 | 4.2 ± 0.9 | 5.1 ± 1.8 |
| gh3.5-1D | Pst DC3000(avrRpt2) | 102.1b ± 11.0 | 12.7 ± 2.2 | 4.4 ± 0.7 | 3.5 ± 1.3 |
| Col-0 | Pst DC3000 | 116.9b ± 12.1 | 12.1 ± 2.0 | 3.0 ± 1.0 | 5.7 ± 0.7 |
| gh3.5-1D | Pst DC3000 | 177.6c ± 22.4 | 22.0 ± 4.7 | 6.8 ± 1.2 | 5.0 ± 1.0 |
GH3.5 Is Induced by Pathogens and SA
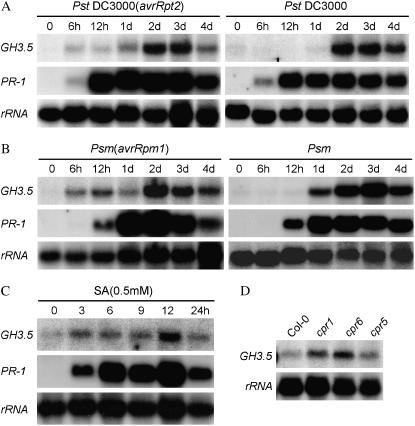
Because GH3.5 plays diverse roles in plant-pathogen interactions, its expression pattern could be indicative of its function. Northern hybridization indicated that the induction of GH3.5 was slightly different in responses to the avirulent and virulent strains in wild-type plants. Induction was detectable as early as 6 h, with a major peak at 48 h after inoculation with both avirulent Pst DC3000(avrRpt2) and Psm(avrRpm1) (Fig. 6, A and B). In contrast, induction was delayed until 24 h, with a peak occurring around 2 dpi by Pst DC3000 and 3 dpi by Psm (Fig. 6B). It was previously shown that GH3.5, also known as AtGH3a, was induced by IAA (Tanaka et al., 2002). We next tested whether GH3.5 could be induced by SA. As shown in Figure 6C, SA induced GH3.5 as early as 3 h, with the peak level at 12 h after treatment. Consistent with these results, GH3.5 expression was up-regulated in cpr1 and cpr6 and slightly increased in cpr5 (Fig. 6D), which are known to contain high levels of SA (Clarke et al., 2000). It should be noted that the degree of GH3.5 induction is much lower by SA than by pathogen (Fig. 6, A and B), suggesting that pathogen-induced expression of the GH3.5 gene might also be mediated by an SA-independent pathway.
Figure 6.
Induction pattern of GH3.5 in response to pathogen and SA. A, GH3.5 expression in leaves of Col-0 at different times after infection with Pst DC3000(avrRpt2) and Pst DC3000 at 107 cfu/mL as detected by northern blot. B, Time-course GH3.5 expression in leaves of Col-0 infected with Psm(avrRpm1) and Psm at 107 cfu/mL. Induction of PR-1 was included as a marker for defense activation (A and B). C, Induction of GH3.5 in Col-0 by SA (0.5 mm). Samples were harvested at the time points indicated. D, Expression of GH3.5 in cpr1, cpr6, and cpr5 mutants and in Col-0 as the control. All experiments were biologically repeated at least once with similar results.
Transcriptional Profiling of gh3.5-1D Plants in Response to Pathogen
To reveal the underlying molecular mechanism of GH3.5 action in modulating the SA and auxin pathways, we performed transcriptional profiling of wild-type Col-0 and gh3.5-1D plants with or without infection by Pst DC3000(avrRpt2) using the Affymetrix Arabidopsis ATH1 GeneChip. With three biological replicates of pairwise experiments (Supplemental Table S1) and high stringent selection criteria (Thilmony et al., 2006), we identified 273 and 831 differentially regulated genes in uninoculated gh3.5-1D and gh3.5-1D infected for 48 h, in comparison with Col-0 plants, respectively, and a total of 1,013 nonredundant genes that exhibited statistically significant (≥2-fold) changes of expression levels (Supplemental Tables S2 and S3).
For more detailed analysis, we first performed hierarchical clustering to study the genes related to auxin biosynthesis and response (Fig. 7; Supplemental Table S4). In uninoculated gh3.5-1D plants, 13 SAUR members and four Aux/IAA genes functioning as repressors in auxin signaling (Tiwari et al., 2001) were down-regulated, consistent with the GH3.5 role in the auxin pathway. A great number of auxin-related genes were either up- or down-regulated in gh3.5-1D infected with pathogens. Among those up-regulated genes were three IAA biosynthesis-related genes (ASA1, CYP79B2, and AAO1), two GH3 genes (DFL1/GH3.6 and GH3.3), and two IAA-amido hydrolase genes (ILR1and ILL5). Up-regulation of the IAA biosynthesis-related and IAA-amido hydrolase genes could contribute to the higher IAA levels observed in gh3.5-1D in response to pathogens (Table I). In contrast, down-regulated genes included 15 SAURs and five Aux/IAAs. Taken together, these results suggest that GH3.5 might regulate auxin signaling by increasing IAA biosynthesis and derepressing the auxin pathway during pathogenesis.
Figure 7.
Hierarchical clustering of auxin-related genes. Auxin-related genes significantly regulated in gh3.5-1D plants as compared to Col-0 plants. Each column represents a single signal log2 ratio of gh3.5-1D versus Col-0 at 0 (uninoculated) and 48 hpi, and each row represents a different gene. Expression scales (log) are illustrated with colors. Descriptions and accession numbers of the genes are indicated (see also Supplemental Table S4).
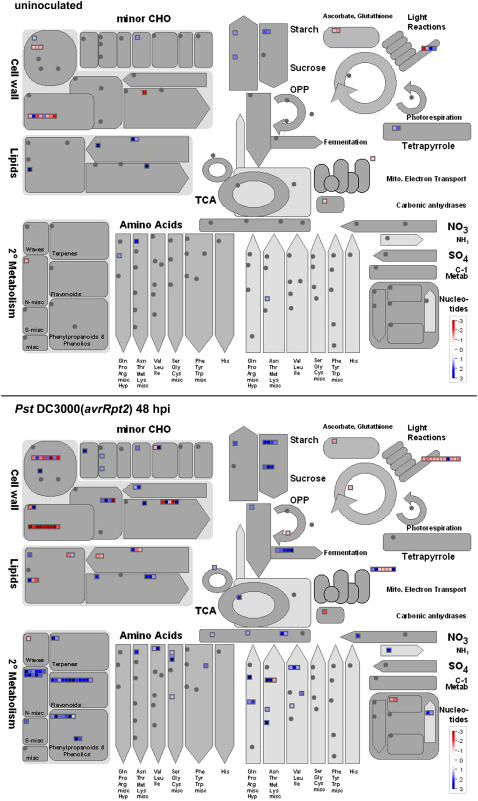
We next profiled metabolic pathways using MAPMAN software (Thimm et al., 2004). The genes involved in the breakdown of starch, Suc, lipids, and nucleotides and the metabolism of amino acids were remarkably up-regulated in gh3.5-1D at 48 hpi (Fig. 8). This suggests high levels of nutrient accumulation during pathogen infection in gh3.5-1D plants. Moreover, MAPMAN analysis revealed that many transporter genes were up-regulated in gh3.5-1D as compared to the wild type at 48 hpi (Supplemental Fig. S6), including several ATP-binding cassette-type transporter genes and genes encoding transporters for oligopeptides, amino acids, and sugars. Strong activation of these genes could result in nutrient efflux to benefit invading pathogens in gh3.5-1D.
Figure 8.
Metabolism changes in gh3.5-1D plants. MAPMAN was used to observe metabolic changes in gh3.5-1D plants either uninoculated or inoculated with Pst DC3000(avrRpt2) at 48 hpi. The average fold change of the three biological replicates is presented as illustrated in the fold change colors in the bottom right of each image (red, repressed; blue, induced).
Consistent with a positive role of gh3.5-1D in the SA-mediated defense pathway, many defense-related genes were more strongly activated by pathogens in gh3.5-1D as compared to wild-type plants (Table II). These genes could be grouped into three main categories: (1) SA-induced genes, such as α-DOX1 (de León, 2002), PAD3, PAL1, and WRKY transcription factor genes; (2) genes involved in cell wall modification known to be a critical aspect of the plant basal defense; and (3) other defense-related genes, such as FRK1 (Asai et al., 2002). WRKY induction was particularly interesting because some WRKYs, such as WRKY18, were recently identified to play important roles in SAR (Wang et al., 2006).
Table II.
Selected defense-related genes that are significantly induced or depressed in gh3.5-1D at 48 hpi with Pst DC3000(avrRpt2)
| AGIa | Description | Fold Changeb |
|---|---|---|
| SA induced | ||
| At3g01420 | α-DOX1 α-dioxygenase | 5.97 |
| At4g34135 | Salicylate-induced glucosyltransferase | 2.73 |
| At2g43820 | Putative glucosyltransferase | 1.77 |
| At1g62300 | WRKY6 | 1.57 |
| At4g31550 | WRKY11 | 1.40 |
| At4g31800 | WRKY18 | 1.97 |
| At5g07100 | WRKY26 | 1.50 |
| At5g13080 | WRKY-like protein | 2.07 |
| At3g26830 | PAD3, phytoalexin deficient 3 | 1.80 |
| At2g37040 | PAL1, Phe ammonia-lyase 1 | 1.70 |
| Cell wall modification | ||
| At5g56870 | β-Galactosidase | 3.27 |
| At2g43570 | Chitinase, putative, similar to chitinase class IV | 1.43 |
| At1g76930 | EXT4, extensin Hyp-rich glycoprotein | 3.80 |
| At1g61810 | Glycosyl hydrolase family 1 protein | 2.27 |
| At4g27830 | Glycosyl hydrolase family 1 protein | 1.63 |
| At3g60130 | Glycosyl hydrolase family 1 protein | 1.67 |
| At4g16260 | Glycosyl hydrolase family 17 protein | 3.13 |
| At3g09260 | PYK10 glycosyl hydrolase family 1 protein | 1.73 |
| At3g60140 | DIN2 glycosyl hydrolase family 1 protein | 6.27 |
| At5g55180 | Glycosyl hydrolase family 17 protein | 1.37 |
| At3g04010 | Glycosyl hydrolase family 17 protein | 2.13 |
| At4g19810 | Glycosyl hydrolase family 18 protein | 2.60 |
| At1g23040 | Hydroxy-Pro-rich glycoprotein family protein | 1.43 |
| At4g22470 | Putative Hyp-rich glycoprotein | 3.33 |
| At5g64120 | Peroxidase | 1.70 |
| At5g05340 | Peroxidase | 2.53 |
| At3g49120 | Peroxidase 34 | 1.37 |
| At2g45220 | Putative pectinesterase | 3.37 |
| Other defense related | ||
| At2g19190 | FRK1 flagellin-induced receptor-like kinase | 1.50 |
| At5g13080 | WRKY75 | 2.07 |
| At4g37990 | Cinnamyl-alcohol dehydrogenase ELI3-2 | 3.37 |
| At3g44880 | ACD1, accelerated cell death 1 | 1.60 |
| At1g33960 | AIG1, avrRpt2-induced gene 1 | 1.67 |
| At4g11650 | OSM34, osmotin-like protein | 4.53 |
| At2g29460 | GSTU4, glutathione transferase 4 | 1.77 |
| At2g29470 | GSTU3, glutathione transferase 3 | 3.97 |
| At1g69930 | GSTU11, glutathione transferase 11 | 2.27 |
| At1g74590 | GSTU10, glutathione transferase 10 | 1.73 |
| At1g65690 | Harpin-induced protein-related | 1.97 |
| At2g39200 | MLO family protein 12, MLO12 | 2.00 |
| At1g72900 | Disease resistance protein (TIR-NBS class), putative, | 1.83 |
| At3g05360 | Disease resistance family protein/LRR family protein | 3.13 |
| At5g44420 | PDF1.2 antifungal protein-like | −3.80 |
Arabidopsis Genome Initiative.
Means of average SLR values (gh3.5-1D 48 hpi versus Col-0 48 hpi) from three independent biological replicates of the reproducibly differently regulated genes.
To confirm the expression patterns of the genes identified by microarray analysis, we next conducted RT-PCR experiments to detect the expression levels of four defense-related and three auxin-related genes in gh3.5-1D and wild-type plants infected with Pst DC3000(avrRpt2). As expected, these genes were differentially induced in gh3.5-1D compared to wild-type plants (Supplemental Fig. S7).
DISCUSSION
GH3.5 Is a Novel Positive Modulator of SA Signaling
SA-mediated SAR has been extensively studied, revealing that SA is necessary and sufficient for induction of SAR. It is well established that the transcription cofactor protein NPR1-regulated expression of PR genes is required for the induction of SAR (Durrant and Dong, 2004). Our current study reveals that GH3.5 is a novel positive modulator in regulating SA signaling during SAR establishment, with several lines of evidence. First, the systemic induction of PR-1 by avirulent strains was increased and associated with higher levels of SA in gh3.5-1D compared with Col-0. Second, systemic PR-1 induction was repressed and SAR was partially compromised in the two GH3.5 insertional mutants. Third, expression of WRK18, a positive regulator of SAR (Wang et al., 2006), was induced in gh3.5-1D compared with the wild type after infection with Pst DC3000(avrRpt2). Therefore, GH3.5 positively regulates SA-mediated SAR most likely by enhancing SA biosynthesis, up-regulating expression of WRKY18, and consequently activating downstream genes.
SA also plays a central role in R-mediated local resistance (Nimchuk et al., 2003). We found that SA levels were significantly elevated in gh3.5-1D than the wild type. Consistent with the increased SA levels in gh3.5-1D after pathogen infection, the Phe ammonia-lyase gene PAL1 was up-regulated. Furthermore, the global expression experiment revealed that many defense-responsive genes were up-regulated in gh3.5-1D as compared to Col-0 after infection with Pst DC3000(avrRpt2), which included four known SA-responsive WRKY genes (Dong et al., 2003; Wang et al., 2006), consistent with the notion that GH3.5 plays a positive role in the SA signaling pathway. Moreover, we also observed up-regulation of the genes involved in cell wall modification and basal defense response. Cell wall modification is recognized as a critical aspect of the plant basal defense, suggesting that GH3.5 is also involved in basal defense. Consistent with this deduction, the basal defense marker gene, FRK1 (Asai et al., 2002), was greatly induced in gh3.5-1D.
GH3.5 Plays an Important Role in Auxin-Elicited Susceptibility
In contrast to disease resistance, little is known about the mechanism of plant susceptibility (Vogel et al., 2002; Nomura et al., 2005). It has long been recognized that many pathogenic microbes can produce IAA, which is proposed to alter host cellular processes to favor pathogen infection (Yamada, 1993; Jameson, 2000; Spaepen et al., 2007). Recent molecular genetics studies and global gene expression experiments have implicated auxin as an important disease susceptibility factor (Navarro et al., 2006; Siemens et al., 2006; Thilmony et al., 2006). Consistent with those studies, our result further reveals an important role for GH3.5, an early auxin-responsive gene, in auxin-elicited susceptibility.
It is intriguing that GH3.5 positively regulates SA signaling but weakens different R-gene-mediated local resistance to diverse avirulent strains in gh3.5-1D. We hypothesized that R-mediated resistance was counteracted by auxin-mediated susceptibility due to GH3.5 overexpression. Indeed, dfl1-D, a similar activation-tagged dwarf mutant that overexpresses GH3.6, showed impaired R-mediated resistance, which was also more susceptible to virulent strains than the wild type. The predicted decrease of basal disease resistance is likely not apparent in gh3.5-1D plants due to counteraction by the simultaneously enhanced SA pathway. This evidence also accounts for the normal R-induced SAR observed in gh3.5-1D plants in which a predicted enhanced SAR response, indicated by increased SA levels and PR-1 induction, was counteracted by susceptibility events.
Regulation of auxin biosynthesis is a key step in the auxin-mediated response. Interestingly, GH3.5 positively regulates IAA accumulation during pathogen infection revealed by the IAA levels, and our microarray assays showed that several genes involved in IAA biosynthesis were more strongly up-regulated in gh3.5-1D than in wild-type plants after pathogen infection. This finding suggests that GH3.5 regulates IAA biosynthesis-related genes to increase IAA accumulation in planta during infection, in addition to IAA generation by P. syringae that harbors the iaaM and iaaH genes encoding enzymes catalyzing the conversion of Trp to IAA via indole-3-acetamide (Glickmann et al., 1998; Buell et al., 2003). Furthermore, microarray assays also showed that GH3.5 overexpression suppresses expression of Aux/IAA genes, thus likely derepressing the auxin signaling pathway, consistent with the previous observation in wild-type Col-0 plants infected with Pst DC3000 and Plasmodiophora brassicae (Siemens et al., 2006; Thilmony et al., 2006). An important outcome of regulation of auxin signaling by GH3.5 in infected gh3.5-1D plants is the up-regulation of genes involved in metabolism and transport of nutrition across the plasma membrane (Fig. 8; Supplemental Fig. S6). How P. syringae, which colonizes the apoplast, modifies the host metabolism to promote nutrient synthesis or release to sustain its growth is a fundamental question that remains to be answered (Alfano and Collmer, 1996). Future research directed toward the detection of metabolism levels during pathogen infection and functional analysis of those transporter genes would be of great interest.
Could GH3.5 Regulate SA and Auxin Homeostasis during Pathogen Infection?
In plants, free SA and IAA accumulate at very low levels and most of the SA and IAA are found in conjugated forms (Sticher et al., 1997; Ljung et al., 2002). SA can form SA-glucoside (SAG) and methyl salicylate in various plants (Sticher et al., 1997). The roles of these conjugated forms of salicylates are diverse: SAG is not active in disease resistance and considered a main storage form of SA (Ryals et al., 1996); methyl salicylate acts as an airborne signal in SAR (Shulaev et al., 1997). SA-Asp, the only known amido derivative of SA, has been reported in grape (Vitis vinifera) and bean (Phaseolus vulgaris; Steffan et al., 1988; Bourne et al., 1991). As GH3.5 exhibited adenylation activity on SA in vitro, it is possible GH3.5 can form SA-amino acid conjugates in vivo. Consistent with this prediction, we have shown that SA-Asp is also present in Arabidopsis and GH3.5 positively regulates the SA-Asp levels in gh3.5-1D. However, we did not observe a lower SA-Asp level in the GH3.5 loss-of-function mutant (Supplemental Fig. S4), suggesting that GH3.5 might not be the only amido synthase for SA-Asp. Because there is no information available on SA-Asp function in planta, it is currently unclear whether SA-Asp is involved in defense responses in gh3.5-1D. Recently, another member of the GH3 family, GH3.12/PBS3, was shown to regulate SA and SAG levels, suggesting its important role in SA metabolism (Jagadeeswaran et al., 2007; Nobuta et al., 2007). Further analysis of SA-Asp might provide insight into the biochemical basis of SA metabolism and signaling.
IAA can form conjugates with sugars, amino acids, and small peptides. It has been shown that IAA conjugation is involved in IAA transport, storage, and metabolism (Ljung et al., 2002). Earlier studies reported that certain members of the GH3 family, including GH3.5, encode IAA-amido synthases that might maintain IAA homeostasis by converting excess auxin into amino acid conjugates that are either inactive or degraded (Staswick et al., 2005). This finding has contributed to our understanding of auxin homeostasis. However, overexpression of GH3.6 in dfl1-D does not alter the IAA level despite increased IAA-Asp accumulation (Staswick et al., 2005). Similar to dfl1-D, gh3.5-1D accumulated the same level of IAA as the wild type. Moreover, IAA levels were significantly elevated after induction by P. syringae. One possible interpretation is that GH3.5 may act differently in regulating IAA homeostasis in different plant physiological processes. In support of this hypothesis, transgenic tobacco (Nicotiana tabacum) overexpressing the iaaL gene, which encodes an IAA-Lys synthase converting IAA to IAA-Lys (Roberto et al., 1990), could decrease, have no effect on, or increase IAA levels depending on the organs and developmental stages of plants (Spena et al., 1991). Unlike in dfl1-D, the amount of IAA-Asp in gh3.5-1D was similar to the wild type even after infection by pathogens. Induction of two IAA-amido hydrolase ILR1 and ILL5 genes by Pst DC3000(avrRpt2) in our microarray assays, which can cleave IAA-amino acid conjugates to free IAA (Bartel and Fink, 1995), may reflect the possibility that IAA-amino acid conjugates transiently form and rapidly hydrolyze in gh3.5-1D, explaining our failure to detect the elevation of three typical IAA-amino acid conjugates in these tissues. On the other hand, gh3.5-1D was less responsive than the wild type, whereas T-DNA knockout mutants became more sensitive to exogenous IAA and naphthylacetic acid (NAA; Supplemental Fig. S1). This suggests that the GH3.5 protein could remove excessive auxin through its activity of amido synthetase.
Dual Roles of GH3.5 in Arabidopsis-P. syringae Interaction
The results presented here reveal that GH3.5 acts as a bifunctional modulator in two distinct signaling pathways during infection by P. syringae: the SA-mediated pathway for disease resistance and the IAA-mediated pathway for disease susceptibility. Intriguingly, two recent papers showed that GH3.12/PBS3 positively regulates SA-dependent disease resistance (Jagadeeswaran et al., 2007; Nobuta et al., 2007). Those results widen our understanding of GH3 functions in planta. A similar activation-tagged mutant of GH3.5, wes1-D, was recently reported to exhibit enhanced disease resistance to Pst DC3000 at the flowering stage with spray inoculation under long-day conditions (Park et al., 2007), which is not observed in our studies. Given the fact that flowering plants are more resistant than young or nonflowering plants, the enhanced resistance observed in wes1-D could result from the flowering mutant plants compared with the nonflowering wild-type plants (Park et al., 2007). Another possibility for this inconsistency might be due to the different inoculation procedures: spray inoculation used in the wes1-D study and injection inoculation used in our study. As plant stomata can act as a barrier against bacterial invasion (Melotto et al., 2006) and GH3.5 is induced by abscisic acid and involved in abscisic acid response, it is also possible that the difference observed in resistance to Pst DC3000 in gh3.5-1D and wes1-D resulted from the potential GH3.5-mediated stomata-based immunity. Those growth conditions and the different flowering times between wes1-D and the wild type might also account for the altered IAA and IAA-Asp levels observed in wes1-D that were not detected in our experiments under short-day conditions with the same flowering time of GH3.5-1D and Col-0 because IAA levels differ greatly with growth times and tissues as previously described (Spena et al., 1991).
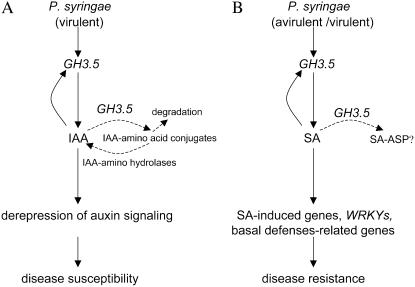
Based on our current data, we propose a functional model for GH3.5 in Arabidopsis-P. syringae interactions (Fig. 9). During virulent pathogen infection in the wild type, bacterial growth requires an increase in nutrition. Pathogens induce GH3.5 expression, whereas GH3.5 modulates the auxin pathway to enhance disease susceptibility by increasing IAA production and derepressing the auxin pathway. At the same time, increased IAA level feedback regulates GH3.5 expression, resulting in an amplifying effect on auxin response (Fig. 9A). To maintain endogenous IAA at an appropriate level, GH3.5 may act as an IAA-amido synthetase to form IAA-amido conjugates, which are either degraded or hydrolyzed to cycle IAA. However, this infection process must pay a penalty: GH3.5 simultaneously modulates the SA pathway to enhance SA accumulation and activate defense-related genes, resulting in increased defense responses (Fig. 9B). There also is a positive feedback loop in this process: SA elevates expression of GH3.5 to amplify plant defense responses. During avirulent pathogen infection, SA accumulation is rapidly induced, whereas IAA does not accumulate; thus, GH3.5 might mainly modulate the SA pathway to enhance disease resistance.
Figure 9.
Function model of GH3.5 in Arabidopsis-P. syringae interactions. A, In the compatible interaction, GH3.5 is activated to modulate the auxin pathway resulting in enhanced disease susceptibility through increasing IAA biosynthesis and derepressing auxin signaling. IAA also induces GH3.5 to enlarge those processes. In addition, GH3.5 might also function as an IAA-amido synthetase to regulate IAA homeostasis. B, In the compatible/incompatible interactions, GH3.5 positively modulates the SA pathway to enhance plant defense response through elevating SA biosynthesis, activating SA-induced genes, WRKYs, and basal defense-related genes. Feedback regulation of GH3.5 by SA amplifies those effects. In this case, GH3.5 might also synthesize SA-Asp with unknown function during the interactions.
The opposing functions of GH3.5 in disease resistance and susceptibility might have an evolutionary benefit, where ancestors of GH3.5 and some other GH3s like GH3.6 function primarily to regulate the auxin pathway to enhance host susceptibility at the early stage of coevolution with pathogens, and GH3.5 evolved to regulate the SA pathway to enhance resistance for host survival. The refined and complex function of GH3.5 in the plant-P. syringae interaction reflects both host defense and pathogen nutrient acquisition strategies.
MATERIALS AND METHODS
Characterization of gh3.5-1D and T-DNA Insertion Mutants and Plant Growth
The gh3.5-1D mutant was isolated from the activation-tagging library that was constructed with the plasmid pSKI015 in ecotype Col-0 as described (Li et al., 2005). A plasmid rescue method was used to clone the GH3.5 gene according to Nakazawa et al. (2001). The rescued genomic DNA was sequenced and compared with the genomic sequence in GenBank. The SALK T-DNA insertion (knockout) mutants, gh3.5-1 (SALK_014376) and gh3.5-2 (SALK_151766), were obtained from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org). The insertional sites for gh3.5-1 and gh3.5-2 were confirmed by sequencing flanking regions. Plants were grown in a growth chamber at 22°C to 23°C, 60% relative humidity, 85 μmol s−1 m−2 fluorescent illumination, with 9-h day/15-h night for pathogen inoculation, and 16-h day/8-h night for physiological and genetic analysis.
Overexpression of GH3.5
Full-length GH3.5 cDNA was generated by RT-PCR with the primers 5′-CTAGACTTCTCTCTTTCTCTTAAAC-3′ and 5′-GGATCCACATTCCATCTTAGTTAC-3′, and was inserted into the expression vector p2300S (provided by Prof. Yinong Yang, University of Arkansas) to generate p35S∷GH3.5. This construct was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis (Arabidopsis thaliana; Col-0) by floral dip with 0.01% Silwett L-77 to produce more than 50 independent GH3.5-overexpressing (p35S∷GH3.5) transgenic plants. Homozygous transgenic plants were selected in the progeny.
Bacterial Strains, Inoculation, and Disease Assessment
The following Pseudomonas syringae strains were used: the virulent strain Pst DC3000 and the avirulent strain Pst DC3000(avrRpt2) (provided by Prof. Andrew Bent), the avirulent strain Pst DC3000(avrRps4) (provided by Prof. Jianmin Zhou), and the virulent strain Psm and the avirulent strain Psm(avrRpm1) (provided by Prof. Frederick Ausubel). Bacterial strains were grown in King's B medium with shaking or on King's B medium agar plates at 28°C containing proper antibiotics. Bacteria were collected and suspended in 10 mm MgCl2. For inoculation, 5-week-old leaves were infiltrated with a bacterial suspension. In parallel, all procedural controls were infiltrated with 10 mm MgCl2 for mock inoculation. Methods for bacterial growth assays were performed as described (Katagiri et al., 2002). Heterozygous gh3.5-1D plants were used for all the experiments in the bacterial growth assay unless otherwise indicated.
SAR Assays
Two lower leaves of 5-week-old plants were inoculated with Psm(avrRpm1) bacterial suspension in 10 mm MgCl2 at 107 cfu/mL. Three upper leaves were infiltrated with Pst DC3000 at 105 cfu/mL 3 d later. Bacterial growth was assayed in the secondary infected leaves. Bacterial growth titers were counted at 0 and 3 dpi.
RNA and Protein Analysis
Total RNA was isolated from leaf tissues using TRIzol reagent according to the manufacturer's instructions (GIBCO-BRL). Ten micrograms of RNA samples were separated on a 1% formaldehyde-agarose gel and then blotted onto Hybond-N+ membranes (Amersham). A 353-bp fragment of the GH3.5 transcript was amplified from genomic DNA using the primers 5′-TAATCAGTATAAGACGCCGAGATGC-3′ and 5′-TCGAGAAAGAGTGATGAGAGTTGGTT-3′ and was labeled with [α-32P]dCTP using a random primer labeling kit (TaKaRa) for hybridization and autoradiography. Northern-blot analysis was also performed to determine the transcript levels of the PR genes PR-1 (At2g14610) and PDF1.2 (At5g44420). The filters were reprobed with a 2.5-kb fragment of 18S Arabidopsis rDNA for loading normalization. The same GH3.5 primers were also used for RT-PCR to detect the GH3.5 transcripts in the T-DNA knockout mutants, with the following cycle conditions: 94°C for 4 min, followed by 32 cycles of 94°C for 20 s, 65°C for 40 s, and 72°C for 30 s, with an elongation step of 72°C for 10 min. Arabidopsis actin cDNA served as an internal control with the primers 5′-TGGCATCAT/CACTTTCTACAA-3′ and 5′-CCACCACTA/G/TAGCACAATGTT-3′. The full-length coding region of GH3.5 (612 amino acids) was ligated into pET-32a to produce the fusion protein in the Escherichia coli strain DE3. The GH3.5 fusion protein was used to immunize rabbit to produce antiserum. Western blotting was performed using the SuperSignal West chemiluminescence kit according to the manufacturer's protocol (Pierce).
SA and SA-Asp Measurement
Leaves of Col-0, gh3.5-1D, gh3.5-2, dfl1-D, and Landsberg erecta (Ler; the wild-type for dfl1-D) plants infected with Psm(avrRpm1) or PstDC3000(avrRpt2) at 107 cfu/mL or with 10 mm MgCl2 (mock) were harvested at 0 (control) and 48 hpi. Free and total SA were extracted from leaf tissues and analyzed by HPLC according to the method described by Dewdney et al. (2000). For SA-Asp measurement, standard SA-Asp was synthesized according to the method described by Bourne et al. (1991) and quantification of SA-Asp was also performed by HPLC accordingly.
Extraction and Quantification of Auxins
Three leaves of 5-week-old Col-0 and gh3.5-1D plants were inoculated with Pst DC3000 and Pst DC3000(avrRpt2) at 105 cfu/mL or with 10 mm MgCl2 (mock). Leaves were harvested 3 dpi and lyophilized. Quantitative analysis of IAA and IAA-amino acid conjugates was performed as previously described (Staswick et al., 2005).
Microarray Analysis
Leaves of plants of 5-week-old wild-type and gh3.5-1D were inoculated with Pst DC3000(avrRpt2) at 105 cfu/mL. Leaves were harvested at 0 (uninoculated) and 48 hpi and stored in liquid nitrogen. Microarray analysis was performed with the oligonucleotide Affymetrix ATH1 chip. RNA preparation and microarray experiments were performed by the Affymetrix-authorized Center (GeneTech Biotechnology Ltd.) following the protocol of Affymetrix GeneChip Expression Analysis Overview with three biological replicates, and data were analyzed according to GeneChip Expression Analysis Data Analysis Fundamentals.
Raw data were analyzed with Affymetrix GeneChip Operating Software (GCOS; version 1.4) using Affymetrix default analysis settings and global scaling as a normalization method. The trimmed mean target intensity of each array was arbitrarily set to 100. Data were then compared between sample chips from the same biological replicate producing a signal log2 ratio (SLR), a change call, and a change P value, which was calculated from the GeneChip fluorescence signal intensity data using the Affymetrix GCOS software. SLRs, change calls, and change P values were determined for each inoculation sample compared with its corresponding mock control.
Reproducibly differentially expressed probe sets were selected from total normalized data (Supplemental Table S1), based on a SLR of at least 1.0, a gene expression change call of I (increase) or MI (marginal increase), and a P value <0.01; or a SLR of at least −1.0, a gene expression change call of D (decrease) or MD (marginal decrease), and a P value >0.99 for all three biological replicate pairs, following stringent selection criteria (Thilmony et al., 2006). Some of the differentially expressed data were selected for RT-PCR analysis.
Hierarchical clustering of auxin-related genes was performed using Cluster, version 2.20, with the default settings with the complete linkage clustering algorithm method selected, and TreeView, version 1.60 (Eisen et al., 1998; http://rana.lbl.gov/EisenSoftware.htm). MAPMAN, version 1.6.0 (Thimm et al., 2004), was used for analysis of the functional classes and metabolic pathways following pathogen inoculation. The complete set of microarray data has been deposited to the National Center for Biotechnology Information (NCBI) GEO database (http://www.ncbi.nlm.nih.gov/geo) in a MIAME-compliant format (GSE6556 and GSM151694–GSM151705).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root growth and auxin responses.
Supplemental Figure S2. Disease symptoms on leaves of Col-0 (left) and gh3.5-1D (right) at 3 dpi with Pst DC3000(avrRpt2) (105 cfu/mL).
Supplemental Figure S3. SA levels in local and systemic leaves of Col-0, gh3.5-1D, and gh3.5-2 plants after infection with Pst DC3000(avrRpt2).
Supplemental Figure S4. SA-Asp levels in local and systemic leaves of Col-0, gh3.5-1D, and gh3.5-2 plants after infection with Psm(avrRpm1).
Supplemental Figure S5. Growth of avirulent strain Pst DC3000(avrRpt2) in Col-0, gh3.5-1, and gh3.5-2 plants.
Supplemental Figure S6. Regulation of transporter genes in gh3.5-1D plants.
Supplemental Figure S7. Expression of seven defense- and auxin-related genes in wild-type and gh3.5-1D plants.
Supplemental Table S1. Summary of normalized SLR expression data from the microarray experiments.
Supplemental Table S2. Summary of 273 reproducibly differentially regulated genes in uninfected gh3.5-1D versus wild-type plants.
Supplemental Table S3. Summary of 831 reproducibly differentially regulated genes in Pst DC3000(avrRpt2)-infected gh3.5-1D versus wild-type plants.
Supplemental Table S4. Regulation of auxin-related genes in gh3.5-1D as compared to wild-type plants.
Supplementary Material
Acknowledgments
We thank Xinnan Dong for the cpr1, cpr5, and cpr6 mutants, and Miki Nakazawa for dfl1-D. We also acknowledge Yiji Xia, Jianming Li, and Hai Huang for critical reading of the manuscript, and Ligeng Ma for suggestions regarding microarray experiments.
This work was supported by grants from the National Basic Research Program of China (2003CB114300), National Natural Science Foundation of China (30730064 and 30421001), and the Shanghai Municipal Science and Technology Commission to Z.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zuhua He (zhhe@sibs.ac.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A (1996) Bacterial pathogens in plants: life up against the wall. Plant Cell 8 1683–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983 [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268 1745–1748 [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265 1856–1860 [DOI] [PubMed] [Google Scholar]
- Bourne DJ, Barrow KD, Milborrow BV (1991) Salicyloylaspartate as an endogenous component in the leaves of Phaseolus vulgaris. Phytochemistry 30 4041–4044 [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Cuzick A, Moeder W, Tang D, Innes RW, Klessig DF, McDowell JM, Kunkel BN (2004) The Pseudomonas syringae type III effector AvrRpt2 functions downstream or independently of SA to promote virulence on Arabidopsis thaliana. Plant J 37 494–504 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- de León IP, Sanz A, Hamberg M, Castresana C (2002) Involvement of the Arabidopsis α-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J 29 61–72 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al (1994) A central role of salicylic acid in plant disease resistance. Science 266 1247–1250 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Shah J, Klessig DF (1999) Salicylic acid and disease resistance in plants. Crit Rev Plant Sci 18 547–575 [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24 205–218 [DOI] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51 21–37 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261 754–756 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20 265–277 [DOI] [PubMed] [Google Scholar]
- Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y (1998) Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 11 156–162 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269 843–846 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49 373–385 [PubMed] [Google Scholar]
- Jagadeeswaran G, Raina S, Acharya BR, Maqbool SB, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R (2007) Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J 51 234–246 [DOI] [PubMed] [Google Scholar]
- Jameson PE (2000) Cytokinins and auxins in plant-pathogen interactions—an overview. Plant Growth Regul 32 369–380 [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0039, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Kunkel BN, Agnew J, Collins JJ, Cohen J, Chen ZY (2005) Molecular genetic analysis of AvrRpt2 activity in promoting virulence of Pseudomonas syringae. In S Tsuyumu, JE Leach, T Shiraishi, T Wolpert, eds, Genomic and Genetic Analysis of Plant Parasitism and Defense. American Phytopathological Society, St. Paul, pp 92–102
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Li ZM, Cao JS, Zhang HK, He ZH (2005) Construction of an activation tagging library of Arabidopsis and cloning for mutant genes. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 31 499–506 [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 49 249–272 [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126 969–980 [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu G-L, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mudgett MB (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56 509–531 [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25 213–221 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF III, Dangl JL (2003) Recognition and response in the plant immune system. Annu Rev Genet 37 579–609 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, Innes RW (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol 144 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Melotto M, He SY (2005) Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol 8 361–368 [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Schmelz EA, Moussatche P, Lund ST, Jones JB, Klee HJ (2003) Susceptible to intolerance—a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J 33 245–257 [DOI] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282 10036–10046 [DOI] [PubMed] [Google Scholar]
- Roberto FF, Klee H, White F, Nordeen R, Kosuge T (1990) Expression and fine structure of the gene encoding Nɛ-(indole-3-acetyl)-L-lysine synthetase from Pseudomonas savastanoi. Proc Natl Acad Sci USA 87 5797–5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM (1997) Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Silverman P, Raskin I (1997) Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385 718–721 [Google Scholar]
- Siemens J, Keller I, Sarx J, Kunz S, Schuller A, Nagel W, Schmulling T, Parniske M, Ludwig-Muller J (2006) Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol Plant Microbe Interact 19 480–494 [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31 425–448 [DOI] [PubMed] [Google Scholar]
- Spena A, Prinsen E, Fladung M, Schulze SC, Van Onckelen H (1991) The indole acetic acid-lysine synthetase gene of Pseudomonas syringae subsp. savastanoi induces developmental alterations in transgenic tobacco and potato plants. Mol Gen Genet 227 205–212 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan H, Ziegler A, Rapp A (1988) N-Salicyloyl-aspartic acid: a new phenolic compound in grapevines. Vitis 27 79–86 [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP (1997) Systemic acquired resistance. Annu Rev Phytopathol 35 235–270 [DOI] [PubMed] [Google Scholar]
- Summermatter K, Sticher L, Métraux JP (1995) Systemic responses in Arabidopsis thaliana infected and challenged with Pseudomonas syringae pv. syringae. Plant Physiol 108 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Mochizuki N, Nagatani A (2002) Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol 43 281–289 [DOI] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46 34–53 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T (1993) The role of auxin in plant-disease development. Annu Rev Phytopathol 31 253–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.