Abstract
Fatty acyl esters of phytosterols are a major form of sterol conjugates distributed in many parts of plants. In this study we report an Arabidopsis (Arabidopsis thaliana) gene, AtSAT1 (At3g51970), which encodes for a novel sterol O-acyltransferase. When expressed in yeast (Saccharomyces cerevisiae), AtSAT1 mediated production of sterol esters enriched with lanosterol. Enzyme property assessment using cell-free lysate of yeast expressing AtSAT1 suggested the enzyme preferred cycloartenol as acyl acceptor and saturated fatty acyl-Coenyzme A as acyl donor. Taking a transgenic approach, we showed that Arabidopsis seeds overexpressing AtSAT1 accumulated fatty acyl esters of cycloartenol, accompanied by substantial decreases in ester content of campesterol and β-sitosterol. Furthermore, fatty acid components of sterol esters from the transgenic lines were enriched with saturated and long-chain fatty acids. The enhanced AtSAT1 expression resulted in decreased level of free sterols, but the total sterol content in the transgenic seeds increased by up to 60% compared to that in wild type. We conclude that AtSAT1 mediates phytosterol ester biosynthesis, alternative to the route previously described for phospholipid:sterol acyltransferase, and provides the molecular basis for modification of phytosterol ester level in seeds.
Plant sterols, known generally as phytosterols, are integral components of the membrane lipid bilayer (Demel and De Kruyff, 1976; Schuler et al., 1991). Unlike animal systems in which cholesterol is most often the lone final product of sterol synthesis, each plant species has its own characteristic distribution of phytosterols, with the three most common phytosterols in nature being β-sitosterol, campesterol, and stigmasterol (Benveniste, 1986, 2004). In addition to free sterol form, phytosterols are also found in the form of conjugates, particularly fatty acyl sterol esters (SEs). SEs can be found in lipid bodies in the cytoplasm and are present at a substantial level in seeds. In canola (Brassica napus), for example, phytosterols constitute about 0.5% of seed oil (Sabir et al., 2003), 35% of which is in the form of SEs (Harker et al., 2003). The biochemical process of sterol acylation is believed to play a role in maintaining the free sterol content of cell membranes at their physiological levels (Schaller, 2004).
The effectiveness of phytosterols as a dietary component to lower serum cholesterol level in humans has been well documented in medical research for more than half a century (Moghadasian et al., 1997, 1999; Hendriks et al., 1999; Sierksma et al., 1999; Piironen et al., 2000; van Rensburg et al., 2000; Awad et al., 2001, 2003; Bouic, 2001). It also has been suggested that the average diet of the western world has a phytosterol intake far below prehistoric levels (Yankah and Jone, 2001). These studies led to recommendations of consumption of phytosterol-enriched food products by the general population and particularly mildly hypercholesterolemic subjects. However, limited quantities of phytosterols are currently a major barrier in satisfying the demands for such functional foods. Based on a firmly established concept that 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) plays a rate-limiting role in the flux control of sterol biosynthesis (Bach, 1986; Gondet et al., 1992, 1994; Chappell et al., 1995), efforts aimed at increasing phytosterol content have been concentrated on enhancing the activity of HMGR through introducing modified HMGR that is resistant to regulation (Chappell et al., 1995; Re et al., 1995; Schaller et al., 1995; Harker et al., 2003; Hey et al., 2006). Nonetheless, evidence suggesting the existence of other regulatory step(s) in the flux control of sterol biosynthesis can be found in some studies, particularly that of Gondet et al. (1994) who studied a sterol-overproducing tobacco (Nicotiana tabacum) mutant LAB1-4, with HMGR levels approximately 3-fold higher than normal (Gondet et al., 1994). LAB1-4 had a 10-fold stimulation of sterol content in calli as compared to wild type, mostly in the form of SEs, but a mere 3-fold stimulation in the regenerated leaves. Intriguingly, both type of tissues had the same stimulation factor of HMGR (3-fold). Therefore, there appears to be other contributing factor(s), independent of HMGR, participating in regulating total sterol content.
Despite the ubiquitous presence of phytosterol esters in plant cells, the metabolic process of sterol acylation remains poorly understood. Sterol esterification is suggested to take place using several acyl donors including phospholipid, diacylglycerol, or triacylglycerol (Zimowski and Wojciechowski, 1981a, 1981b; Dyas and Goad, 1993). A recent report on the phospholipid:sterol O-acyltransferase gene from Arabidopsis (Arabidopsis thaliana; AtPSAT) established a route of transacylation between phospholipids and sterols in plant SE synthesis (Banaś et al., 2005). AtPSAT is a lecithin:cholesterol acyltransferase protein family member homologous to human soluble lecithin:cholesterol acyltransferase. In this study, we took a functional complementation approach using a yeast (Saccharomyces cerevisiae) strain in which two sterol O-acyltransferase-encoding genes, ARE1 and ARE2, are disrupted. We identified a novel Arabidopsis sterol O-acyltransferase AtSAT1, which is structurally related to the acyl-CoA cholesterol acyltransferase (ACAT) in animal systems. We present evidence that overexpression of AtSAT1 alters SE synthesis in Arabidopsis, providing elevated levels of commercially desirable phytosterols.
RESULTS
Cloning of AtSAT1 from Arabidopsis
Taking a yeast functional complementation strategy (Bach and Benveniste, 1997), we focused our search of a plant sterol O-acyltransferase to the superfamily of membrane-bound O-acyltransferases (MBOATs; Hofmann, 2000), because the common biochemical functionality of the MBOAT family proteins is to mediate the transfer of organic acids in ester or thioester forms to hydroxyl groups of membrane-embedded acyl acceptors.
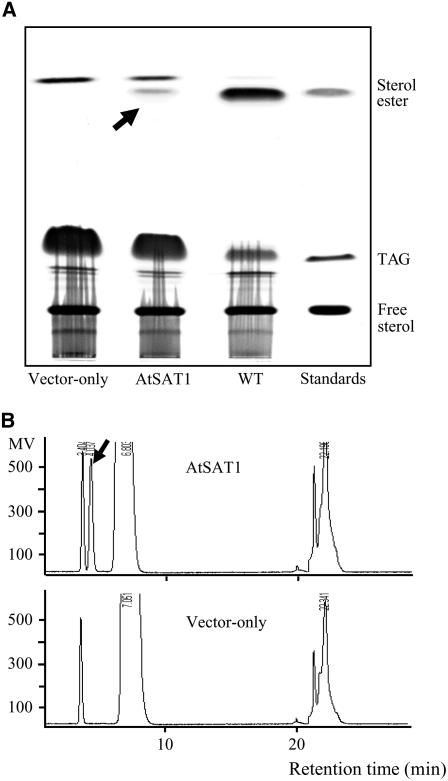
The yeast mutant SCY059 produces only a residual amount of SE because the two principle sterol O-acyltransferase genes, ARE1 and ARE2, were interrupted (Yang et al., 1996). Employing this strain, we introduced members of the Arabidopsis MBOAT family to test functional complementation. Fifteen cDNAs corresponding to MBOAT family candidate genes (listed in “Materials and Methods”) were cloned into the yeast expression vector pYES2.1 under the control of GAL1 promoter. The yeast transformants were subjected to GAL1 induction condition and harvested for neutral lipid analysis. When separated by thin-layer chromatography (TLC), SCY059 harboring At3g51970 produced an additional band in the neutral lipid extraction (Fig. 1A). HPLC analysis also revealed similar results from these cells (Fig. 1B). Both the TLC Rf value and the HPLC retention time of the novel products were consistent with that of SE.
Figure 1.
TLC and HPLC separation of neutral lipids from SCY059 harboring control vector (vector-only), SCY059 expressing AtSAT1 (AtSAT1), and a parental strain SCY062 (WT). A, Neutral lipids separated through TLC developed in hexane/diethyl ether/acetic acid (40/10/1.5). B, Normal-phase HPLC separation of neutral lipids extracted from SCY059 expressing AtSAT1 (AtSAT1) and SCY059 harboring the vector (vector-only). Arrows denote the novel product found in SCY059 expressing AtSAT1.
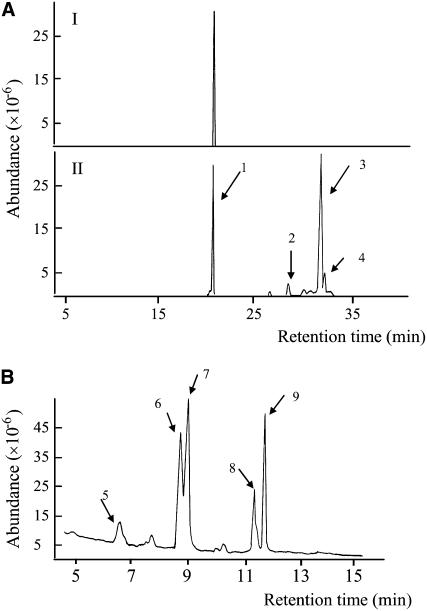
To verify the identity of the product, the HPLC fraction was collected and subjected to alkaline hydrolysis (saponification). The saponification extract was derivatized with N,O-bis(trimethysily)-trifluoroacetamide + 1% trimethylchlorosilane (BSTFA + 1% TMCS) and detected as trimethylsilyl (TMS) derivatives by gas chromatography (GC)-mass spectrometry (MS). GC profile revealed four major peaks from the sterol fraction (Fig. 2A). All the GC fractions were identified by searching the National Institute of Standards and Technology (NIST) 2.0 mass spectra library. The first peak, also present in the control strain, corresponded to squalene. The other three peaks displayed mass spectra identical to that of ergosterol, lanosterol, and 4,4-dimethyl-8,24-cholestadienol (also known as 4,4-dimethylzymosterol), respectively (Fig. 2A). The five peaks from the fatty acid fraction were found to be 14:0, 16:1, 16:0, 18:1, and 18:0 fatty acids (Fig. 2B). These results confirmed that expression of At3g51970 conferred SCY059 an ability to produce SE and At3g51970 was then tentatively designated as AtSAT1.
Figure 2.
Analysis of TMS derivatives of sterols and fatty acids of SE. A, GC profiles of sterols saponified from SE of yeast strain SCY059 harboring control vector (I) and SCY059 expressing AtSAT1 (II). B, GC profile of TMS derivatives of fatty acids released from SE of AtSAT1 yeast transformant. The TMS derivatives of peaks in the profiles were subsequently subjected to mass spectrum analysis and the mass spectra were found as being identical to: 1, Squalene; 2, ergosterol; 3, lanosterol; 4, 4,4-dimethyl-8,24-cholestadienol; 5, myristic acid (14:0); 6, palmitoleic acid (16:1); 7, palmitic acid (16:0); 8, oleic acid (18:1); and 9, stearic acid (18:0).
AtSAT1 Is Structurally Related to Acyl-CoA Sterol Acyltransferases of Yeast and Animal Origins
AtSAT1 is predicted to be an integral membrane protein with eight transmembrane domains, and has a putative signal peptide with a cleavage site at the 19th amino acid from the N terminus, which was proposed to direct nascent proteins into the secretory pathway, including the endoplasm reticulum (Nielsen et al., 1997). Deduced amino acid sequence alignment shows that AtSAT1 is structurally related to acyl-CoA sterol acyltransferase from yeast and animal systems (Fig. 3), among which ACATs have been extensively studied. The RxWNxxVxxxLxxxVY motif present in all ACATs, commonly believed to be crucial for fatty acyl-CoA binding (Guo et al., 2001), is also found in AtSAT1. A His residue (His245 in AtSAT1) located within a long stretch of hydrophobic amino acids is present at the C terminus of AtSAT1. This conserved His residue was suggested to be a part of the active site (Hofmann, 2000). However, when compared with other ACATs, AtSAT1 protein is a shorter polypeptide, lacking the extended N-terminal region found in other sterol acyltransferases. Moreover, two conservative motifs, FYxDWWN and H/YSF previously suggested as being crucial for ACAT activity (Cao et al., 1996; Oelkers et al., 1998), were also absent in AtSAT1.
Figure 3.
Sequence alignment of AtSAT1 with other reported sterol acyltransferases. Alignment was performed with CLUSTALV from the DNASTAR package run with default multiple alignment parameters (gap opening penalty: 10, gap extension penalty: 10). Accession numbers of proteins are: AtSAT1 (Arabidopsis, AAQ65159); HsACAT1 (Homo sapiens, NP_003092); HsACAT2 (H. sapiens, NP_003569); MmACAT1 (Mus musculus, Q61263); MmACAT2 (M. musculus, NP_666176); RnACAT1 (Rattus norvegicus, O70536); RnACAT2 (R. norvegicus, NP_714950); ScARE1 (yeast, P25628); and ScARE2 (yeast, P53629). Residues boxed in dashed or solid lines were previously reported as conserved functional motifs in ACATs. The black bar on the bottom marks a shared domain, and the arrow indicates the His likely to be an active-site residue (Hofmann, 2000).
Heterologous Expression of AtSAT1 in Yeast Produces Mainly Lanosterol Esters
We estimated SE content of the yeast strains by quantifying sterols released through alkaline hydrolysis. As reported previously (Yang et al., 1996), the vector-only transformant of SCY059 still produced a trace amount of SE (0.027 μmol/g dry weight [DW] of yeast cells). The same strain heterologously expressing AtSAT1 increased SE production by 10-fold to 2.7 μmol/g DW, though still lower than 9.4 μmol/g DW of parental wild-type strain SCY062. The levels of free sterols in SCY059, SCY059/AtSAT1, and SCY062 were found at 18.1 μmol/g DW, 14.9 μmol/g DW, and 13.2 μmol/g DW, respectively.
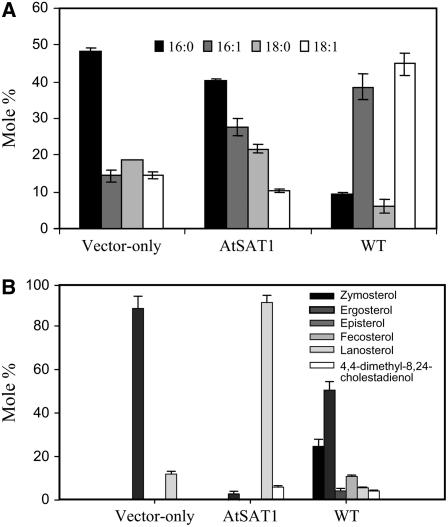
Considering SCY059 still retained a low level of SE synthesis capacity, there was the possibility of AtSAT1 being a facilitator, enhancing the residual SE synthesis activity in the mutant strain. If this were true, SCY059 harboring AtSAT1 would likely produce SE with a profile of sterol or fatty acid moiety similar to that of the vector-alone transformant. This was experimentally proven untrue. As shown in Figure 4A, the molar composition of the fatty acid species saponified from the SE collected from the vector-only strain and the SEs produced by SCY059/AtSAT1 were different. Likewise, the sterol composition of the SE also displayed major differences due to the expression of AtSAT1 (Fig. 4B). The residual SE in SCY059/empty vector consisted of mainly ergosterol. On the other hand, expression of AtSAT1 resulted in production of SE containing chiefly lanosterol, which contributed to 88.1% of total sterol moiety. Thus, AtSAT1 appeared to preferentially acylate lanosterol when expressed in yeast.
Figure 4.
Fatty acid profile and sterol composition of SE in yeast strains. A, Mol % of fatty acid species released from SE of SCY059 expressing AtSAT1 in comparison to that of the vector-only control and the parental strain (WT). B, Profiles of the sterol moieties of SE. The SE was saponified in methanolic-KOH for 2 h at 80°C. The fatty acids were transmethylated with methanolic HCl and free sterols were derivatized with BSTFA (1% TMCS).
Substrate Preference of AtSAT1
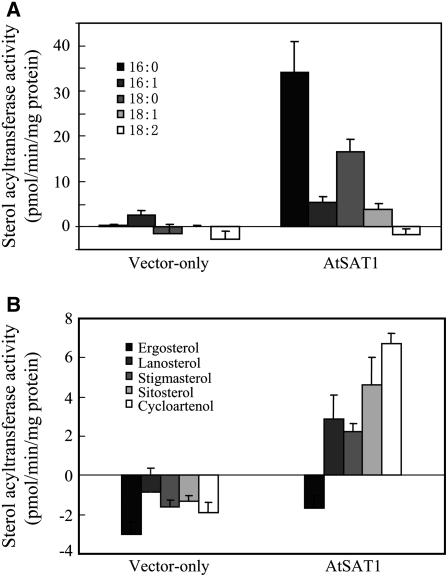
In light of substrate preferences of ACAT-related enzymes reported to date, we examined fatty acyl donor preferences of AtSAT1 using cell-free lysate of yeast with various fatty acyl-CoAs. Lipids extracted from in vitro reactions were separated by TLC and the incorporation of [3H]lanosterol into SE was measured. Because the cell-free lysate was expected to contain certain amount of acyl-CoA, reactions without added acyl-CoA were set as blank controls for enzyme activity calculation. As shown in Figure 5A, AtSAT1 had a substrate preference in the order of 16:0 > 18:0 > 16:1 > 18:1. 18:2 was found to negatively affect sterol O-acyltransfease activity.
Figure 5.
Substrate preference assessment of AtSAT1. A, Fatty acyl-CoA selectivity of AtSAT1. Cell-free lysates were assayed for acylation of [3H]lanosterol. Fatty acyl-CoAs used for the assay were palmitoyl-CoA (16:0), palmitoleoyl-CoA (16:1), stearoyl-CoA (18:0), oleoyl-CoA (18:1), and linoleoyl-CoA (18:2). B, Sterol preference of AtSAT1. Cell-free lysates were assessed for acylation of ergosterol, lanosterol, stigmasterol, β-sitosterol, and cycloartenol in the presence of [14C]palmitoyal-CoA. Reactions without exogenous sterol (or fatty acyl-CoA) served as control. Enzyme activity was calculated based on the difference of sterol acylation between reactions with added sterol (or fatty acyl-CoAs) substrates and reactions without sterols (or fatty acyl-CoAs). The negative values in some reactions were caused by high background sterol acylation in the control in the absence of exogenous sterol substrates.
We used stigmasterol, β-sitosterol, and cycloartenol as well as two sterols of yeast origin, lanosterol and ergosterol, to assess [14C]16:0-CoA acylation. Assessment of sterol substrate preferences entailed a methyl-β-cyclodextrin wash to extract free sterols from the preparations (Rodal et al., 1999), due to the concern that endogenous sterols may interfere with substrate specificity assessment. However, there apparently was still a substantial amount of endogenous sterols remaining, because the background value in the absence of added sterol substrate was nonetheless high. We also attempted acetone extraction of the membrane fractions to reduce the endogenous sterol levels. Unfortunately the acetone treatment essentially eliminated the enzyme activity (data not shown). This led us to use reactions without exogenous sterol additions as control experiments. Under our assay conditions, the yeast strain harboring empty vector displayed a reduced 16:0 acylation activity in the presence of exogenous sterol. Addition of ergosterol to the yeast lysate expressing AtSAT1 also gave rise to a negative value on SE formation. Such a negative effect of exogenous sterol addition on sterol O-acyltransferase activity was previously reported for animal ACAT (Tavani et al., 1982; Yang et al., 1997) and the Arabidopsis AtPSAT1 assays (Banaś et al., 2005). Despite these results, we could consistently detect SE synthesis activity with most of the sterol substrates when AtSAT1 was expressed. The highest activity was found with cycloartenol, followed by β-sitosterol, lanosterol, and stigmasterol (Fig. 5B).
Because phosphatidylcholine (PC) has been previously implicated as an acyl donor for SE synthesis (Banaś et al., 2005), the ability of AtSAT1 to use this lipid as substrate was assessed using dipalmitoyl-PC with [14C]label in both the sn-1 and sn-2 fatty acyl moieties. We found that, while the PC could indeed sustain SE synthesis above the background of the control yeast strain, it was nonetheless at a level one-fiftieth of that when 16:0-CoA was provided. We also expressed AtSAT1 in the are1are2lro1dga1 quadruple yeast mutant devoid of triacylglycerol (Sandager et al., 2002). While the formation of triacylglycerols was not detectable, the quadruple yeast mutant expressing AtSAT1 was still capable of producing SE. This result was significant in two respects: first, it suggested that AtSAT1 did not require triacyglycerols (TAGs) as a fatty acyl donor for SE biosynthesis, and second, it could not acylate diacylglycerol to produce TAG.
Expression Profile of AtSAT1
Based on transcript profile obtained from Genevestigator at https://www.genevestigator.ethz.ch/, AtSAT1 is expressed in all tissues examined, but particularly abundant in the elongation zone of roots followed by developing microspore and germinating seedlings. Previous published works (Ting et al., 1998; Hernández-Pinzón et al., 1999) have shown the accumulation of SE in developing pollen and seed. AtSAT1 expression level increased as pollen development proceeded from uninucleate microspore to bicellular pollen, tricellular pollen, and mature pollen (Honys and Twell, 2004). Increased expression of AtSAT1 was also an evident trend during seed development.
Overexpression of AtSAT1 in Arabidopsis Leads to Cycloartenol Ester Overproduction
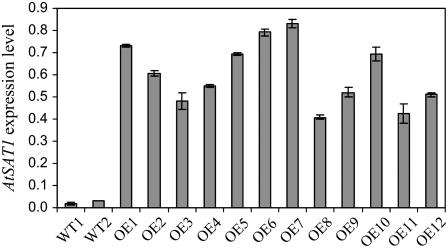
To further confirm that AtSAT1 is involved in SE biosynthesis in plants, we cloned AtSAT1 cDNA into a plant expression vector under the control of a seed-specific napin promoter (Josefsson et al., 1987). The transgenic plants showed no apparent developmental difference from wild-type plants. Real-time quantitative (qRT)-PCR revealed that AtSAT1 was approximately 20-fold overexpressed in developing siliques of most AtSAT1 transgenic lines (Fig. 6).
Figure 6.
qRT-PCR analysis of transcript levels of AtSAT1. Each sample contained RNA extracted from pooled developing siliques. qRT-PCR was performed using Applied Biosystem StepOne Real-Time PCR system. CT was automatically determined by software StepOne (version 1.0). AtSAT1 expression levels were calculated through normalization with β-Actin-8 by the formula 2−(CTAtSAT1 − CTβ-actin). Data represent the mean and sd of three replicates.
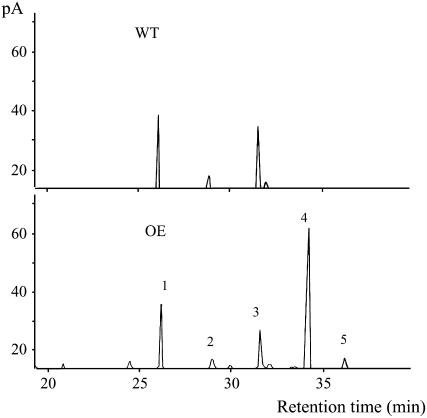
Seeds harvested from four wild-type plants and 12 transgenic plant (T1) lines, raised simultaneously in the same growth chamber, were selected for sterol analysis. AtSAT1 overexpression led to increases in SE by more than 2-fold and total sterol content by 64% in some transgenic lines (Table I). Overexpression of AtSAT1 in seed, as exemplified by one transgenic line shown in Fig. 7, also drastically altered the SE-sterol profile. In wild-type seeds, the two major SE species, β-sitosterol ester and campesterol ester, accounted for 80.4% and 17.5% of total SE, respectively. Cycloartenol ester was detected as a minor component at a distant third with 2.1%. Strikingly, the AtSAT1 transgenic seeds produced cycloartenol ester as the most prominent component, representing up to an average of 64.1% of total SE. On the other hand, the esters of β-sitosterol and campesterol were reduced to averages of 25.2% and 5.2%, respectively. We also found that 24-methylene cycloartenol was increased from nondetectable level in wild-type seeds to 5.6% in the transgenic line seeds. The enhanced SE overproduction also caused reduction of free sterol from 58.8% in wild type to 28.3% of total sterols in transgenic plants.
Table I.
Content of phytosterol esters and free sterols in wild-type and AtSAT1 transgenic seeds
Arabidopsis (Columbia) plants were transformed with vector pSE129∷AtSAT1, in which AtSAT1 was under the control of a napin promoter. The four wild-type (WT) controls and 12 AtSAT1 overexpression (OE) lines were raised under identical conditions. Lipids were extracted in chloroform/methanol (2/1) from WT and transgenic seeds, followed by separation via normal-phase HPLC. SE fractions were collected and saponified in 7.5% methanolic KOH. Released free sterols were derivatized with BSTFA containing 1% TMCS and quantified by GC. The data were presented as averages of two analyses. Camp, Campesterol; Sito, sitosterol; Cyclo, cycloartenol; MethyCyclo, 24-methylene cycloartenol; nd, nondetectable.
| Lines | SE Composition
|
Total SE Sterol | Free Sterol Composition
|
Total Free Sterol | Total Sterol | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Camp | Sito | Cyclo | Methy-Cyclol | Camp | Sito | Cyclo | Methy Cyclo | ||||
| % | % | % | % | μg/mg | % | % | % | % | μg/mg | μg/mg | |
| WT1 | 16.3 | 79.8 | 4.0 | nd | 1.7 | 12.1 | 74.9 | 1.3 | 11.8 | 2.1 | 3.8 |
| WT2 | 18.2 | 78.9 | 2.9 | nd | 1.6 | 10.3 | 69.7 | 0.0 | 20.1 | 2.6 | 4.2 |
| WT3 | 17.3 | 82.7 | 0.0 | nd | 1.8 | 11.8 | 66.4 | 0.6 | 21.2 | 2.9 | 4.7 |
| WT4 | 18.1 | 80.2 | 1.7 | nd | 1.6 | 14.8 | 69.8 | 0.9 | 14.6 | 2.1 | 3.7 |
| OE1 | 2.7 | 18.0 | 73.0 | 6.3 | 4.5 | 16.3 | 74.2 | 1.7 | 7.7 | 1.2 | 5.7 |
| OE2 | 5.3 | 27.8 | 61.5 | 5.5 | 3.5 | 12.5 | 69.3 | 2.5 | 15.7 | 2.1 | 5.6 |
| OE3 | 7.3 | 34.5 | 52.3 | 5.8 | 3.4 | 11.6 | 70.3 | 1.1 | 17.1 | 1.8 | 5.2 |
| OE4 | 5.3 | 26.4 | 63.7 | 4.7 | 4.0 | 11.0 | 66.4 | 0.9 | 21.8 | 1.8 | 5.9 |
| OE5 | 4.6 | 22.0 | 67.7 | 5.7 | 4.6 | 11.1 | 63.9 | 2.1 | 22.9 | 1.7 | 6.3 |
| OE6 | 4.1 | 19.7 | 70.1 | 6.1 | 5.0 | 12.1 | 64.0 | 1.9 | 22.1 | 1.8 | 6.7 |
| OE7 | 3.7 | 19.4 | 71.8 | 5.2 | 5.2 | 14.2 | 68.1 | 2.2 | 15.6 | 1.4 | 6.6 |
| OE8 | 7.0 | 30.5 | 57.6 | 5.0 | 3.1 | 16.7 | 67.2 | 2.6 | 13.5 | 1.5 | 4.6 |
| OE9 | 5.6 | 27.8 | 60.9 | 5.8 | 3.8 | 18.7 | 73.7 | 3.4 | 4.3 | 1.2 | 5.0 |
| OE10 | 4.5 | 20.6 | 69.5 | 5.4 | 4.3 | 15.1 | 67.9 | 2.5 | 14.5 | 1.4 | 5.7 |
| OE11 | 6.9 | 31.2 | 56.1 | 5.9 | 3.2 | 12.3 | 67.1 | 1.9 | 18.8 | 2.0 | 5.2 |
| OE12 | 5.0 | 24.3 | 64.5 | 6.3 | 3.8 | 12.7 | 68.7 | 1.5 | 17.1 | 1.3 | 5.1 |
Figure 7.
GC profiles of sterols saponified from SE extracted from seeds of wild type (WT) and one representative AtSAT1 overexpression line. Lipids were extracted from seeds with chloroform:methanol (2:1, v/v) and separated though normal-phase HPLC. SE fractions were collected and saponified with 7.5% KOH in 95% methanol. The resulting free sterols were derivatized with BSTFA:pyridine (1:1, v/v). Identification of mass spectra and assignation of GC/MS peaks was carried out by a library search (NIST, version 2.0). 1, Cholesterol (internal standard); 2, campesterol; 3, β-sitosterol; 4, cycloartenol; and 5, 24-methylene cycloartenol.
We further analyzed the profile of the fatty acid component of the SEs in seeds overexpressing AtSAT1 (Table II). In comparison to wild-type seeds, there was a drastic increase in the molar percentage of 16:0, from less than 15% to approaching 30%. Significant increases were also apparent in the composition of very long-chain fatty acids 20:0 and 20:1. The overall changes in fatty acid component of the SEs in the transgenic lines can be summarized as saturated and long-chain fatty acids replacing unsaturated fatty acids in wild-type seeds, particularly 18:2 and 18:3. The fatty acid composition of SE in AtSAT1-overexpressed seeds was consistent with the in vitro fatty-CoA specificity results.
Table II.
Fatty acid composition of SEs in wild-type (WT) and AtSAT1 overexpression (OE) plants
WT and transgenic seeds were from the same lines as that in Table I. SE purified through HPLC was transmethylated in methanolic HCl. The resultant fatty acid methyl ester products were quantified by GC for fatty acid mole ratio calculation. The data were presented as averages of two analyses.
| Lines | 16:0 | 16:1 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 |
|---|---|---|---|---|---|---|---|---|
| WT1 | 9.0 | 3.1 | 3.0 | 6.6 | 50.7 | 23.3 | 0.1 | 4.3 |
| WT2 | 16.0 | 3.6 | 7.9 | 11.2 | 38.6 | 17.3 | 2.4 | 3.0 |
| WT3 | 13.5 | 3.1 | 4.0 | 11.6 | 33.9 | 22.7 | 2.1 | 8.3 |
| WT4 | 14.0 | 7.3 | 4.7 | 9.1 | 41.0 | 20.8 | 0.1 | 3.1 |
| OE1 | 28.3 | 2.6 | 14.5 | 11.5 | 9.1 | 4.1 | 8.1 | 21.9 |
| OE2 | 29.3 | 2.3 | 9.2 | 9.0 | 13.1 | 6.0 | 8.3 | 22.6 |
| OE3 | 24.7 | 2.4 | 8.0 | 9.4 | 17.9 | 8.7 | 7.5 | 21.4 |
| OE4 | 22.6 | 6.8 | 8.6 | 14.4 | 15.8 | 8.3 | 6.0 | 17.6 |
| OE5 | 32.1 | 2.2 | 8.7 | 8.8 | 11.6 | 4.3 | 9.1 | 23.2 |
| OE6 | 29.1 | 1.6 | 8.5 | 9.8 | 10.8 | 4.6 | 9.0 | 26.5 |
| OE7 | 28.5 | 2.1 | 8.3 | 8.7 | 12.3 | 5.0 | 9.3 | 25.7 |
| OE8 | 29.1 | 2.9 | 8.3 | 9.0 | 11.9 | 4.5 | 9.2 | 24.9 |
| OE9 | 25.3 | 2.3 | 10.2 | 10.9 | 15.8 | 6.8 | 7.2 | 21.5 |
| OE10 | 27.6 | 2.4 | 9.7 | 8.4 | 11.9 | 5.6 | 9.4 | 25.0 |
| OE11 | 24.4 | 0.9 | 7.6 | 9.8 | 18.3 | 8.1 | 8.4 | 22.6 |
| OE12 | 23.7 | 2.2 | 8.8 | 11.6 | 15.2 | 6.3 | 8.2 | 24.0 |
DISCUSSION
In this study we identified AtSAT1 as a sterol O-acyltransferase from Arabidopsis that belongs to the MBOAT family. Our conclusion on the functional identity of AtSAT1 was based on the following lines of evidence: (1) heterologous expression of AtSAT1 enabled a yeast SE-deficient mutant SCY059 to synthesize a considerable amount of SE; (2) AtSAT1 mediated the synthesis of SE with sterol and fatty acid profiles distinctively different from that of the trace amount of SE synthesized in the yeast mutant strain; and (3) transgenic overexpression of AtSAT1 in Arabidopsis substantially increased SE content in seeds. Furthermore, since we did not find detectable levels of TAG when expressing AtSAT1 in the are1are2lro1dga1 quadruple yeast mutant, it appears that AtSAT1 could not acylate diacylglycerol for TAG biosynthesis.
Expression of AtSAT1 in yeast strain SCY059 resulted in the accumulation of SE consisting mainly of lanosterol, rather than the most abundant sterol being ergosterol, the end product of the sterol biosynthesis pathway in yeast (Zweytick et al., 2000), suggesting that AtSAT1 possesses a certain degree of sterol substrate specificity. The SE composition resulting from expression of AtSAT1 was clearly different from reports of overexpressing the ARE1 and ARE2 genes in SCY059, which produced SE enriched in zymosterol and erogosterol (Jensen-Pergakes et al., 2001). Thus, it can also be inferred that sterol preference of AtSAT1 is distinctively different from that of the yeast sterol O-acyltransferases. AtSAT1 also appeared to possess substrate preference toward fatty acyl donors, displaying the highest activity when saturated fatty acyl-CoAs (16:0 and 18:0) were provided. This is in contrast to the Arabidopsis phospholipid sterol O-acyltransferase AtPSAT, which discriminates against saturated fatty acids and was suggested to play a minor role in the synthesis of SE with saturated fatty acids (Banaś et al., 2005). On this basis, it is important to note that previously published work has established that SE in some tissues of Brassica and Arabidopsis are highly enriched in saturated fatty acids (Hernández-Pinzón et al., 1999).
Data obtained from transgenic plants were generally in line with results of enzyme property assessment performed through heterologous expression of AtSAT1 in yeast. Consistent with the high enzyme activity detected with cycloartenol in our in vitro assays, enhanced AtSAT1 expression drastically advanced accumulation of cycloartenol esters in Arabidopsis seeds. Likewise, for the fatty acid components of the SEs, we observed increases in the molar ratio of saturated fatty acids, including 16:0, 18:0, and 20:0. Elevated sterol acylation with the long-chain monounsaturated eicosenoic acid was also evident. On the other hand, the polyunsaturated fatty acids were proportionally reduced.
Previous biochemical studies suggested that there are several potential acyl donors for SE biosynthesis in plants (Garcia and Mudd, 1978a, 1978b, 1978c; Zimowski and Wojciechowski, 1981a, 1981b), including acyl-CoA, phospholipids, diacylglycerol, and triglyceride. In the case of AtSAT1, triglyceride can be ruled out because TAG-deficient are1are2lro1dga1 quadruple yeast mutant expressing AtSAT1 produced SE. When PC was supplied as an acyl donor the detected sterol acylation was above residual level of the control, but far below that in the presence of acyl-CoA. It is unclear whether PC served as an immediate acyl donor or indirectly through mobilization of acyl group from PC to the fatty acyl-CoA pool. We suggest that AtSAT1 is an acyl-CoA:sterol acyltransferase. However, since the specific activity in our assay was lower than that of other reported ACATs (Macauley et al., 1986; Yang et al., 1997), and our in vitro assays with yeast cell lysate was complicated by a high background of sterol acylation, this supposition should be approached with caution. In light of the fact that AtSAT1 belongs to a large family of proteins in which some have promiscuous substrate preference, the substrate specificity of AtSAT1 requires further study.
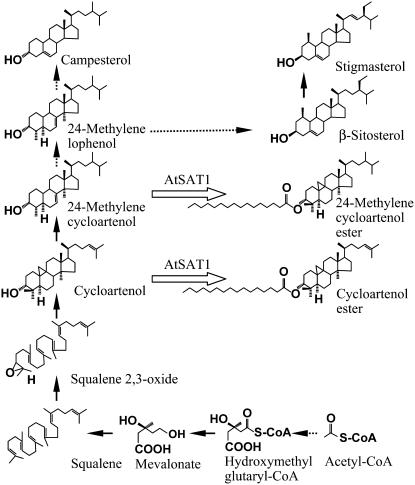
SE has been long proposed to be the storage and transport form of sterols for their intracellular movement (Kemp et al., 1967; Janiszowska and Kasprzyk, 1977) and movement between tissues (Holmer et al., 1973). Radiolabeling experiments in Apium graveolens suspension cell culture indicated SE pool is under rapid turnover with esterification and hydrolysis process occurring concomitantly (Dyas and Goad, 1993). It was also proposed that esterification and hydrolysis should occur alternatively to newly synthesized sterol intermediates for orderly structural modification until end product 4-desmethylsterols were formed. In our study, seed-specific overexpression of AtSAT1 in Arabidopsis resulted in the accumulation of cycloartenol esters, which otherwise occur only as a minor sterol component in wild-type seeds. Taken together with the in vitro sterol substrate preference results, the simplest explanation for such a metabolic outcome is that AtSAT1 selectively channeled cycloartenol into the ester pool (Fig. 8). Cycloartenol is the first tetracyclic sterol precursors in plant sterol biosynthesis pathway, and is considered an intermediate in the phytosterol biosynthesis pathway equivalent to lanosterol in yeast and animal systems (Benveniste 1986; Lovato et al., 2000; Kolesnikova et al., 2006; Suzuki et al., 2006). After cycloartenol, several major biochemical steps including methyltransferase, demethylation, and desaturation reactions need to take place before the final products of β-sitosterol and campesterol can be formed (Benveniste, 2004). The elevated expression of AtSAT1 could enhance the esterification of cycloartenol, thereby removing cycloartenol from the mainstream of sterol biosynthetic pathway into the formation of esters. This is consistent with our results in the transgenic seeds where the contents of β-sitosterol and campesterol, regardless if in the form of free or esterified forms, were reduced when compared to wild type. A question remains as to the physiological significance of such a role of AtSAT1 in plant metabolism and development. While highly speculative, we suggest that because cycloartenol has much better micellar solubility than the sterol end products, a role for AtSAT1 is to synthesize cycloartenol esters, which may serve as a significant form of transported sterol conjugates in certain developmental stages.
Figure 8.
A proposed role for AtSAT1 in the sterol biosynthesis pathway. The diagram accentuates the section where the conversion of squalene to sterol end products occurs. Solid lines denote single-step enzymatic reactions and dashed lines represent multiple steps. The open arrows suggest the roles of AtSAT1 in sterol biosynthesis.
Incorporating phytosterols into the diet may be an effective approach to lower total and LDL cholesterol levels in humans, but free phytosterols are difficult to incorporate into commercial foods because of their low solubility. Fortunately, phytosterol esters can be dissolved in vegetable oil at a concentration 10 times higher than that of the free phytosterols. In this study, we found that overexpression of AtSAT1 could lead to enhanced total sterol content, primarily due to the increase in cycloartenol ester level. Cycloartenol is particularly enriched in oryzanol derived from rice (Oryza sativa) bran oil (Rukmini and Raghuram, 1991). It has been suggested that cycloartenol is the effective ingredient in rice bran oil for lowering plasma non-HDL cholesterol level (Wilson et al., 2007). We believe that the engineering outcome of the current research is likely to be repeated in crops species, particularly canola, offering a novel genetic engineering target for enhanced phytosterol content in vegetable oil. We must note, although the increase in seed oil SE content was significant, increase in total sterol content was still modest. Further studies on the regulation of phytosterol biosynthesis are clearly required to improve phytosterol content in seeds.
MATERIALS AND METHODS
TOPA TA Cloning and Yeast Complementation
RT-PCR of the open reading frames of Arabidopsis (Arabidopsis thaliana) MBOAT family genes At1g12460, At1g34490, At1g34500, At1g34520, At1g57600, At3g08930, At3g51970, At5g01460, At5g55320, At5g55330, At5g55340, At5g55350, At5g55360, At5g55370, and At5g55380 was performed with primer pairs designed based on sequences of gene annotation available on The Arabidopsis Information Resource Web site. The primer pair for At3g51970 cDNA was 5′-CCATGGCGAGTTTCATCAAGGCAT-3′ (forward primer) and 5′-GGCAGGGTTAAAAAAGATATGCGGTCAGT-3′ (reverse primer). The cDNA was cloned into vector pYES2.1 using pYES2.1 TOPO TA cloning kit according to the manufacturer's protocol (Invitrogen), and subsequently introduced into yeast (Saccharomyces cerevisiae) strain SCY059 (MATα, his 3-11,15 leu 2-3, 112 trp 1-1, ura 3-1, kan 1-100, ade2 met14Δare1∷HIS3 are2∷LEU2), kindly provided by Dr. Stephen Sturley (Columbia University), and a quadruple mutant strain (are1 are2 lro1 dgat1) from Dr. Sten Stymne (Swedish University of Agricultural Sciences).
Yeast Growth, Lipid Extraction, and HPLC Analysis
Single colonies of vector-only or AtSAT1 transformed yeast were inoculated in 10 mL of synthetic complete medium with Leu, His, and Ura omitted, and grown overnight at 28°C. Twenty-milliliters of fresh medium was then added and the cultures were further maintained overnight. Neutral lipid fraction was extracted following an established protocol (Folch et al., 1957) and separated through normal-phase HPLC on Agilent HPLC 1100 system by the method detailed by Moreu et al. (1996).
Qualitative Analysis of Sterol and Fatty Acid Released from SE
SEs were saponified for 2 h in 1 mL of 7.5% methanolic KOH at 80°C. The released sterol was extracted twice with hexane and dried under N2 stream. The aqueous layer was neutralized with HCl and extracted twice with hexane to obtain free fatty acids. The fatty acid fraction was derivatized with BSTFA containing 1% TMCS and sterol derivatives were obtained in a mixture of BSTFA and pyridine (1:1, v/v) at room temperature for 1 h, and separated on a Hewlett-Packard 6890 n GC with a capillary column DB-5 (Hewlett-Packard HP5, 35 m length × 0.25 mm diameter × 0.25 μm thickness). GC-MS analysis was accomplished using an Agilent 5973 mass selective detector coupled to an Agilent 6890 n gas chromatograph. The mass selective detector was run under standard electron impact conditions (70 eV), scanning an effective mass-to-charge ratio range of 40 to 700 at 2.26 scans/s. The mass spectra were compared with entries in the NIST mass-spectral database, version 2.0.
Quantitative Analysis of SE
After HPLC separation of neutral lipid, SE fraction was collected, dried under N2 stream, and saponified in 7.5% KOH in 95% methanol. Separation and quantification of sterol species was performed on a Hewlett-Packard 6890 series GC with a capillary column DB-5 and flame ionization detector. The peaks of each sterol species were compared to the cholesterol (internal standard) peak to determine the amount of each sterol present. Fatty acid methyl esters were separated on GC with DB23 column. The quantification was performed in triplicate with three different batches of yeast cultures. The content of SE was expressed as micromoles of sterol moiety saponified from SE on a dry yeast cell weight basis.
Sterol O-Acyltransferase Activity Assay Using Cell-Free Lysates
AtSAT1 expression induction in yeast cells was performed according to manufacturer's protocol for pYES2.1 TOPO TA expression kit (Invitrogen). The cells, resuspended in 1 mL cell wall-breaking buffer, were shaken vigorously in the presence of acid-washed glass beads. The resultant homogenate was centrifuged at 1,500g for 5 min at 4°C. The supernatant was removed, aliquoted, and stored at −76°C. For sterol substrate specificity determination, the supernatant was washed twice with 80 mm methyl-β-cyclodextrin (Sigma) for removal of free sterols embedded in endoplasmic reticulum membrane (Rodal et al., 1999). Protein concentration was measured using Bio-Rad Protein Assay kit for final AtSAT1 activity calculation. Substrate specificity was determined by a previously described method (Yang et al., 1997) with slight modification. For fatty acid-CoA substrate specificity, a reaction mixture containing 200 μg microsomal protein, 10 μg unlabeled lanosterol, 200 μg bovine serum albumin, and 30 μg reduced glutathione in a final volume of 400 μL of 0.1 m potassium phosphate buffer, pH 7.4, was sonicated for 5 min. One microliter of [3H]lanosterol (1 μCi/μL, 10 mCi/μmol, American Radiolabeled Chemicals) was added and incubated at 30°C with 100 rpm shaking for 30 min to allow incorporation of sterols into the membrane. Sterols were suspended in reaction buffer with the help of Triton-100 at a ratio of 30:1 (Tritus/sterol, w/w). The reaction was initiated by adding 50 mL 0.4 mm acyl-CoA (C16:0, C16:1, C18:0, C18:1, and C18:2, respectively). Reaction without exogenous acyl-CoA was employed as control. The reaction mixture for sterol substrate preference determination was essentially the same as that for acyl-CoA specificity assays except that [14C]palmitoyl-CoA (50 mL, 10nCi/nmol, 180 mm) was utilized to measure SE production. Reactions were terminated by the addition of 3 mL of chloroform/methanol (2:1, v/v). Cholesterol oleate was added as a carrier. The chloroform layer containing lipid was removed and dried under a N2 stream, redissolved in 100 μL chloroform, and then applied to a J.T. baker Si250 PA TLC silica plate gel. After visualization with iodine, the bands corresponding to SE were scraped off and subjected to scintillation counting. The specific activity of AtSAT1 activity was determined as picomoles of acyl-CoA or lanosterol converted to SE per milligram protein per minute (pmol mg−1 min−1).
l-α-dipalmitoyl PC (dipalmitoyl-1-α-[14C]PC, 55 mCi/mmol, 0.1 mCi/mL) was used to examine if AtSAT1 was capable of using PC as fatty acyl donor. The reaction mixture was essentially the same as that for sterol substrate specificity analysis, except that 2 μL of the 14C-labeled PC in ethanol:toluene (1:1) solution was added to the reaction mixture. A similar assay with [14C]16:0-CoA (55mCi/mmol, 0.1 mCi/mL) as acyl donor was also performed in parallel for comparison.
Generation of Transgenic Plants
A cDNA of AtSAT1 was PCR amplified with primers FP-XbaI (5′-CAAGAATCTAGAATGGCGAGTTTCATCAAGGCA-3′) and RP-KpnI (5′-GATCACTCAAGTTACCACACACGGCAGGGTTA-3′), and ligated into the XbaI and KpnI sites of pSE129 vector. The AtSAT1 sequence and the construct integrity were verified by sequencing. The resulting gene expression cassette in the transformation vector contained a canola (Brassica napus) seed-specific napin promoter and Agrobacterium NOS terminator flanking the 5′ and 3′ ends of the AtSAT1 cDNA, respectively. A single colony of Agrobacterium (GV3101) carrying napin:AtSAT1 transformation vector was cultured in Luria-Bertani medium containing 50 mg mL−1 kanamycin and 25 mg mL−1 gentamycin. Plant transformation was performed through vacuum infiltration. Selection of T0 transgenic seeds was carried on media containing one-half Murashige and Skoog Gamborg medium, 0.8% (w/v) phytagar, 3% (w/v) Suc, 50 mg mL−1 kanamycin, and 50 mg mL−1 timentin.
qRT-PCR and Data Analysis
To quantify transcript levels, total RNA was extracted from siliques of wild-type and transgenic T2 plants using Qiagen NEasy Plant Mini kit. One microgram of DNase I-treated RNA was used as template for cDNA synthesis (Invitrogen superscript First-Strand Synthesis kit). qRT-PCR was performed on Applied Biosystem StepOne Real-Time PCR system in triplicates. The β-actin-8 gene (AT1G49240) was used as endogenous standard for expression level calculation. The cDNA samples were 50 times diluted for amplication of SYBR green-labeled PCR fragments (Power SYBR Green PCR Master mix, Applied Biosystems) by using gene-specific primers (forward 5′-CTTGAGATCATCCTTGCCGCCACAGC-3′ and reverse 5′-AGTGATGTCGCTAGGTACGGCTTGTTG-3′) for AtSAT1. The expression of β-actin-8 was assayed using forward primer 5′-AACAGCAGAACGGGAAATTGTGAGA-3′ and reverse primer 5′-TGGAGGAGCTGGTTTTCGAGGT-3′. The corresponding amplicon size of β-actin-8 and AtSAT1 are 101 and 99 bp, respectively. The cycling parameters included an initial DNA denaturation step at 95°C for 10 min, followed by 40 cycles of PCR with DNA denaturation at 95°C for 15 s, and primer annealing and extension at 60°C for 1 min. qRT-PCR reactions with RNA templates (minus RT) were used as controls to check for DNA contamination or amplification of nonspecific products. Controls with no added template were included for each primer pairs to ensure primer dimmer was not interfering with amplification detection. qRT-PCR results were captured and analyzed using software StepOne (ver. 1.0). The cycle number at which the fluorescence passed the cycle threshold (CT), automatically determined by the software StepOne (version 1.0), was used for AtSAT1 expression level quantitation. AtSAT1 relative expression levels in each cDNA sample were obtained through normalization to β-actin-8 by the formula 2−(CTAtSAT1 − CTβ-actin-8).
Seed Sterol Extraction and Quantification
Six to 8 mg of mature seeds from wild-type and AtSAT1 transgenic lines were suspended in 2 mL of chloroform:methanol (2:1, v/v) containing 20 μg cholesterol and 20 μg cholesteryl myristate as internal standards. The suspension was probe sonicated, washed with 0.9% NaCl, and then vigorously vortexed. The bottom chloroform layer was transferred into a new solvent-washed tube. The aqueous layer was extracted twice with 2 mL of chloroform:methanol (2:1, v/v), and the organic layers were combined and dried down under N2 stream. The residue was dissolved in 200 μL hexane. SE and free sterol were separated and collected through HPLC. The SE fraction was saponified in 2 mL 7.5% KCl in 95% methanol and the resultant free sterols were repeatedly extracted with hexane. Sterol-TMS derivatives were qualitatively and quantitatively analyzed following the same method for yeast sterol analysis. Campesterol, β-sitosterol, cycloartenol, and methylenecycloartenol were identified by searching NIST 2.0 mass spectrum library or standard chemicals.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_115056.
Acknowledgments
We thank Dr. Stephen L. Sturley for providing the yeast strain SCY059 and Dr. Sten Stymne for providing the quadruple disrupted yeast strain devoid of TAG synthesis. We thank Mike Giblin and Darwin Reed for technical guidance in HPLC separation of SEs, and Ms. Kim Bryce, project manager for the National Research Council Canada-Crops for Enhanced Human Health Program. Helpful discussion with Drs. Sten Stymne, Pat Covello, and Mark Smith are acknowledged. This is National Research Council Canada publication number 48407.
This work was supported by the National Research Council Canada-Crops for Enhanced Human Health Program and a Natural Science and Engineering Research Council of Canada grant (grant no. NSERC RGPIN 327217–06 to J.Z.). Q.C. is a recipient of the Post-Doctoral Fellowship for Visiting Government Laboratories from the Natural Science and Engineering Research Council of Canada.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jitao Zou (jitao.zou@nrc-cnrc.gc.ca).
Open Access articles can be viewed online without a subscription.
References
- Awad AB, Roy R, Fink CS (2003) β-sitosterol, a plant sterol, induces apoptosis and activates key caspases in MDA-MB-231 human breast cancer cells. Oncol Rep 10 497–500 [PubMed] [Google Scholar]
- Awad AB, Smith AJ, Fink CS (2001) Plant sterols regulate rat vascular smooth muscle cell growth and prostacyclin release in culture. Prostaglandins Leukot Essent Fatty Acids 64 323–330 [DOI] [PubMed] [Google Scholar]
- Bach TJ (1986) Hydroxymethyl glutaryl-CoA reductase, a key enzyme in phytosterol synthesis? Lipids 21 82–88 [DOI] [PubMed] [Google Scholar]
- Bach TJ, Benveniste P (1997) Cloning of cDNAs or genes encoding enzymes of sterol biosynthesis from plants and other eukaryotes: heterologous expression and complementation analysis of mutations for functional characterization. Prog Lipid Res 36 197–226 [DOI] [PubMed] [Google Scholar]
- Banaś A, Carlsson AS, Huang B, Lenman M, Banas W, Lee M, Noiriel A, Benveniste P, Schaller H, Bouvier-Nave P, et al (2005) Cellular sterol ester synthesis in plants is performed by an enzyme (phospholipid:sterol acyltransferase) different from the yeast and mammalian acyl-CoA:sterol acyltransferases. J Biol Chem 280 34626–34634 [DOI] [PubMed] [Google Scholar]
- Benveniste P (1986) Sterol biosynthesis. Annu Rev Plant Physiol 37 275–308 [Google Scholar]
- Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55 429–457 [DOI] [PubMed] [Google Scholar]
- Bouic PJ (2001) The role of phytosterols and phytosterolins in immune modulation: a review of the past 10 years. Curr Opin Clin Nutr Metab Care 4 471–475 [DOI] [PubMed] [Google Scholar]
- Cao G, Goldstein JL, Brown MS (1996) Complementation of mutation in acyl-CoA: cholesterol acyltransferase (ACAT) fails to restore sterol regulation in ACAT-defective sterol-resistant hamster cells. J Biol Chem 271 14642–14648 [DOI] [PubMed] [Google Scholar]
- Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C (1995) Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol 109 1337–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demel RA, De Kruyff B (1976) The function of sterols in membranes. Biochim Biophys Acta 457 109–132 [DOI] [PubMed] [Google Scholar]
- Dyas L, Goad LJ (1993) Steryl fatty acyl esters in plants. Phytochemistry 34 17–29 [Google Scholar]
- Folch J, Lees M, Sloane-Stanley GH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226 497–509 [PubMed] [Google Scholar]
- Garcia RE, Mudd JB (1978. a) Partial characterization of steryl ester biosynthesis in spinach leaves. Plant Physiol 61 354–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia RE, Mudd JB (1978. b) Fatty acid and sterol specificity in the biosynthesis of sterol ester by enzyme preparations from spinach leaves. Arch Biochem Biophys 190 315–321 [DOI] [PubMed] [Google Scholar]
- Garcia RE, Mudd JB (1978. c) Identification of an acyl donor in sterol ester biosynthesis by enzyme preparations from spinach leaves. Plant Physiol 62 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondet L, Bronner R, Benveniste P (1994) Regulation of sterol content in membranes by subcellular compartmentation of sterol-esters accumulating in a sterol-overproducing tobacco mutant. Plant Physiol 105 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondet L, Weber T, Maillot-Vernier P, Benveniste M, Bach TJ (1992) Regulatory role of microsomal 3-hydroxy-3-methyl-glutaryl-Coenzyme A reductase in a tobacco mutant that overproduces sterols. Biochem Biophys Res Commun 186 878–893 [DOI] [PubMed] [Google Scholar]
- Guo Z, Cromley D, Billheimer JJ, Sturley SL (2001) Identification of potential substrate-binding sites in yeast and human acyl-CoA sterol acyltransferases by mutagenesis of conserved sequences. J Lipid Res 42 1282–1291 [PubMed] [Google Scholar]
- Harker M, Holmberg N, Clayton JC, Gibbard CL, Wallace AD, Rawlins S, Hellyer SA, Lanot A, Safford R (2003) Enhancement of seed phytosterol levels by expression of an N-terminal truncated Hevea brasiliensis (rubber tree) 3-hydroxy-3-methylglutaryl-CoA reductase. Plant Biotechnol J 1 113–121 [DOI] [PubMed] [Google Scholar]
- Hendriks HF, Weststrate JA, van Vliet T, Meijer GW (1999) Spreads enriched with three different levels of vegetable oil sterols and the degree of cholesterol lowering in normocholesterolaemic and mildly hypercholesterolaemic subjects. Eur J Clin Nutr 53 319–327 [DOI] [PubMed] [Google Scholar]
- Hernández-Pinzón I, Ross JH, Barnes KA, Damant AP, Murphy DJ (1999) Composition and role of tapetal lipid bodies in the biogenesis of the pollen coat of Brassica napus. Planta 208 588–598 [DOI] [PubMed] [Google Scholar]
- Hey SJ, Powers SJ, Beale MH, Hawkins ND, Ward JL, Halford NG (2006) Enhanced seed phytosterol accumulation through expression of a modified HMG-CoA reductase. Plant Biotechnol J 4 219–229 [DOI] [PubMed] [Google Scholar]
- Hofmann K (2000) A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem Sci 25 111–112 [DOI] [PubMed] [Google Scholar]
- Holmer G, Ory RL, Hoy CE (1973) Changes in lipid composition of germinating barley embryo. Lipids 8 277–283 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszowska W, Kasprzyk Z (1977) Intracellular distribution and origin of sterols in Calendula officinalis leaves. Phytochemistry 16 473–476 [Google Scholar]
- Jensen-Pergakes K, Guo Z, Giattina M, Sturley SL, Bard M (2001) Transcriptional regulation of the two sterol esterification genes in the yeast Saccharomyces cerevisiae. J Bacteriol 183 4950–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson LG, Lenman M, Ericson ML, Rask L (1987) Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J Biol Chem 262 12196–12201 [PubMed] [Google Scholar]
- Kemp RJ, Goad LJ, Mercer EI (1967) Changes in the level and composition of the esterified and unesterified sterols of maize seedling during germination. Phytochemistry 6 1609–1615 [Google Scholar]
- Kolesnikova MD, Xiong Q, Lodeiro S, Hua L, Matsuda SP (2006) Lanosterol biosynthesis in plants. Arch Biochem Biophys 447 87–95 [DOI] [PubMed] [Google Scholar]
- Lovato MA, Hart EA, Segura MJ, Giner JL, Matsuda SP (2000) Functional cloning of an Arabidopsis thaliana cDNA encoding cycloeucalenol cycloisomerase. J Biol Chem 275 13394–13397 [DOI] [PubMed] [Google Scholar]
- Macauley SK, Billheimer JT, Ritter KS (1986) Sterol substrate specificity of acyl coenzyme A:cholesterol acyltransferase from the corn earworm, Heliothis zea. J Lipid Res 27 64–71 [PubMed] [Google Scholar]
- Moghadasian MH, McManus BM, Godin DV, Rodrigues B, Frohlich JJ (1999) Proatherogenic and antiatherogenic effects of probucol and phytosterols in apolipoprotein E-deficient mice: possible mechanisms of action. Circulation 99 1733–1739 [DOI] [PubMed] [Google Scholar]
- Moghadasian MH, McManus BM, Pritchard PH, Frohlich JJ (1997) “Tall oil” derived phytosterols reduce atherosclerosis in ApoE-deficient mice. Arterioscler Thromb Vasc Biol 17 119–126 [DOI] [PubMed] [Google Scholar]
- Moreu RA, Powell MJ, Hicks KB (1996) Extraction and quantitative analysis of oil from commercial corn fiber. J Agric Food Chem 44 2149–2154 [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Heijne GV (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10 1–6 [DOI] [PubMed] [Google Scholar]
- Oelkers P, Behari A, Cromley D, Billheimer JT, Sturley SL (1998) Characterization of two human genes encoding acyl coenzyme A:cholesterol acyltransferase related enzymes. J Biol Chem 273 26765–26771 [DOI] [PubMed] [Google Scholar]
- Piironen V, Lindsay D, Miettinen TA, Toivo J, Lampi AM (2000) Plant sterols: biosynthesis, biological function and their importance to human nutrition. J Sci Food Agric 80 939–966 [Google Scholar]
- Re EB, Jones D, Learned RM (1995) Co-expression of native and introduced genes reveals cryptic regulation of HMG CoA reductase expression in Arabidopsis. Plant J 7 771–784 [DOI] [PubMed] [Google Scholar]
- Rodal SK, Skretting G, Garred Ø, Vilhardt F, van Deurs B, Sandvig K (1999) Extraction of cholesterol with methyl-β-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell 10 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukmini C, Raghuram TC (1991) Nutritional and biochemical aspects of the hypolipidemic action of rice bran oil: a review. J Am Coll Nutr 10 593–601 [DOI] [PubMed] [Google Scholar]
- Sabir SM, Hayat I, Gardezi DA (2003) Estimation of sterols in edible fats and oils. Pakistan J Nutr 2 178–181 [Google Scholar]
- Sandager L, Gustavsson MH, Stahl U, Dahlqvist A, Wiberg E, Banaś A, Lenman M, Ronne H, Stymne S (2002) Storage lipid synthesis is non-essential in yeast. J Biol Chem 277 6478–6482 [DOI] [PubMed] [Google Scholar]
- Schaller H (2004) New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol Biochem 42 465–476 [DOI] [PubMed] [Google Scholar]
- Schaller H, Grausem B, Benveniste P, Chye ML, Tan CT, Song YH, Chua NH (1995) Expression of the Hevea brasiliensis (H.B.K.) Mull. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiol 109 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler I, Milon A, Nakatani Y, Ourisson G, Albrecht A, Benveniste P, Hartman M (1991) Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phospatidylcholine bilayers. Proc Natl Acad Sci USA 88 6926–6930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierksma A, Weststate JA, Meijer GW (1999) Spreads enriched with plant sterols, either esterified 4,4-dimethylsterols or free 4-desmethylsterols, and plasma total and LDL cholesterol concentrations. Br J Nutr 82 273–282 [PubMed] [Google Scholar]
- Suzuki M, Xiang T, Ohyama K, Seki H, Saito K, Muranaka T, Hayashi H, Katsube Y, Kushiro T, Shibuya M, et al (2006) Lanosterol synthase in dicotyledonous plants. Plant Cell Physiol 47 565–571 [DOI] [PubMed] [Google Scholar]
- Tavani DM, Nes WR, Billheimer JT (1982) The sterol substrate specificity of acyl CoA: cholesterol acyltransferase from rat liver. J Lipid Res 23 774–781 [PubMed] [Google Scholar]
- Ting JTL, Wu SSH, Ratnayake C, Huang AHC (1998) Constituents of the tapetosomes and elaioplasts in Brassica campestris and their degradation and retention during microsporogenesis. Plant J 16 541–551 [DOI] [PubMed] [Google Scholar]
- van Rensburg SJ, Daniels WM, van Zyl JM, Taljaard JJ (2000) A comparative study of the effects of cholesterol, β-sitosterol, β-sitosterol glucoside, dehydroepiandrosterone sulphate and melatonin on in vitro lipid peroxidation. Metab Brain Dis 15 257–265 [DOI] [PubMed] [Google Scholar]
- Wilson TA, Nicolosi RJ, Woolfrey B, Kritchevsky D (2007) Rice bran oil and oryzanol reduce plasma lipid and lipoprotein cholesterol concentrations and aortic cholesterol ester accumulation to a greater extent than ferulic acid in hypercholesterolemic hamsters. J Nutr Biochem 18 105–112 [DOI] [PubMed] [Google Scholar]
- Yang H, Bard M, Bruner DA, Gleeson A, Deckelbaum RJ, Aljinovic G, Pohl TM, Rothstein R, Sturley SL (1996) Sterol esterification in yeast: a two-gene process. Science 272 1353–1356 [DOI] [PubMed] [Google Scholar]
- Yang H, Cromley D, Wang H, Billheimer JT, Sturley SL (1997) Functional expression of a cDNA to human acyl-coenzyme A:cholesterol acyltransferase in yeast: species-dependent substrate specificity and inhibitor sensitivity. J Biol Chem 272 3980–3985 [DOI] [PubMed] [Google Scholar]
- Yankah VV, Jone RJH (2001) Phytosterols and health implications—efficacy and nutritional aspects. Inform 12 899–903 [Google Scholar]
- Zimowski J, Wojciechowski ZA (1981. a) Acyl donors for sterol esterification by cell-free preparations from Sinapis alba root. Phytochemistry 20 1795–1798 [Google Scholar]
- Zimowski J, Wojciechowski ZA (1981. b) Partial purification and specificity of triacylglycerol: sterol acyltransferase from Sinapis alba. Phytochemistry 20 1799–1803 [Google Scholar]
- Zweytick D, Leitner E, Kohlwein SD, Yu C, Rothblatt J, Daum G (2000) Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur J Biochem 267 1075–1082 [DOI] [PubMed] [Google Scholar]