Abstract
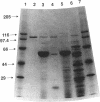
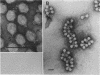
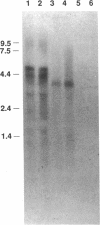
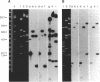
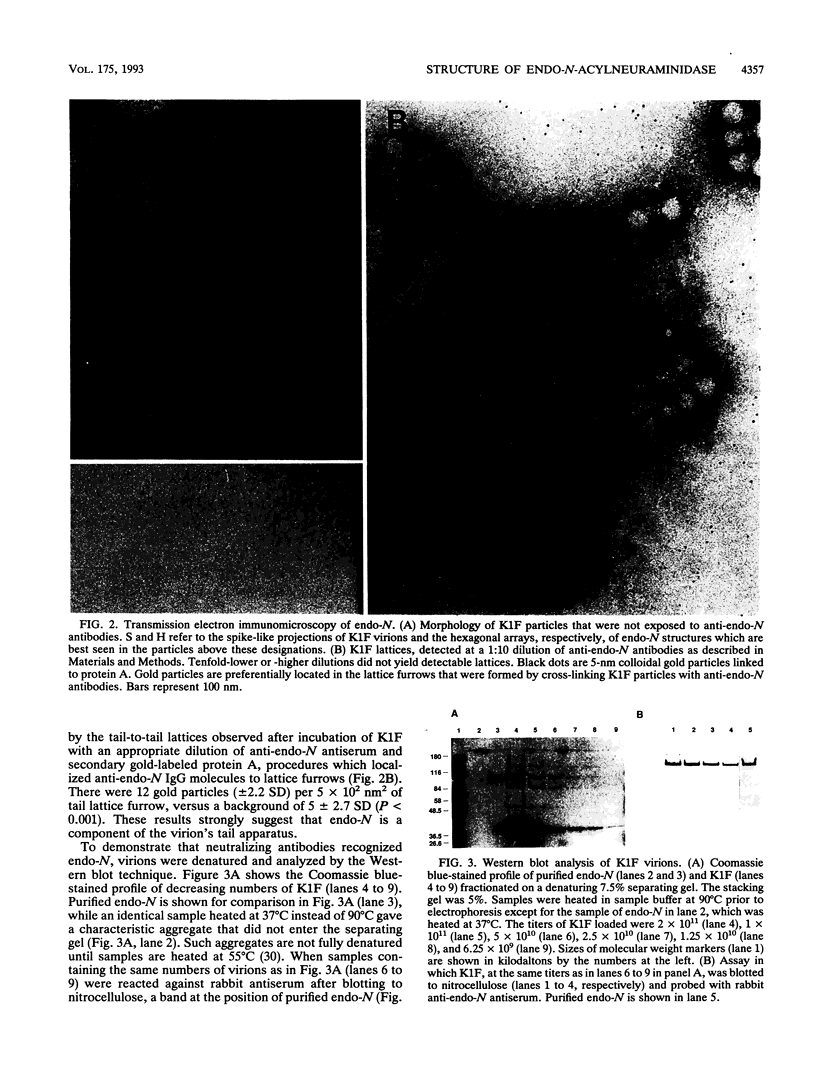
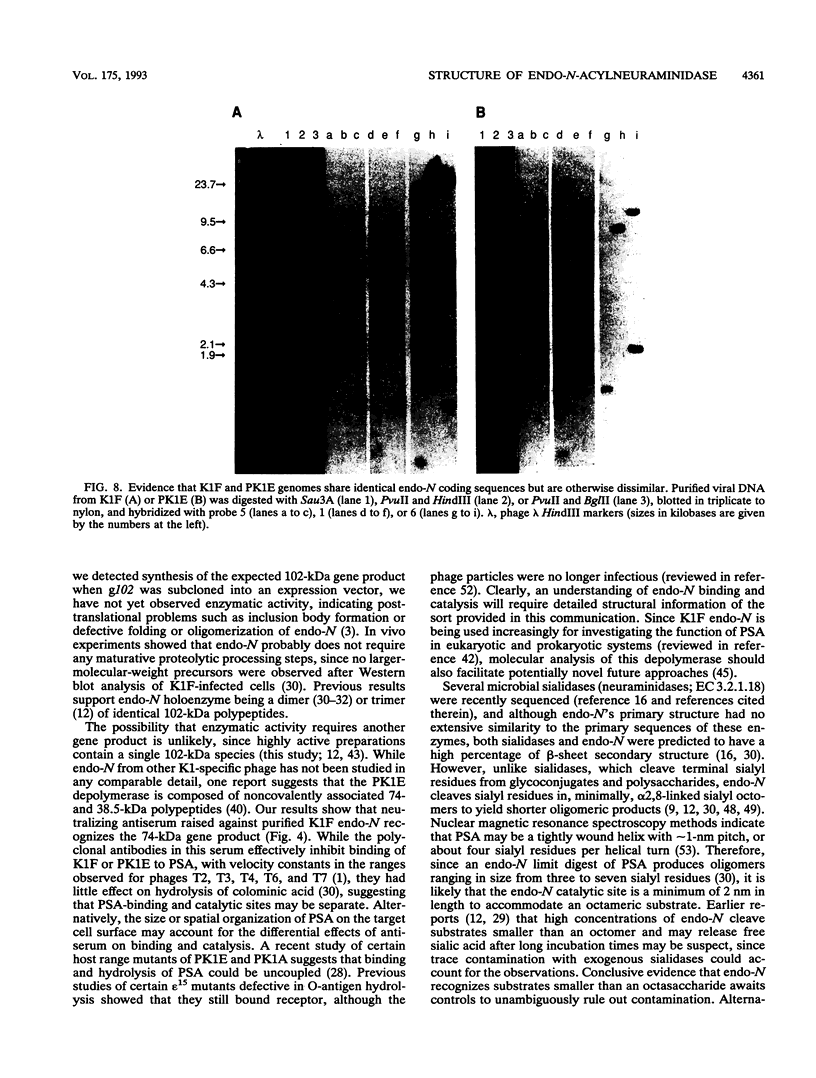
Endo-N-acylneuraminidase (endo-N) is a phage-encoded depolymerase that degrades the alpha (2-8)-linked polysialic acid chains of K1 serotypes of Escherichia coli and vertebrate neural cell adhesion molecules. We have determined the DNA sequence of the bacteriophage K1F tail protein structural gene, which codes for a polypeptide of 920 residues. Purification of the tail protein yields a 102-kDa species upon denaturing gel electrophoresis and detection by Western immunoblot analysis. An identical polypeptide was detected by Western blot analysis of K1F virions. Peptide sequencing confirmed that the open reading frame determined by nucleotide sequencing encodes endo-N. Immunoelectron microscopy with neutralizing antibodies raised against the depolymerase confirmed that endo-N is a component of the K1F tail apparatus. Antibodies in the serum cross-reacted with endo-N from another K1-specific phage, PK1E, demonstrating the presence of shared epitopes. Homology between K1F and PK1E endo-N was confirmed by Southern, Northern (RNA), and Western blot analyses. The endo-N amino-terminal domain is homologous to the amino termini of phage T7 and T3 tail proteins, indicating by analogy that this domain functions in attachment of endo-N to the K1F virion's head. A central domain of 495 residues has weak similarity to sea urchin aryl sulfatase, suggesting that this region may contain the endo-N catalytic site. Failure to detect homology between the PK1E homolog and the carboxy-terminal domain of K1F endo-N is consistent with the central domain's involvement in binding and catalysis of polysialic acid. These results provide the initial molecular and genetic description of polysialic acid depolymerase, which has so far been detected only in K1-specific phage.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradley D. E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967 Dec;31(4):230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Finne J., Mäkelä P. H. Cleavage of the polysialosyl units of brain glycoproteins by a bacteriophage endosialidase. Involvement of a long oligosaccharide segment in molecular interactions of polysialic acid. J Biol Chem. 1985 Jan 25;260(2):1265–1270. [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggård-Ljungquist E., Halling C., Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992 Mar;174(5):1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenbeck P. C., Vimr E. R., Yu F., Bassler B., Troy F. A. Purification and properties of a bacteriophage-induced endo-N-acetylneuraminidase specific for poly-alpha-2,8-sialosyl carbohydrate units. J Biol Chem. 1987 Mar 15;262(8):3553–3561. [PubMed] [Google Scholar]
- Hendrix R. W. Purification and properties of groE, a host protein involved in bacteriophage assembly. J Mol Biol. 1979 Apr 15;129(3):375–392. doi: 10.1016/0022-2836(79)90502-3. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer L. L., Hamilton A. C., Steenbergen S. M., Vimr E. R. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol Microbiol. 1992 Apr;6(7):873–884. doi: 10.1111/j.1365-2958.1992.tb01538.x. [DOI] [PubMed] [Google Scholar]
- Hoyer L. L., Roggentin P., Schauer R., Vimr E. R. Purification and properties of cloned Salmonella typhimurium LT2 sialidase with virus-typical kinetic preference for sialyl alpha 2----3 linkages. J Biochem. 1991 Sep;110(3):462–467. doi: 10.1093/oxfordjournals.jbchem.a123603. [DOI] [PubMed] [Google Scholar]
- Häyrinen J., Bitter-Suermann D., Finne J. Interaction of meningococcal group B monoclonal antibody and its Fab fragment with alpha 2-8-linked sialic acid polymers: requirement of a long oligosaccharide segment for binding. Mol Immunol. 1989 Jun;26(6):523–529. doi: 10.1016/0161-5890(89)90003-5. [DOI] [PubMed] [Google Scholar]
- Kornblum J. S., Projan S. J., Moghazeh S. L., Novick R. P. A rapid method to quantitate non-labeled RNA species in bacterial cells. Gene. 1988;63(1):75–85. doi: 10.1016/0378-1119(88)90547-1. [DOI] [PubMed] [Google Scholar]
- Krüger D. H., Schroeder C. Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol Rev. 1981 Mar;45(1):9–51. doi: 10.1128/mr.45.1.9-51.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski B., Boschek B., Thiele H., Stirm S. Substrate specificity of two bacteriophage-associated endo-N-acetylneuraminidases. J Virol. 1983 Jan;45(1):367–374. doi: 10.1128/jvi.45.1.367-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L., Dahm L., Tang J. C., Rutishauser U. Polysialic acid as a regulator of intramuscular nerve branching during embryonic development. Neuron. 1990 May;4(5):655–667. doi: 10.1016/0896-6273(90)90193-j. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lu H. S., Klein M. L., Lai P. H. Narrow-bore high-performance liquid chromatography of phenylthiocarbamyl amino acids and carboxypeptidase P digestion for protein C-terminal sequence analysis. J Chromatogr. 1988 Aug 26;447(2):351–364. [PubMed] [Google Scholar]
- Manfioletti G., Schneider C. A new and fast method for preparing high quality lambda DNA suitable for sequencing. Nucleic Acids Res. 1988 Apr 11;16(7):2873–2884. doi: 10.1093/nar/16.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierendorf R. C., Percy C., Young R. A. Gene isolation by screening lambda gt11 libraries with antibodies. Methods Enzymol. 1987;152:458–469. doi: 10.1016/0076-6879(87)52054-7. [DOI] [PubMed] [Google Scholar]
- Pelkonen S., Aalto J., Finne J. Differential activities of bacteriophage depolymerase on bacterial polysaccharide: binding is essential but degradation is inhibitory in phage infection of K1-defective Escherichia coli. J Bacteriol. 1992 Dec;174(23):7757–7761. doi: 10.1128/jb.174.23.7757-7761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkonen S., Pelkonen J., Finne J. Common cleavage pattern of polysialic acid by bacteriophage endosialidases of different properties and origins. J Virol. 1989 Oct;63(10):4409–4416. doi: 10.1128/jvi.63.10.4409-4416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U., Acheson A., Hall A. K., Mann D. M., Sunshine J. The neural cell adhesion molecule (NCAM) as a regulator of cell-cell interactions. Science. 1988 Apr 1;240(4848):53–57. doi: 10.1126/science.3281256. [DOI] [PubMed] [Google Scholar]
- Rutishauser U., Jessell T. M. Cell adhesion molecules in vertebrate neural development. Physiol Rev. 1988 Jul;68(3):819–857. doi: 10.1152/physrev.1988.68.3.819. [DOI] [PubMed] [Google Scholar]
- Rutishauser U., Watanabe M., Silver J., Troy F. A., Vimr E. R. Specific alteration of NCAM-mediated cell adhesion by an endoneuraminidase. J Cell Biol. 1985 Nov;101(5 Pt 1):1842–1849. doi: 10.1083/jcb.101.5.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Yamada K., Akasaka K., Kawasaki H., Suzuki K., Saito A., Sato M., Shimada H. cDNA cloning, nucleotide sequence and expression of the gene for arylsulfatase in the sea urchin (Hemicentrotus pulcherrimus) embryo. Eur J Biochem. 1988 Oct 15;177(1):9–13. doi: 10.1111/j.1432-1033.1988.tb14338.x. [DOI] [PubMed] [Google Scholar]
- Steenbergen S. M., Vimr E. R. Mechanism of polysialic acid chain elongation in Escherichia coli K1. Mol Microbiol. 1990 Apr;4(4):603–611. doi: 10.1111/j.1365-2958.1990.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Steven A. C., Trus B. L., Maizel J. V., Unser M., Parry D. A., Wall J. S., Hainfeld J. F., Studier F. W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988 Mar 20;200(2):351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- Tomlinson S., Taylor P. W. Neuraminidase associated with coliphage E that specifically depolymerizes the Escherichia coli K1 capsular polysaccharide. J Virol. 1985 Aug;55(2):374–378. doi: 10.1128/jvi.55.2.374-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy F. A., 2nd Polysialylation: from bacteria to brains. Glycobiology. 1992 Feb;2(1):5–23. doi: 10.1093/glycob/2.1.5. [DOI] [PubMed] [Google Scholar]
- Troy F. A., Hallenbeck P. C., McCoy R. D., Vimr E. R. Detection of polysialosyl-containing glycoproteins in brain using prokaryotic-derived probes. Methods Enzymol. 1987;138:169–185. doi: 10.1016/0076-6879(87)38014-0. [DOI] [PubMed] [Google Scholar]
- Varki A., Hooshmand F., Diaz S., Varki N. M., Hedrick S. M. Developmental abnormalities in transgenic mice expressing a sialic acid-specific 9-O-acetylesterase. Cell. 1991 Apr 5;65(1):65–74. doi: 10.1016/0092-8674(91)90408-q. [DOI] [PubMed] [Google Scholar]
- Vimr E. R., Aaronson W., Silver R. P. Genetic analysis of chromosomal mutations in the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1989 Feb;171(2):1106–1117. doi: 10.1128/jb.171.2.1106-1117.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R., McCoy R. D., Vollger H. F., Wilkison N. C., Troy F. A. Use of prokaryotic-derived probes to identify poly(sialic acid) in neonatal neuronal membranes. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1971–1975. doi: 10.1073/pnas.81.7.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R. Selective synthesis and labeling of the polysialic acid capsule in Escherichia coli K1 strains with mutations in nanA and neuB. J Bacteriol. 1992 Oct;174(19):6191–6197. doi: 10.1128/jb.174.19.6191-6197.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R., Troy F. A. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol. 1985 Nov;164(2):845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki R., Bacon B. Three-dimensional structural analysis of the group B polysaccharide of Neisseria meningitidis 6275 by two-dimensional NMR: the polysaccharide is suggested to exist in helical conformations in solution. Biochemistry. 1991 Jan 22;30(3):851–857. doi: 10.1021/bi00217a039. [DOI] [PubMed] [Google Scholar]