Abstract
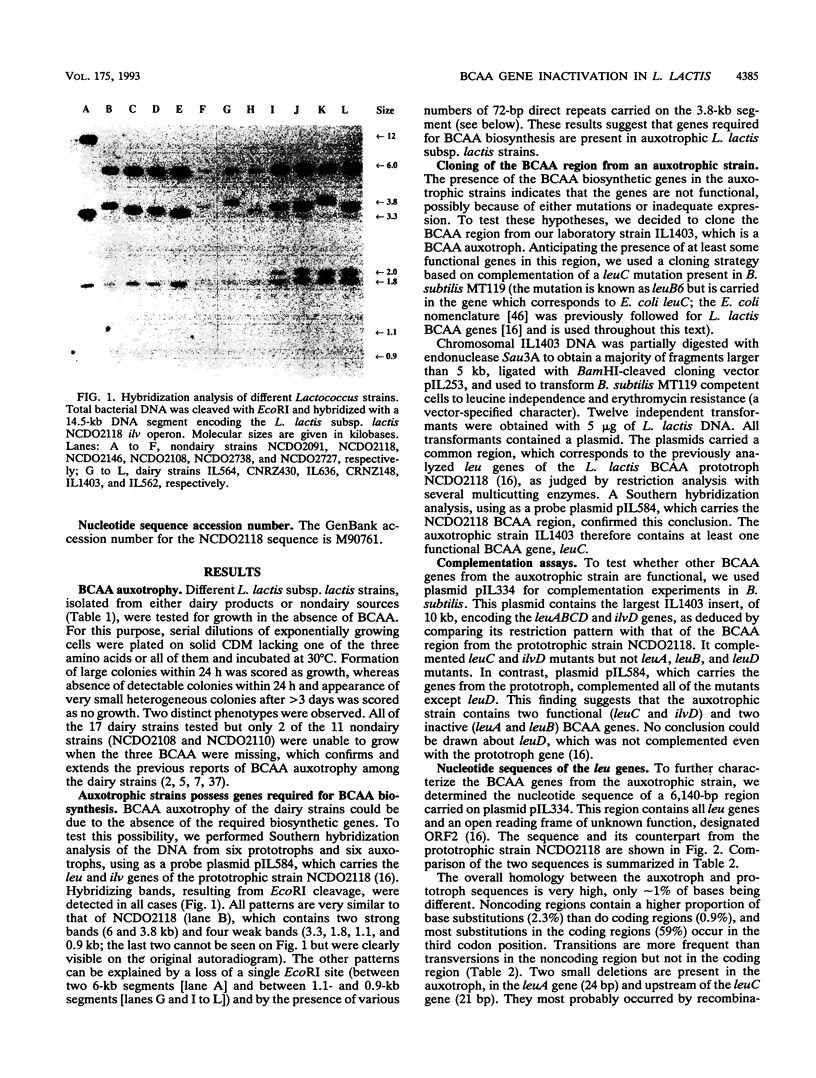
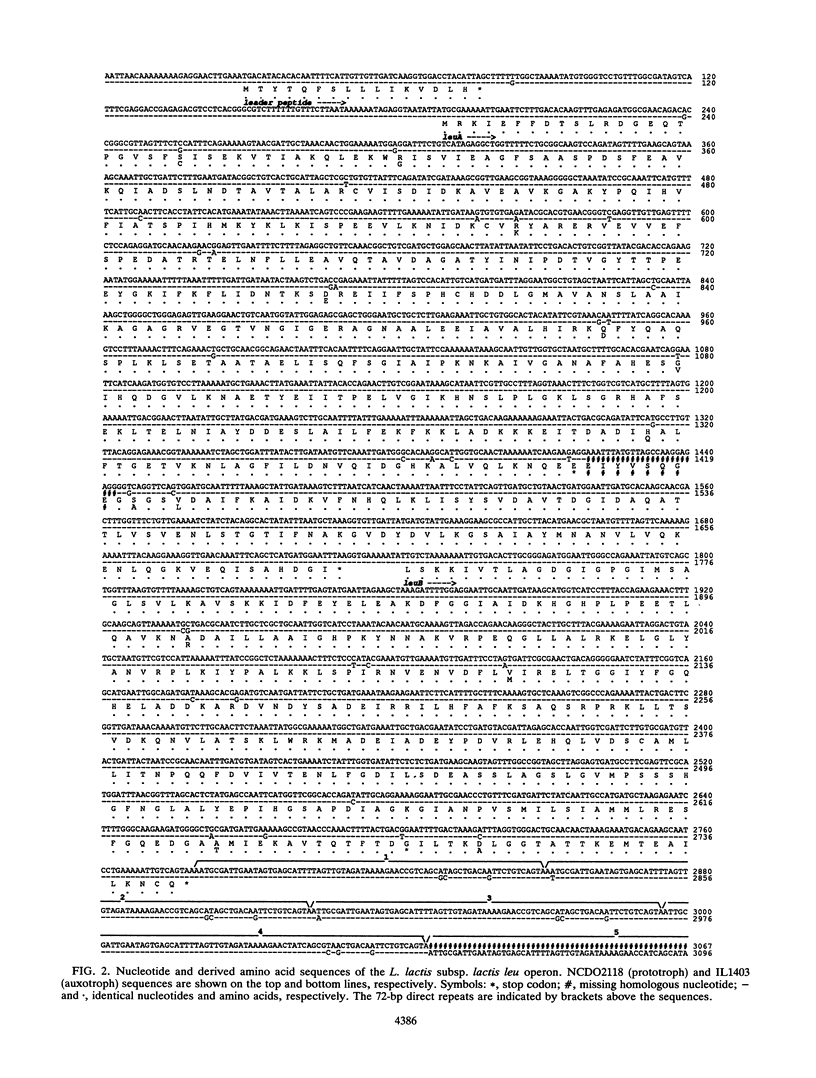
The Lactococcus lactis subsp. lactis strains isolated from dairy products are auxotrophs for branched-chain amino acids (leucine, isoleucine, and valine), while most strains isolated from nondairy media are prototrophs. We have cloned and sequenced the leu genes from one auxotroph, IL1403. The sequence is 99% homologous to that of the prototroph NCDO2118, which was determined previously. Two nonsense mutations and two small deletions were found in the auxotroph sequence, which might explain the branched-chain amino acid auxotrophy. Nevertheless, the leu genes from the auxotroph appear to be transcribed and regulated similarly to those from the prototroph.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carifo K., Catlin B. W. Neisseria gonorrhoeae auxotyping: differentiation of clinical isolates based on growth responses on chemically defined media. Appl Microbiol. 1973 Sep;26(3):223–230. doi: 10.1128/am.26.3.223-230.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- De Vos W. M., Gasson M. J. Structure and expression of the Lactococcus lactis gene for phospho-beta-galactosidase (lacG) in Escherichia coli and L. lactis. J Gen Microbiol. 1989 Jul;135(7):1833–1846. doi: 10.1099/00221287-135-7-1833. [DOI] [PubMed] [Google Scholar]
- Delorme C., Ehrlich S. D., Renault P. Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6571–6579. doi: 10.1128/jb.174.20.6571-6579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme C., Godon J. J., Ehrlich S. D., Renault P. Gene inactivation in Lactococcus lactis: histidine biosynthesis. J Bacteriol. 1993 Jul;175(14):4391–4399. doi: 10.1128/jb.175.14.4391-4399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Ingraham L. MEIOTROPHIC MUTANTS OF Pasteurella Pestis AND THEIR USE IN THE ELUCIDATION OF NUTRITIONAL REQUIREMENTS. Proc Natl Acad Sci U S A. 1957 May 15;43(5):369–372. doi: 10.1073/pnas.43.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. A. Lactose hydrolysing enzymes in Streptococcus lactis and Streptococcus cremoris and also in some other species of streptococci. J Appl Bacteriol. 1980 Dec;49(3):493–503. doi: 10.1111/j.1365-2672.1980.tb04724.x. [DOI] [PubMed] [Google Scholar]
- Glatron M. F., Rapoport G. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54(10):1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- Godon J. J., Chopin M. C., Ehrlich S. D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Yokoyama S., Calhoun D. H. Role of cryptic genes in microbial evolution. Mol Biol Evol. 1983 Dec;1(1):109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- Juni E., Heym G. A. Studies of some naturally occurring auxotrophs of Neisseria gonorrhoeae. J Gen Microbiol. 1980 Nov;121(1):85–92. doi: 10.1099/00221287-121-1-85. [DOI] [PubMed] [Google Scholar]
- Kandler O. Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek. 1983 Sep;49(3):209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- Koch A. L. Enzyme evolution. I. The importance of untranslatable intermediates. Genetics. 1972 Oct;72(2):297–316. doi: 10.1093/genetics/72.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):15–42. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Calhoun D. H., Adams C. W., Hauser C. A., Gray J., Hatfield G. W. Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1981 Feb;78(2):922–925. doi: 10.1073/pnas.78.2.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Calhoun D. H., Gray J., Adams C. W., Hauser C. A., Hatfield G. W. DNA sequence fine-structure analysis of ilvG (IlvG+) mutations of Escherichia coli K-12. J Bacteriol. 1982 Jan;149(1):294–298. doi: 10.1128/jb.149.1.294-298.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey C. J., Zahler S. A. Insertion of bacteriophage SP beta into the citF gene of Bacillus subtilis and specialized transduction of the ilvBC-leu genes. J Bacteriol. 1982 Sep;151(3):1222–1229. doi: 10.1128/jb.151.3.1222-1229.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson M. D., Yanofsky C. Naturally occurring sites within the Shigella dysenteriae tryptophan operon severely limit tryptophan biosynthesis. J Bacteriol. 1976 May;126(2):668–678. doi: 10.1128/jb.126.2.668-678.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzari G., Yanofsky C. Naturally occurring promoter down mutation: nucleotide sequence of the trp promoter/operator/leader region of Shigella dysenteriae 16. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5580–5584. doi: 10.1073/pnas.75.11.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Deguchi Y., Yajima M., Sakurai T., Yura T. Multiple nutritional requirements of lactobacilli: genetic lesions affecting amino acid biosynthetic pathways. J Bacteriol. 1981 Oct;148(1):64–71. doi: 10.1128/jb.148.1.64-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L. L., Hall B. G. Mechanisms of activation of the cryptic cel operon of Escherichia coli K12. Genetics. 1990 Mar;124(3):473–482. doi: 10.1093/genetics/124.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle L. E., Finch L. R. A physical map of the genome of Mycoplasma mycoides subspecies mycoides Y with some functional loci. Nucleic Acids Res. 1988 Jul 11;16(13):6027–6039. doi: 10.1093/nar/16.13.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Rak B. IS5: a mobile enhancer of transcription in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1244–1248. doi: 10.1073/pnas.89.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Smid E. J., Konings W. N. Relationship between utilization of proline and proline-containing peptides and growth of Lactococcus lactis. J Bacteriol. 1990 Sep;172(9):5286–5292. doi: 10.1128/jb.172.9.5286-5292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. Restriction of plasmid-mediated transformation in Bacillus subtilis 168. Mol Gen Genet. 1979 Sep;175(2):235–237. doi: 10.1007/BF00425542. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Ward J. B., Jr, Zahler S. A. Genetic studies of leucine biosynthesis in Bacillus subtilis. J Bacteriol. 1973 Nov;116(2):719–726. doi: 10.1128/jb.116.2.719-726.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamenhof S., Eichhorn H. H. Study of microbial evolution through loss of biosynthetic functions: establishment of "defective" mutants. Nature. 1967 Nov 4;216(5114):456–458. doi: 10.1038/216456a0. [DOI] [PubMed] [Google Scholar]