Abstract
Plant cell wall invertases and fructan exohydrolases (FEHs) are very closely related enzymes at the molecular and structural level (family 32 of glycoside hydrolases), but they are functionally different and are believed to fulfill distinct roles in plants. Invertases preferentially hydrolyze the glucose (Glc)-fructose (Fru) linkage in sucrose (Suc), whereas plant FEHs have no invertase activity and only split terminal Fru-Fru linkages in fructans. Recently, the three-dimensional structures of Arabidopsis (Arabidopsis thaliana) cell wall Invertase1 (AtcwINV1) and chicory (Cichorium intybus) 1-FEH IIa were resolved. Until now, it remained unknown which amino acid residues determine whether Suc or fructan is used as a donor substrate in the hydrolysis reaction of the glycosidic bond. In this article, we present site-directed mutagenesis-based data on AtcwINV1 showing that the aspartate (Asp)-239 residue fulfills an important role in both binding and hydrolysis of Suc. Moreover, it was found that the presence of a hydrophobic zone at the rim of the active site is important for optimal and stable binding of Suc. Surprisingly, a D239A mutant acted as a 1-FEH, preferentially degrading 1-kestose, indicating that plant FEHs lacking invertase activity could have evolved from a cell wall invertase-type ancestor by a few mutational changes. In general, family 32 and 68 enzymes containing an Asp-239 functional homolog have Suc as a preferential substrate, whereas enzymes lacking this homolog use fructans as a donor substrate. The presence or absence of such an Asp-239 homolog is proposed as a reliable determinant to discriminate between real invertases and defective invertases/FEHs.
The nonreducing disaccharide Suc (α-d-Glc-(1 → 2)-β-d-Fru; Fig. 1A) is, besides starch, one of the most common reserve carbohydrates in plants, playing a key role in plant metabolism and development (Sturm and Tang, 1999). Suc is synthesized in source leaves during photosynthesis and is thereof transported to sink tissues. Upon arrival into sink cells, Suc can serve as a source of carbon for the synthesis of polysaccharides, such as starch and fructans, or as a source of energy through respiration. Metabolic use of Suc as a carbon-building block or energy source and its channeling into the sink metabolism require the cleavage of the α1-β2 glycosidic bond. In plants, this reaction can be catalyzed by two distinct enzymes: Suc synthase (EC 2.4.1.13) and invertases (EC 3.2.1.26). Suc synthase is a glycosyltransferase catalyzing the UDP-dependent reversible cleavage of Suc into UDP-Glc and Fru, whereas invertase irreversibly hydrolyzes Suc into Glc and Fru. Because the substrate and reaction products of invertases are not only important nutrients, but also regulators of different classes of genes, invertases are fundamental enzymes in the control of cell differentiation and plant development (Koch, 1996).
Figure 1.
Schematic structure of Suc (A) and 1-kestose (B).
Based on their subcellular localization, pH optima, solubility, and isoelectric point, three different types of invertase isoenzymes could be distinguished: vacuolar, cell wall-bound, and cytoplasmic invertases (Tymowska-Lananne and Kreis, 1998). Cell wall invertases are key metabolic enzymes involved in the regulation of Suc partitioning (Eschrich, 1980; Sturm, 1999; Sherson et al., 2003; Roitsch and Gonzalez, 2004), developmental processes such as pollen and seed development (Cheng et al., 1996; Goetz et al., 2001), and the response to wounding and pathogen infection (Sturm and Crispeels, 1990; Benhamou et al., 1991; Fotopoulos et al., 2003; Ditt et al., 2006). In Arabidopsis (Arabidopsis thaliana), two vacuolar, six putative cell wall, and nine cytoplasmic invertase genes are present (Ji et al., 2005). However, two of the putative cell wall invertase genes turned out to encode fructan exohydrolases (FEHs), enzymes that hydrolyze terminal Fru moieties from fructans but not from Suc (De Coninck et al., 2005).
Both vacuolar and cell wall invertases are grouped together with fructan biosynthetic and degrading enzymes and microbial β-fructosidases in family 32 of glycoside hydrolases (GH32; www.cazy.org). Family GH32 can be combined in the clan GH-J together with the related family GH68 (Naumoff, 2001), harboring bacterial invertases, levansucrases, and inulosucrases. These enzymes use Suc as the preferential donor and transfer the fructosyl moiety to a variety of acceptors, including water (Suc hydrolysis), Suc, and fructan (levan or inulin). Within this GH-J clan, three-dimensional (3-D) structures of six enzymes have been determined, namely, the levansucrases from Bacillus subtilis (Meng and Fütterer, 2003; Protein Data Bank [PDB] code 1OYG) and Gluconacetobacter diazotrophicus (Martinez-Fleites et al., 2005; PDB code 1W18), an exoinulinase from Aspergillus awamori (Nagem et al., 2004; PDB code 1Y4W), a β-fructosidase from Thermotoga maritima (Alberto et al., 2004; PDB code 1UYP; Alberto et al., 2006; PDB code 1W2T), a fructan 1-exohydrolase (1-FEH) from chicory (Cichorium intybus; Verhaest et al., 2005b; PDB code 1ST8), and, most recently, a cell wall invertase from Arabidopsis (Verhaest et al., 2006; PDB code 2AC1). All these proteins show a common fold; they consist of an N-terminal five-bladed β-propeller domain (GH32 and GH68) followed by a C-terminal domain formed by two β-sheets (only in GH32). Superposition of these structures indicates that the active sites are located in the β-propeller domain. The active site is characterized by the presence of three highly conserved acidic groups that play a crucial role in the catalytic mechanism for the hydrolysis of the glycosidic bond (Reddy and Maley, 1990, 1996; Batista et al., 1999; Yanase et al., 2002).
A phylogenetic tree containing deduced amino acid sequences from vacuolar and cell wall invertases and fructan biosynthetic and breakdown enzymes revealed that FEHs are more related to cell wall invertases than to fructan biosynthetic enzymes, which, in turn, are more related to vacuolar invertases. FEHs and cell wall invertases are believed to have evolved from a common ancestor (Van den Ende et al., 2000). FEHs and invertases are both hydrolases but use different preferential donor substrates. Suc is the preferential donor substrate for cell wall invertases, but other donor substrates, such as 1-kestose (Fig. 1B), raffinose, and stachyose, were also reported to be degraded to a certain extent (Tymowska-Lananne and Kreis, 1998; De Coninck et al., 2005). It was reported that the side activity against raffinose is determined by a single amino acid (Goetz and Roitsch, 1999). Plant FEHs seem to be unifunctional enzymes, only degrading fructans but not Suc (Van Laere and Van den Ende, 2002), in contrast with microbial FEHs that can degrade Suc as well. Depending on the linkage type that is hydrolyzed, 1-FEH or inulinase (β-2,1 linkages) and 6-FEH or levanase (β-2,6 linkages) can be distinguished.
To gain insight into (1) which amino acids determine the preference of Glu-Fru linkage (Suc, raffinose, stachyose) hydrolysis versus Fru-Fru linkage (1-kestose and higher degree of polymerization [DP] fructans) hydrolysis and, more particularly, (2) which amino acids are important for the binding and degradation of Suc, site-directed mutagenesis studies were performed focusing on the identification of putative important residues as determined by sequence and 3-D structure comparisons. To our knowledge, this is the first report specifically considering the difference between GH32 hydrolases only differing in donor substrate specificity (Suc versus fructan). All studies were performed on Arabidopsis cell wall Invertase1 (AtcwINV1; Arabidopsis gene code At3g13790), a genuine cell wall invertase (De Coninck et al., 2005).
RESULTS AND DISCUSSION
The Active Site of AtcwINV1 Consists of Three Highly Conserved Amino Acids
By multiple-sequence alignments of members of GH32, GH68, and GH43 (harboring β-xylanases, β-xylosidases, α-l-arabinases, and α-l-arabinofuranosidases), and GH62 (harboring some α-l-arabinofuranosidases), three highly conserved amino acids, at an equivalent position in all the enzymes, can be identified (Pons et al., 2004). The general GH reaction mechanism requires at least two critical acidic residues, a proton donor, and a nucleophile. The reaction involves two major mechanisms resulting in either an overall retention (GH32 and GH68) or inversion (GH43) of the anomeric configuration (Davies and Henrissat, 1995). Several mutation experiments on enzyme members of these families and on a plant invertase in particular (Goetz and Roitsch, 2000), revealed a crucial role for Asp and Glu residues in substrate hydrolysis (Reddy and Maley, 1990, 1996; Batista et al., 1999; Nurizzo et al., 2002; Yanase et al., 2002; Meng and Fütterer, 2003; Ozimek et al., 2004; Altenbach et al., 2005; Ritsema et al., 2006). Recently, the 3-D structures of chicory 1-FEH IIa/AtcwINV1 were resolved, and the acidic residues Asp-22/Asp-23, Asp-147/Asp-149, and Glu-201/Glu-203 were considered as nucleophile, transition-state stabilizer, and acid/base catalyst, respectively (Verhaest et al., 2005b, 2006). These residues belong to the conserved NDPNG, RDP, and EC regions within the GH32 family (Fig. 2; Naumoff, 2001). Because the Asp of the RDP motif is considered to be too distant to take part in the catalytic process (Meng and Fütterer, 2003), we focused on Asp-23 and Glu-203 from AtcwINV1 and mutated them into Ala residues. The total activity, measured as the release of Fru from Suc, of wild-type and mutant enzymes, were compared and their kinetic parameters determined (Table I). Compared to the wild-type enzyme, the kcat for both mutant enzymes drastically decreased (600- to 900-fold), whereas the Km was modified to a lesser extent (8-fold). The large reduction in kcat values for both D23A and E203A mutants confirms that these residues are essential for catalysis in AtcwINV1. In accordance with previous studies on yeast (Saccharomyces cerevisiae) invertase (Reddy and Maley, 1996) in which the nucleophile and proton donor were identified, we conclude that the Asp-23 and Glu-203 residues in AtcwINV1 act as the nucleophile and proton donor in the hydrolysis reaction of Suc.
Figure 2.
Multiple sequence alignment (using EMBL-EBI ClustalW) of three conserved regions of some plant GH32 enzymes. The following sequences were used for the alignment: AtcwINV1, chicory vacuolar invertase, 1-FEH IIa, 1-SST, 1-FFT, and an invertase from yeast. The three crucial catalytic residues are shown in bold. Arrows indicate the residues subjected to mutagenesis. Numbering refers to AtcwINV1.
Table I.
Kinetic parameters for the hydrolysis of Suc of purified AtcwINV1 wild-type, D23A mutant, and E203A mutant proteins
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm−1s−1 | |
| Wild type | 0.35 ± 0.05 | 59 ± 4 | 168.6 |
| D23A | 2.4 ± 0.4 | 10 × 10−2 ± 0.1 × 10−2 | 42 × 10−3 |
| E203A | 2.7 ± 0.3 | 7 × 10−2 ± 0.5 × 10−2 | 26 × 10−3 |
Presence of an Additional Asp/Asn Residue Is Important for Binding and Hydrolysis of Suc in AtcwINV1
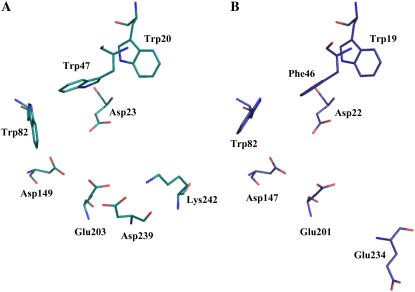
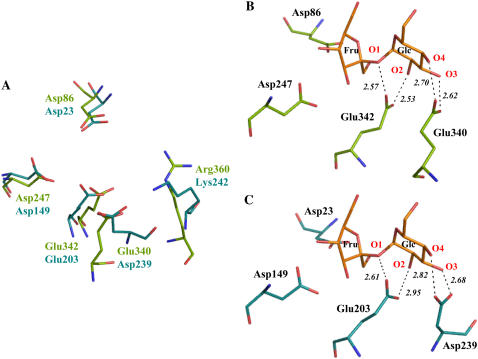
A comparison of the AtcwINV1 and chicory 1-FEH IIa structures reveals some marked differences in their active site regions (Fig. 3). In AtcwINV1, an extra acidic residue (Asp-239) is observed in the vicinity of the conserved acid/base catalyst Glu-203 (Fig. 3A). An acidic amino acid is also found in 1-FEH IIa (Glu-234), but the acidic chain is twisted over 180° and oriented away from the active site (Fig. 3B). This different orientation is most probably due to the presence of a double deletion in the vicinity of this residue (Fig. 4). The presence of an Asp-239 homolog is highly conserved in cell wall invertases, a noticeable exception being Arabidopsis cell wall invertase 5 (Fig. 4). Besides cell wall invertases, vacuolar invertases in general also share this feature (data not shown). It can be hypothesized that the presence of an Asp-239 homolog contributes to the specificity of Suc recognition of plant acid invertases. By contrast, analogous to chicory 1-FEH IIa, all other known FEHs are generally characterized by the absence of such a structural Asp-239 homolog. In some cases, an acidic residue is present, but, analogous to chicory 1-FEH IIa, always surrounded by deletions (Fig. 4). Taking into account the orientation of Glu-234 in chicory 1-FEH IIa away from the active site, most probably due to a double deletion, it can be assumed that the presence of deletions in this region might be a reason for the absence of a structural Asp-239 homolog in FEHs.
Figure 3.
Overview of the 3-D structure of the active site of AtcwINV1 (PDB code 2AC1; A) and chicory 1-FEH IIa (PDB code 1ST8; B). Figures were prepared with PyMol (Delano, 2002). The sequence motifs containing the indicated residues are shown in Figures 2, 4, and 7: Asp-23/Asp-22 (DPN region), Asp-149/Asp-147 (RDP region), Glu-203/Glu-201 (EC region), Asp-239/Glu-234, Lys-242 (LDDTKH/FEG--H region), Trp-19/Trp-20 (WMN region), Trp-47/Phe-46 (WGN/FGD region), and Trp-82/Trp-82 (WSGSAT region). [See online article for color version of this figure.]
Figure 4.
Multiple sequence alignment of the Asp-239/Lys-242 region (shown in bold) in cell wall invertases and all known plant FEHs. The following sequences were used for the alignment: Arabidopsis cell wall invertase 1, Solanum tuberosum cell wall invertase 1, Nicotiana tabacum cell wall invertase, S. tuberosum cell wall invertase 2, S. tuberosum cell wall invertase 3, tomato cell wall invertase Lin5, Vicia faba cell wall invertase, Pisum sativum cell wall invertase, V. faba cell wall invertase 2, Chenopodium rubrum cell wall invertase, Arabidopsis cell wall invertase 2, Arabidopsis cell wall invertase 4, Daucus carota cell wall invertase 1, Oryza sativa cell wall invertase, wheat cell wall invertase, Zea mays cell wall invertase 1, Z. mays cell wall invertase 2, Z. mays cell wall invertase 3, Arabidopsis cell wall invertase 5, Arabidopsis 6-FEH (ancient cell wall invertase 3), Beta vulgaris 6-FEH, chicory 1-FEH I, chicory 1-FEH IIa, chicory 1-FEH IIb, Campanula rapunculoides 1-FEH, Arabidopsis 6&1-FEH (ancient cell wall invertase 6), wheat 1-FEH w1, wheat 1-FEH w2, wheat 6&1-FEH, Hordeum vulgare 1-FEH, wheat 6-KEH w1, wheat 6-KEH w2, Lolium perenne 1-FEH, and wheat 6-FEH. Functionally characterized enzymes are marked with an asterisk.
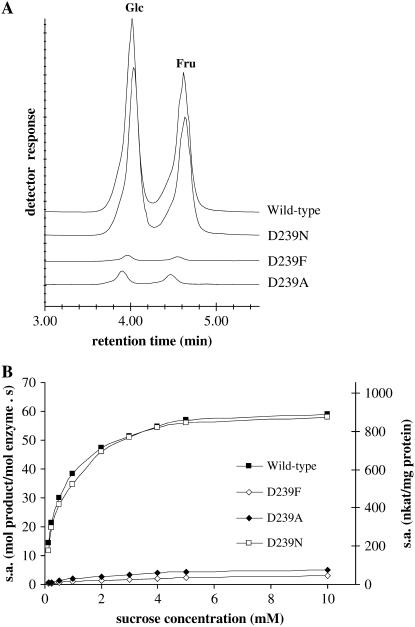
Because polar forces (i.e. the formation of H-bonds between the side chains of polar amino acids and the hydroxyl groups of the carbohydrates) can play an important role in the recognition and binding of carbohydrate substrates in the active site, the possible role of Asp-239 in the binding and hydrolysis of Suc was further investigated by site-directed mutagenesis studies on AtcwINV1. Several mutants were constructed in which Asp-239 was changed into an Ala (D239A), a Phe (D239F), or an Asn (D239N). A comparison of the invertase activities of wild-type and mutant enzymes is presented in Figure 5. The kinetic parameters of the purified mutant enzymes were determined and compared with those obtained for the wild-type invertase (Table II). In both D239A and D239F mutants, Km increased 6- to 11-fold, respectively, inferring an important role for the Asp-239 residue in substrate binding. The lower substrate affinity of D239F compared to D239A can result from the more extended steric hindrance of the bulky Phe residue. A similar tendency is observed on kcat values, which decrease 10- to 20-fold, suggesting that Asp-239 is important for efficient catalysis as well. In contrast, Km and kcat values of the D239N mutant differ only slightly from the wild type, convincingly demonstrating that an acidic group is not essential at this position and can be replaced by an Asn. This is not surprising because both Asp and Asn, containing a carbonyl on their side chain, can form H-bonds with the substrate in almost exactly the same way. Conclusively, these data show that the presence of an additional Asp or Asn residue, adjacent to the Glu-203 proton donor, is important for optimal binding and efficient catalysis of Suc.
Figure 5.
A, Chromatographic pattern of AtcwINV1 wild-type and Asp-239 mutant enzymatic activities. Reaction conditions were as follows: incubation of 50 ng purified enzyme with 10 mm Suc in 50 mm acetate buffer, pH 5.0, during 30 min at 30°C. B, Comparison of specific enzymatic activities [specific (enzymatic) activity in (mol Fru) × (mol enzyme)−1 (s)−1 and (nkat) × (mg protein)−1] of AtcwINV1 wild-type and Asp-239 mutant purified proteins in function of increasing substrate concentrations.
Table II.
Kinetic parameters for the hydrolysis of Suc of purified AtcwINV1 wild-type, D239A mutant, D239F mutant, D239N mutant, and K242L mutant proteins
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm−1s−1 | |
| Wild type | 0.35 ± 0.05 | 59 ± 4 | 168.6 |
| D239A | 2.1 ± 0.2 | 6 ± 0.3 | 2.9 |
| D239F | 4.5 ± 0.5 | 3 ± 0.2 | 0.7 |
| D239N | 0.6 ± 0.07 | 61 ± 5 | 101.7 |
| K242L | 3.7 ± 0.4 | 29 ± 1 | 7.8 |
Remarkably, Asp-239 interacts very strongly with the Lys-242 residue (Fig. 3A), which can be considered as the structural homolog of the Arg-360 residue in B. subtilis levansucrase. It was demonstrated that Arg-360 fulfills a crucial role in the transfructosylation process and interacts via H-bonds with the bound substrate (Chambert and Petit-Glatron, 1991; Meng and Fütterer, 2003). Modeling studies (data not shown) indicate that the Asp-Lys or Asp-Arg interaction might occur in all cell wall invertases and is more probable than a Lys substrate interaction homologous to the Arg substrate interaction observed in levansucrase. Indeed, at the Lys-242 position, a Lys or Arg residue is invariably observed among all cell wall invertases, except for Arabidopsis cwINV5 (Fig. 4). To investigate whether the preceding results on the Asp-239 mutants were due to the removal of this Asp residue itself, or indirectly caused by the disturbance of the Asp-239-Lys-242 interaction, a K242L mutant was constructed and the kinetic properties of the purified enzyme were determined (Table II). The K242L mutant enzyme Km increased with a comparable factor as those of the Asp-239 mutants. However, kcat value only decreased 2-fold in comparison with the wild-type enzyme. According to these data, a crucial role for the Lys-242 residue itself is excluded. However, regarding the conserved interaction between Asp-239 and Lys-242, it is plausible that Lys-242 is necessary to keep Asp-239 in the right orientation toward the active site.
A superposition of the B. subtilis levansucrase and AtcwINV1 structures strongly suggests that Glu-340 in levansucrase can be considered as the functional (but not structural) homolog of Asp-239 in AtcwINV1 (Fig. 6A). The presence of a Glu or Gln besides the proton donor Glu-342 is strictly conserved in levansucrases, implying a possible important role for this residue in binding and/or degradation of Suc in these enzymes. Indeed, it was shown that Glu-340 forms strong H-bridges with the Glc moiety of Suc (O3 and O4 groups) in the active site of B. subtilis levansucrase (Fig. 6B; Meng and Fütterer, 2003). As a consequence of these interactions, we can presume that Glu-342 is not hindered by these groups in its proton donation to the glycosidic oxygen. Regarding this superposition, it can be assumed that the Glu-340 residue is important for both Suc binding and degradation in B. subtilis levansucrase, although this was not checked by site-directed mutagenesis in this system. By analogy with these findings, our results on the Asp-239 AtcwINV1 mutants could be explained (Fig. 6C). Although the exact position of Suc in the active site of AtcwINV1 is unknown and the structure of the AtcwINV1-Suc complex remains to be determined, the orientation of Suc in AtcwINV1 can be predicted (Fig. 6C) based on the position of Suc in the active site of B. subtilis levansucrase because the position of the terminal fructosyl at the −1 subsite is equal for all GH32 and GH68 enzymes (Verhaest et al., 2007). An Asp-239/Glu-340 functional homolog can also be found in G. diazotrophicus levansucrase (Gln-399) and T. maritima invertase (Glu-188). On the contrary, chicory 1-FEH IIa lacks this additional residue. Like all other known plant FEHs, 1-FEH IIa is unable to degrade Suc. Moreover, Suc is a strong inhibitor of most FEHs, including 1-FEH IIa (De Roover et al., 1999). Unlike plant FEHs, microbial FEHs are able to degrade Suc to a certain extent (Nagem et al., 2004). Although A. awamori exoinulinase lacks a structural Asp-239 homolog, the function of this residue could probably be partly taken over by Trp-335.
Figure 6.
A, Superposition of the active sites of AtcwINV1 (turquoise) and B. subtilis levansucrase (green; PDB code 10YG) showing similarities in the catalytic region. B, Details of Suc (orange) in the active site of B. subtilis levansucrase Glu-342A mutant (PDB code 1PT2) in which Ala-342 was replaced in silico by Glu-342 (wild-type orientation; PDB code 1OYG). C, Illustration of the putative position of Suc in the active site of AtcwINV1 based on the known structural complex of B. subtilis levansucrase with Suc. Putative Suc-protein interactions are indicated with dashed lines. Figures were prepared with PyMol (Delano, 2002).
Taken together, our data strongly suggest that the presence of an extra Asp/Asn/Glu/Gln group in GH32 and GH68 Suc-metabolizing enzymes might be important for optimal binding and efficient catalysis of Suc. Overall, the presence or absence of a carbonyl-containing residue adjacent to the acid/base catalyst could be an important determinant of differences in substrate specificity between 1-FEH IIa and fungal exoinulinase (fructan as preferential donor), on one hand, and invertase and levansucrase (Suc as preferential donor) on the other hand. However, this general hypothesis should be further tested by additional site-directed mutagenesis work on a wider array of enzymes also including microbial invertases, fructan hydrolases, and levansucrases.
Conserved Trp Residues Are Important for Stable Substrate Binding
Besides polar forces that are involved in the carbohydrate recognition of various enzymes, it has been reported that other interactions can play a distinctive role. Depending on the stereochemistry of the carbohydrate monomers, the presence of a number of apolar C-H groups (aromatic residues) can play an important role by stabilizing the sugar-enzyme complexes by means of interactions between the aromatic and sugar rings through van der Waals contacts and CH-π stackings (Van der Veen et al., 2001; Muraki, 2002; Chavez et al., 2005).
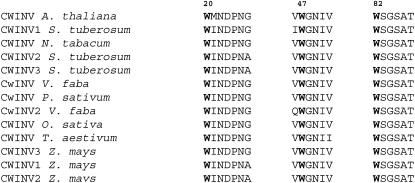
At the rim of the AtcwINV1 active site, there is a prominent presence of three Trp residues, namely, Trp-20, Trp-47, and Trp-82, together forming an aromatic zone (Fig. 3A). These residues belong to the conserved (among invertases) WMNDPN, WGN, and WSGSAT regions (Fig. 7). Mutagenesis experiments were performed in which these Trp residues were changed into a Leu. Determination of the kinetic parameters of the purified mutant proteins and comparison with the wild-type enzyme revealed an important role for these hydrophobic residues in substrate binding (Table III). Km values increased 200, 600, and 120 times for the W20L, W47L, and W82L mutants, respectively, in comparison with the wild-type enzyme. The especially high value of the W47L mutant is not surprising because this residue is very close to the active site and the hydrophobic ring has a good orientation for interacting with the substrate. As a consequence of the very unstable substrate binding in all three mutants, the release of the Glc and Fru residues out of the active site probably takes place at a higher velocity, giving rise to an increase of the kcat values in two of the three cases.
Figure 7.
Multiple sequence alignment of the three conserved hydrophobic residues (shown in bold) of some cell wall invertases: Arabidopsis cell wall invertase 1, S. tuberosum cell wall invertase 1, N. tabacum cell wall invertase, S. tuberosum cell wall invertase 2, S. tuberosum cell wall invertase 3, V. faba cell wall invertase, P. sativum cell wall invertase, V. faba cell wall invertase 2, O. sativa cell wall invertase, wheat cell wall invertase, Z. mays cell wall invertase 3, Z. mays cell wall invertase 1, and Z. mays cell wall invertase 2.
Table III.
Kinetic parameters for the hydrolysis of Suc of purified AtcwINV1 wild-type, W20L mutant, W47L mutant, and W82L mutant proteins
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm−1s−1 | |
| Wild type | 0.35 ± 0.05 | 59 ± 4 | 168.6 |
| W20L | 72 ± 8 | 89 ± 6 | 1.2 |
| W47L | 212 ± 20 | 175 ± 15 | 0.8 |
| W82L | 43 ± 4 | 39 ± 4 | 0.9 |
Taken together, these data strongly suggest that the presence of an aromatic zone in the active site is important for optimal and stable binding of Suc in invertases. Interestingly, most FEHs lack a Trp-47 homolog at this position, but often contain a Phe, which has a less hydrophobic character compared to a Trp residue. A structural Trp-47 homolog can also be detected in T. maritima invertase (Trp-41) and A. awamori exoinulinase (Trp-65). On the contrary, such a homolog is absent in all levansucrases. However, in G. diazotrophicus levansucrase, His-172 might possibly fulfill an equal function because it is present at a similar position in the active site (Martinez-Fleites et al., 2005). Although the active sites of both B. subtilis levansucrase and G. diazotrophicus levansucrase show quite similar architecture, a His-172 homolog is absent in B. subtilis levansucrase. It has been reported that the ratio between high DP fructan and oligosaccharide formation differs significantly between B. subtilis levansucrase, on one hand, and G. diazotrophicus levansucrase, on the other hand (Hernandez et al., 1995). In this context, further research on the absence or presence of a His-172 homolog could be interesting.
1-Kestose as a Substrate for AtcwINV1: The D239A Mutant Acts as a FEH
Cell wall invertases can, besides Suc, degrade other substrates like 1-kestose, raffinose, and stachyose, although to a much lesser extent. Because 1-kestose is the preferential substrate of chicory 1-FEH IIa (De Roover et al., 1999) and an Asp-239 homolog is missing in 1-FEH IIa (Fig. 3B), we investigated whether the breakdown of 1-kestose by AtcwINV1 was also affected by mutagenesis on the Asp-239 residue. The kinetic parameters for the degradation of 1-kestose were determined for the wild-type, D239A, and D239F mutant proteins (Table IV). There was no marked difference in Km and kcat values between the wild-type and D239F mutant proteins. D239A was seriously affected in its ability to degrade Suc (Fig. 5; Table II), but retained its ability to degrade 1-kestose, supporting the idea that plant FEHs lacking invertase activity can evolve from cell wall invertases by a minimal number of mutations or deletions. Similarly, degradation of raffinose and stachyose was affected to the same extent as observed for Suc (data not shown). Higher DP inulin-type fructans were found to be very poor substrates for AtcwINV1 (De Coninck et al., 2005) and for the mutant D239A enzyme (data not shown), so the mutant D239A enzyme can in fact be considered as a special 1-FEH preferentially degrading 1-kestose, the smallest inulin-type fructan (a 1-kestose exohydrolase [1-KEH]). Enzymes that preferentially degrade fructan trisaccharides were recently discovered in fructan plants. Specific 6-kestosidase (6-KEH) enzymes have been purified and cloned from wheat (Triticum aestivum; Van den Ende et al., 2005) and 1-KEH activities were reported in onion (Allium cepa; Benkeblia et al., 2005).
Table IV.
Kinetic parameters for the hydrolysis of 1-kestose of purified AtcwINV1 wild-type, D239A mutant, and D239F mutant proteins
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm−1s−1 | |
| Wild type | 1 ± 0.1 | 21 ± 2 | 21.0 |
| D239A | 0.6 ± 0.03 | 21 ± 3 | 35.0 |
| D239F | 1.2 ± 0.1 | 20 ± 2 | 16.7 |
Strikingly, the D239A mutant showed superior affinity (Km 600 μm) for 1-kestose compared to the naturally occurring FEHs from chicory (1-FEH IIa: 58 mm; De Roover et al., 1999) and wheat (1-FEH w1 and w2: 7 mm; Van den Ende et al., 2003a). However, compared to chicory 1-FEH IIa, the D239A mutant has a similar kcat (11 s−1). The difference in Km between invertases and FEHs is not surprising because cell wall invertases and FEHs fulfill different functions in plants. Cell wall invertases fulfill crucial roles in phloem unloading, carbohydrate partitioning, and sugar signaling, determining normal plant growth and development. It is reasonable to assume that high affinity for Suc is absolutely essential for this purpose. FEHs play a role in remobilization of large amounts of fructans (accumulating up to 20% on a fresh weight basis) and very high affinity of FEHs for their substrates might be less critical for this function. The active site of AtcwINV1 is well designed to bind and hydrolyze Suc in a very efficient way. This appears intrinsically linked to efficient 1-kestose binding and degradation as well. The fact that the D239A mutant has superior affinity for 1-kestose (especially when compared to naturally occurring FEHs) shows that site-directed mutagenesis in the active region of GH32 family members can lead to the development of new enzymes with changed substrate specificities and better kinetics. This allows further exploration of the biotechnological potential of interfering with invertase function in plants. Indeed, it is not unthinkable that further manipulation of AtcwINV1 will result in the development of enzymes with even lower Km values (higher affinities). Such enzymes might be used in the future to potentially increase sink strength and increase yield, enhance fruit quality, or secondary product formation in sink organs of crop plants (Sonnewald et al., 1997). As an example, it has recently been illustrated that a natural cell wall invertase isoform with superior kinetics determines fruit quality in a particular tomato (Lycopersicon esculentum) cultivar (Fridman et al., 2004).
Asp-239 as a Reliable Determinant for Prediction of Functionality within the Group of Plant Cell Wall Invertases/FEHs
Without exception, all functionally characterized FEHs lack an Asp-239 homolog and are unable to break down Suc (Van den Ende et al., 2000, 2001, 2003a, 2003b, 2005; De Coninck et al., 2005; Van Riet et al., 2006). On the contrary, the presence of such an Asp-239 homolog seems to be a general feature of cell wall invertases. Unfortunately, only four so-called cell wall invertases are functionally characterized as real invertases (Sturm and Crispeels, 1990; Roitsch et al., 1995; Fridman et al., 2004; De Coninck et al., 2005). Functional characterization and reliable modeling studies (data not shown) now revealed that three of six of the previously termed cell wall invertase genes of Arabidopsis do not encode real invertases, but defective invertases/FEHs with different substrate specificities (De Coninck et al., 2005). Based on mutagenesis data, extensive functional characterization, and modeling studies on many FEHs, Asp-239 can be proposed as a reliable marker to predict whether a new, uncharacterized member within the cell wall invertase/FEH group encodes a real invertase or a defective invertase/FEH.
CONCLUSION
The GH32 family harbors β-fructosidases, invertases, FEHs, and various types of fructosyltransferases, differing only in their donor and acceptor substrate specificities. High levels of amino acid sequence similarities between these enzymes reveal evolutionary relationships. Fructan biosynthesizing enzymes are closely related to vacuolar invertases (Vijn and Smeekens, 1999; Francki et al., 2006; Ritsema et al., 2006), whereas FEHs are closely related to cell wall invertases, suggesting that they both evolved from a common β-fructosidase ancestor (Van den Ende et al., 2000). Site-directed mutagenesis-based data on AtcwINV1 demonstrate an important role for the Asp-239 residue in both binding and hydrolysis of Suc. A hydrophobic zone at the rim of the active site further stabilizes the optimal binding of Suc. Moreover, an D239A mutant acted as a 1-FEH, preferentially degrading 1-kestose. In general, GH32 family enzymes containing an Asp-239 functional homolog have Suc as a preferential donor, whereas enzymes lacking this homolog use fructans as their donor substrate. Asp-239 is proposed as a reliable determinant for the identification of noncharacterized members within the group of cell wall invertases/FEHs.
MATERIALS AND METHODS
Cloning and Site-Directed Mutagenesis
Arabidopsis (Arabidopsis thaliana) AtcwINV1 (gene accession no. At3g13790) was cloned into the pPICZαA vector as described by De Coninck et al. (2005). Single amino acid substitutions were generated following the Quick Change protocol (Stratagene), using the pPICZαA-AtcwINV1 construct as a template. For site-directed mutagenesis, the following forward oligonucleotide primers (and complementary reverse primers) were used: 5′-GGATGAACGCTCCTAATGGG-3′ (D23A), 5′-GGAAGTGGAATGTGGGCATGTCCTGATTTTTTC-3′ (E203A), 5′-GAAAATAAGTTTGGCCGACACGAAACATG-3′ (D239A), 5′-GTTGAAAATAAGTCTGTTCGACACGAAACATG-3′ (D239F), 5′-GTTGAAAATAAGTCTGAACGACACGAAACATG-3′ (D239N), 5′-GTTTGGACGACACGCTACATGATTATTAC-3′ (K242L), 5′-CCCCCAAAAATTTGATGAACGATCC-3′ (W20L), 5′-GGAGCCGTGTTGGGTAACATC-3′ (W47L), and 5′-CAACGGATGCCTGTCCGGTTCAG-3′ (W82L; mutations are in bold). After DpnI digestion, an additional purification step was performed (QiaQuick PCR purification kit; Qiagen) and 3 μL of the purified plasmids were used for transformation of Escherichia coli TOP10 cells. Mutations were confirmed by sequence analysis.
Expression and Purification
The methylotrophic yeast Pichia pastoris was used for extracellular gene expression as described in De Coninck et al. (2005). Purification of the protein from the culture supernatant was achieved by 80% ammonium sulfate precipitation. After centrifugation (8,500g, 30 min at 4°C), the pellet was redissolved in 50 mm sodium acetate buffer, pH 5.0. The solution was further centrifuged (10,000g, 10 min at 4°C) to spin down undissolved material. The supernatant was subsequently dialyzed in 50 mm sodium acetate buffer, pH 5.0, for 4 h at 4°C. As a final purification step, the dialyzed solution was loaded onto a Mono S column as described in Verhaest et al. (2005a). The purity of the protein was checked by SDS-PAGE and a single band could be observed after gel staining with Coomassie Blue. Concentration measurements of purified enzymes were performed using the Bradford method with bovine serum albumin as a standard (Sedmak and Grossberg, 1977). The amount of pure protein yielded from one P. pastoris expression culture varied depending on the P. pastoris clone and ranged between 50 and 200 μg.
Enzyme Assays
For kinetic determination, appropriate aliquots of enzyme were mixed with Suc or 1-kestose (final concentration ranging from 250 μm to 1 m) in 50 mm sodium acetate buffer, pH 5.0. Reaction mixtures were incubated at 30°C for different time periods. Sodium azide (0.02% [v/v]) was added to prevent microbial growth. Total enzyme activity was determined by measuring the amount of released Fru by anion-exchange chromatography with pulsed amperometric detection (Van den Ende and Van Laere, 1996). For all reactions, different time points were analyzed and only data from the linear range were used. The experiments were repeated three times with consistent results. Kinetic parameters (Km and kcat) were determined using the linear Hanes plot.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: AY079422 (Arabidopsis cell wall invertase 1), AJ419971 (chicory [Cichorium intybus] vacuolar invertase), AJ295033 (chicory 1-FEH IIa), U81520 (chicory 1-SST), U84398 (chicory 1-FFT), P10594 (yeast [Saccharomyces cerevisiae] invertase), Q43171 (Solanum tuberosum cell wall invertase 1), X81834 (Nicotiana tabacum cell wall invertase), Q9M4K7 (S. tuberosum cell wall invertase 2), Q9M4K8 (S. tuberosum cell wall invertase 3), AJ272304 (tomato [Lycopersicum esculentum] cell wall invertase Lin5), Z35162 (Vicia faba cell wall invertase), AK118343 (Arabidopsis cell wall invertase 2), AB049617 (Arabidopsis cell wall invertase 4), X69321 (Daucus carota cell wall invertase 1), AF063246 (Pisum sativum cell wall invertase), Z35163 (V. faba cell wall invertase 2), X81792 (Chenopodium rubrum cell wall invertase), AB073749 (Oryza sativa cell wall invertase), AF030420 (wheat [Triticum aestivum] cell wall invertase), AF050129 (Zea mays cell wall invertase 1), AF050128 (Z. mays cell wall invertase 2), AF050631 (Z. mays cell wall invertase 3), AB029310 (Arabidopsis 6-FEH [ancient cell wall invertase 3]), BAB01929 (Arabidopsis cell wall invertase 5), AJ508534 (Beta vulgaris 6-FEH), AJ242538 (chicory 1-FEH), AJ295034 (chicory 1-FEH IIb), AJ509808 (Campanula rapunculoides 1-FEH), AY060533 (Arabidopsis 6&1-FEH [ancient cell wall invertase 6]), AJ516025 (wheat 1-FEH w1), AJ508387 (wheat 1-FEH w2), AB089269 (wheat 6&1-FEH), AJ605333 (Hordeum vulgare 1-FEH), AB089271 (wheat 6-KEH w1), AB089270 (wheat 1-FEH w2), DQ073968 (Lolium perenne 1-FEH), and AM075205 (wheat 6-FEH).
Acknowledgments
We thank Rudy Vergauwen for his outstanding technical assistance. We are also very grateful to Ed Etxeberria and Marc De Maeyer for critical reading of the manuscript.
This work was supported by the Fund for Scientific Research (grants to W.V.d.E. and A.R.) and the Institute for the Promotion of Innovation through Science and Technology in Flanders (grant to B.D.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wim Van den Ende (wim.vandenende@bio.kuleuven.be).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M (2004) The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J Biol Chem 279 18903–18910 [DOI] [PubMed] [Google Scholar]
- Alberto F, Jordi E, Henrissat B, Czjzek M (2006) Crystal structure of inactivated Thermotoga maritima invertase in complex with the trisaccharide substrate raffinose. Biochem J 395 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach D, Nüesch E, Ritsema T, Boller T, Wiemken A (2005) Mutational analysis of the active center of plant fructosyltransferases: Festuca 1-SST and barley 6-SFT. FEBS Lett 579 4647–4653 [DOI] [PubMed] [Google Scholar]
- Batista FR, Hernandez L, Fernandez JR, Arrieta J, Menendez C, Gomez R, Tambarra Y, Pons T (1999) Substitution of Asp-309 by Asn in the Arg-Asp-Pro (RDP) motif of Acetobacter diazotrophicus levansucrase affects sucrose hydrolysis, but not enzyme specificity. Biochem J 337 503–506 [PMC free article] [PubMed] [Google Scholar]
- Benhamou N, Grenier J, Crispeels MJ (1991) Accumulation of β-fructosidase in the cell walls of tomato roots following infection by a fungal wilt pathogen. Plant Physiol 97 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkeblia N, Ueno K, Onodera S, Shiomi N (2005) Variation of fructooligosaccharides and their metabolizing enzymes in onion bulb (Allium cepa L. cv. Tenshin) during long-term storage. J Food Sci 70 S208–S214 [Google Scholar]
- Chambert R, Petit-Glatron MF (1991) Polymerase and hydrolase activities of Bacillus subtilis levansucrase can be separately modulated by site-directed mutagenesis. Biochem J 279 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez MI, Andreu C, Vidal P, Aboitiz N, Freire F, Groves P, Asensio JL, Asensio G, Muraki M, Canada FJ, et al (2005) On the importance of carbohydrate-aromatic interactions for the molecular recognition of oligosaccharides by proteins: NMR studies of the structure and binding affinity of AcAMP2-like peptides with non-natural naphthyl and fluoroaromatic residues. Chem Eur J 11 7060–7074 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS (1996) The miniature1 seed locus of maize encodes a cell wall invertase required for the normal development of endosperm and maternal cells in the pedicel. Plant Cell 8 971–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Henrissat B (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3 853–859 [DOI] [PubMed] [Google Scholar]
- De Coninck B, Le Roy K, Francis I, Clerens S, Vergauwen R, Halliday AM, Smith SM, Van Laere A, Van den Ende W (2005) Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities. Plant Cell Environ 28 432–443 [Google Scholar]
- De Roover J, Van Laere A, De Winter M, Timmermans JW, Van den Ende W (1999) Purification and properties of a second fructan exohydrolase from the roots of Cichorium intybus. Physiol Plant 106 28–34 [Google Scholar]
- Delano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific, Palo Alto, CA
- Ditt RF, Kerr KF, de Figueiredo P, Delrow J, Comai L, Nester EW (2006) The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol Plant Microbe Interact 19 665–681 [DOI] [PubMed] [Google Scholar]
- Eschrich W (1980) Free space invertase, its possible role in phloem unloading. Ber Dtsch Bot Ges 93 363–378 [Google Scholar]
- Fotopoulos V, Gilbert MJ, Pittman JK, Marvier AC, Buchanan AJ, Sauer N, Hall JL, Williams LE (2003) The monosaccharide transporter gene, AtSTP4, and the cell wall invertase, Atβfruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol 132 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki MG, Walker E, Forster JW, Spangenberg G, Appels R (2006) Fructosyltransferase and invertase genes evolved by gene duplication and rearrangements: rice, perennial ryegrass, and wheat gene families. Genome 49 1081–1091 [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305 1786–1789 [DOI] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Guivarc'h A, Kahmann U, Chirqui D, Roitsch T (2001) Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci USA 98 6522–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Roitsch T (1999) The different pH optima and substrate specificities of extracellular and vacuolar invertases from plants are determined by a single amino-acid substitution. Plant J 20 707–711 [DOI] [PubMed] [Google Scholar]
- Goetz M, Roitsch T (2000) Identification of amino acids essential for enzymatic activity of plant invertases. J Plant Physiol 157 581–585 [Google Scholar]
- Hernandez L, Arrieta J, Menendez C, Vazquez R, Coego A, Suarez V, Selman G, Petit-Glatron M, Chambert R (1995) Isolation and enzymatic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium associated with sugar cane. Biochem J 309 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Van den Ende W, Van Laere A, Cheng S, Bennett J (2005) Structure, evolution and expression of the two invertase gene families of rice. J Mol Evol 60 615–634 [DOI] [PubMed] [Google Scholar]
- Koch K (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47 509–540 [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites C, Ortiz-Lombardia M, Pons T, Tarbouriech N, Taylor EJ, Arrieta JG, Hernandez L, Davies GJ (2005) Crystal structure of levansucrase from the Gram-negative bacterium Gluconacetobacter diazotrophicus. Biochem J 390 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Fütterer K (2003) Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat Struct Biol 10 935–941 [DOI] [PubMed] [Google Scholar]
- Muraki M (2002) The importance of CH/pi interactions to the function of carbohydrate binding proteins. Protein Pept Lett 9 195–209 [DOI] [PubMed] [Google Scholar]
- Nagem RAP, Rojas AL, Gobulev AM, Korneeva OS, Eneyskaya EV, Kulminskaya AA, Neutroev KN, Polikarpov I (2004) Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition. J Mol Biol 344 471–480 [DOI] [PubMed] [Google Scholar]
- Naumoff DG (2001) β-Fructosidase superfamily: homology with some α-l-arabinases and β-d-xylanases. Proteins 42 66–76 [DOI] [PubMed] [Google Scholar]
- Nurizzo D, Turkenburg JP, Charnock SJ, Roberts SM, Dodson EJ, McKie VA, Taylor EJ, Gilbert HJ, Davies GJ (2002) Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold. Nat Struct Biol 9 665–668 [DOI] [PubMed] [Google Scholar]
- Ozimek LK, van Hijum SAFT, van Koningsveld GA, van der Maarel MJEC, van Geel-Schutten GH, Dijkhuizen L (2004) Site-directed mutagenesis study of the three catalytic residues of the fructosyltransferases of Lactobacillus reuteri 121. FEBS Lett 560 131–133 [DOI] [PubMed] [Google Scholar]
- Pons T, Naumoff DG, Martinez-Fleites C, Hernandez L (2004) Three acidic residues are at the active site of a beta-propeller architecture in glycoside hydrolase families 32, 43, 62 and 68. Proteins 54 424–432 [DOI] [PubMed] [Google Scholar]
- Reddy A, Maley F (1990) Identification of an active-site residue in yeast invertase by affinity labeling and site-directed mutagenesis. J Biol Chem 265 10817–10820 [PubMed] [Google Scholar]
- Reddy A, Maley F (1996) Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J Biol Chem 271 13953–13957 [DOI] [PubMed] [Google Scholar]
- Ritsema T, Hernandez L, Verhaar A, Altenbach D, Boller T, Wiemken A, Smeekens S (2006) Developing fructan-synthesizing capability in a plant invertase via mutations in the sucrose-binding box. Plant J 48 228–237 [DOI] [PubMed] [Google Scholar]
- Roitsch T, Bittner M, Godt DE (1995) Induction of apoplastic invertase of Chenopodium rubrum by D-glucose and a glucose analog and tissue-specific expression suggest a role in sink-source regulation. Plant Physiol 108 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T, Gonzalez MC (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci 9 606–613 [DOI] [PubMed] [Google Scholar]
- Sedmak JJ, Grossberg SE (1977) Rapid, sensitive and versatile assay for protein using Coomassie Brilliant Blue G250. Anal Biochem 79 544–552 [DOI] [PubMed] [Google Scholar]
- Sherson SM, Alford HL, Wallace G, Smith SM (2003) Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J Exp Bot 54 525–531 [DOI] [PubMed] [Google Scholar]
- Sonnewald U, Hajirezaei MR, Kossmann J, Heyer A, Trethewey RN, Willmitzer L (1997) Increased potato tuber size resulting from apoplastic expression of a yeast invertase. Nat Biotechnol 5 794–797 [DOI] [PubMed] [Google Scholar]
- Sturm A (1999) Invertases: primary structures, functions and roles in plant development and sucrose partitioning. Plant Physiol 121 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Crispeels MJ (1990) cDNA cloning of a carrot extracellular β-fructosidase and its expression in response to wounding and bacterial infection. Plant Cell 2 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Tang GQ (1999) The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci 4 401–407 [DOI] [PubMed] [Google Scholar]
- Tymowska-Lananne Z, Kreis M (1998) The plant invertases: physiology, biochemistry and molecular biology. Adv Bot Res 28 71–117 [Google Scholar]
- Van den Ende W, Clerens S, Vergauwen R, Van Riet L, Van Laere A, Yoshida M, Kawakami A (2003. a) Fructan 1-exohydrolases: β-(2,1)-trimmers during graminan biosynthesis stems of wheat? Purification, characterization, mass mapping and cloning of two fructan 1-exohydrolase isoforms. Plant Physiol 131 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W, De Coninck B, Clerens S, Vergauwen R, Van Laere A (2003. b) Unexpected presence of fructan exohydrolases (6-FEHs) in non-fructan plants: characterization, cloning, mass mapping and functional analysis of a novel “cell-wall invertase-like” specific 6-FEH from sugar beet (Beta vulgaris L.). Plant J 36 697–710 [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, De Roover J, Verhaert P, Van Laere A (2000) Cloning and functional analysis of chicory root fructan-1-exohydrolase I (1-FEH I): a vacuolar enzyme derived from a cell-wall invertase ancestor? Mass fingerprint of the 1-FEH I enzyme. Plant J 24 447–456 [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, Van Wonterghem D, Clerens SP, De Roover J, Van Laere A (2001) Defoliation induces fructan 1-exohydrolase II in witloof chicory roots: cloning and purification of two isoforms, fructan 1-exohydrolase IIa and fructan 1-exohydrolase IIb. Mass fingerprint of the fructan exohydrolase II enzymes. Plant Physiol 126 1186–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W, Van Laere A (1996) Fructan synthesizing and degrading activities in chicory roots (Cichorium intybus L.) during growth, storage and forcing. J Plant Physiol 149 43–50 [Google Scholar]
- Van den Ende W, Yoshida M, Clerens S, Vergauwen R, Kawakami A (2005) Cloning, characterization and functional analysis of novel 6-kestose exohydrolases (6-KEHs) from wheat (Triticum aestivum). New Phytol 166 917–932 [DOI] [PubMed] [Google Scholar]
- Van der Veen BA, Leemhuis H, Kralj S, Uitdehaag JC, Dijkstra BW, Dijkhuizen L (2001) Hydrophobic amino acid residues in the acceptor binding site are main determinants for reaction mechanism and specificity of cyclodextrin-glycosyltransferase. J Biol Chem 276 44557–44562 [DOI] [PubMed] [Google Scholar]
- Van Laere A, Van den Ende W (2002) Inulin metabolism in dicots: chicory as a model system. Plant Cell Environ 25 803–813 [Google Scholar]
- Van Riet L, Nagaraj V, Van den Ende W, Clerens S, Wiemken A, Van Laere A (2006) Purification, cloning and functional characterization of a fructan 6-exohydrolase from wheat (Triticum aestivum L.). J Exp Bot 57 213–223 [DOI] [PubMed] [Google Scholar]
- Verhaest M, Lammens W, Le Roy K, De Ranter CJ, Van Laere A, Rabijns A, Van den Ende W (2007) Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor. New Phytol 174 90–100 [DOI] [PubMed] [Google Scholar]
- Verhaest M, Lammens W, Le Roy K, De Ranter CJ, Van Laere A, Van den Ende W, Rabijns A (2006) X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr D Biol Crystallogr 62 1555–1563 [DOI] [PubMed] [Google Scholar]
- Verhaest M, Le Roy K, Sansen S, De Coninck B, Lammens W, De Ranter CJ, Van Laere AV, Van den Ende W, Rabijns A (2005. a) Crystallization and preliminary x-ray diffraction study of a cell wall invertase from Arabidopsis thaliana. Acta Crystallogr Sect F Struct Biol Cryst Commun 61 766–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaest M, Van den Ende W, Le Roy K, De Ranter CJ, Van Laere AV, Rabijns A (2005. b) X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. Plant J 41 400–411 [DOI] [PubMed] [Google Scholar]
- Vijn I, Smeekens S (1999) Fructans: more that a reserve carbohydrate? Plant Physiol 120 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanase H, Maeda M, Hagiwara E, Yagi H, Taniguchi K, Okamoto K (2002) Identification of functionally important amino acid residues in Zymomonas mobilis levansucrase. J Biol Chem 132 565–572 [DOI] [PubMed] [Google Scholar]