Abstract
PDX3 and SALT OVERLY SENSITIVE4 (SOS4), encoding pyridoxine/pyridoxamine 5′-phosphate oxidase and pyridoxal kinase, respectively, are the only known genes involved in the salvage pathway of pyridoxal 5′-phosphate in plants. In this study, we determined the phenotype, stress responses, vitamer levels, and regulation of the vitamin B6 pathway genes in Arabidopsis (Arabidopsis thaliana) plants mutant in PDX3 and SOS4. sos4 mutant plants showed a distinct phenotype characterized by chlorosis and reduced plant size, as well as hypersensitivity to sucrose in addition to the previously noted NaCl sensitivity. This mutant had higher levels of pyridoxine, pyridoxamine, and pyridoxal 5′-phosphate than the wild type, reflected in an increase in total vitamin B6 observed through HPLC analysis and yeast bioassay. The sos4 mutant showed increased activity of PDX3 as well as of the B6 de novo pathway enzyme PDX1, correlating with increased total B6 levels. Two independent lines with T-DNA insertions in the promoter region of PDX3 (pdx3-1 and pdx3-2) had decreased PDX3 activity. Both also had decreased activity of PDX1, which correlated with lower levels of total vitamin B6 observed using the yeast bioassay; however, no differences were noted in levels of individual vitamers by HPLC analysis. Both pdx3 mutants showed growth reduction in vitro and in vivo as well as an inability to increase growth under high light conditions. Increased expression of salvage and some of the de novo pathway genes was observed in both the pdx3 and sos4 mutants. In all mutants, increased expression was more dramatic for the salvage pathway genes.
Pyridoxine/pyridoxamine 5′-P (PNP/PMP) oxidase and pyridoxal (PL) kinase, encoded in Arabidopsis (Arabidopsis thaliana) by the PDX3 (Sang et al., 2007) and the SALT OVERLY SENSITIVE4 (SOS4; Shi and Zhu, 2002) genes, respectively, are key enzymes involved in the biosynthetic pathway leading to pyridoxal 5′-P (PLP). PLP is the active coenzyme product of the vitamin B6 pathway and is essential in many biochemical reactions, including decarboxylation, transamination, deamination, racemization, and trans-sulfuration reactions associated primarily with amino acid synthesis (Drewke and Leistner, 2001). PLP is also involved in enzymes that catalyze some steps in carbohydrate and lipid metabolism and is a cofactor for aminocyclopropane-1-carboxylate synthase. Moreover, vitamin B6 has been linked to resistance to oxidative and environmental stresses and is an efficient singlet oxygen quencher and antioxidant (Shi et al., 2002; Denslow et al., 2005, 2007). It is required for postembryonic root development and root hair development in plants (Shi and Zhu, 2002; Chen and Xiong, 2005; Denslow et al., 2005, 2007; Titiz et al., 2006).
Organisms can synthesis PLP de novo or by a salvage pathway. Escherichia coli and other proteobacteria synthesize PLP de novo by the well-studied pdxA/pdxJ pathway (Roa et al., 1989; Hill et al., 1996; Man et al., 1996; Laber et al., 1999). In this pathway, PNP, a direct biosynthetic intermediate leading to PLP, is synthesized by the condensation of 4-(phosphohydroxy)-l-Thr and 1-deoxy-d-xylulose-5-P, catalyzed by the enzymes PdxA and PdxJ. PNP is then oxidized by a PNP/PMP oxidase encoded by pdxH to form PLP (Laber et al., 1999). An alternative de novo PLP biosynthesis pathway that involves two different proteins, PDX1 and PDX2, has also been characterized and is present in the majority of organisms that can produce vitamin B6 de novo, including plants, fungi, archeabacteria, and most bacteria (Ehrenshaft et al., 1999; Osmani et al., 1999; Ehrenshaft and Daub, 2001; Mittenhuber, 2001). In this pathway, proteins encoded by the genes PDX1 and PDX2 form a complex that has Gln amidotransferase activity (Belitsky, 2004). This enzyme complex catalyzes a reaction where Gln plus either Rib 5-P or ribulose 5-P and either dihydroxyacetone phosphate or glyceraldehyde 3-P are used to produce PLP, the first B6 vitamer synthesized in this pathway (Burns et al., 2005; Raschle et al., 2005). PDX1 and PDX2 were recently isolated and characterized in tobacco (Nicotiana tabacum; Denslow et al., 2005) and Arabidopsis (Chen and Xiong, 2005; Tambasco-Studart et al., 2005; Titiz et al., 2006; Wagner et al., 2006; Denslow et al., 2007). Two PDX1 sequences were recovered from tobacco, suggesting either two copies or two alleles; one copy of PDX2 was identified. In Arabidopsis, there is a single copy of PDX2 on chromosome 5 and three homologs of PDX1 located on chromosomes 2, 3, and 5, referred to as PDX1.1, PDX1.2, and PDX1.3, respectively.
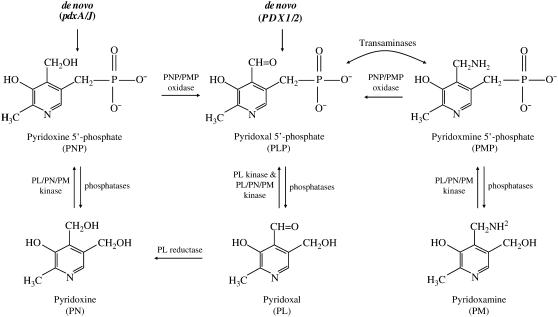
Many organisms, including those that cannot synthesize PNP or PLP de novo, contain a salvage pathway that interconverts between the different vitamers (Fig. 1). In this pathway, the nonphosphorylated forms of vitamin B6 are converted to PLP by the action of kinases that phosphorylate pyridoxine (PN), pyridoxamine (PM), and PL to form PNP, PMP, and PLP, respectively, and an oxidase that catalyzes the oxidation of PNP and PMP to form PLP. Alternatively, PMP oxidation can also be catalyzed by transaminases (Mittenhuber, 2001). The phosphorylated forms can be hydrolyzed by phosphatases to restore the free vitamers and maintain PLP homoeostasis (McCormick and Chen, 1999; Mittenhuber, 2001). Additionally, a PL reductase that reduces PL to PN has been suggested to be involved in the vitamin B6 salvage pathway (Guirard and Snell, 1988; Nakano et al., 1999; Morita et al., 2004).
Figure 1.
Vitamin B6 salvage pathway. PLP is produced both via the de novo pathway in organisms containing PDX1 and PDX2 as well as by conversion from PNP or PMP via the PNP/PMP oxidase. PMP oxidation can also be catalyzed by transaminases. PN, PM, and PL are phosphorylated by the action of a PL/PN/PM kinase or by an additional PL-specific kinase for PLP. Phosphorylated derivatives are dephosphorylated by phosphatases. PL can also be converted to PN by a PL reductase. This pathway is based on data from E. coli, yeast, B. subtilis, and mammals.
The vitamin B6 salvage pathway enzymes and genes have been characterized in several organisms. In E. coli, two kinases with different substrate specificity have been identified. PN/PM/PL kinase (PdxK) phosphorylates PN, PM, and PL to form PNP, PMP, and PLP, respectively, whereas PL kinase (PdxY) specifically phosphorylates PL (Yang et al., 1996, 1998). The oxidation of PNP and PMP to form PLP is carried out by PdxH, the same oxidase involved in the de novo pathway (Lam and Winkler, 1992). Kinases and oxidases with sequence and functional homology to PdxK and PdxH have also been identified and thoroughly characterized in mammals (Kazarinoff and McCormick, 1975; Kwok and Churchich, 1980; Choi et al., 1987; Hanna et al., 1997). However, little is known about the vitamin B6 genes involved in the salvage pathway in organisms that synthesize PLP de novo by the PDX1/PDX2 pathway. A kinase (ThiD) involved in thiamine biosynthesis in Bacillus subtilis was recently characterized and shown also to have PN, PM, and PL kinase activity (Park et al., 2004). A PL kinase has also been identified and characterized in plants (Lum et al., 2002; Shi et al., 2002; Wang et al., 2004). This enzyme, identified in Arabidopsis by its ability to complement E. coli pdxK mutants (Shi et al., 2002), is encoded by the SOS4 gene associated with NaCl tolerance and root hair development in Arabidopsis (Shi et al., 2002; Shi and Zhu, 2002) and wheat (Triticum aestivum; Wang et al., 2004). A PNP/PMP oxidase, PDX3, has been also identified and characterized in yeast (Saccharomyces cerevisiae; Loubbardi et al., 1995), and recently in Arabidopsis by complementation of a yeast pdx3 mutant (Sang et al., 2007).
In this study, we confirm, by complementation of pdxH E. coli mutants, the recently published identity (Sang et al., 2007) of the Arabidopsis locus At5g49970 as the gene encoding PDX3, the PNP/PMP oxidase. In addition, we report the effects on phenotype, vitamer levels, stress responses, enzyme activity, and regulation of salvage and de novo pathway genes, resulting from mutations in pdx3 and sos4. In addition to the previously reported sensitivity of the sos4 mutant to salt, we show that this mutant has root-specific sensitivity to Suc. Analysis of vitamer levels in the sos4 mutant identified an unexpected increase in PLP levels along with predicted increases in PM and PN. The elevated B6 levels correlated with increased enzyme activity of both PDX1 and PDX3, as well as increased expression of the encoding genes. By contrast, pdx3 mutant lines have decreased total vitamin B6 levels, which correlate with decreased PDX1 enzyme activity but not with the increased PDX1 and SOS4 gene expression seen in the pdx3 mutants. Gene expression cannot explain the differential effect on B6 vitamer levels between the mutants.
RESULTS
Arabidopsis PDX3 Gene Complements pdxH
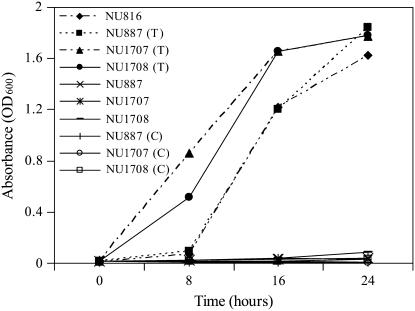
Identity of At5g49970 as encoding a PNP/PMP oxidase was determined by complementation of E. coli pdxH mutants. As shown in Figure 2, wild-type growth was restored in all pdxH E. coli mutants (NU887, NU1707, and NU1708). Only the mutants transformed with PDX3 were capable of growing on minimal medium without PL and had comparable growth to the parental strain NU816. These results confirm the report of Sang et al. (2007) and also show that PDX3 from Arabidopsis is functional in E. coli.
Figure 2.
Growth of E. coli pdxH mutants, mutants transformed with Arabidopsis PDX3, and the parental strain on minimal medium. NU887, NU1706, and NU1708, pdxH E. coli mutants; (T), pdxH E. coli mutants transformed with the Arabidopsis PDX3 gene; NU816, parental strain; (C), pdxH E. coli mutants transformed with vector control. Mutants were grown on minimal medium (liquid EM plus 0.01 mm FeSO4).
Recovery of pdx3 Homozygous T-DNA Insertion Mutant Lines
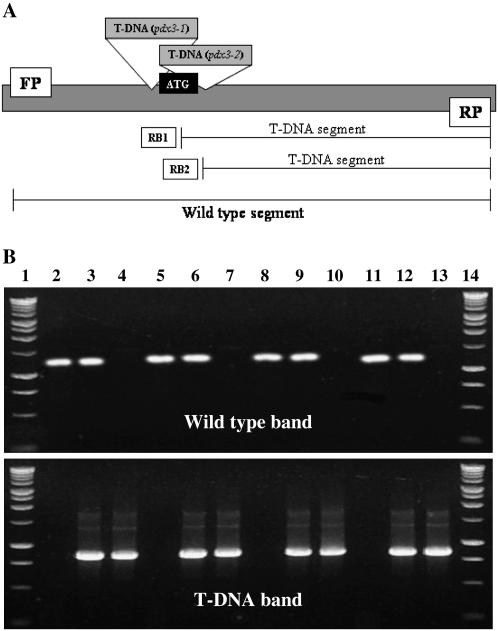
Plants from Arabidopsis ecotype Columbia (Col-0) with T-DNA insertions located in two different sites of the PDX3 promoter region (SALK_060749 and SALK_149382, named pdx3-1 and pdx3-2, respectively; Fig. 3A) were obtained from the Arabidopsis Biological Resource Center (ABRC). Homozygous mutant plants were differentiated from wild-type and heterozygous plants by the presence of a single band of approximately 800 bp (Fig. 3B), compared to the presence of only a single 1,200-bp band in wild-type plants and the presence of both bands in heterozygous plants. The difference in distance between the T-DNA insertion in the pdx3-1 and pdx3-2 mutant lines is only 38 bp; therefore, the bands amplified in the pdx3-1 and pdx3-2 homozygous lines are indistinguishable in the gel (Fig. 3B).
Figure 3.
A, Diagram of the location of the T-DNA insertions in the pdx3 mutant lines (pdx3-1 and pdx3-2) within the PDX3 gene. FP and RP indicate forward and reverse primers used to amplify the 1,200-bp wild-type PDX3 segment. RB1 and RB2 are the T-DNA right-border primers used with RP to amplify the approximately 800-bp segments for the T-DNA insertion mutants. B, Screening of T-DNA pdx3 insertion lines by PCR. DNA from plants segregating for the T-DNA insertion was amplified with the FP and RP primers (top) to amplify the 1,200-bp wild-type sequence or with the RB1 or RB2 primers plus the RP primer (bottom) to amplify the approximately 800-bp sequence resulting from the T-DNA insertions. Plants lacking T-DNA insertions are identified by the amplification of only the 1,200-bp band; homozygous T-DNA mutants have only the 800-bp band and heterozygous plants have both bands. Lanes 1 and 14, 1-kb ladder; lanes 2, 5, 8, and 11, plants lacking the T-DNA insertion (wild type); lanes 3 and 6, pdx3-1 heterozygous plants; lanes 9 and 12, pdx3-2 heterozygous plants; lanes 4 and 7, pdx3-1 homozygous mutants; lines 10 and 13, pdx3-2 homozygous mutants. PCR products were visualized on 1% agarose gels. The difference in distance between the T-DNA insertion in the pdx3-1 and pdx3-2 mutant lines is only 38 bp, resulting in bands that are not distinguishable on the gel.
PDX3 Gene Expression and Enzyme Activity in Insertion Mutants
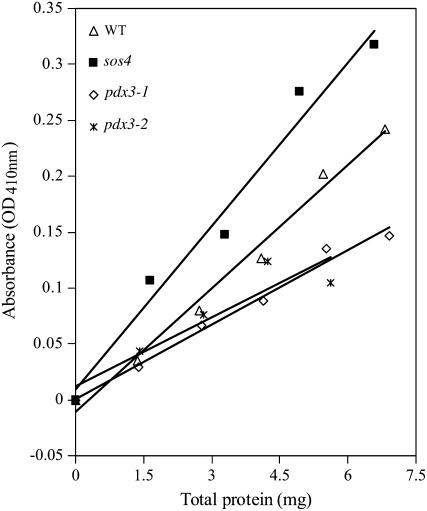
Expression of PDX3 in the pdx3-1 and pdx3-2 mutant lines was assayed by quantitative real-time PCR (qRT-PCR). No decrease was seen in PDX3 expression in the pdx3-1 mutant, but the pdx3-2 mutant showed a 70% decrease in PDX3 expression. PDX3 enzyme activity was then assayed in both mutant lines. We were unable to detect activity in the wild-type strain using the assay developed by Sang et al. (2007) for the purified Arabidopsis enzyme; however, we were able to measure activity using methods developed for PNP/PMP oxidase enzymes from E. coli and rabbit liver and brain tissues (Wada and Snell, 1961; Zhao and Winkler, 1995). Results are shown in Figure 4. Both pdx3 mutants had lower oxidase activity than the wild type, with approximately one-half the activity at the highest amount of protein assayed. In spite of the different expression measured by qRT-PCR, oxidase activities in the two insertion lines were indistinguishable from each other. As a comparison, PDX3 activity was also assayed in the sos4 mutant. This mutant showed higher oxidase activity than the wild type or either of the pdx3 mutants.
Figure 4.
PDX3 activity in Arabidopsis pdx3 and sos4 mutant lines and Arabidopsis ecotype Col-0 (WT). The activity of PDX3 was measured in crude extracts of the mutant lines and WT by determining the amount of PLP formed with a colorimetric assay after a 1-h incubation of enzyme reactions containing increasing amounts of total protein. PDX3 activity data were analyzed with a regression line adjusted to the absorbance values obtained for the increasing amounts of total protein.
Phenotypic Analysis of Mutant Lines
Phenotypes of the pdx3 and sos4 mutant lines were assessed both in soil and in vitro. In soil, pdx3 mutant lines did not show a distinct phenotype in contrast to sos4 mutant plants, which were chlorotic and had reduced plant size. The first flower buds were visible in sos4 mutant plants before those in wild-type and pdx3 mutant plants; however, there were no significant differences in the number of days to the first open flower between any of the mutant lines and the wild type (Table I). Only pdx3-1 mutant plants, and not those of pdx3-2, showed significant reduction in the number of seeds per silique as compared to the wild type.
Table I.
Phenotypic characterization of Arabidopsis pdx3 and sos4 mutants compared to wild-type Arabidopsis ecotype Col-0 (WT)
| First Flower Buds Visible (Days)ab | First Flower Open (Days)ab | No. of Seeds/Siliqueab | Percentage of Seed Germination | |
|---|---|---|---|---|
| WT | 30.3 b | 45.2 a | 34.5 a | 95.8% |
| sos4 | 31.8 a | 47.0 a | 31.2 bc | 96.5% |
| pdx3-1 | 30.3 b | 43.9 a | 30.4 c | 95.1% |
| pdx3-2 | 30.3 b | 45.6 a | 34.0 ab | 95.5% |
Plants were monitored from seed germination to senescence.
Means followed by the same letter are not significantly different at P = 0.05 according to the Waller-Duncan k-ratio t test.
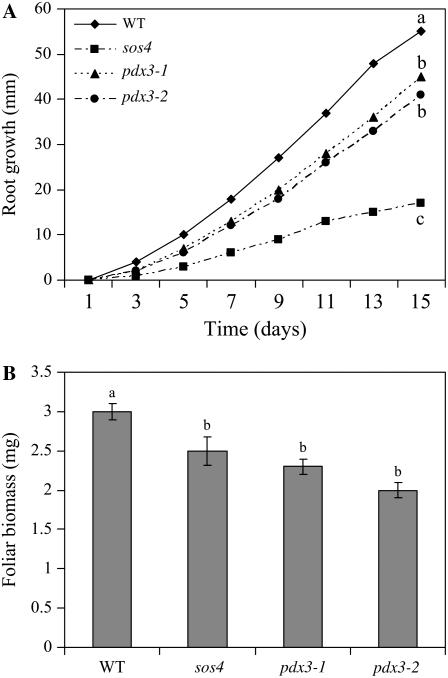
Seed germination and early seedling growth were assessed in vitro. None of the mutant lines showed differences in percent seed germination (Table I). Seedlings of both pdx3 and sos4 mutants had reduced root growth and foliar biomass compared to the wild type (Fig. 5). At day 15, pdx3-1 and pdx3-2 mutant lines showed, respectively, statistically significant 19% and 25% reduction in root growth and 23% and 33% reduction in foliar biomass. The sos4 mutant plants showed the greatest reduction in root growth of over 69% at day 15. Foliar biomass in the sos4 mutants was also significantly reduced (17%).
Figure 5.
In vitro root growth and foliar biomass of Arabidopsis pdx3 and sos4 mutant lines and Arabidopsis ecotype Col-0 (WT). A and B, Root growth (A) and foliar biomass (B) of plants grown on Murashige and Skoog medium. Root length was measured every other day for 15 d. Biomass (wet weight) was determined at 15 d. Means followed by the same letter are not significantly different at P = 0.05 according to the Waller-Duncan k-ratio t test.
Suc, NaCl, and Mannitol Sensitivity
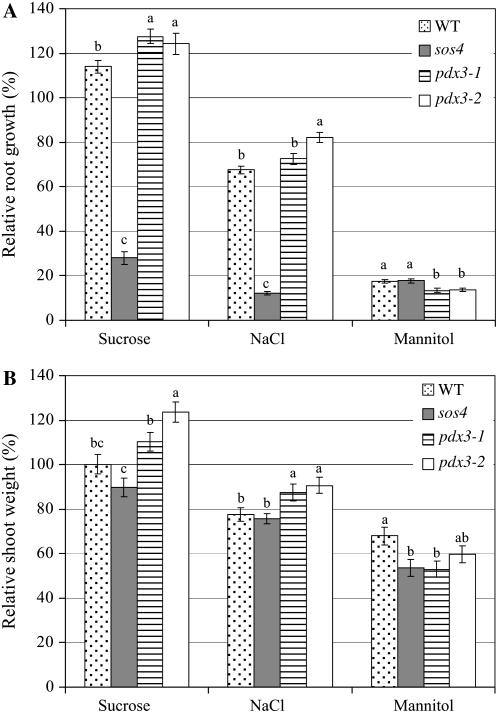
Seedlings of the pdx3 and sos4 mutant lines and the wild type were assayed for growth on Murashige and Skoog medium supplemented with 100 mm Suc, NaCl, or mannitol. Addition of Suc to the medium increased root growth in the pdx3 mutant lines and the wild type (Fig. 6A), with greater increases seen with the pdx3-1 and pdx3-2 mutant lines (28% and 24% increases, respectively) as compared with the wild type (14%). By contrast, sos4 mutant plants were inhibited by Suc, showing 72% reduction in growth compared to growth on medium lacking Suc. Effects of Suc on shoot weight were less dramatic than on root growth. Suc increased shoot weight in pdx3-1 and pdx3-2 mutant lines (10% and 24% increases, respectively), but had no effect on the wild type and did not significantly reduce shoot growth of the sos4 mutant as compared to the wild type (Fig. 6B).
Figure 6.
Sensitivity of Arabidopsis pdx3 and sos4 mutant lines and Arabidopsis ecotype Col-0 (WT) to Suc, NaCl, and mannitol. A and B, Relative root growth (A) and relative shoot weight (B) of plants grown on Murashige and Skoog medium amended with 100 mm of Suc, NaCl, or mannitol; values shown are relative to the growth of plants of the same line on Murashige and Skoog medium alone. Roots were measured every other day for 15 d. Shoot weight was determined at 15 d. Means followed by the same letter are not significantly different at P = 0.05 according to the Waller-Duncan k-ratio t test.
In agreement with the report of Shi et al. (2002), we also found that root growth of the sos4 mutant plants was strongly inhibited by NaCl (88% reduction; Fig. 6A). Shoot growth of sos4, however, was not significantly different from that of the wild type on NaCl (Fig. 6B). NaCl reduced root growth and shoot weight of the pdx3 and wild-type plants, with the pdx3-1 and pdx3-2 mutant lines showing less reduction in root growth (27% and 18% reduction, respectively) and shoot weight (12% and 9% reduction, respectively) than the wild type (32% and 22% reduction, respectively, in root growth and shoot weight). Mannitol reduced root growth and shoot weight in pdx3 and sos4 mutants and in the wild type (Fig. 6, A and B). Reductions in root growth were greater than in shoot weight, both in the mutant lines and in the wild type.
Response to High Light, Chilling, and Drought
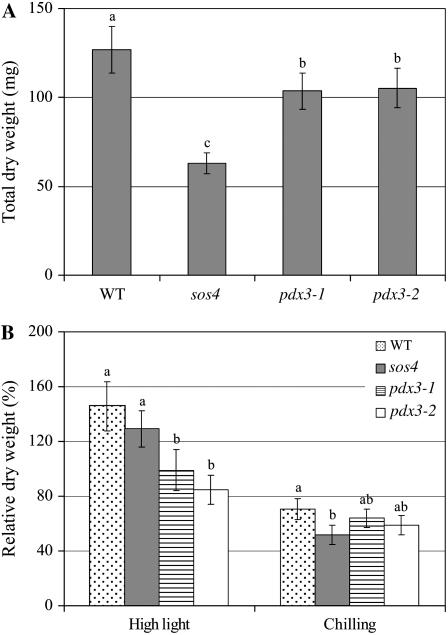
Eight- to 9-week-old plants of the pdx3 and sos4 mutant lines and wild type grown in soil were assayed for their response to high light, chilling, and drought. Plants of all three mutant lines showed a reduction in biomass when grown under control conditions (20°C, 8-h photoperiod at 200 μmol s−1 m−2) as compared to the wild type (Fig. 7A). The sos4 mutant plants showed higher reduction in biomass (50%) than the pdx3-1 and pdx3-2 mutant lines (18% and 17%, respectively). All plants (pdx3 and sos4 mutant lines and wild type) growing under high light conditions showed dark red pigmentation due to accumulation of anthocyanins (data not shown). However, high light conditions increased growth in wild-type and sos4 plants, whereas pdx3 mutant plants did not have increased growth under high light as compared to control conditions (Fig. 7B). An evident reduction in plant size under chilling conditions was observed in the mutant lines and the wild type, which was then reflected in a reduction in dry weight of these plants (Fig. 7B). sos4 plants were more tolerant to drought than wild-type and pdx3 plants, a phenotype also seen with plants mutant in the de novo pathway gene PDX1.3 (Supplemental Fig. S1).
Figure 7.
Response of Arabidopsis pdx3 and sos4 mutant lines and Arabidopsis ecotype Col-0 (WT) to high light and chilling conditions. A, Total dry weight of mutant and WT plants grown under control conditions (20°C, 8-h photoperiod, 200 μmol s−1 m−2 light) for 3 weeks. B, Relative dry weight of mutant and WT plants exposed to high light (20°C, 8-h photoperiod, 1,000 μmol s−1 m−2) and chilling (5°C, 8-h photoperiod, 200 μmol s−1 m−2 light) conditions. Dry weight is relative to the dry weight of plants of the same line grown under control conditions. Means followed by the same letter are not significantly different at P = 0.05 according to the Waller-Duncan k-ratio t test.
Analysis of Vitamin B6 Levels by HPLC Analysis and Yeast Bioassay
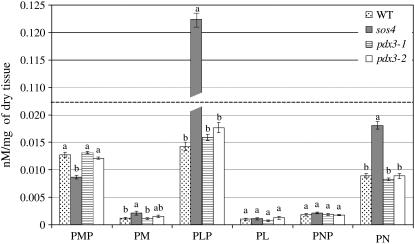
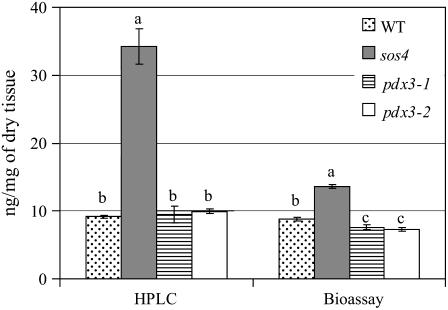
The sos4 mutant showed significant shifts in vitamer concentration compared to the wild type (Fig. 8). Higher levels of PM, PLP, and PN, with a 2-fold increase in PM and PN and an almost 9-fold increase in PLP, were observed in sos4 mutant plants. The increase in these vitamers was reflected in a 3.7- and a 1.5-fold increase in total vitamin B6 measured using HPLC analysis and yeast bioassay, respectively (Figs. 8 and 9). The sos4 mutant also showed 69% reduction in PMP levels compared to the wild type (Fig. 8).
Figure 8.
Levels of vitamin B6 vitamers in Arabidopsis pdx3 and sos4 mutant plants and Arabidopsis ecotype Col-0 (WT) determined by HPLC analysis. Values represent the average concentration in plant extracts obtained from two independent sets of plants. Means followed by the same letter are not significantly different at P = 0.05 according to the Waller-Duncan k-ratio t test.
Figure 9.
Comparison of total vitamin B6 in Arabidopsis pdx3 and sos4 mutant lines and Arabidopsis ecotype Col-0 (WT) determined by yeast bioassay and HPLC analysis. HPLC values represent the sum of PMP, PM, PLP, PL, PNP, and PN. Total vitamin B6 values represent the average concentration in plant extracts obtained from two independent sets of plants. Means followed by the same letter are not significantly different at P = 0.05 according to the Waller-Duncan k-ratio t test.
Total vitamin B6 concentration in pdx3 mutant lines was significantly lower than in the wild type, measured using yeast bioassay (Fig. 9). Although pdx3 mutant lines did not show significant changes in total vitamin B6 concentration as assayed by HPLC, slight shifts were observed in some vitamer levels (Fig. 8).
Regulation of B6 Pathway Genes in Mutant Lines
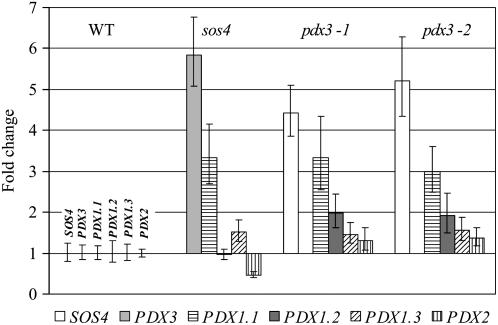
Expression of salvage and de novo pathway genes in wild-type and mutant plants was assayed by qRT-PCR. The pdx3 and sos4 mutants had increased expression of the other salvage gene (SOS4 and PDX3, respectively; Fig. 10). PDX3 was up-regulated almost 6-fold in the sos4 mutant and the expression of SOS4 was increased 4.4- and 5.2-fold in the pdx3-1 and pdx3-2 mutant lines, respectively, as compared to the wild type. Up-regulation of PDX3 in the sos4 mutant correlates with the increased PDX3 expression seen in the enzyme assay (Fig. 4). Expression of the de novo pathway PDX1 genes also was affected in the pdx3 and sos4 mutant lines (Fig. 10). PDX1.1 was the most up-regulated, with 3-fold increases in expression in the sos4 and pdx3 mutants. PDX1.2 was up-regulated 2-fold in the pdx3 mutants, but not in sos4. PDX1.3 was up-regulated less than 2-fold in all mutants. Expression of the de novo pathway gene PDX2 was not dramatically affected, with very slight increases in the pdx3 mutant lines and down-regulation in the sos4 mutant (Fig. 10).
Figure 10.
Gene expression of de novo and salvage pathway genes in Arabidopsis pdx3 and sos4 mutant lines and Arabidopsis Col-0 (WT). Lines are indicated across the top of the figure and fold changes in expression level for each gene are shown by the bars. Data represent the average of three replications. Genes were amplified by qRT-PCR using the primers listed in Table II and gene expression was normalized to UBQ10 expression.
PDX1 Activity in Mutant Lines
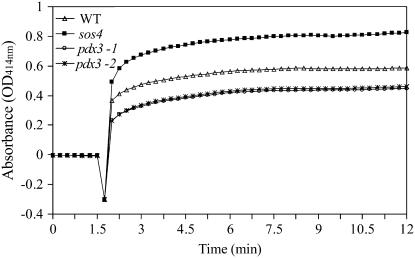
sos4 and pdx3 mutants differ significantly in PLP levels. This difference may be explained by increases in the de novo pathway; however, gene regulation studies showed similar expression of the de novo pathway genes. To further investigate this observation, we assayed PDX1 enzyme activity in wild-type and mutant lines using an assay developed for measuring PDX1 activity in tobacco (Herrero and Daub, 2007). In agreement with the PLP levels, but in contrast to gene expression data, the sos4 mutant showed an increase in PDX1 activity compared to the wild type (Fig. 11), whereas pdx3 mutant lines showed lower PDX1 activity than the wild type. Thus, PLP levels of the sos4 and pdx3 mutants correlate with PDX1 activity, but do not correlate with expression of the PDX1 genes.
Figure 11.
PDX1 activity in Arabidopsis pdx3 and sos4 mutant lines and Arabidopsis ecotype Col-0 (WT). The activity of PDX1 was measured in crude extracts of the mutant lines and WT by monitoring the formation of PLP in enzyme reactions containing the same amount of total protein for the mutant lines and WT. Before the addition of the substrate, a 1.5-min baseline was established. Absorbance values represent the average of three replications.
DISCUSSION
Arabidopsis plants mutant in the vitamin B6 salvage pathway PDX3 and SOS4 genes showed phenotypic changes, altered vitamin B6 vitamer concentrations, and changes in enzyme activity and regulation of salvage and de novo pathway genes. pdx3 and sos4 mutant lines also showed different in vitro and in vivo responses to stress conditions compared to wild-type plants. PMP/PNP oxidase (PDX3) and PL kinase (SOS4) are key enzymes involved in the vitamin B6 salvage pathway in Arabidopsis (Shi et al., 2002; Sang et al., 2007), where the nonphosphorylated forms of vitamin B6 are converted to PLP, the active cofactor of vitamin B6, essential in many aspects of primary metabolism. Additionally, vitamin B6 has been linked to stress responses in plants (Shi et al., 2002; Denslow et al., 2005, 2007; Sang et al., 2007) and is essential for root development (Shi and Zhu, 2002; Chen and Xiong, 2005; Titiz et al., 2006). Therefore, it is not surprising that Arabidopsis pdx3 and sos4 plants are affected in their growth and development under normal growth and stress conditions.
As previously reported by Shi et al. (2002), Arabidopsis sos4 plants grown under in vitro conditions show distinct reduction in root growth under normal conditions as well as root hypersensitivity to NaCl, attributed to higher accumulation of Na+ and lower retention of K+ compared to wild-type plants. Under their conditions, shoot growth was not affected under normal growth conditions but was reduced on medium containing NaCl. Our results support their root growth findings. However, in our studies, we found that shoot growth of sos4 mutant plants was reduced under normal conditions, but was not significantly different from the wild type when grown on NaCl. In addition to NaCl sensitivity, sos4 mutant plants also showed in vitro root hypersensitivity to Suc, as well as chlorosis and a visibly smaller in vivo shoot size when grown under normal conditions, reflected in the reduction of dry weight compared to wild-type plants. Similar changes in phenotype, including chlorosis and root hypersensitivity to Suc, were also observed in experiments conducted in our laboratory with Arabidopsis plants mutant for the PDX1.3 gene involved in the vitamin B6 de novo pathway (E. Rueschhoff and M. Daub, unpublished data). Interestingly, sos4 plants showed resistance to drought compared to wild type. However, plants mutant in the de novo pathway gene PDX1.3, which produce only 33% of total B6 levels of the wild type (E. Rueschhoff and M. Daub, unpublished data), showed the same level of drought tolerance (Supplemental Fig. S1). Thus, drought tolerance is unrelated to total B6 levels and may be due to the smaller and more compact rosette morphology of both of the sos4 and pdx1.3 mutants as compared to rosettes of the pdx3 mutant lines and the wild type.
Although pdx3 mutant plants showed a decrease in root and shoot growth and in total vitamin B6 compared to wild-type plants, these mutant lines were not as debilitated as sos4 mutant plants. The growth of pdx3 mutant plants was not affected in the presence of Suc and NaCl. However, pdx3 mutant plants were incapable of increased growth under high light conditions and showed decreased PDX1 activity as compared to both wild-type and sos4 mutant plants. The less dramatic phenotype may be due to the fact that these mutants are only reduced in PDX3 activity and are not completely deficient in this enzyme.
Our results demonstrate alterations in total B6 levels and in levels of the different vitamers in the sos4 and pdx3 mutant plants. pdx3 mutant plants showed significantly lower overall levels of total B6 when measured by yeast bioassay. Levels of each individual vitamer measured by HPLC, however, were not significantly different from the wild type. By contrast, the level of total B6 in the sos4 mutant was significantly greater than the wild type, with significant shifts in levels of individual vitamers. The sos4 mutant had significantly higher PM and PN levels with a corresponding decrease in PMP. Studies on the activity of the Arabidopsis (Lum et al., 2002) and wheat (Wang et al., 2004) PL kinases, SOS4 and TaPdxK, respectively, have only tested for PL kinase activity. However, Shi et al. (2002) confirmed the activity of SOS4 by complementation of E. coli mutants deficient in pdxK, which encodes a PL/PN/PM kinase, and concluded that SOS4 is functionally homologous with PdxK. The increased levels in PN and PM showed by sos4 mutant plants in the HPLC analysis support the hypothesis that SOS4 can also phosphorylate PN and PM, consistent with SOS4 encoding a PL/PN/PM kinase.
An unexpected result of the vitamer analysis was the increase in PLP levels in the sos4 mutant, a result contrary to predictions. One hypothesis that would explain this observation is increased activity of the de novo pathway. Gene regulation experiments did not support this hypothesis. Both the pdx3 and sos4 mutant lines showed similar increased expression of PDX1.1 and PDX1.3, and expression of PDX1.2 and PDX2 was actually less in sos4 as compared to the pdx3 mutants. However, PDX1 enzyme activity did correlate with PLP levels, with the sos4 plants having the highest PDX1 activity followed by wild-type and then the pdx3 plants. The sos4 mutant also had higher PDX3 activity than the wild type, indicating greater activity of both salvage and de novo pathways in this mutant. Overall, increased expression of both the salvage and the de novo genes and altered activity of PDX1 and PDX3 in the mutants confirm that plants mutant in PDX3 and SOS4 affect the normal pathway of vitamin B6 biosynthesis.
The increase in PLP in sos4 mutant plants suggests the presence of an additional kinase in Arabidopsis that would preferentially phosphorylate PL, hence the increase in PN and PM. In E. coli, an alternate kinase (PdxY) is involved in the vitamin B6 salvage pathway (Yang et al., 1998). This kinase was shown to have specific activity for PL and its function is confined to the salvage pathway. Additionally, in B. subtilis, ThiD, a kinase involved in thiamine biosynthesis, was recently shown to have activity for PL, PM, and PN (Park et al., 2004). We have identified two Arabidopsis genes encoding predicted proteins with homology to ThiD and PdxY. At5g58730 is predicted to encode a protein with 27% identity and 48% similarity to E. coli PdxY, and At1g22940 (also known as THI1 and involved in thiamine biosynthesis; Machado et al., 1997) encodes a predicted protein with 31% identity and 51% similarity to B. subtilis ThiD. Future studies are needed to confirm that these loci encode enzymes involved in the B6 salvage pathway and to determine whether they are responsible for the increased PLP found in sos4 mutant plants.
As mentioned before, the PMP/PNP oxidase in E. coli, PdxH, is essential for PLP biosynthesis because this enzyme converts the PNP synthesized in the de novo pathway into PLP (Laber et al., 1999) and pdxH E. coli mutants are incapable of growth in minimal medium (Lam and Winkler, 1992). In plants, however, PLP is the first vitamer synthesized via the de novo pathway (Tambasco-Studart et al., 2005); thus, mutations in PDX3 in plants would not be expected to be lethal. Our mutants resulted from insertions in the promoter region of the gene and still retained some enzyme activity; thus, it is not known whether this gene is essential for viability. The pdx3 mutant lines had approximately half the PDX3 activity of the wild type, resulting in only slightly less total B6 and no significant differences in levels of individual vitamers. However, these changes had measurable effects on phenotype, including less shoot growth and an inability to increase growth under high light conditions. Gene regulation studies also indicated that reduced PDX3 activity resulted in increased expression of both the de novo and salvage pathway genes, which may have allowed for the near-normal production of B6.
Although sos4 mutant plants produce elevated levels of B6, they show significant reduction in root and shoot growth and increased sensitivity to NaCl and Suc, suggesting that elevated B6 levels may be debilitating rather than advantageous. This hypothesis is supported by studies in animals and microorganisms, where elevated levels of vitamin B6 have been shown to be problematic. Large doses of vitamin B6 can cause sensory neuropathy syndrome in animals (Schaumburg et al., 1983), and overproducing microorganisms excrete excess amounts of vitamin B6 into the medium (Argoudelis, 1999; Chumnantana et al., 2001) and show delayed recovery from the stationary phase (Rodríguez-Navarro et al., 2002). In our studies to overexpress the B6 de novo pathway genes in tobacco (Herrero and Daub, 2007), 50% of the transgenic lines constitutively expressing the two de novo genes, PDX1 and PDX2, showed delays in seed germination and early seedling growth, whereas none of the transgenic plants expressing either PDX1 or PDX2 alone showed germination and growth delays. These results suggest that elevated B6 levels may be harmful to cells.
In summary, we have shown that mutations in the Arabidopsis genes encoding PMP/PNP oxidase and PL kinase result in alterations in B6 levels, growth, and response to stress conditions. Mutations in both loci result in increases in gene expression of both de novo and salvage pathway genes and in activity of the de novo pathway PDX1 enzyme, with enzyme activity but not gene expression correlating with B6 levels. Our results also suggest the presence of additional genes involved in the salvage pathway in Arabidopsis because vitamer shifts in sos4 mutants were contrary to predictions based on the function of PL kinase. We are currently conducting experiments to determine the presence of an alternate kinase as well as to identify and characterize a putative PL reductase involved in the vitamin B6 salvage pathway in Arabidopsis.
MATERIALS AND METHODS
Cloning of the Arabidopsis PDX3 Gene
To clone PDX3, total RNA was extracted from leaf tissue of wild-type Arabidopsis (Arabidopsis thaliana) ecotype Col-0 plants and reversed transcribed as described previously (Denslow et al., 2005). PDX3 was amplified from cDNA by PCR using primers 5′-TTTGGGTCATAAAGAGACTACCAATT-3′ and 5′-AATTTGGCCCCATGACTTCTACTCA-3′ and a mix of Taq DNA polymerase (Promega) and Pfu Ultra HF DNA polymerase (Stratagene) in a ratio of 10:1. The amplified gene was cloned into vector pCR 4-TOPO and was used to transform Escherichia coli One Shot TOP10 chemically competent cells using TOPO TA cloning technology (Invitrogen). Transformants were selected on Luria-Bertani medium containing 50 μg/mL kanamycin. The resulting construct, extracted from the E. coli transformants with the Wizard Plus Minipreps DNA purification system (Promega), was used to transform freshly prepared E. coli pdxH-competent cells of mutants NU877, NU1707, and NU1708, kindly provided by Dr. Malcolm E. Winkler (University of Indiana). These strains are mutant in pdxH and are therefore unable to grow on minimal medium lacking PL (Lam and Winkler, 1992). Competent cells were prepared as described previously by Miller and Nickoloff (1995). E. coli pdxH mutants were also transformed with vector pCR 4-TOPO containing a control PCR template following the same procedure described above.
Complementation of pdxH in E. coli
Complementation of pdxH E. coli mutants (NU887, NU1707, and NU1708) was determined by comparing the growth of mutants transformed with PDX3 from Arabidopsis to the growth of the mutants, the parental strain NU816, and the vector-transformed control. Strains were grown overnight at 37°C with shaking at 250 rpm in 5 mL of liquid Vogel-Bonner 1XE medium (EM) containing 0.01 mm FeSO4 and 1 μm PL. Overnight cultures were centrifuged for 10 min at 1,000 relative centrifugal force (rcf) at 4°C. Pellets were washed twice by resuspending them in 10 mL of sterile deionized water and centrifuging them for 10 min at 1,000 rcf at 4°C. Finally, clean pellets were resuspended in sterile deionized water and the density of the cell suspension was determined using a Beckman DU 600 spectrophotometer at OD600. An aliquot from the cell suspensions containing 5 × 106 bacterial cells was used to inoculate 5 mL of liquid EM plus 0.01 mm FeSO4. Cultures were incubated at 37°C with shaking at 250 rpm and growth was determined at 0, 8, 16, and 24 h by measuring the absorbance at OD600.
Arabidopsis pdx3 T-DNA Insertion Mutant Lines
Plants from Arabidopsis ecotype Col-0 with T-DNA insertions located in two different sites of the PDX3 promoter region (SALK_060749 and SALK_149382; Alonso et al., 2003; later named pdx3-1 and pdx3-2, respectively; Fig. 3A) obtained from the ABRC were screened by PCR to verify the T-DNA insertion and identify homozygous mutant plants. Seeds of SALK_060749 and SALK_149382 and the wild type were surface sterilized in a 50% commercial bleach solution (6.15% NaOCl) plus 0.01% Triton X-100 for 10 min and rinsed two to three times with sterile deionized water. Seeds were suspended in 0.1% agarose and plated onto Murashige and Skoog medium (Caisson Laboratories) with 1.2% agar, pH 5.7. Plates were placed at 4°C for 48 h to synchronize germination and then incubated at 22°C and 8-h photoperiod for about 3 weeks. Plants were transferred to Arabidopsis growing medium PM-15-13 AIS mix (Lehle Seeds) and placed at room temperature (approximately 24°C) under an 8-h photoperiod until seeds were collected. Total DNA was extracted using the Quick DNA Prep for PCR protocol described by Weigel and Glazebrook (2002). PDX3 primers (forward primer, 5′-TCCTCATTTTATCTTAGGATATTC-3′; and reverse primer, 5′-CCACCAAACCATCACCACCAT-3′) designed using the SIGnal T-DNA verification primer design tool, powered by the Genome Express Browser Server (http://signal.salk.edu/tdnaprimers.2.html), were used to amplify an approximate 1,200-bp DNA segment in PDX3 using the following program: 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, 56°C for 1 min, 72°C for 1.5 min, and 72°C for 7 min. To confirm the T-DNA insertion in the PDX3 gene, a DNA segment of about 800 bp was amplified using the PDX3 reverse primer and the right-border primer of the T-DNA insertion (5′-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3′), following the program described above with a 1-min extension at 72°C.
Phenotypic Analysis of pdx3 and sos4 Mutant Lines
Plants of the pdx3-1 and pdx3-2 mutant lines, Arabidopsis ecotype Col-0, and the sos4-1 mutant, previously generated by Shi et al. (2002), were monitored from seed germination to senescence (ready for seed harvest), to determine differences in phenotype, days to first flower buds visible, days to first flower open, and the number of seeds produced per silique. Seeds collected from homozygous pdx3-1 and pdx3-2 mutant lines, seeds from the sos4 mutants (kindly provided by Dr. Jian-Kang Zhu), and seeds from Arabidopsis Col-0 obtained from ABRC were sown into 32-pot plastic trays containing PM-15-13 AIS mix and grown at room temperature (approximately 24°C) under an 8-h photoperiod. After 2 weeks, plants were thinned to one plant per pot and grown until senescence. Thirty-two plants per mutant and wild type were analyzed. Additionally, 138 seeds harvested from the plants were plated on Murashige and Skoog medium with 1.2% agar and pH 5.7 to determine the percentage of seed germination. In vitro root growth and foliar biomass were also determined. Seeds of the mutant lines and wild type were surface sterilized and plated on square plates containing Murashige and Skoog medium as described above for 6 d. Plates were oriented vertically for in vitro assays. Six 6-d-old seedlings of each mutant and the wild type were transferred to new square plates containing the same medium, and root length was measured using a ruler every other day for 15 d. All plates were run in triplicate and the experiment was repeated one time.
Stress Responses
The in vitro sensitivity of the pdx3 and sos4 mutants and wild type to Suc, mannitol, and NaCl was tested based on a previously described method with some modifications (Wu et al., 1996). Seeds were surface sterilized as above and plated on square plates containing Murashige and Skoog medium with 1.2% agar and pH 5.7. Plates were placed at 4°C for 48 h to synchronize germination and then incubated vertically at 22°C under an 8-h photoperiod. To test Suc, mannitol, and NaCl sensitivity, six 6-d-old seedlings were transferred to square plates containing the same medium alone or supplemented with 100 mm Suc, mannitol, or NaCl. Three replicate plates were used for each treatment and assay plates were oriented vertically. Root length was measured with a ruler every other day for 15 d and the experiment was repeated one time. Response of the mutant lines and wild type to high light, chilling, and drought was tested in growth chambers following a protocol described by Denslow et al. (2007). Plants mutant in the PDX1.3 de novo gene were also included in the drought experiment as a comparison; these plants produce 33% of the total B6 levels of wild-type plants (E. Rueschhoff and M. Daub, unpublished data). For these experiments, 3-week-old seedlings of each mutant and the wild type were transferred to 32-pot plastic trays containing PM-15-13 AIS mix. The plants to be tested for high light and chilling were randomly arranged in a controlled-environment growth chamber at 20°C under an 8-h photoperiod at 200 μmol s−1 m−2 light (control chamber). After 3 weeks, one-third of the plants were moved to a growth chamber at 20°C and an 8-h photoperiod at 1,000 μmol s−1 m−2 light (high light conditions), and another one-third to a growth chamber at 5°C and an 8-h photoperiod at 200 μmol s−1 m−2 light (chilling conditions). The remaining one-third of the plants was kept in the control chamber. Plants were monitored for phenotypic changes for 2 weeks, after which all plants were harvested and dry weights were determined. The experiment was repeated one time. For the drought experiment, 32 plants of each mutant line and the wild type were randomly arranged in a growth chamber at 20°C and watered once a week for 3 weeks. After this period, watering was stopped in one-half of the plants, and all plants were monitored for phenotypic changes for the next 3 weeks.
Extraction of Vitamin B6
Vitamin B6 was extracted from leaf tissue from pdx3-1, pdx3-2, and sos4 mutant lines and Arabidopsis ecotype Col-0 obtained from two independent sets of plants. Seeds were sown into 48-pot plastic trays containing PM-15-13 AIS mix, randomly arranged, and grown at room temperature (approximately 24°C) under an 8-h photoperiod. After 2 weeks, plants were thinned to one plant per pot and grown for 6 to 8 more weeks. Leaf tissue from all plants of each mutant and wild type was bulked, lyophilized, and ground in liquid nitrogen. The finely ground tissue was divided into three samples and vitamin B6 was extracted by adding 8 mL/g of ground tissue of 5% (w/v) TCA (HPLC grade; LabChem). Samples were homogenized with a Sorvall omni-mixer (Du Pont) in a 50-mL stainless steel chamber for 5 min at power setting 5. Homogenates were centrifuged for 15 min at 10,000 rcf at 4°C. Supernatant was transferred to a clean tube and clarified by centrifugation as above. Extracts were kept on ice and in darkness at all times. Leaf extracts were stored in the dark at −20°C until use.
Yeast Bioassay
Levels of total vitamin B6 in the leaf extracts were determined by a yeast bioassay that quantifies growth of a yeast strain (ATCC 9080) auxotrophic for vitamin B6. Leaf extracts were prepared using a modified protocol from Denslow et al. (2005). Concentrated leaf extracts were diluted 4-fold with sterile deionized water, and pH was adjusted to 4.8 with sodium acetate. To deglycosylate and dephosphorylate vitamin B6 vitamers, freshly prepared β-glycosidase (5 mg in 1 mL of water) and acid phosphatase (75 mg in 2 mL of water) were added to 50-mL tubes containing the diluted plant extracts. Tubes were incubated overnight at 37°C with shaking at 70 rpm. Treated leaf extracts were filter sterilized using a 50-mL steriflip unit (0.22 μm; Millipore) and stored in the dark at −20°C until use. Yeast cells were grown on malt agar medium overnight at 30°C. Cells were washed twice with 10 mL of sterile deionized water and pelleted by centrifuging at 4°C and 1,000 rcf for 10 min. Pellets were resuspended in 2 mL of sterile deionized water, and the concentration of the cell suspension was determined using a hemacytometer. For the assay, 14-mL polystyrene tubes containing 5 mL of yeast extract mannitol medium (Becton Dickinson) were inoculated with 5 × 106 yeast cells and 8 μL of treated leaf extract. Standards contained 0, 1, 2, 3, 4, 5, 6, 7, 8, and 9 ng of total PN per 5-mL volume. All samples and standards were run in triplicate. Tubes were incubated at 30°C with shaking at 250 rpm. Growth was measured at 17 and 24 h with a spectrophotometer at OD540. Total vitamin B6 in the leaf extracts was determined based on comparison to the standard curve.
HPLC Analysis
Levels of all six vitamin B6 vitamers (PN, PM, PL, PNP, PMP, and PLP) were determined by HPLC. A reverse-phase HPLC method described by Valls et al. (2001) was used with some modifications. The HPLC system consisted of a Waters 616 pump, a Rheodyne 7125 injector equipped with a 100-μL sample loop, a Shimadzu RF-10AXL fluorescence detector, and a Waters 600S controller. The chromatographic column was a 250- × 4.6-mm i.d. HyperClone 5-μm BDS C18 (Phenomenex) equipped with a 4- × 3-mm i.d. SecurityGuard C18 (ODS; Phenomenex) guard column. The guard and analytical column were mounted in a column heater set at 35°C. The mobile phase consisted of two components: solution A (0.05 m phosphoric acid in water) and solution B (solution A containing 1% acetonitrile). Both solutions were adjusted to pH 3.2 with 10 n potassium hydroxide prior to and after degassing. Separation was carried out using a mixture of 50% solution A and 50% solution B at a flow rate of 1 mL/min. The detector excitation and emission wavelengths were set at 290 and 395 nm, respectively. Reference standards of PN (PN hydrochloride), PL (PL hydrochloride; Sigma grade), PM (PM dihydrochloride, 98% purity), PMP (98%), and PLP (98%) were obtained from Sigma-Aldrich. PNP was kindly provided by Dr. Christos Argoudelis (University of Illinois). HPLC-grade phosphoric acid, acetonitrile, and water were from Fisher Scientific. Stock solutions of 100 ng/μL of all standards were stored at 4°C and in darkness. Spiking of the leaf extracts with reference standards of PN, PNP, PM, PMP, PL, and PLP was used to confirm peak identity. Six-point standard curves were prepared and run daily before running the leaf extract samples, by injection of standards in the following concentration ranges: PN, 2.5 to 49.1 pmol; PNP, 2.1 to 33.2 pmol; PL, 0.3 to 9.8 pmol; PMP, 4.0 to 80.6 pmol; PM, 0.3 to 8.3 pmol; and PLP, 12.1 to 809.3 pmol. Concentrated leaf extracts were filter sterilized using a 50-mL steriflip unit (0.22 μm; Millipore) and diluted 4-fold with water (HPLC grade) before injection. Concentrations of vitamin B6 vitamers were determined based on comparison to the standard curve, which was obtained by fitting a linear curve to the average peak area of the different known concentrations for each vitamer. To prevent changes in retention time due to changes in retention caused by the use of a 99% aqueous mobile phase, at the beginning of each day and after every leaf extract injection, the column was flushed with acetonitrile using a solvent gradient starting with 50% A and 50% B and moving to 15% A, 15% B, and 70% acetonitrile for 40 min, followed by 10 min with 15% A, 15% B, and 70% acetonitrile, returning to 50% A and 50% B in a linear gradient for 20 min, and finally 10 min with 50% A and 50% B.
PDX1 and PDX3 Enzyme Assays
PDX1 activity was measured in crude extracts of pdx3-1, pdx3-2, and sos4 mutant lines and Arabidopsis ecotype Col-0 based on an enzyme assay previously described by Herrero and Daub (2007) with some modifications. Crude extracts were obtained by grinding 100 mg of leaf tissue in 400 μL of 50 mm Tris-HCl, pH 8.0. PDX1 activity was determined by monitoring the formation of PLP in a 100-μL reaction containing 80 to 90 μg of total protein from the crude extracts, 25 mm NH4SO2, and 10 mm glyceraldehyde-3-P. Before the addition of glyceraldehyde-3-P, a 1.5-min baseline was established in the presence of the crude extract and NH4SO2. The reaction progress was monitored spectrophotometrically at 414 nm. The activity of PDX3 was also measured in crude extracts of pdx3-1, pdx3-2, and sos4 mutant lines and Arabidopsis ecotype Col-0 with a colorimetric procedure previously described for E. coli (Zhao and Winkler, 1995) with some modifications. Crude extracts were obtained from 500 mg of lyophilized tissue (obtained from the same samples used for the HPLC analysis) homogenized in 1 mL of 200 mm Tris-HCl and 200 mm KPO4, pH 8.5, with a Silamat S5 (Ivoclar Vivadent) for 20 s. PMP oxidase activity in 1 to 7 mg of total protein from the crude extracts was determined in 0.5-mL reaction mixtures containing 200 mm Tris-HCl, 200 mm KPO4, pH 8.5, and 0.2 mm PMP. After 1 h of incubation at 37°C, reactions were stopped by addition of 50 μL of chilled 50% (w/v) TCA. Precipitated protein was pelleted by centrifuging at 4°C and 1,500 rcf for 10 min, and supernatant transferred to a clean tube. PLP formation was measured following addition of 2% (w/v) phenylhydrazine in 10 n H2SO4 and incubation on ice for 30 min (Wada and Snell, 1961) by determining the absorbance at OD410. PDX3 activity data were analyzed with a regression line adjusted to the absorbance values obtained for increasing amounts of total protein in the enzyme reactions conducted for each mutant and the wild type. Total protein concentration in the crude extracts was determined with the Bradford protein assay kit (Bio-Rad Laboratories) with bovine serum albumin as the standard, following the protocol recommended by the manufacturer. All enzyme reactions were repeated three times.
qRT-PCR
Expression of the de novo pathway genes PDX1.1, PDX1.2, PDX1.3, and PDX2 (Denslow et al., 2007) and the salvage genes SOS4 and PDX3 was determined in pdx3-1, pdx3-2, and sos4 mutant lines and Arabidopsis ecotype Col-0 using qRT-PCR. Total RNA was extracted from leaf tissue and reverse transcribed as described previously (Denslow et al., 2005). The qRT-PCR reactions consisted of 2 μL of cDNA, reverse transcribed from 1 μg of RNA, 5 μL of 2× SYBR Green master mix (Applied Biosystems), and gene-specific primers at a concentration of 0.9 pmol/μL (Table II). All primers were designed using Primer Express (Applied Biosystems) and cDNA sequences were obtained from The Arabidopsis Information Resource. Reactions were adjusted to a total volume of 10 μL with sterile deionized water. All genes were compared to expression of the genes encoding UBQ10 and ACTIN2. The primers used to amplify these genes were at a concentration of 300 nm and are shown in Table II. All reactions were run in triplicate on a DNA Engine Opticon 2 System (Bio-Rad Laboratories) using the following program: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, and the experiment was repeated three times. Only gene expression data normalized to the UBQ10 gene are shown as these data were the most constant among repetitions.
Table II.
Forward and reverse primers used for amplification of cDNA from Arabidopsis mutants for qRT-PCR
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| SOS4 | 5′-CGCTCTGTAAACCCGAATCTTAC-3′ | 5′-GTTGTATGTGCCTGAAGAACTGG-3′ |
| PDX3 | 5′-AACTGATCCTACGGTACAGTTTCG-3′ | 5′-GAGAAACAAATGCTATGGCTTTGT-3′ |
| PDX1.1 | 5′-TCTCCCTTCTCCGTGAAAGTTG-3′ | 5′-GCGTTGACGACATCCATGATT-3′ |
| PDX1.2 | 5′-AGGTCGGATTAGCTCAGGTACTTC-3′ | 5′-CGGATTCAGCGAGCTTAGCTT-3′ |
| PDX1.3 | 5′-TTTGCGGTTGCCGGAAT-3′ | 5′-ATCATCGCCGCACCTTCA-3′ |
| PDX2 | 5′-GTTCATACGTGCTCCAGCTGT-3′ | 5′-TTGATGGGACGGGATAATCC-3′ |
| UBQ10 | 5′-CACACTCCACTTGGTCTTGCG T-3′ | 5′-TGGTCTTTCCGGTGAGAGTCTTCA-3′ |
| ACTIN2 | 5′-TCCCTCAGCACATTCCAGCAGAT-3′ | 5′-AACGATTCCTGGACCTGCCTCATC-3′ |
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM124376 (Arabidopsis PDX3), M92351 (E. coli pdxH), and AF400125 (Arabidopsis SOS4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Response of Arabidopsis sos4 and pdx1.3 mutant lines and Arabidopsis ecotype Col-0 (wild type) to drought conditions.
Supplementary Material
Acknowledgments
We thank Dr. Malcolm Winkler, University of Indiana, for providing the E. coli pdxH mutants; Dr. Jian-Kang Zhu, University of California, Riverside, for the sos4-1 mutant seeds; and Dr. Christos Argoudelis, University of Illinois, for the pyridoxine 5′-P standard.
This work was supported by the National Science Foundation (grant no. MCB–0322562).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Margaret E. Daub (margaret_daub@ncsu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanove AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Argoudelis CJ (1999) Identification of the vitamers of vitamin B6 excreted by a yeast mutant growing in a glucose minimal culture medium. J Chromatogr B Analyt Technol Biomed Life Sci 721 21–29 [DOI] [PubMed] [Google Scholar]
- Belitsky BR (2004) Physical and enzymological interaction of Bacillus subtilis proteins required for de novo pyridoxal 5′-phosphate biosynthesis. J Bacteriol 186 1191–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Xiang Y, Kinsland CL, McLafferty FW, Begley TP (2005) Reconstitution and biochemical characterization of a new pyridoxal-5′-phosphate biosynthetic pathway. J Am Chem Soc 127 3682–3683 [DOI] [PubMed] [Google Scholar]
- Chen H, Xiong L (2005) Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stress. Plant J 44 396–408 [DOI] [PubMed] [Google Scholar]
- Choi SY, Churchich JE, Zaiden E, Kwok F (1987) Brain pyridoxine-5-phosphate oxidase. J Biol Chem 262 12013–12017 [PubMed] [Google Scholar]
- Chumnantana R, Hirose K, Baba H, Yagi T (2001) Production of pyridoxal phosphate by a mutant strain of Schizosaccharomyces pombe. Biosci Biotechnol Biochem 65 1789–1795 [DOI] [PubMed] [Google Scholar]
- Denslow SA, Reuschhoff EE, Daub ME (2007) Regulation of the Arabidopsis thaliana vitamin B6 biosynthesis genes by abiotic stress. Plant Physiol Biochem 45 152–161 [DOI] [PubMed] [Google Scholar]
- Denslow SA, Walls AA, Daub ME (2005) Regulation of biosynthetic genes and antioxidant properties of vitamin B6 vitamers during plant defense responses. Physiol Mol Plant Pathol 66 244–255 [Google Scholar]
- Drewke C, Leistner E (2001) Biosynthesis of vitamin B6 and structurally related derivatives. In G Litwack, T Begley, eds, Vitamins and Hormones. Advances in Research and Applications, Vol 61. Academic Press, San Diego, pp 121–125 [DOI] [PubMed]
- Ehrenshaft M, Biliski P, Li MY, Chignell CF, Daub ME (1999) A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA 96 9374–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenshaft M, Daub ME (2001) Isolation of PDX2, a second novel gene in the pyridoxine biosynthesis pathway of eukaryotes, archaebacteria, and a subset of eubacteria. J Bacteriol 183 3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirard BM, Snell EE (1988) Physical and kinetic properties of pyridoxal reductase purified from bakers' yeast. Biofactors 1 187–192 [PubMed] [Google Scholar]
- Hanna MC, Turner AJ, Kirkness EF (1997) Human pyridoxal kinase. J Biol Chem 272 10756–10760 [DOI] [PubMed] [Google Scholar]
- Herrero S, Daub ME (2007) Genetic manipulation of vitamin B-6 biosynthesis in tobacco and fungi uncovers limitations to up-regulation of the pathway. Plant Sci 172 609–620 [Google Scholar]
- Hill RE, Himmeldirk K, Kennedy IA, Pauloski RM, Sayer BG, Wolf E, Spencer ID (1996) The biogenetic anatomy of vitamin B6. J Biol Chem 271 30426–30435 [DOI] [PubMed] [Google Scholar]
- Kazarinoff MN, McCormick DB (1975) Rabbit liver pyridoxamine (pyridoxine) 5′-phosphate oxidase. Purification and properties. J Biol Chem 250 3436–3442 [PubMed] [Google Scholar]
- Kwok F, Churchich JE (1980) Interaction between pyridoxal kinase and pyridoxine-5-P oxidase, two enzymes involved in the metabolism of vitamin B6. J Biol Chem 255 882–887 [PubMed] [Google Scholar]
- Laber B, Maurer W, Scharf S, Stepusin K, Schmidt FS (1999) Vitamin B6 biosynthesis: formation of pyridoxine 5′-phosphate from 4-(phosphohydroxy)-L-threonine and 1-deoxy-D-xylulose-5-phosphate by PdxA and PdxJ protein. FEBS Lett 449 45–48 [DOI] [PubMed] [Google Scholar]
- Lam HM, Winkler ME (1992) Characterization of the complex pdxH-tyrS operon of Escherichia coli K-12 and pleiotropic phenotypes caused by pdxH insertion mutations. J Bacteriol 171 6033–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubbardi A, Marcireau C, Karst F, Guilloton M (1995) Sterol uptake induced by an impairment of pyridoxal phosphate synthesis in Saccharomyces cerevisiae: cloning and sequencing of the PDX3 gene encoding pyridoxine (pyridoxamine) phosphate oxidase. J Bacteriol 177 1817–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum HK, Kwok F, Lo SCL (2002) Cloning and characterization of Arabidopsis thaliana pyridoxal kinase. Planta 215 870–879 [DOI] [PubMed] [Google Scholar]
- Machado CR, Praekelt UM, deOliveira RC, Barbosa ACC, Byrne KL, Meacock PA, Menck CFM (1997) Dual role for the yeast THI4 gene in thiamine biosynthesis and DNA damage tolerance. J Mol Biol 273 114–121 [DOI] [PubMed] [Google Scholar]
- Man TK, Zhao GS, Winkler ME (1996) Isolation of a pdxJ point mutation that bypasses the requirement for the PdxH oxidase in pyridoxal 5′-phosphate coenzyme biosynthesis in Escherichia coli K-12. J Bacteriol 178 2445–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick DB, Chen H (1999) Update on interconversions of vitamin B-6 with its coenzyme. J Nutr 129 325–327 [DOI] [PubMed] [Google Scholar]
- Miller EM, Nickoloff JA (1995) Escherichia coli electrotransformation. In JA Nickoloff, ed, Methods in Molecular Biology, Vol 47. Humana Press, Totowa, NJ, p 105 [DOI] [PubMed]
- Mittenhuber G (2001) Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J Mol Microbiol Biotechnol 3 1–20 [PubMed] [Google Scholar]
- Morita T, Takegawa K, Yagi T (2004) Disruption of the plr1+ gene encoding pyridoxal reductase of Schizosaccharomyces pombe. J Biol Chem 135 225–230 [DOI] [PubMed] [Google Scholar]
- Nakano M, Morita T, Yamamoto T, Sano H, Ashiuchi M, Masui R, Kuramitsu S, Yagi T (1999) Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. J Biol Chem 274 23185–23190 [DOI] [PubMed] [Google Scholar]
- Osmani A, May G, Osmani S (1999) The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J Biol Chem 274 23565–23569 [DOI] [PubMed] [Google Scholar]
- Park JH, Burns K, Kinsland C, Begley TP (2004) Characterization of two kinases involved in thiamine pyrophosphate and pyridoxal phosphate biosynthesis in Bacillus subtilis: 4-amino-5-hydroxymethyl-2-methylpyrimidine kinase and pyridoxal kinase. J Bacteriol 186 1571–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle T, Amrhein N, Fitzpatrick TB (2005) On the two components of pyridoxal 5′-phosphate synthase from Bacillus subtilis. J Biol Chem 280 32291–32300 [DOI] [PubMed] [Google Scholar]
- Roa BB, Connolly DM, Winkler ME (1989) Overlap between pdxA and ksgA in the complex pdxA-ksgA-apaG-apaH operon of Escherichia coli K-12. J Bacteriol 171 4767–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S, Llorente B, Rodríguez-Manzaneque MT, Ramne G, Uber D, Marchesan B, Dujon E, Herrero E, Sunnerhagen P, Pérez-Ortín JE (2002) Functional analysis of yeast gene families involved in metabolism of vitamins B1 and B6. Yeast 19 1261–1276 [DOI] [PubMed] [Google Scholar]
- Sang Y, Barbosa JM, Wu H, Locy RD, Singh NK (2007) Identification of a pyridoxine (pyridoxamine) 5′-phosphate oxidase from Arabidopsis thaliana. FEBS Lett 581 344–348 [DOI] [PubMed] [Google Scholar]
- Schaumburg H, Kaplan J, Windebank A, Vick N, Rasmus S, Pleasure D, Brown MJ (1983) Sensory neuropathy from pyridoxine abuse. A new megavitamin syndrome. N Engl J Med 309 445–448 [DOI] [PubMed] [Google Scholar]
- Shi H, Xiong L, Stevenson B, Lu T, Zhu JK (2002) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhu JK (2002) SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol 129 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB (2005) Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA 102 13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, Fitzpatrick TB (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J 48 933–946 [DOI] [PubMed] [Google Scholar]
- Valls F, Sancho MT, Fernandez-Muino MA, Checa MA (2001) Determination of vitamin B6 in cooked sausages. J Agric Food Chem 49 38–41 [DOI] [PubMed] [Google Scholar]
- Wada H, Snell EE (1961) Enzymatic oxidation of pyridoxine and pyridoxamine phosphates. J Biol Chem 236 2089–2095 [PubMed] [Google Scholar]
- Wagner S, Bernhardt A, Leuendorf JE, Drewke C, Lytovchenko A, Mujahed N, Gurgui C, Frommer WB, Leistner E, Fernie AR, et al (2006) Analysis of the Arabidopsis rsr4-1/pdx1-3 mutant reveals the critical function of the PDX1 family in metabolism, development, and vitamin B6 biosynthesis. Plant Cell 18 1722–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu D, Liu C, Zhang A (2004) The pyridoxal kinase gene TaPdxK from wheat complements vitamin B6 synthesis-defective Escherichia coli. J Plant Physiol 161 1053–1060 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Quick DNA prep for PCR. In D Weigel, J Glazebrook, eds, Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 168–169
- Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Tsui HC, Man TK, Winkler ME (1998) Identification and function of the pdxY gene, which encodes a novel pyridoxal kinase involved in the salvage pathway of pyridoxal 5′-phosphate biosynthesis in Escherichia coli K-12. J Bacteriol 180 1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhao G, Winkler ME (1996) Identification of the pdxK gene that encodes pyridoxine (vitamin B6) kinase in Escherichia coli K-12. FEMS Microbiol Lett 141 89–95 [DOI] [PubMed] [Google Scholar]
- Zhao G, Winkler ME (1995) Kinetic limitation and cellular amount of pyridoxine (pyridoxamine) 5′-phosphate oxidase of Escherichia coli K-12. J Bacteriol 177 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.