Abstract
To clarify the functions of the Arabidopsis thaliana REV1 (AtREV1) protein, we expressed it in Escherichia coli and purified it to near homogeneity. The deoxynucleotidyl transferase activity of the recombinant AtREV1 was examined in vitro using a primer extension assay. The recombinant AtREV1 transferred one or two nucleotides to the primer end. It efficiently inserted dCMP regardless of the opposite base. AtREV1 also inserted a dCMP opposite an apurinic/apyrimidinic site, which is physiologically generated or induced by various DNA-damaging agents. In contrast, AtREV1 had no insertion activities against UV-inducible DNA lesions as reported in yeast or mammalian system. Although the substrate specificity of AtREV1 was rather narrow in the presence of magnesium ion, it widened in the presence of manganese ion. These results suggest that AtREV1 serves as a deoxycytidyl transferase in plant cells.
Plants are continuously exposed to harmful UV-B (290–320 nm) as well as the photosynthetic light (400–700 nm). Therefore, an increase of UV-B is predicted to inhibit plant growth in a large area, causing the disruption of ecosystems. UV-B radiation induces various lesions on plant DNA. The vast majority of UV-B-induced damage is cyclobutane pyrimidine dimer (CPD), which corresponds to 75% to 78% of total damage (Mitchell, 1995; Cadet et al., 2005). The 6-4 photoproducts (6-4PP) and Dewar isomer, a derivative of 6-4PP, correspond to most of the remainder (20%–24%; Mitchell, 1995; Cadet et al., 2005). Other types of DNA damage, including 8-oxoG, pyrimidine hydrate, thymine glycol, DNA single-strand break, etc., are generated by UV-B radiation at very low yield (<0.1%–2%; Mitchell, 1995; Gao and Murphy, 2001; Song et al., 2002; Cadet et al., 2005; Friedberg et al., 2006). On the other hand, apurine/apyrimidine (AP) sites are among the most abundant DNA lesions generated spontaneously (Boiteux and Guillet, 2004). For example, approximately 10,000 AP sites arise spontaneously in a mammalian cell per day (Nakamura et al., 1998). The AP site also is generated when exposed to the various DNA-damaging agents, such as ionizing radiation and alkylating chemicals, through the reaction of DNA N-glycosylases that remove the damaged base (Friedberg et al., 2006). The AP sites are rarely formed after UV irradiation directly (Song et al., 2002) or as a consequence of a cellular process removing the damaged base (Friedberg et al., 2006). Such lesions can lead to incorporation of the wrong base or can inhibit DNA replication and transcription, resulting in mutations and cell death (Britt, 1999). To prevent the effects of UV and induced DNA lesions, plants have many DNA repair mechanisms, such as photorepair by the CPD and 6-4 photolyases (Landry et al., 1997; Nakajima et al., 1998). In addition, they have nucleotide excision repair (Gallego et al., 2000; Liu et al., 2001, 2003), base excision repair (García-Ortiz et al., 2001), and recombination repair (Osakabe et al., 2006).
Translesion synthesis (TLS) is one of the damage-tolerant mechanisms prevalent in both prokaryotes and eukaryotes (Prakash et al., 2005). In TLS, a DNA lesion is bypassed by the action of specialized DNA polymerases (Broomfield et al., 2001; Friedberg et al., 2005). DNA synthesis during TLS often results in mutations because of the intrinsic nature of TLS-type polymerases (Friedberg et al., 2000). The TLS-type polymerases include DNA polymerases (Pols) ζ, ι, η, REV1, and other specialized polymerases (Prakash et al., 2005). The REV1 gene was first identified in yeast through a screening for reversionless mutants (Lemontt, 1971). Subsequent analyses showed that REV1 is a Y-family DNA polymerase. In vitro analyses showed that yeast Rev1 inserts a C opposite the AP site (Nelson et al., 1996a), and then the mismatched end is efficiently extended by Pol ζ (Nelson et al., 1996b; Lin et al., 1999). Thus, Rev1 and DNA Pol ζ are thought to be major components of the error-prone TLS (Nelson et al., 1996a, 1996b).
Little is known about the proteins that are involved in TLS in plants. We recently described an Arabidopsis (Arabidopsis thaliana) homolog of Pol ζ, which is a heterodimer of family-B DNA polymerase REV3 (Sakamoto et al., 2003) and the regulatory subunit REV7 (Takahashi et al., 2005). The Arabidopsis genome has three Y-family polymerases, which are most similar to REV1, DinB1, and Rad30, respectively (Ohmori et al., 2001). The homolog of DinB1 (AtPolK) was recently characterized and its polymerase activity was analyzed in vitro (García-Ortiz et al., 2004). The homolog of Rad30 (AtPolH) was shown to suppress the UV sensitivity of the Saccharomyces cerevisiae rad30 strain (Santiago et al., 2006). However, it is still unknown whether these Y-family polymerases perform TLS and whether they are involved in damage tolerance in plants. We recently showed that a mutant with disrupted AtREV1 had increased sensitivity to UV-B and other DNA-damaging agents (Takahashi et al., 2005), suggesting that AtREV1 has a role in the damage tolerance in Arabidopsis.
Here, we show that recombinant AtREV1 has deoxynucleotidyl transferase activity. The recombinant AtREV1 clearly inserted a nucleotide opposite AP sites in vitro. Our results suggest that AtREV1 has a role in providing tolerance to DNA damage through the TLS pathway.
RESULTS
Overproduction and Purification of Recombinant AtREV1 Protein
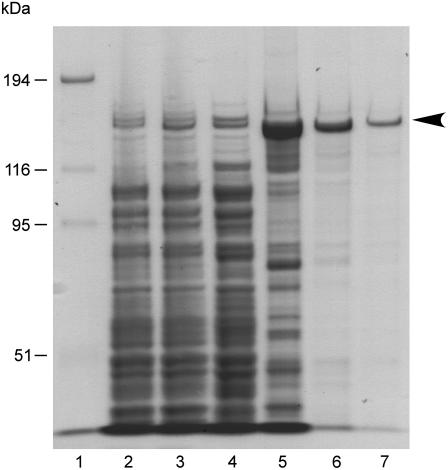
To prepare soluble and active AtREV1 protein, the AtREV1 cDNA was inserted into the pET32a vector that carries a thioredoxin-tag fusion protein, which increases the solubility of the expressed protein (Fig. 1). Under optimized growth conditions of Escherichia coli cells (see “Materials and Methods”), the recombinant AtREV1 protein was successfully expressed on a large scale. SDS-PAGE analysis showed the presence of a thick band that migrated between the 116-kD and 194-kD size markers after induction with isopropyl β-d-1-thiogalactopyranoside (IPTG; Fig. 2, lane 3). Since the size of the band (approximately 140 kD) was consistent with the predicted molecular mass of the recombinant AtREV1 fusion protein and since the band cross-reacted with the anti-His tag antibody (data not shown), we concluded that the band corresponded to AtREV1. After sonicating the E. coli cells, the recombinant AtREV1 as well as some degradation products were detected in the soluble fraction (clear lysate; Fig. 2, lane 4). The recombinant AtREV1 proteins were then fractionated by their affinity for nickel (Ni)-chelated Sepharose and successfully concentrated to a major band (Fig. 2, lane 5). To exclude the other remaining proteins derived from E. coli, the fraction was purified by heparin column and anion-ion-exchange column chromatography (Fig. 2, lanes 6 and 7). The activity of AtREV1 was monitored in each purification step. The fractions eluted from the heparin column and the anion-exchange column-purified fraction were examined by a primer extension assay using a primer template containing G and dCTP (template 30G; see “Materials and Methods”). As a result, the dCMP insertion activity was detected in both fractions (data not shown). Thus, the anion-exchange column-purified recombinant AtREV1 protein, represented as a single band (Fig. 2, lanes 6 and 7), was used to assay deoxynucleotidyl transferase activity.
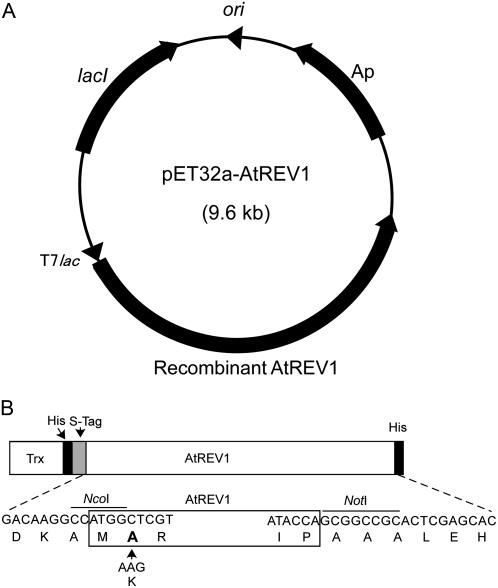
Figure 1.
Structure of the recombinant AtREV1 expression plasmid. A, AtREV1 cDNA was modified to include restriction sites on both sides and inserted into the pET32a(+) vector. The pET32a(+) vector includes a T7lac promoter (T7lac), an ampicillin-resistant gene (Ap), a lacI coding sequence (lacI), and pBR322 origin (ori). B, Schematic representation of the recombinant AtREV1 protein. The recombinant protein contains a thioredoxin tag (Trx), a His tag (His), and an S protein-binding tag (S-tag) at the N terminus, and another His tag at the C terminus. The bottom DNA and amino acid sequences represent the restriction sites (NcoI and NotI) induced for subcloning. The K (Lys) of the native AtREV1 amino acid sequence was changed to A (Ala; in boldface) in the recombinant AtREV1.
Figure 2.
Purification of recombinant AtREV1 proteins. Overproduction and purification of recombinant AtREV1 protein in E. coli. Fractions in each purification step were separated on a 7.5% SDS-polyacrylamide gel, then stained with Coomassie Brilliant Blue. Lane 1, Mr marker (Invitrogen); lanes 2 and 3, total E. coli proteins with (lane 3) or without (lane 2) IPTG induction; lane 4, clear lysate; lane 5, fraction eluted from Ni-affinity column; lane 6, fraction from a heparin affinity column; lane 7, purified fraction from HiTrap Q HP column. Molecular weight is shown on the left.
In Vitro Assay of Deoxynucleotidyl Transferase Activity of Recombinant AtREV1
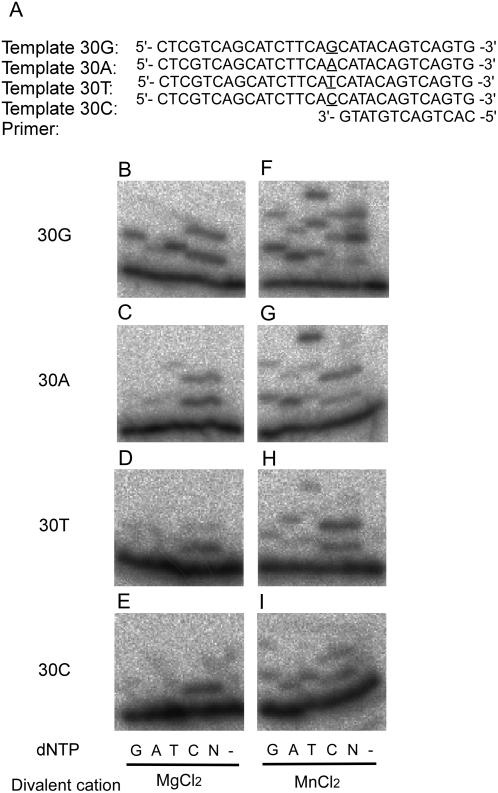
To examine the substrate specificity of the recombinant AtREV1 protein, we performed a primer extension assay using four different primer templates (Fig. 3A) in the presence of dGTP, dATP, dTTP, or dCTP individually, or all four dNTPs together. At first, the activity was examined in the presence of 2 mm magnesium ion, based on previous observations (Masuda and Kamiya, 2002). The recombinant AtREV1 protein efficiently inserted a dCMP regardless of the opposite base (Fig. 3, B–E). The protein sometimes inserted additional dCMPs, for example, when the first template base was G, A, or T (Fig. 3, B–D). AtREV1 also inserted dTMP or dGMP when the opposite base was G (Fig. 3B). Although the efficiency was low, we detected dGMP insertions opposite G, T, and C, dAMP insertions opposite G, A, and T, and dTMP insertion opposite A (Fig. 3, B–E). These results indicate that the recombinant AtREV1 has an ability to transfer a deoxynucleotide at the end of the primer. Comparing the nucleotide transferase activities to the four templates, we found that the recombinant AtREV1 seems to prefer template G to other templates. This is consistent with the character of yeast and human REV1s (Haracska et al., 2002; Zhang et al., 2002). Although the recombinant AtREV1 incorporated two to four kinds of nucleotides depending on the template base, a robust dCMP incorporation activity was detected at all four templates we examined. Thus, we concluded that the recombinant AtREV1 has deoxycytidyl transferase activity.
Figure 3.
Deoxynucleotidyl transferase activity of the recombinant AtREV1 protein. The primer P13 was 32P-labeled at the 5′ end and annealed with each of the templates, preparing four template/primer pairs: 30G (B and F), 30A (C and G), 30T (D and H), and 30C (E and I). The nucleotide sequences around the primer terminus are shown in A. The underlined bases represent the variation of templates. One hundred and sixty nanograms of AtREV1 recombinant protein and the indicated template/primer were incubated with 0.1 mm of a single dNTP (G, A, T, or C), 0.1 mm each of all four dNTPs (N), or no dNTP (−) under standard reaction conditions with 2 mm magnesium chloride (B–E) or 1 mm manganese chloride (F–I) at 30°C for 10 min. The reaction products were resolved in 20% polyacrylamide gels containing 8 m urea and visualized by autoradiography.
Effects of Divalent Cations on Activity of Recombinant AtREV1
To examine the effects of divalent cations on the transferase activity of AtREV1, the assay was performed in the presence of 1 mm manganese chloride (Fig. 3, F–I). The substrate specificity of the recombinant AtREV1 was weaker in the manganese buffer than in the magnesium buffer. Namely, in the manganese buffer, the recombinant AtREV1 transferred almost all four dNTPs regardless of the opposite base (Fig. 3, F–I). Also, up to three nucleotides were inserted in the manganese buffer, while a maximum of two nucleotides were inserted in the magnesium buffer. These results suggest that the cations chelated in the active site of AtREV1 affect the substrate specificity of this protein, as reported in other polymerases (Doublie et al., 1998; Steitz, 1998).
Insertion of Nucleotide Opposite the AP Site by Recombinant AtREV1
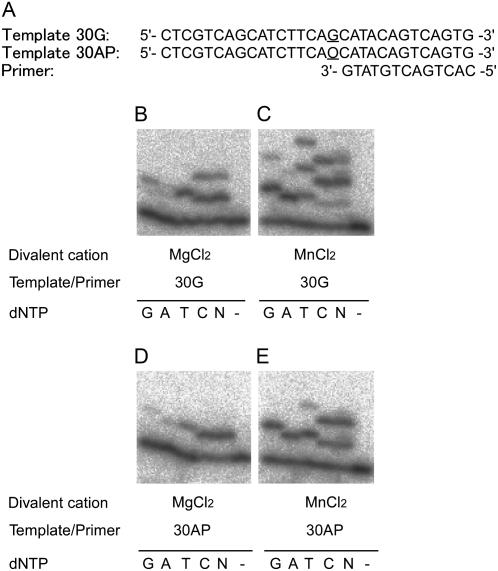
To examine whether AtREV1 recombinant protein can insert a nucleotide opposite an AP site, we performed the in vitro transferase assay with template 30AP (Fig. 4A). In the presence of magnesium ion, the recombinant AtREV1 protein efficiently inserted only one nucleotide opposite the AP site (Fig. 4D). Among four deoxynucleotides, one dCMP was preferentially incorporated opposite the AP site when compared to the other three deoxynucleotides. Other nucleotides, dTMP and dAMP, were also inserted opposite AP, but activity of incorporation was lower than that of the dCMP insertion. In the presence of manganese chloride, the recombinant AtREV1 protein inserted one or two deoxynucleotides opposite the AP site (Fig. 4E).
Figure 4.
Deoxynucleotidyl transferase activity of the recombinant AtREV1 protein opposite the AP site. The primer P13 was 32P-labeled at the 5′ end and annealed with the template 30G or 30AP. The nucleotide sequences are shown in A. The underlined bases represent the variation of templates; O indicates the AP site in template 30AP. One hundred and sixty nanograms of AtREV1 recombinant protein and the indicated template/primer were incubated with 0.1 mm of a single dNTP (G, A, T, or C), 0.1 mm each of all four dNTPs (N), or no dNTP (−) under standard reaction conditions with 2 mm magnesium chloride (B and D) or 1 mm manganese chloride (C and D) at 30°C for 10 min. The reaction products were resolved in 20% polyacrylamide gels containing 8 m urea, and then visualized by autoradiography.
No Nucleotide Insertion Activity of AtREV1 Opposite the UV Damage
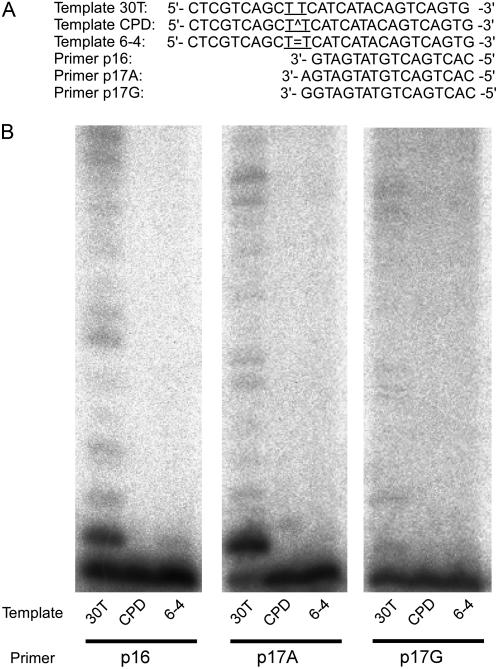
To examine whether the AtREV1 recombinant protein can insert a nucleotide opposite the typical UV-induced damage, we performed an in vitro transferase assay with a template containing CPD or 6-4PP (Fig. 5A). The template CPD or 6-4PP and a primer (P16) were annealed and transferase activity was measured in the presence of all four dNTPs. As a result, no insertion activity was detected even in a prolonged (up to 30 min) incubation (Fig. 5B). In contrast, the presence of ladder bands over the primer band with template 30T indicates that the recombinant AtREV1 can replicate this template unless UV damage is present. When the templates were annealed with P17A primer that possesses a correct nucleotide (A) opposite the 3′T of CPD or 6-4PP, AtREV1 did not extend this primer end (Fig. 5B). Naturally, no nucleotide insertion activity was observed when the templates were annealed with P17G that possesses an incorrect nucleotide (G) opposite the 3′T of CPD or 6-4PP, although AtREV1 extended this mismatch end if there was no UV damage. The absence of nucleotide insertion activity of AtREV1 against the UV damage is consistent with several reports about human or yeast REV1s (Haracska et al., 2002; Zhang et al., 2002). However, the absence of nucleotide insertion activity conflicts with the finding that an AtREV1-disrupted plant is sensitive to UV-B (Takahashi et al., 2005).
Figure 5.
Deoxynucleotidyl transferase activity of the recombinant AtREV1 protein opposite sites of UV damage. The primer P16, P17A, or P17G was 32P-labeled at the 5′ end and annealed with template 30T, CPD, or 6-4 (A). The nucleotide sequences around the primer terminus and position of UV damage (T^T for CPD, T = T for 6-4PP) are shown in A. Four hundred nanograms of AtREV1 recombinant protein and the indicated template/primer were incubated with 0.1 mm of each dNTPs under standard reaction conditions with 2 mm magnesium chloride at 30°C for 30 min. The reaction products were resolved in 20% polyacrylamide gels containing 8 m urea and visualized by autoradiography.
DISCUSSION
REV1 is known as a DNA template-dependent dCMP transferase in yeast, human, and mouse (Nelson et al., 1996a; Masuda et al., 2002; Zhang et al., 2002). We previously identified AtREV1, an Arabidopsis homolog of the REV1 gene (Takahashi et al., 2005), but did not investigate its biochemical characteristics. In this study, we successfully purified an active recombinant AtREV1 protein and examined its deoxynucleotidyl transferase activity using a primer extension assay. The recombinant AtREV1 protein incorporated nucleotides, especially cytosines and thymines, onto the primer end, regardless of the opposite template residue. In addition, the AtREV1 incorporated dCTP or dTTP opposite of the AP sites. The substrate specificity of AtREV1 is similar to the substrate specificities of yeast, human, and mouse REV1 proteins (Haracska et al., 2002; Masuda and Kamiya, 2002; Masuda et al., 2002). These results indicate that AtREV1 is a deoxycytidyl transferase.
Nair et al. (2005) determined the crystal structure of yeast Rev1 protein when it is bound to the template G and ready for an incoming dCTP. In the template/REV1 complex, an incoming dCTP binds to Arg-324 in the N-digit, a unique subdomain in REV1, rather than to the G in the template (Nair et al., 2005). At the same time, the G residue in the template is evicted from the DNA helix and makes a hydrogen bond with Leu-325 in the N-digit. This hydrogen bond is not made if the G is replaced with an A, T, or C residue in the template. This result supports the previous observation that human REV1 has a high kcat/Km value for the template G. The Arg and the Leu residues in the N-digit of yeast, mouse, and human REV1s (Masuda et al., 2001, 2002) are conserved in AtREV1 (Takahashi et al., 2005). In addition, AtREV1 more efficiently incorporated nucleotides into template G than into template A, T, or C. These results suggest that AtREV1 transfers a dCMP opposite G in a REV1-specific manner, and not through Watson-Crick pairing between dG and dCTP.
The substrate specificity of AtREV1 was reduced by replacing the magnesium ion in the buffer with manganese ion. The specificity of human REV1 was also reduced by replacing magnesium ions with manganese ions (Masuda and Kamiya, 2002). Vaisman et al. (2005) analyzed the effect of metal ions on the activity of a Y-family polymerase from Sulfolobus solfataricu Dpo4 based on its crystal structure and kinetics. They suggested that four conditions are required to form an active catalytic center in Dpo4: (1) 3′-OH of the primer end, (2) matching of the template base and the incoming nucleotide, (3) three catalytic carboxylates, and (4) two metal ions in correct coordination. Replacing the magnesium ion with manganese ion, which has a relaxed coordination requirement, dramatically increased the catalytic efficiency of misincorporation by Dpo4. Therefore, magnesium ions are probably essential for limiting misincorporations by AtREV1.
The recombinant AtREV1 protein inserted one or two nucleotides opposite the AP site, but did not extend from the mispaired termini. It is generally believed that the mispaired termini caused by the action of REV1 are extended by another polymerase (Nelson et al., 1996a; Zhang et al., 2002; Guo et al., 2004). In fact, an oligonucleotide including an AP site was efficiently replicated in vitro if the reaction mixture included both REV1 and Pol ζ (Haracska et al., 2001; Acharya et al., 2006). Does such a two-step replication involving AtREV3 occur in Arabidopsis? We do not know the properties of AtREV3 because it is too large to express as a recombinant protein. However, recent evidence strongly suggests that AtREV1 and AtREV3 play roles in a common pathway (Takahashi et al., 2005). First, the sensitivities of double-knockout rev1rev3 plants to UV-B, cisplatin, and γ-rays were similar to those of AtREV3-disrupted plants (Takahashi et al., 2005). Second, when we measured the UV-induced reversion frequencies by using the point-mutated GUS (uidA) reporter system (Kovalchuk et al., 2000), the AtREV1- and AtREV3-disrupted plants showed a similar reversion frequency on somatic cells (M. Nakagawa, S. Takahashi, and A.N. Sakamoto, unpublished data). These facts suggest that the DNA Pol ζ and AtREV1 might cooperate to bypass some classes of DNA damage in Arabidopsis.
We previously showed that the AtREV1-disrupted plants are more sensitive to UV-B and γ-ray irradiation than wild-type plants (Takahashi et al., 2005). It is known that AP sites are generated by various DNA-damaging agents, including ionizing radiation (Boiteux and Guillet, 2004). Thus, it is reasonable that the γ-ray sensitivity of AtREV1-disrupted plants is due to the loss of the dCMP insertion activity against the AP site (Takahashi et al., 2005). On the other hand, the majority of UV-induced DNA damage consists of CPDs, which are approximately 75% to 78%, while the 6-4PPs and Dewar isomers account for most of the remainder (Mitchell, 1995; Cadet et al., 2005). The monomeric damage, such as 8-oxoG or cytosine hydrate, which could be turned to AP sites by the action of DNA N-glycosylase, is generated at very low level (Cadet et al., 2005; Friedberg et al., 2006). Based on these facts, it is presumable that the UV sensitivity of AtREV1-disrupted plants is caused by loss of activity to bypass CPDs and/or 6-4PPs rather than the AP site. However, in this study, the recombinant AtREV1 protein showed no insertion or extension activity either on CPD- or 6-4PP-containing oligonucleotides in vitro. There are at least two possible explanations for this contradictory result. One of the possibilities is that the presence of the AP site is quite toxic for plants, even though the yield after UV irradiation is low, and that the inability of bypassing the AP site caused inhibition of growth. However, this hypothesis is unlikely because the AP site is abundantly generated even under no exogenous agent (Nakamura et al., 1998; Friedberg et al., 2006). Thus, if the inability of bypassing the AP site were essential for plants, the REV1-disrupted plant would show the severe growth defect even under natural growth condition. The indistinguishable growth of rev1-1 plants from wild-type plants without UV treatment (Takahashi et al., 2005) suggests that this hypothesis is not appropriate to explain the UV sensitivity of REV1-deficient plants.
The alternate possibility is that AtREV1 has another function(s) independent of dCMP transferase activity, which contributes the UV tolerance. Ross et al. (2005) showed that the human REV1 protein with point mutations in its polymerase domain completely suppresses the UV sensitivity of REV1-disrupted chicken cells. This result indicates that transferase activity is not directly required for damage tolerance at least in vertebrates. Moreover, the yeast strain involving the rev1-1 mutation, which was disrupted in the BRCT domain, could not bypass 6-4PPs on the plasmid (Nelson et al., 2000), although the REV1-1p protein with the same mutation showed normal dCMP transferase activity in vitro. These results support the idea that the BRCT domain of REV1s has a distinct function in addition to its dCMP transferase activity. Thus, by analogy, it is conceivable that the BRCT domain of AtREV1 plays some important roles in UV tolerance in Arabidopsis.
Recently, it was reported that the proliferating cell nuclear antigen (PCNA) plays a major role in polymerase switching, which is regulated by the posttranslational modification (Stelter and Ulrich, 2003; Lehmann et al., 2007). Especially, the monoubiquitination of the Lys-164 residue increases the affinity of PCNA to TLS-type polymerases, promoting TLS at the replication fork (Parker et al., 2007). Consistently, all the Y-family polymerases have conserved ubiquitin-binding sequences (UBM or UBZ), which are thought to provide an interface interacting with monoubiquitinated PCNA (Bienko et al., 2005). In yeast, the damage-bypass activity of REV1 is enhanced in the presence of ubiquitinated PCNA in vitro (Garg and Burgers, 2005), and this enhancement needs one of two UBM motifs in the C-terminal region of REV1 (Wood et al., 2007). A mutation in the UBM motif diminished the tolerance of yeast against UV, methylmethane sulfate, and 4-nitroquinolene-1-oxide to the same level as the REV1-knockout mutant (Guo et al., 2006; Wood et al., 2007). Similarly, the mouse REV1 gene that has a mutation in the UBM failed to suppress the UV or cisplatin tolerance of REV1-deficient chicken cells (Guo et al., 2006). These results indicate that the interaction of REV1 to the monoubiquitinated PCNA is commonly important for the function of REV1. The presence of putative UBMs in the C terminus of AtREV1 (Supplemental Fig. S1) strongly suggests the involvement of AtREV1 with monoubiquitinated PCNA in Arabidopsis.
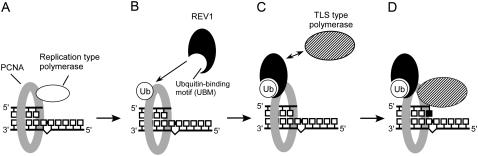
Murakumo et al. showed that human REV1 interacts with the REV7 subunit of Pol ζ through its C-terminal domain (Murakumo et al., 2001). Also, the C-terminal region of mouse REV1 interacts with Pol η and Pol ι, and Pol κ (Guo et al., 2003). From these facts, it is further proposed that the REV1 protein bound to the monoubiquitinated PCNA serves as a platform of polymerase switching to mediate recruitment of the appropriate polymerase to the damage site (Tissier et al., 2004; Lehmann et al., 2007). Taken together, the protein-protein interactions through the BRCT domain, UBMs, and/or C-terminal domain of REV1 seem to be indispensable for polymerase switching and for damage-tolerance activity. The AtREV1 protein putatively contains both a BRCT domain (Takahashi et al., 2005) and UBMs (Supplemental Fig. S1). Thus, it is conceivable that these multiple protein-protein interactions through the distinct domains of AtREV1 play an important role in UV tolerance in plant, although the exact position of the C-terminal polymerase-interacting domain remains to be identified. Based on this hypothesis, we propose a model for a possible role of AtREV1 and polymerase switching at the replication fork (Fig. 6). The replication fork is stalled when it encounters DNA damage (Fig. 6A). A signal from stalled replication triggers ubiquitination of PCNA, which leads to dissociation of the replication-type polymerase and attracts AtREV1 (Fig. 6B). TLS-type polymerases are recruited to the DNA damage through interaction with AtREV1 (Fig. 6C). TLS is accomplished by the action of one or more specialized polymerases (Fig. 6D).
Figure 6.
Possible role of AtREV1 in polymerase switching. A, Replication-type polymerase (white oval) bound to PCNA (gray ring) is stalled at the damage. B, PCNA is monoubiquitinated (Ub), which increases its affinity for AtREV1 (black oval with pocket that represents the UBM). C, TLS-type polymerase (hatched oval) interacts with the PCNA-bound AtREV1. D, A nucleotide (black square) is inserted by the action of the TLS-type polymerase. See the text for more detail.
Structure-function analysis of AtREV1, in which the BRCT, UBM, or C-terminal region is mutated, will be necessary to evaluate the function(s) of AtREV1 protein in plant damage tolerance.
MATERIALS AND METHODS
Construction of a Recombinant AtREV1 Expression Plasmid
To express the His-tagged AtREV1 protein, the AtREV1 coding fragment, prepared previously (Takahashi et al., 2005), was reamplified by PCR using the REV1-pET-1B (5′-GCCCATGGCTCGTAGCTTGGGTTCAAATTC-3′) and REV1-pET-2B (5′-GCGCGGCCGCTGGTATACTCAAGCTTCCTC-3′) primers, which contain NcoI and NotI restriction sites (underlined), respectively. To insert the NcoI site at the N terminus of AtREV1, the second codon was changed from K to A (Fig. 1B). For sequencing, the amplified fragment was ligated into the pGEM-T easy vector (Promega). The fragment contained two base substitutions that were introduced by PCR, but neither of them caused an amino acid change. The plasmid was digested with NcoI and NotI, and inserted into the corresponding sites of the pET32a(+) vector (Novagen), which is located downstream of the T7lac promoter under the control of lacI. The resulting plasmid was named pET32a-AtREV1 (Fig. 1A).
Overexpression and Purification of Recombinant AtREV1
Escherichia coli BL21(DE3) cells (Novagen) carrying pET32a-AtREV1 were inoculated into 3 mL of Luria-Bertani (LB) broth containing 100 μg mL−1 carbenicillin and were grown at 37°C for 18 h. One milliliter of this preculture was added into 50 mL of fresh LB broth containing 100 μg mL−1 carbenicillin and further grown at the same temperature for 6 h. All of the culture was transferred to 0.9 L of fresh LB broth containing 100 μg mL−1 carbenicillin and grown at 37°C until OD600 reached 0.6. To optimize the recovery of recombinant protein, expression was induced with IPTG for 3 h at 30°C or 22°C, or overnight at 15°C. At 30°C and 22°C, recombinant AtREV1 was predominantly accumulated in inclusion bodies. However, the recombinant protein was detected in the soluble fraction when the expression was induced at 15°C (data not shown). Therefore, the cells were grown at 37°C until OD600 reached 0.6, transferred to 15°C, induced with 0.8 mm IPTG (final concentration), and cultured for additional 20 h. One liter of cell culture was centrifuged at 4°C, and the cell paste was washed once with 100 mL of buffer A (20 mm Na2HPO4, pH 7.4, 0.5 m NaCl, 10% glycerol, 1 mm dithiothreitol) and then stored at −80°C. Cell paste from 3 L of culture (approximately 11.5 g wet weight) was resuspended in 100 mL of buffer A containing lysozyme and the Protease inhibitor cocktail (Sigma-Aldrich), and lysed by sonication. The cell lysate was incubated at 4°C in 0.1% Triton X-100 for 30 min, centrifuged at 12,000g for 30 min, and then the supernatant (clear lysate) was recovered. The His-tagged recombinant AtREV1 was separated with a Ni-affinity column. First, 10 mm imidazole was added to the clear lysate, then the lysate was mixed with 3 mL of Ni2+-charged Chelating Sepharose Fast Flow (GE Healthcare) equilibrated in buffer B (20 mm Na2HPO4, pH 7.4, 0.5 m NaCl, 10% glycerol, 10 mm 2-mercaptoethanol) containing 10 mm imidazole. The mixture was centrifuged at 500g, washed with 5 volumes of buffer B, and then packed in a PD-10 empty column (GE Healthcare). Bound proteins were eluted with 3 mL of buffer B containing 30, 60, 100, and 300 mm imidazole and collected in 3-mL fractions. The 300 mm imidazole fraction, containing AtREV1 proteins, was collected (Ni fraction). Six milliliters of the Ni fraction was desalted by 5-mL HiTrap Desalting column (GE Healthcare), in which the buffer was replaced with buffer C (20 mm Na2HPO4, pH 7.4, 300 mm NaCl, 10% glycerol, 1 mm dithiothreitol). The desalted Ni fraction was then applied onto a 1-mL HiTrap heparin HP column (GE Healthcare) equilibrated with buffer C. After washing the column with 10 mL of buffer C, proteins were eluted with 3.5 mL of 20 mm Na2HPO4 buffer, pH 7.4, supplemented with 400, 450, and 500 mm NaCl. The fractions with 450 and 500 mm NaCl were collected (heparin fraction).
The heparin fraction (7 mL) was desalted as described above; the buffer was replaced with buffer D (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10% glycerol, 1 mm dithiothreitol). The fraction was then applied to a 1-mL HiTrap Q HP column (GE Healthcare) equilibrated with buffer D. After the column was washed with 10 mL of buffer D, proteins were eluted with 3.5 mL of buffer D supplemented with 350 or 400 mm NaCl. The purified AtREV1 protein, eluted in 350 mm NaCl, was collected. The total volume was approximately 1.5 mL and concentration was 80 μg mL−1. Aliquots of the purified AtREV1 were dialyzed against buffer E (5 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 50% glycerol, 1 mm dithiothreitol) overnight and stored at −20°C.
Primer Extension Assay
Deoxynucleotidyl transferase activity of the purified recombinant AtREV1 was assayed according to Masuda and Kamiya (2002). Oligonucleotide template 5′-CTCGTCAGCATCTTCAXCATACAGTCAGTG-3′ (X = G, 30G; A, 30A; T, 30T; C, 30C) and the primer 5′-CACTGACTGTATG-3′ (P13) were purchased from Sigma Genosys Japan. The template with AP sites (same sequence as above template except for X = AP) was purchased from Midland Certified Reagent Company. Templates with UV damage, 5′-CTCGTCAGCTTCATCATACAGTCAGTG-3′ (TT = CPD or 6-4PP), were kindly provided by Dr. Shigenori Iwai, Osaka University. The primers 5′-CACTGACTGTATCATG-3′ (P16), 5′-CACTGACTGTATCATGA-3′ (P17A), and 5′-CACTGACTGTATCATGG-3′ (P17G) were purchased from Sigma Genosys. To prepare primer templates, the primers were labeled using T4 polynucleotide kinase (Takara BIO) and [γ-32P]ATP (GE Healthcare) and annealed to templates. The reaction mixture (20 μL) contained 40 mm Tris-HCl buffer, pH 8.0, 10% glycerol, 0.1 mg mL−1 bovine serum albumin, 5 mm dithiothreitol, 0.1 mm of a single dNTP or 0.1 mm each of all four dNTPs, 25 nm primer template, 2 mm MgCl2 or 1 mm MnCl2 as divalent cations, and 2 μL (approximately 160 ng) of AtREV1. After incubation at 30°C for 10 min, the reactions were terminated with 10 μL of stop solution (30 mm EDTA, 94% formamide, 0.05% bromphenol blue, 0.05% xylene cyanol) and the products were resolved on a 20% polyacrylamide gel containing 8 m urea. The gel was dried and used to expose an imaging plate (Fuji Photo Film) for about 40 h. The radioactivity was visualized using Bio-Imaging Analyzer BAS1800II (Fuji Photo Film).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB187523.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Conserved UBM sequences in the REV1 proteins.
Supplementary Material
Acknowledgments
We thank Dr. Shigenori Iwai at Osaka University for his kind gift of UV damage-containing oligonucleotides, and Dr. Motoshi Suzuki at Nagoya University for his technical advice on in vitro transferase assays. We also thank Ms. Chihiro Suzuki at JAEA for her technical assistance; and Drs. Katsuya Sato, Satoshi Kitamura, Yutaka Oono, and Ms. Satomi Ishii at JAEA and Mr. Youichirou Matuo at Osaka University for their technical advice on protein expression and purification. We are grateful to Drs. James Raymond and Alan Clark for careful review of the manuscript.
This work was supported in part by the Ministry of Education, Science, Sports and Culture of Japan (grants-in-aid 15201010 and 19570049 for scientific research).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ayako N. Sakamoto (sakamoto.ayako@jaea.go.jp).
The online version of this article contains Web-only data.
References
- Acharya N, Johnson RE, Prakash S, Prakash L (2006) Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol 26 9555–9563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudorf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310 1821–1824 [DOI] [PubMed] [Google Scholar]
- Boiteux S, Guillet M (2004) Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 3 1–12 [DOI] [PubMed] [Google Scholar]
- Britt AB (1999) Molecular genetics of DNA repair in higher plants. Trends Plant Sci 4 20–25 [DOI] [PubMed] [Google Scholar]
- Broomfield S, Hryciw T, Xiao W (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res 486 167–184 [DOI] [PubMed] [Google Scholar]
- Cadet J, Douki T, Pouget J-P, Ravanat J-L (2005) UVB and UVA induced formation of photoproducts within cellular DNA. In E Sage, R Drouin, M Rouabhia, eds, From DNA Photolesions to Mutations, Skin Cancer and Cell Death. RSC Publishing, London, pp 1–14
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391 251–257 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Feaver WJ, Gerlach VL (2000) The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci USA 97 5681–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RPP (2005) Trading places: How do DNA polymerases switch during translesion DNA synthesis? Mol Cell 18 499–505 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA Repair and Mutagenesis, Ed 2. American Society of Microbiology Press, Washington, DC
- Gallego F, Fleck O, Li A, Wyrzykowska J, Tinland B (2000) AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. Plant J 21 507–518 [DOI] [PubMed] [Google Scholar]
- Gao MJ, Murphy TM (2001) Alternative forms of formamidopyrimidine-DNA glycosylase from Arabidopsis thaliana. Photochem Photobiol 73 128–134 [DOI] [PubMed] [Google Scholar]
- García-Ortiz MV, Ariza RR, Hoffman PD, Hays JB, Roldán-Arjona TR (2004) Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J 39 84–97 [DOI] [PubMed] [Google Scholar]
- García-Ortiz MV, Ariza RR, Roldán-Arjona T (2001) An OGG1 orthologue encoding a functional 8-oxoguanine DNA glycosylase/lyase in Arabidopsis thaliana. Plant Mol Biol 47 795–804 [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM (2005) Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc Natl Acad Sci USA 102 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC (2003) Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J 22 6621–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC (2006) Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol 26 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Xie Z, Shen H, Zhao B, Wang Z (2004) Translesion synthesis of acetylaminofluorene-dG adducts by DNA polymerase ζ is stimulated by yeast Rev1 protein. Nucleic Acids Res 32 1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, Johansson E, Burgers PMJ, Prakash S, Prakash L (2001) Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev 15 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L (2002) Yeast Rev1 protein is G template-specific DNA polymerase. J Biol Chem 277 15546–15551 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Hohn B (2000) Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J 19 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Stapleton AE, Lim J, Hoffman P, Hays J, Walbot V, Last R (1997) An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc Natl Acad Sci USA 94 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM (2007) Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 6 891–899 [DOI] [PubMed] [Google Scholar]
- Lemontt JF (1971) Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Xu H, Zhang Y, Wu X, Yuan F, Wang Z (1999) The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res 27 4468–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hall JD, Mount DW (2001) Arabidopsis UVH3 gene is a homolog of the Saccharomyces cerevisiae RAD2 and human XPG DNA repair genes. Plant J 26 329–338 [DOI] [PubMed] [Google Scholar]
- Liu J, Hong SW, Escobar M, Vierling E, Mitchell D, Mount DW, Hall JD (2003) Arabidopsis UVH6, a homolog of human XPD and yeast RAD3 DNA repair genes, functions in DNA repair and is essential for plant growth. Plant Physiol 132 1405–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Kamiya K (2002) Biochemical properties of the human REV1 protein. FEBS Lett 520 88–92 [DOI] [PubMed] [Google Scholar]
- Masuda Y, Takahashi M, Fukuda S, Sumii M, Kamiya K (2002) Mechanisms of dCMP transferase reactions catalyzed by mouse Rev1 protein. J Biol Chem 277 3040–3046 [DOI] [PubMed] [Google Scholar]
- Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K (2001) Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J Biol Chem 276 15051–15058 [DOI] [PubMed] [Google Scholar]
- Mitchell DL (1995) DNA damage and repair. In WM Horspool, P-S Song, eds, CRC Handbook of Organic Photochemistry and Photobiology. CRC Press, New York, pp 1326–1331
- Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M (2001) Interaction in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem 276 35644–35651 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2005) Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309 2219–2222 [DOI] [PubMed] [Google Scholar]
- Nakajima S, Sugiyama M, Iwai S, Hitomi K, Otoshi E, Kim ST, Jiang CZ, Todo T, Britt A, Yamamoto K (1998) Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res 26 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA (1998) Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological condition. Cancer Res 58 222–225 [PubMed] [Google Scholar]
- Nelson JR, Gibbs PEM, Nowicka AM, Hinkle DC, Lawrence CW (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol 37 549–554 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC (1996. a) Deoxicytidil transferase activity of yeast REV1 protein. Nature 382 729–731 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC (1996. b) Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272 1646–1649 [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al (2001) The Y-family of DNA polymerases. Mol Cell 8 7–8 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yoshioka T, Osakabe Y, Todoriki S, Ichikawa H, Hohn B, Toki S (2006) Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant J 48 827–842 [DOI] [PubMed] [Google Scholar]
- Parker JL, Bielen AB, Dikic I, Ulrich HD (2007) Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res 35 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74 317–353 [DOI] [PubMed] [Google Scholar]
- Ross A-L, Simpson LJ, Sale JE (2005) Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res 33 1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Lan VTT, Hase Y, Shikazono N, Matsunaga T, Tanaka A (2003) Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and γ-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 15 2042–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago MJ, Alejandre-Durán E, Ruiz-Rubio M (2006) Analysis of UV-induced mutation spectra in Escherichia coli by DNA polymerase from Arabidopsis thaliana. Mutat Res 601 51–60 [DOI] [PubMed] [Google Scholar]
- Song JM, Milligan JR, Sutherland BM (2002) Bistranded oxidized purine damage clusters: induced in DNA by long-wavelength ultraviolet (290-400 nm) radiation? Biochemistry 41 8683–8688 [DOI] [PubMed] [Google Scholar]
- Steitz TA (1998) DNA polymerases: structural diversity and common mechanisms. J Biol Chem 274 17395–17398 [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425 188–191 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Sakamoto A, Sato S, Kato T, Tabata S, Tanaka A (2005) Roles of Arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiol 138 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck M-P, Lehmann AR, Fuchs RPP, Cordonnier A (2004) Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol η and REV1 protein. DNA Repair (Amst) 3 1503–1514 [DOI] [PubMed] [Google Scholar]
- Vaisman A, Ling H, Woodgate R, Yang W (2005) Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophospholysis. EMBO J 24 2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Garg P, Burgers MJ (2007) A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential function interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J Biol Chem 282 20256–20263 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Rechkoblit O, Geacintov E, Taylor JS, Wang Z (2002) Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res 30 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.