Abstract
Gibberellins (GA) are known to influence phase change in Arabidopsis (Arabidopsis thaliana) as well as the development of trichomes, which are faithful epidermal markers of shoot maturation. They modulate these developmental programs in part by antagonizing DELLA repressors of growth, GIBBERELLIC ACID INSENSITIVE (GAI) and REPRESSOR OF ga1-3 (RGA). In this study, we have probed the relative roles played by RGA, GAI, and two homologs, RGA-LIKE1 (RGL1) and RGL2, in these processes and investigated molecular mechanisms through which they influence epidermal differentiation. We found that the DELLAs act collectively to regulate trichome initiation on all aerial organs and that the onset of their activity is accompanied by the repression of most genes known to regulate trichome production. These effects are consistent with the results of genetic analysis, which conclusively place theses genes downstream of the DELLAs. We find that repression of trichome regulatory genes is rapid, but involves an indirect, rather than a direct, molecular mechanism, which requires de novo protein synthesis. DELLA activity also influences postinitiation events and we show that GAI is a major repressor of trichome branching, a role in which it is antagonized by RGL1 and RGL2. Finally, we report that, in contrast to most other effects, the repression by GA applications of flower trichome initiation is not dependent on RGA, GAI, RGL1, or RGL2. In summary, our data show that DELLA proteins are central to trichome development in Arabidopsis and that their effect can be largely explained by their transcriptional influence on trichome initiation activators.
The initiation of trichomes, which are large unicellular epidermal structures on the aerial organs of many plant species, has long been a model for the study of cell-fate determination in plants. In Arabidopsis (Arabidopsis thaliana), trichomes decorate stems, sepals, and leaves, where they first appear on the adaxial side, early in leaf development (Larkin et al., 1994; Hülskamp et al., 1999). Genetic analyses have identified at least three regulatory proteins that promote trichome initiation: GLABROUS1 (GL1), an R2R3-Myb-type transcription factor; TRANSPARENT TESTA GLABRA1 (TTG1), a protein containing WD-40 repeats; and GLABRA3 (GL3), a basic helix-loop-helix-type transcription factor. Loss-of-function mutations in the GL1 and TTG1 genes nearly abolish trichome initiation on leaves (Larkin et al., 1994; Walker et al., 1999; Payne et al., 2000). GL3 has been shown to self-associate and assemble with GL1 and TTG1 in yeast (Saccharomyces cerevisiae) two-hybrid experiments, but GL1 and TTG1 are not known to interact (Larkin et al., 1994; Walker et al., 1999; Payne et al., 2000; Zhang et al., 2003).
Trichome initiation is dependent on GA signaling in Arabidopsis: The GA biosynthesis mutant ga1-3 is almost completely glabrous and GA applications stimulate initiation in both ga1-3 and wild-type plants (Chien and Sussex, 1996; Telfer et al., 1997; Perazza et al., 1998). GAs also influence trichome morphology, promoting the formation of supernumerary branches on leaf trichomes (Perazza et al., 1998). Trichome formation is therefore a useful system for dissecting how GA signaling controls cellular differentiation in plants. In addition, the appearance of trichomes on the abaxial side of rosette leaves marks the transition between juvenile and adult vegetative phases, which, like flowering, is modulated by GA signaling (Wilson et al., 1992; Telfer et al., 1997). We have recently shown that the effects of GA on inflorescence initiation and shoot maturation are influenced in Arabidopsis by redundant transcription factors GLABROUS INFLORESCENCE STEMS (GIS), ZINC FINGER PROTEIN8 (ZFP8), and GIS2 (Gan et al., 2006, 2007). All three proteins are positive regulators of initiation and both GIS and GIS2 have been shown to activate GL1 expression (Gan et al., 2006, 2007). Whereas the molecular mechanisms through which these three genes are regulated are unclear, much is known of upstream steps in the GA response pathway in plants. GA signaling initiates with the hormones binding to soluble receptors GIBBERELLIN INSENSITIVE DWARF1 (OsGID1) and OsGID1-like (Ueguchi-Tanaka et al., 2005; Hartweck and Olszewski, 2006; Cao et al., 2006), which triggers the degradation of plant growth repressor DELLA proteins (DELLAs) via the 26S proteasome pathway (Silverstone et al., 2001; Fu et al., 2002; Itoh et al., 2002; Hussain et al., 2005). This degradation process is mediated by GA-specific F-box proteins OsGID2 (Sasaki et al., 2003) and AtSLY1 (McGinnis et al., 2003; Dill et al., 2004; Fu et al., 2004), which remove the restraint on growth exerted by the DELLAs (Harberd, 2003; Cao et al., 2006). In Arabidopsis, the DELLAs are encoded by a family of five genes: GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), and three RGA-LIKE genes (RGL1, RGL2, and RGL3; Peng et al., 1997; Silverstone et al., 1998; Lee et al., 2002; Tyler et al., 2004). These repressors have overlapping, but distinct, influences on plant growth and development (Dill and Sun, 2001; King et al., 2001; Lee et al., 2002; Wen and Chang, 2002; Cheng et al., 2004). GAI and RGA play a predominant role in repressing stem elongation (Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; King et al., 2001), whereas RGA, RGL2, and RGL1 influence flower development by restricting petal and stamen morphogenesis (Cheng et al., 2004; Tyler et al., 2004; Yu et al., 2004). RGL2 is central to the repression of seed germination, a process in which RGL1, GAI, and RGA also have limited involvement (Wen and Chang, 2002; Lee et al., 2002; Tyler et al., 2004; Cao et al., 2005, 2006). The role of RGL3 is less clear (Tyler et al., 2004).
RGA and GAI are known to repress trichome formation on leaves because loss-of-function mutations in RGA and GAI can rescue leaf trichome initiation in ga1-3 mutants (Dill and Sun, 2001). The repressors also have a profound influence on vegetative and reproductive phase change. However, nothing is known of how this is achieved at the molecular level or of the role played by other DELLA proteins in these processes.
In this study, we have used a combination of DELLA loss-of-function mutants and inducible overexpressors to examine these issues. We found that the different repressors act synergistically in the control of trichome development, but that specific DELLA proteins play predominant roles in the control of either initiation or branching. We also found that the effect of DELLA repression is associated with profound changes in the expression of trichome initiation regulators, which are an indirect, rather than a direct, consequence of DELLA action.
RESULTS
DELLA Proteins Act as Repressors of Most Known Positive Regulators of Trichome Initiation in Arabidopsis
In GA-treated plants, higher trichome production is associated with the induction of GIS, ZFP8, and GIS2 and with an increase in the expression of GL1, which is itself dependent on GIS activity. In addition, GL1 and GIS are strongly down-regulated in the ga1-3 mutant (Perazza et al., 1998; Gan et al., 2006, 2007). We therefore sought to determine whether variations in GA signaling have a similar effect on other trichome regulators and assess the role played by DELLA proteins in this response.
We first compared the levels of TTG1, GL3, ZFP8, and GIS2 expression in ga1-3 and in wild-type plants. We found that decreased GA levels in the mutant resulted in the down-regulation of GL3, ZFP8, and GIS2, but that the expression of TTG1 was not affected (Supplemental Fig. S1). This response indicated that reductions in GA signaling negatively impact the expression of multiple genes encoding regulators of trichome initiation.
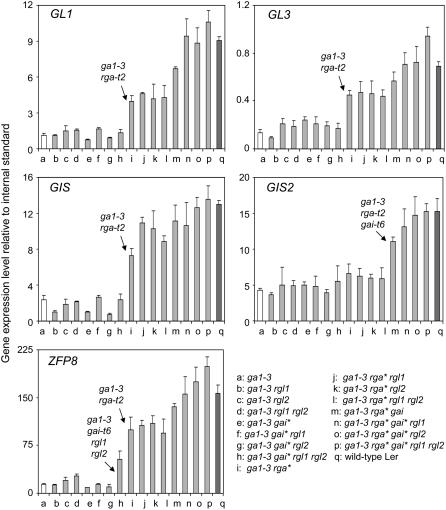
To assess the role played by DELLA proteins in this transcriptional response, we examined the effects of loss-of-function mutations in RGA, GAI, RGL1, and RGL2 on the expression of GL1, GL3, GIS, ZFP8, and GIS2 in the ga1-3 background (Fig. 1). We found that, in every case, loss of all DELLA function restored gene expression to wild-type levels or higher, which indicated that, collectively, DELLA repressors play a prominent role in the regulation of these genes by GA. Whereas the impact of DELLA activity on gene expression was generally similar to its effects on trichome initiation, DELLA proteins differed in their overall influence and in their effects on individual genes. For example, RGA loss of function strongly stimulated the expression of GL1, GL3, GIS, and ZFP8, but had little effect on GIS2 expression. GIS2 was only significantly induced when the functions of both RGA and GAI were abolished (Fig. 1). Similarly, the combination of gai-t6, rgl1, and rgl2 mutations strongly promoted ZFP8 expression, but not expression of other genes. The influence of RGL1 and RGL2 on gene expression was clearly detectable in the case of GL1 and ZFP8, although it was most marked when the function of other DELLA proteins had been abolished. Similarly, loss-of-function mutations in both RGL1 and RGL2 boosted GL3 expression above wild-type levels, but only if both RGA and GAI had been knocked out. In all other cases, the influence of individual DELLA proteins was limited and dependent on the activity of all the others (Fig. 1).
Figure 1.
Expression of GL1, GL3, GIS, GIS2, and ZFP8 in DELLA-defective ga1-3 mutants. Transcript levels of the different trichome initiation regulatory genes in various DELLA mutants were measured by real-time PCR. Values represent the average and sd from three measurements and are relative to the expression of the UBQ10 gene. Gene expression was measured in young developing inflorescences. Mutants presenting a significant change in gene expression are indicated by an arrow. gai*, gai-t6; rga*, rga-t2; Ler, Landsberg erecta ecotype control.
In summary, our findings indicated that DELLA proteins are essential in the control by GA of the expression of genes encoding activators of trichome initiation. They also suggested that, whereas RGA generally plays a predominant role in this process, GAI, RGL1, and RGL2 also participate, sometimes in a critical way, in the regulation of downstream gene expression.
DELLA Proteins Rapidly, But Indirectly, Target Trichome Initiation Regulators
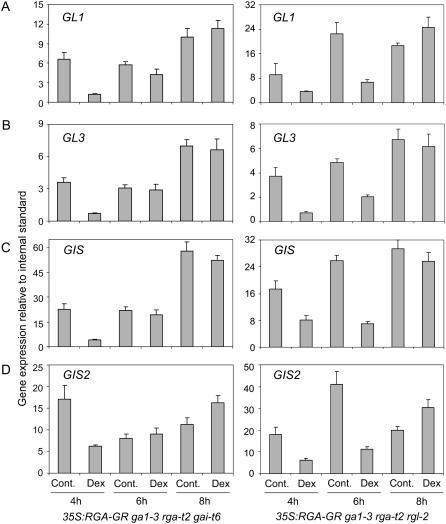
To further investigate regulation by DELLA proteins of downstream regulators and, in particular, assess whether their role in this process is direct or indirect, we examined the expression of GL1, GL3, GIS, GIS2, and ZFP8 in DELLA mutants that overexpress RGA-GR, which encodes a functional fusion of RGA to the receptor domain of the rat glucocorticoid receptor. In these plants, RGA (RGA-GR) activity is inducible by dexamethasone (DEX; Yu et al., 2004). Through tight control of RGA-GR activity, we aimed to determine the kinetics of induced gene expression changes as well as assess the possible requirement for secondary regulators. To cross-validate our results, we analyzed gene expression in response to RGA-GR activity in two different mutant backgrounds: rga-t2 rgl2 ga1-3 and rga-t2 gai-t6 ga1-3 (Yu et al., 2004). The effects of RGA-GR induction were determined at different time points following DEX treatment.
We found that, in most cases, RGA-GR strongly and rapidly repressed the expression of trichome regulatory genes and that this effect was detectable as early as 2 h after DEX application (Fig. 2). In the case of ZFP8, repression was faster in 35S:RGA-GR rga-t2 rgl2 ga1-3 than in 35S:RGA-GR rga-t2 gai-t6 ga1-3 plants, possibly because the amplitude of the response was more limited in the latter background. In all cases, the repression of gene expression by RGA-GR was found to be temporary and was no longer detectable 8 h after treatment (Fig. 2).
Figure 2.
Short-term effect of RGA activity on the expression of GL1, GL3, GIS, and GIS2. Time course of GL1 (A), GL3 (B), GIS (C), and GIS2 (D) expression in DEX-treated ga1-3 rga-t2 gai-t6 (left) or ga1-3 rga-t2 rgl2 (right) plants overexpressing RGA-GR and in mock-treated plants (Cont.). Transcript levels were measured in young developing inflorescence by real-time PCR 4, 6, and 8 h after treatment. Values represent the average and sd from three measurements and are relative to the expression of the UBQ10 gene.
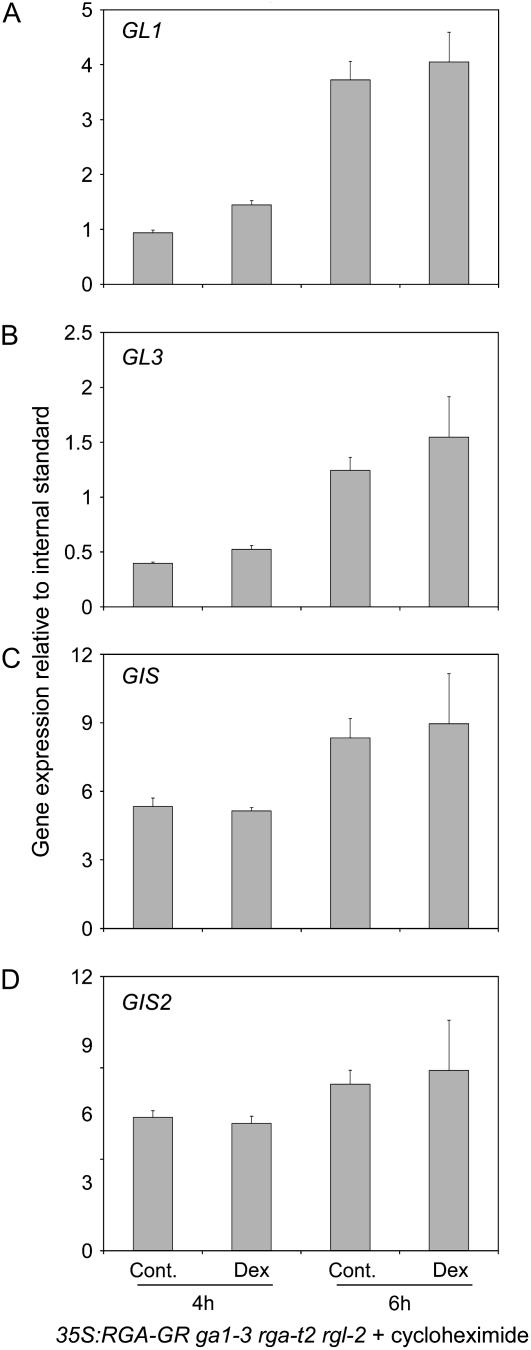
Because DEX induction of RGA-GR activity does not require protein synthesis, it is possible to determine whether its effects are direct or indirect by examining the influence of protein synthesis inhibitors on induced gene expression changes. To determine whether RGA-GR directly or indirectly represses trichome regulators, we repeated DEX applications to 35S:RGA-GR rga-t2 rgl2 ga1-3 in the presence of the protein synthesis inhibitor cycloheximide. We found that cycloheximide strongly inhibited the repression by DEX of all genes (Fig. 3). The transcriptional effect of RGA-GR activity was abolished or strongly attenuated and we never measured down-regulation in the first 6 h following application.
Figure 3.
GL1, GL3, GIS, and GIS2 are indirect targets of RGA. Time course of GL1 (A), GL3 (B), GIS (C), and GIS2 (D) expression in DEX-treated 35S:RGA-GR ga1-3 rga-t2 rgl2 plants and in mock-treated plants (Cont.). All plants were treated with the protein synthesis inhibitor cycloheximide before DEX applications. Transcript levels were measured in young developing inflorescence by real-time PCR 4 and 6 h after treatment. Values represent the average and sd from three measurements and are relative to the expression of the UBQ10 gene.
Based on the above results, we concluded that, although RGA-GR activity rapidly triggers repression of trichome initiation activators, the corresponding genes are indirect, rather than direct, targets and repression is therefore likely to require the production of additional, yet unidentified, factors.
RGA, GAI, RGL1, and RGL2 Differentially, But Synergistically, Regulate Trichome Initiation
To investigate whether the roles of RGA, GAI, RGL1, and RGL2 in repressing trichome activators reflect their influence on trichome initiation throughout the plant, we examined the effect of DELLA loss-of-function mutations in the absence of GA in the ga1-3 background.
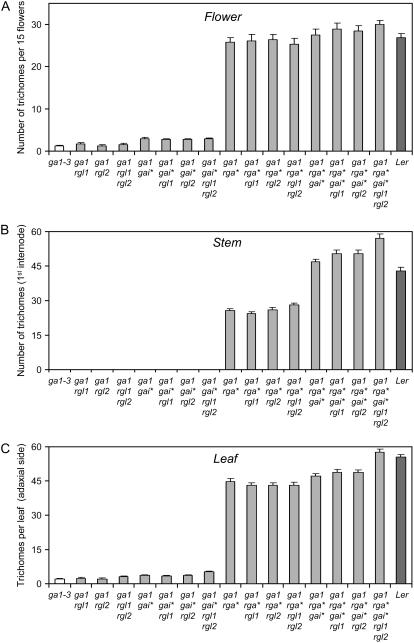
Among all single DELLA mutants (in ga1-3), rgl1-1 ga1-3 and rgl2-1 ga1-3 displayed the lowest trichome density. Despite the importance of RGL1 in flower development (Cheng et al., 2004; Tyler et al., 2004; Yu et al., 2004), loss of RGL1 function had only a limited effect on flower trichome initiation (Fig. 4A). The rgl2-1 ga1-3 mutant was, like ga1-3 control plants, nearly glabrous. In contrast, loss of RGA function had a dramatic impact on trichome production in ga1-3. Initiation was not only restored on the abaxial side of rga-t2 ga1-3 rosette leaves, as previously observed (Silverstone et al., 1997; Dill and Sun, 2001), but also on the adaxial side and on all inflorescence organs. In fact, the effect was strongest on flowers, where the epidermal phenotype was most similar to wild type. On stems as on the abaxial side of rosette leaves, no trichome initiation was visible in the ga1-3 background unless RGA function was abolished (Fig. 4, B and C).
Figure 4.
Influence of DELLA loss of function on trichome initiation. Trichome production of different DELLA mutants in the ga1-3 background on sepals (A), stems (B), and the adaxial side of the second adult leaf (C). Sepal trichome counts are the total number of trichomes for about 15 flowers per plant. Stem values represent the total number of trichomes on the first internode of the main stem. Leaf values represent an estimate of the total number of adaxial trichomes on the second adult leaf based on a small area and average leaf surfaces for each genotype. gai*, gai-t6; rga*, rga-t2; Ler, Landsberg erecta ecotype control. Values represent averages and se for 20 plants.
GAI loss of function also had a significant, although lesser, impact on trichome initiation: gai-t6 ga1-3 mutants were glabrous, but gai-t6 rga-t2 ga1-3 produced more trichomes than rga-t2 ga1-3 mutants on stems and, to a lesser degree, on leaves (P = 0.07; Fig. 4, B and C). The gai-t6 mutation had the greatest effect on stems of rga-t2 ga1-3, where it nearly doubled trichome production (Fig. 4B). Interestingly, the quadruple DELLA rga-t2 gai-t6 rgl1 rgl2 ga1-3 mutant produced significantly more leaf and stem trichomes than gai-t6 rga-t2 ga1-3 mutants. These observations indicated that RGL1 and RGL2 also play a significant role in the regulation of trichome initiation. Their influence was, however, greatest when the repression exerted by other redundant DELLA proteins had been lifted. We also observed that rga-t2 gai-t6 rgl1-1 rgl2-1 ga1-3 produces a higher number of trichomes than wild-type plants on both vegetative and inflorescence organs (Fig. 4). This phenotype suggested that four DELLA proteins exert a repressive effect on trichome initiation even in wild-type plants.
In summary, our data indicated that GAI, RGA, RGL1, and RGL2 are all involved, albeit to different degrees, in regulation by GA of trichome initiation and that they act collectively in their repressive role. The gene expression and phenotypic data together suggest that the DELLAs are major gatekeepers in the control by GA of trichome initiation acting mainly through a repressive transcriptional mechanism.
GAI Is the Major Repressor of Trichome Branching, But the Effects of GAI Derepression Are Suppressed by Loss-of-Function Mutations in RGL1 and RGL2
GA treatments are known to stimulate trichome branching on leaves and the trichomes of spy mutants, which display a constitutive GA response, are more highly branched than those of wild-type plants (Perazza et al., 1998, 1999; Gan et al., 2006). Because RGA and, to a lesser extent, GAI, play an important role in trichome initiation, we investigated their involvement and that of RGL1 and RGL2 in the control of trichome branching. To this end, we examined the branching pattern of trichomes of leaves and stems of plants in which the functions of RGA, GAI, RGL1, and/or RGL2 were impaired.
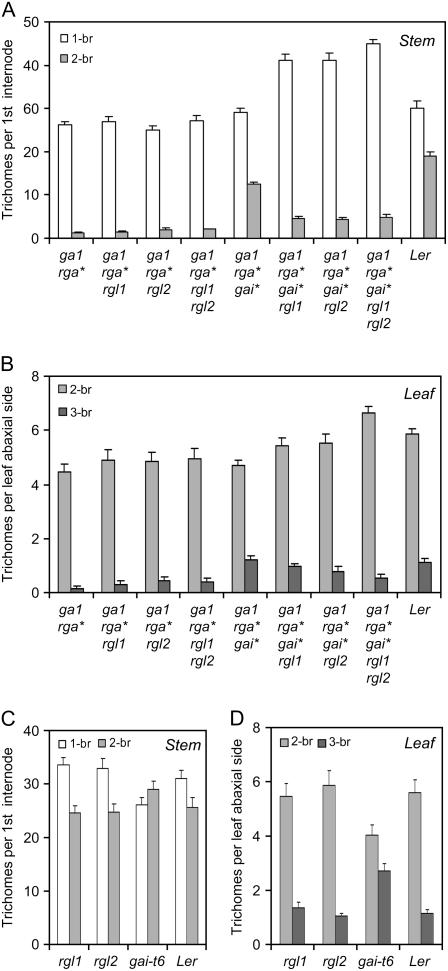
Our first observation was that, whereas RGA loss of function restored trichome initiation in ga1-3 mutants, rga-t2 ga1-3 was strongly deficient in branching. For example, mutant stems harbored only 4.4% branched trichomes, as compared to 38.8% in wild-type plants (Fig. 5A). The ga1-3 rga-t2 rgl1 and ga1-3 rga-t2 rgl2 mutants presented a similar phenotype, indicating that lifting the repression exerted by RGA, RGL1, and/or RGL2 repression is not sufficient to restore branching. In contrast, we found that GAI loss of function strongly stimulated branching, restoring the number of branched trichomes to about 30% of the total (Fig. 5A). A similar situation was found in leaves: Whereas the rga-t2, rgl1, and rgl2 mutations had little influence on branching in the ga1-3 background, the gai-t6 mutation restored branching to wild-type levels or above (Fig. 5B).
Figure 5.
Influence of DELLA activity on trichome branching. Branching phenotype of stem (A and C) and leaf trichomes (B and D) of DELLA mutants in the ga1-3 (A and B) or wild-type (C and D) background. All trichomes were counted on the first internode of the main stem or on the abaxial side of the second adult leaf. “1-br,” “2-br,” and “3-br,” one-branched, two-branched, and three-branched trichomes. gai*, gai-t6; rga*, rga-t2; Ler, Landsberg erecta ecotype control. Values represent averages and se for 20 plants.
Interestingly, loss of RGL1 and/or RGL2 function antagonized the effect of the gai-t6 mutation, an effect that was most dramatic in the quintuple mutant. This negative effect of rgl1 and rgl2 in the ga1-3 rga-t2 gai-t6 background was visible both on leaves and on stems (Fig. 5, A and B). The positive influence of RGL1 and RGL2 on branching was dependent on GAI activity because we could not detect any decrease in trichome branching in rgl1 and rgl2 (Fig. 5C) or in the ga1-3 rga-t2 rgl1 and ga1-3 rga-t2 rgl2 mutants. In contrast, the single gai-t6 mutant displayed significantly more branched trichomes on leaves and stems, which indicated that the stimulation of trichome branching by GAI loss of function is not dependent on other DELLA proteins (Fig. 5C).
In summary, our analysis revealed an essential role for GAI in trichome branching and indicated that the repressors are antagonized by RGL1 and RGL2 in this process.
DELLA-Dependent Derepression of Trichome Initiation Requires Downstream Regulator GIS2
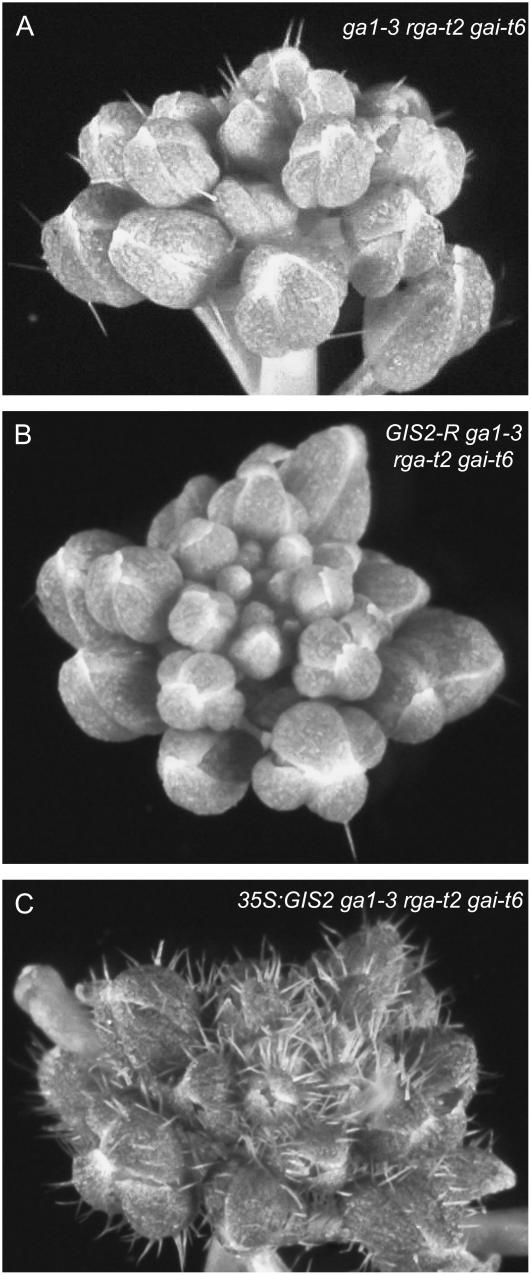
We previously reported that overexpressing GIS is not sufficient for restoring trichome initiation in the gai or ga1-3 mutants even though GIS, GIS2, and ZFP8 are required in the positive regulation of trichome initiation by GA (Gan et al., 2006). We therefore sought to determine whether members of the GIS clade are, conversely, required for trichome initiation when the repression exerted by the DELLA proteins has been removed. To answer this question, we used RNAi to specifically down-regulate GIS2, which plays a central role in the regulation of trichome initiation on flowers, in the ga1-3 rga-t2 gai-t6 mutant (Gan et al., 2007). In this mutant, loss of RGA and GAI function restores flower trichome initiation to normal levels (Fig. 4A). We found that, in nine independent transformants, trichome production was greatly decreased on sepals as compared to controls (Table I; Fig. 6B). In contrast, GIS2 overexpression caused a marked increase in trichome production (Fig. 6C), a phenotype that we also observed when GIS or ZFP8 was overexpressed in this mutant (Supplemental Fig. S2, B and C). A similar result was obtained when overexpression experiments were repeated in the ga1-3 rga-t2 rgl2 mutant (data not shown).
Table I.
Trichome initiation on flowers of DELLA mutants in which GIS2 was silenced by RNAi
Trichome counts are the total number of trichomes for about 15 flowers per plant. ga1-3 rga-t2 gai-t6 values represent averages and ses for 16 plants. grg, ga1-3 rga-t2 gai-t6; g2R, GIS2-RNAi.
| Control Line | Silenced Lines | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ga1-3 | grg | grg | grg | grg | grg | grg | grg | grg | grg |
| rga-t2 | g2R1 | g2R2 | g2R3 | g2R4 | g2R5 | g2R6 | g2R7 | g2R8 | g2R9 |
| gai-t6 | |||||||||
| 29.9 (1.1) | 4 | 7 | 2 | 9 | 0 | 2 | 1 | 3 | 3 |
Figure 6.
Effects of GIS2 silencing and overexpression on flower trichome initiation in ga1-3 rga-t2 gai-t6. Trichome initiation on flowers of ga1-3 rga-t2 gai-t6 control (A), GIS2-silenced (GIS2-R; B), and GIS2-overexpressing (C) plants. GIS2 overexpression causes the proliferation of trichomes on sepals, whereas silencing inhibits trichome initiation.
These observations therefore indicated that GIS2 is required for trichome initiation in the absence of DELLA repression. They also indicated that GIS2 (GIS/ZFP8) activity is limiting to trichome initiation when repression by RGA and GAI or RGL2 is lifted and that derepression is sufficient to restore the ability of GIS clade members to induce trichome proliferation in the ga1-3 background. These findings provided conclusive evidence that GIS2 and, in all likelihood, GIS and ZFP8 act downstream of the DELLA proteins.
RGL1 and RGL2 Act to Delay Vegetative and Reproductive Phase Change
In Arabidopsis, GAs are required for the transition between juvenile and adult stages and for flowering under short days, processes in which RGA and GAI play a major role (Wilson et al., 1992; Chien and Sussex, 1996; Telfer et al., 1997; Perazza et al., 1998; Dill and Sun, 2001).
Our observations confirmed that only RGA loss of function can restore abaxial trichome production in ga1-3 and that GAI loss of function further accelerates both vegetative and reproductive phase change, although the gai-t6 mutation alone has no effect (data not shown). Similarly, the effects of the rgl1 and rgl2 mutations were not detectable unless the function of other DELLA proteins was compromised. Loss of RGA function was required for the influence of RGL1 and RGL2 to be measurable and the effect was most pronounced when the activities of both RGA and GAI were abolished (Supplemental Fig. S3). Loss of RGL1 and/or RGL2 function in the rga-t2 gai-t6 ga1-3 background also resulted in earlier adult leaf production and flowering than in wild-type plants. In comparison, the combination of rga-t2 and gai-t6 mutations only restored a normal shoot maturation program in the ga1-3 mutant (Supplemental Fig. S3).
The above findings indicated that both RGL1 and RGL2 play a role in the regulation by GAs of vegetative phase change and flowering and that their influence is conditional on RGA and GAI function. Interestingly, there was, overall, a strong correlation between the effects of DELLA loss of function on vegetative and reproductive phase change, which suggested that the mechanisms through which they control the two processes may be similar (Supplemental Fig. S3).
GAI, RGA, RGL1, and RGL2 Are Not Implicated in the Repression by GA of Trichome Initiation on Flowers
Exogenous GA application counteracts the repression by DELLA proteins of floral development and stem elongation (Peng and Harberd, 1997; Dill and Sun, 2001; King et al., 2001; Lee and Schiefelbein, 2001; Cheng et al., 2004; Tyler et al., 2004; Yu et al., 2004). To assess whether the effects of loss of DELLA function on trichome initiation are equivalent to those resulting from increases in GA levels, we compared the effects of applying GA to ga1-3, wild-type, and ga1-3 rga-t2 gai-t6 rgl1 rgl2 plants. We found that the trichome phenotype of GA-treated ga1-3 leaves and stems is very similar to that of the wild type and the untreated quintuple mutant. Interestingly, GA applications to flowers, which inhibit trichome initiation in Columbia-0 plants (Gan et al., 2007), had a similarly negative effect on initiation in the wild-type Landsberg erecta (Ler) and quintuple mutant backgrounds (Table II). This observation suggested that the four DELLA proteins are not implicated in the repression of trichome initiation occurring on upper inflorescence organs when high GA doses are applied.
Table II.
Effect of GA applications on trichome initiation and vegetative phase change in ga1-3 rga-t2 gai-t6 rgl1 rgl2
Trichome counts are the total number of trichomes for about 15 flowers per plant. Values represent averages and ses for 20 plants.
| Sepals (∼15 Flowers) | Stems | Leaf (Abaxial) | ||||
|---|---|---|---|---|---|---|
| GA | 0 μm | 100 μm | 0 μm | 100 μm | 0 μm | 100 μm |
| Ler | 27.0 (1.7) | 20.1 (1.3) | 49.2 (1.7) | 48.8 (2.3) | 7.0 (0.3) | 7.5 (0.2) |
| ga1-3 | 1.2 (0.3) | 23.2 (1.2) | 0.0 (0.0) | 48.0 (2.6) | 0.0 (0.0) | 7.3 (0.4) |
| Five mutations | 26.3 (1.3) | 18.3 (0.8) | 49.8 (1.2) | 48.9 (1.6) | 7.2 (0.3) | 7.8 (0.2) |
| First Adult Leaf
|
||||||
| GA | 0 μm | 100 μm | ||||
| Ler | 4.9 (0.1) | 4.2 (0.1) | ||||
| ga1-3 | n.a. | 6.3 (0.1) | ||||
| Five mutations | 3.1 (0.1) | 3.0 (0.0) | ||||
DISCUSSION
In this study, we have shown that GA-mediated control over trichome initiation in Arabidopsis involves repression of most known trichome initiation activators by DELLA proteins RGA, GAI, RGL1, and RGL2. The onset of DELLA activity results in the rapid, but indirect, down-regulation of the corresponding genes, in particular of GL1, GL3, GIS, GIS2, and ZFP8. Consistently with these observations, we find that RGA, GAI, RGL1, and RGL2 collectively repress trichome initiation in Arabidopsis, RGA having the largest influence on this process. In contrast to its secondary role in initiation, GAI is the key repressor of trichome branching, antagonized in this process by RGL1 and RGL2. In addition to their role in epidermal differentiation, GAI, RGL1, and RGL2 act synergistically with RGA to repress vegetative and reproductive phase change. Finally, we show that GA applications generally recapitulate the effects of RGA/GAI/RGL1/RGL2 loss of function, except for their negative effect on flower trichome initiation, which suggests that other developmental signals and alternative regulators are implicated in this response.
DELLA Proteins Play an Indirect Role in Repressing the Expression of Genes Encoding Activators of Trichome Initiation
GAs are known to influence GL1 expression and, upstream, the expression of GIS, GIS2, and ZFP8 (Gan et al., 2007). Our results indicate that the influence of the phytohormones on the transcription of trichome initiation regulatory genes also extends to other members of the trichome initiation complex, in particular GL3. We have found that the control by GA of this process is mediated by the DELLAs, as DELLA derepression (loss of function) in a GA biosynthetic mutant restores the expression of GL1, GL3, GIS, GIS2, and ZFP8. These observations are consistent with the results of our genetic analysis and place the DELLA proteins upstream of all of these regulators (Fig. 7). Strikingly, we observed that the relative influence of each DELLA on trichome initiation generally mirrors their impact on the expression of the different trichome regulatory genes. This result highlights the importance of transcriptional regulation in modulating the influence of GA signaling and underlines the critical role played by transcription factors in this process (Perazza et al., 1998; Gan et al., 2006, 2007). Interestingly, decreases in GA levels have little effect on TTG1 expression; increases in GA signaling have, however, a positive effect (Supplemental Fig. S1). These observations suggest that TTG1 may be less responsive to GA signaling than other regulators of trichome production, but that, although its expression may not be limiting when GA signaling is decreased, its induction may promote initiation when DELLA repression is lifted.
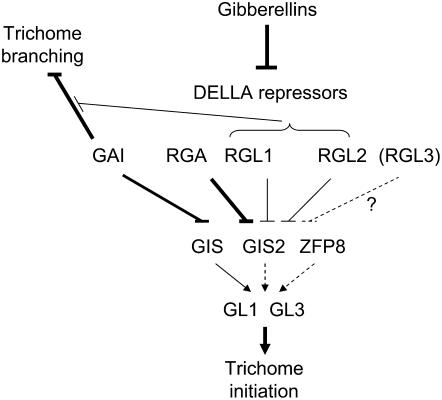
Figure 7.
Model of DELLA-mediated control of trichome initiation and branching. Dotted arrows indicate relationships that have not been fully characterized. Thicker lines indicate a more predominant effect in the pathway.
Whereas the repressive effects on GL1, GL3, GIS, and GIS2 of the DELLA proteins are clear, the results of our experiments with RGA-GR argue against a direct interaction between the repressors and their downstream targets. This situation is reminiscent of the interaction between RGA and homeotic genes, which is also indirect (Yu et al., 2004). An alternative interpretation of our results could be that cycloheximide treatments inhibited all responses in treated tissues and that the absence of a transcriptional effect on trichome regulatory genes was unspecific. We discount this possibility, although we cannot rule it out, on the basis that APETALA1 could be activated by LEAFY factor under similar conditions in similar tissues (Wagner et al., 1999). Regardless, it will be interesting to define the transcriptional networks acting immediately downstream of the DELLAs in the epidermis to start delineating the pathway leading from DELLA repression to trichome initiation.
DELLA Proteins Play a Central, Yet Contradictory, Role in Trichome Branching
Whereas GA applications are known to promote trichome branching (Perazza et al., 1998), the mechanism of GA action in this process has not been defined. Here, we show that the DELLAs play a key role in repressing branching. Surprisingly, their relative influences in branch formation and trichome initiation are very different. In particular, GAI plays a predominant role in the control of branching, but only plays a secondary role in trichome production. This is not only clear in stems, where GAI exerts the largest influence on initiation (Fig. 4B) and other processes (Dill and Sun, 2001), but also in leaves. RGA, in contrast, has little influence on branching despite playing a major role in initiation throughout the plant (Figs. 4, 5, and 7). The unique influence of GAI on this process is also reflected in the branching phenotype of single gai-t6 mutants, where all the other DELLAs are active. Possibly, GAI has specificity for downstream regulators of branching or other DELLA proteins are not produced at high enough levels in developing trichomes to have a clear influence.
The antagonism between RLG1/RGL2 and GAI with respect to branching is also surprising because the three proteins repress, although to differing degrees, the initiation process. One possible interpretation is that the mode of GAI action during trichome development is unusual and opposite to that of RGL1 and RGL2. Alternatively, a minimal level of DELLA activity may be required for branching to occur.
Roles of RGL1 and RGL2 in the Control of Trichome Initiation and Shoot Maturation
Previous studies have indicated that RGA plays a central role in leaf trichome initiation, flowering, and postgerminative growth (Silverstone et al., 1997; Dill and Sun, 2001). Our results confirm and indicate that RGA is also essential for trichome initiation throughout the life cycle and that GAI also plays an important, yet secondary, role. Interestingly, RGL1 and RGL2, which are mostly known for their involvement in flowering and germination (Lee et al., 2002; Wen and Chang, 2002; Tyler et al., 2004; Cao et al., 2005, 2006), also influence trichome initiation, although this influence is mainly incremental to that of the other DELLAs (Fig. 7). Surprisingly, despite its importance in flower morphogenesis, RGL2 only plays a minor role in flower trichome initiation. It is striking that the relative influences of the DELLAs on trichome initiation, vegetative and reproductive phase change are similar and that RGA and, to a lesser extent, GAI are predominant in all these processes. Our results therefore further support the central role of RGA in the control by GA of shoot maturation and epidermal differentiation and indicate that the synergy between the DELLAs extends to shoot maturation throughout the plant.
RGA, GAI, RGL1, and RGL2 Are Not Implicated in the Repression by GA of Trichome Initiation on Flowers
Loss of function of the four DELLA proteins generally results in trichome and shoot maturation phenotypes that are similar to those obtained with GA treatments. It does not, however, inhibit trichome formation on flowers, an effect that is observed when high doses of GA are applied, or in spy mutants (Greenboim-Wainberg et al., 2005; Gan et al., 2007). Furthermore, GA applications have the same inhibitory effect on flower trichome initiation in wild-type plants and in DELLA mutants, which rules out a role for the four repressors in mediating the GA response. In exerting their negative influence on flower trichome initiation, GAs therefore act through alternative regulators. One possibility is that they act through RGL3, although we consider it unlikely as RGL3 is expressed at low levels in reproductive organs (Tyler et al., 2004). Another possibility is that GAs impact on inflorescence trichome initiation through their effect on cytokinin signaling (Greenboim-Wainberg et al., 2005; Gan et al., 2007). Under this scenario, GA may directly influence cytokinin signaling without involving the DELLAs. A prime candidate in mediating this response would be SPY (Greenboim-Wainberg et al., 2005), although a role for alternative, upstream regulators cannot be ruled out.
CONCLUSION
Our study highlights the importance of repressive mechanisms modulating the action of transcriptional activators in the control by GAs of epidermal differentiation in Arabidopsis. We show that these mechanisms are complex and are likely to involve multiple steps, including the intervention of growth repressors of the DELLA family. These regulatory proteins play a central role in this developmental process as part of their larger control over shoot maturation, regulating both trichome initiation and development. Their influence may, however, be more limited in trichome production on flowers, where other regulators are likely to play a central role. It is expected that elucidating the molecular steps linking the DELLAs to downstream activators and the role played by other direct and indirect regulators will be facilitated by multifaceted approaches drawing from genetics, gene expression profiling, and protein biochemistry.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) mutants used in this study are in the Ler background unless otherwise stated. The DELLA mutant combinations in the ga1-3 background have been described before (Lee et al., 2002; Cao et al., 2005). The ga1-3 and gai mutants were obtained from the Nottingham Arabidopsis Stock Centre (NASC). To break dormancy, all ga1-3 mutants were imbibed with 100 μm GA at 4°C for 2 d, and then rinsed thoroughly with water before sowing. For all phenotypic analyses, plants were grown under 16-h-light (95 μmol cm−2 s−2, 21°C) and 8-h-dark (18°C) cycles. Inflorescence organs were harvested when the main stem had reached approximately 3 to 4 cm in length. Leaf trichome initiation was evaluated by counting trichomes in an 0.36 cm−2 area of the leaf midsection on both the adaxial and abaxial sides. Whole-leaf trichome production was extrapolated from these counts using average leaf areas. Stem trichome initiation and branching were monitored by counting all trichomes of each type on the first internode of main stems. Trichome production on the flower was measured by counting trichomes on all the sepals of 10 to 15 flowers. Unless specified otherwise, a minimum of 20 plants was used for trichome analysis for each treatment × genotype combination. Differences in average values were judged significant if they produced P < 0.05 in a one-sided paired t test. All experiments were repeated at least once.
Molecular Biology
Plasmid Constructs
ga1-3 rga-t2 gai-t6 35S:RGA-GR lines were produced by transforming ga1-3 rga-t2 gai-t6 plants with an RGA-GR overexpression construct (Yu et al., 2004). Transgenic plants containing 35S:RGA-GR were selected for their resistance to glufosinate (Basta) and the ability of RGA-GR to complement the mutant phenotype was tested by applying DEX. We focused on a particular overexpressing line containing only one copy of the transgene and with a phenotype that closely resembles that of ga1-3 gai-t6 after treatment with DEX.
Analysis of Gene Expression
Plant RNA was extracted from young inflorescence tissue using the TRIzol reagent (Invitrogen) according to the manufacturer's conditions. Pooled tissue samples from at least eight soil-grown plants were used for RNA extractions. The following gene-specific primer sequences were used for real-time PCR analysis: GL1, 5′-CGACTCTCCACCGTCATTGTT-3′ and 5′-TTCTCGTAGATATTTTCTTGTTGATGATG-3′; GL3, CCAGCAAGATCCGATTATCACA and ACTGAACATAGGCGCGTAAATCTC; TTG1, CCGTCTTTGGGAAATTAACGAA and GCTCGTTTTGCTGTTGTTGAGA; GIS, 5′-TTCATGAACGTCGAATCCTTCTC-3′ and 5′-ACGAATGGGTTTAGGGTTCTTATCT-3′; ZFP8, 5′-AAGCCGCCATTATTCGTCTCT-3′ and 5′-CTGCGGATAAGTTGTCGGAGTT-3′; GIS2, 5′-ACCGCCAACAAAACCACATT-3′ and 5′-CGCGTCGTTGATTTGAACAG-3′; and UBQ10, 5′-GGTTCGTACCTTTGTCCAAGCA-3′ and 5′-CCTTCGTTAAACCAAGCTCAGTATC-3′. Quantitative PCR primer design and reaction conditions were as previously described (Gan et al., 2006). UBQ10, whose expression is stable between different tissues and treatments (Gan et al., 2005), was used as an internal control for normalizing expression of the other genes. All reverse transcription-PCR experiments were performed twice.
GA and DEX Treatments
GA3 (Sigma) was used in all experiments that involved exogenous GA treatments and GA solutions were applied twice per week by spraying as previously described (Gan et al., 2006, 2007). DEX and cycloheximide treatments were performed according to Yu et al. (2004).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Influence of GA signaling and DELLA activity on the expression of genes encoding transcriptional activators of trichome initiation.
Supplemental Figure S2. Effects of overexpressing GIS or ZFP8 on flower trichome initiation in the ga1-3 rga-t2 gai-t6 background.
Supplemental Figure S3. Effect of DELLA mutations on reproductive and vegetative phase change.
Supplementary Material
Acknowledgments
We thank Nicholas Welsby and Katchan Thacker for their technical assistance.
This work was supported by a grant from the Garfield Weston Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pierre Broun (pierre.broun@rdto.nestle.com).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Cao D, Cheng H, Peng J, Wu W, Soo H, Peng J (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain H, Cheng H, Peng J (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 104–113 [DOI] [PubMed] [Google Scholar]
- Cheng H, Qin LJ, Lee SC, Fu XD, Richards DE, Cao DN, Luo D, Harberd NP, Peng JR (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064 [DOI] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun TP (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Ait-ali T, Hynes LW, Ougham H, Peng J, Harberd NP (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222 1–13 [DOI] [PubMed] [Google Scholar]
- Gan Y, Kumimoto R, Liu C, Ratcliffe OJ, Yu H, Broun P (2006) GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18 1383–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Liu C, Yu H, Broun P (2007) Integration of cytokinin and gibberellin signalling by redundant transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate in Arabidopsis. Development 134 2073–2081 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP (2003) Relieving DELLA restraint. Science 299 1853–1854 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Olszewski NE (2006) Rice GIBBERELLIN INSENSITIVE DWARF1 is a gibberellin receptor that illuminates and raises questions about GA signaling. Plant Cell 18 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Schnittger A, Folkers U (1999) Pattern formation and cell differentiation: trichomes in Arabidopsis as a genetic model system. Int Rev Cytol 186 147–178 [DOI] [PubMed] [Google Scholar]
- Hussain A, Cao D, Cheng H, Wen Z, Peng J (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J 44 88–99 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Lloyd AM, Paparozzi ET, Marks MD (1994) Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell 6 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (2001) Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128 1539–1546 [DOI] [PubMed] [Google Scholar]
- Lee SC, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng JR (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JR, Harberd NP (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113 1051–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hulskamp M, Brown S, Dorne AM, Bonneville JM (1999) Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M (1998) Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol 117 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kiitano H, Ashikari M, et al (2003) Accumulation of phosphorylated repressors for gibberellin signaling in an F-box mutant. Science 299 1896–1898 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun TP (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martinez EC, Sun TP (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 645–654 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698 [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville C (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM (2004) Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA 101 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 4859–4869 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.