Abstract
Two genes encoding Arabidopsis (Arabidopsis thaliana) DEAD-box RNA helicases were identified in a functional genomics screen as being down-regulated by multiple abiotic stresses. Mutations in either gene caused increased tolerance to salt, osmotic, and heat stresses, suggesting that the helicases suppress responses to abiotic stress. The genes were therefore designated STRESS RESPONSE SUPPRESSOR1 (STRS1; At1g31970) and STRS2 (At5g08620). In the strs mutants, salt, osmotic, and cold stresses induced enhanced expression of genes encoding the transcriptional activators DREB1A/CBF3 and DREB2A and a downstream DREB target gene, RD29A. Under heat stress, the strs mutants exhibited enhanced expression of the heat shock transcription factor genes, HSF4 and HSF7, and the downstream gene HEAT SHOCK PROTEIN101. Germination of mutant seed was hyposensitive to the phytohormone abscisic acid (ABA), but mutants showed up-regulated expression of genes encoding ABA-dependent stress-responsive transcriptional activators and their downstream targets. In wild-type plants, STRS1 and STRS2 expression was rapidly down-regulated by salt, osmotic, and heat stress, but not cold stress. STRS expression was also reduced by ABA, but salt stress led to reduced STRS expression in both wild-type and ABA-deficient mutant plants. Taken together, our results suggest that STRS1 and STRS2 attenuate the expression of stress-responsive transcriptional activators and function in ABA-dependent and ABA-independent abiotic stress signaling networks.
As a sessile organism, a plant's ability to adapt to abiotic stresses such as heat, cold, drought, and high salinity is crucial for its survival. Plant responses to abiotic stresses involve a complex variety of tolerance mechanisms (Bray et al., 2000; Tester and Davenport, 2003; Wang et al., 2003) that are activated and integrated by the expression of thousands of genes (Chen et al., 2002; Kreps et al., 2002; Seki et al., 2002). These genes encode proteins involved in numerous biological processes as well as a large number of proteins of unknown function. Furthermore, the expression of many genes with regulatory functions, such as transcription factors, RNA-binding proteins, calcium-binding proteins, kinases, phosphatases, etc., is altered by stress. These genes are probably involved not only in regulating downstream stress responses but also in stress perception and signaling (for review, see Xiong et al., 2002b; Zhu, 2002; Shinozaki et al., 2003; Bartels and Sunkar, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006).
In recent years, much progress has been made in identifying and characterizing components of stress signaling networks in Arabidopsis (Arabidopsis thaliana). Promoter analyses of cold- and dehydration-responsive genes such as RD29A have revealed two cis-acting elements mediating stress-induced expression, the DRE/CRT and ABRE elements (Yamaguchi-Shinozaki and Shinozaki, 1994; Shinozaki et al., 2003). The DRE/CRT element, which has the core sequence CCGAC, is essential for regulating gene expression in response to cold and hyperosmotic stresses, and this control is independent of the plant hormone abscisic acid (ABA; Yamaguchi-Shinozaki and Shinozaki, 1994). Members of the DRE-binding protein (DREB)/C-repeat-binding factor (CBF) family of transcription factors specifically bind the DRE/CRT element and activate transcription of downstream stress-inducible genes in response to cold, osmotic, and salt stresses (Stockinger et al., 1997; Liu et al., 1998). Ectopic or inducible expression of DREB/CBF genes leads to enhanced expression of downstream stress-inducible genes and increased tolerance to freezing, drought, and salt stresses (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). The cold-induced expression of at least one of the DREB/CBF genes, as well as that of other cold-responsive transcription factors, is controlled by the constitutively expressed transcription factor INDUCER OF CBF EXPRESSION1 (ICE1), the most upstream transcription factor in the cold stress signaling subnetwork identified to date (Chinnusamy et al., 2003; Lee et al., 2005).
Control of ABA-regulated gene expression is mediated via the ABRE cis-acting element PyACGTGGC, which is bound by bZIP transcription factors known as ABRE-binding (AREB) proteins or AREB factors (ABFs; Choi et al., 2000; Uno et al., 2000). Expression of genes encoding several of these proteins are up-regulated by ABA, drought, and high salinity, and the proteins themselves can act as transcriptional activators in protoplast transient expression assays (Uno et al., 2000; Fujita et al., 2005). Overexpression of ABF3 and AREB/ABF4 confers ABA-hypersensitive germination and seedling phenotypes, enhanced expression of ABA-regulated genes, and tolerance to drought (Kang et al., 2002). Several other cis-elements also act in ABA-dependent expression. For instance, RD22 expression is dependent on ABA for its drought-inducible induction (Abe et al., 1997), but the gene contains no ABRE element in its promoter. Instead, drought-inducible RD22 expression is mediated by the AtMYC2 (RD22BP1) and AtMYB2 transcription factors that can bind MYC and MYB cis-elements, respectively, that are present in the RD22 promoter (Abe et al., 2003). These two transcription factors are synthesized after ABA accumulates and cooperatively activate RD22 expression.
Posttranscriptional and posttranslational control of stress gene expression is increasingly being recognized as playing a major role in regulating plant stress responses. For example, in unstressed Arabidopsis lines overexpressing the Na+/H+ antiporter SOS1, levels of SOS1 transcript are similar to wild-type levels. Only under salt stress does SOS1 transcript accumulate to high levels in the overexpressors, suggesting that SOS1 mRNA is unstable in unstressed conditions (Shi et al., 2003). Evidence is beginning to accumulate that small RNAs, such as microRNAs and short interfering RNAs (siRNAs), may regulate gene expression in response to environmental stresses. The expression of a substantial number of small RNAs is altered in response to abiotic stress (Sunkar and Zhu, 2004), and at least one siRNA is involved in regulating a salt stress response (Borsani et al., 2005). Phosphorylation/dephosphorylation also appears to be important in stress responses. In ABA signaling, for instance, phosphorylation of AREB/ABFs by ABA-activated SNF1-related protein kinases may activate these transcription factors (Furihata et al., 2006). Control of stress-responsive gene expression at the level of protein degradation had also been demonstrated. HOS1, a negative regulator of cold responses, is a RING finger protein that has E3 ligase activity and can mediate ubiquitination of ICE1 (Dong et al., 2006). Moreover, degradation of ICE1 is induced by cold stress and requires HOS1 activity.
The upstream stress-responsive transcription factors such as ICE1, the DREB/CRT family, the AREB/ABFs, ATMYC2, and AtMYB2, as well as proteins mediating posttranscriptional regulation of gene expression, ultimately control the expression of many downstream stress-responsive genes involved in the response to multiple abiotic stresses (e.g. Maruyama et al., 2004; Sakuma et al., 2006). Therefore, in an effort to identify new upstream components of abiotic stress signaling networks, we devised a novel, functional, genomics-based screen for identifying regulatory genes that control Arabidopsis responses to multiple abiotic stresses (P. Kant, M. Gordon, S. Kant, G. Zolla, O. Davydov, Y.M. Heimer, V. Chalifa-Caspi, R. Shaked, and S. Barak, unpublished data). Here, we present the characterization of two mutants that exhibited greater tolerance than wild type to multiple abiotic stresses and showed more highly induced expression of genes encoding stress-responsive transcription factors and their downstream target genes. Both mutants were defective in different DEAD-box RNA helicase proteins, and expression of their respective genes, designated STRESS RESPONSE SUPPRESSOR1 (STRS1) and STRS2, in wild-type plants was down-regulated by salt, drought, and heat stress, but not cold stress. Expression of the STRS genes was reduced by ABA, but the STRSs regulate both ABA-dependent and -independent stress signaling subnetworks. This study not only identifies STRS1 and STRS2 as upstream negative regulators of Arabidopsis responses to multiple abiotic stresses but also illustrates the growing importance of RNA metabolism in the control of stress-responsive gene expression.
RESULTS
Identification of the strs Mutants
A functional genomics-based screen was performed to identify genes that may function as upstream regulators of multiple abiotic stress responses (P. Kant, M. Gordon, S. Kant, G. Zolla, O. Davydov, Y.M. Heimer, V. Chalifa-Caspi, R. Shaked, and S. Barak, unpublished data). In brief, a microarray analysis of early Arabidopsis heat stress-responsive genes was performed, and the resulting data were combined in a “stress gene” database with data from published microarray analyses examining Arabidopsis responses to a variety of abiotic stresses. The database was queried for a set of regulatory genes whose expression was affected early by multiple abiotic stresses, and Arabidopsis T-DNA insertion mutants defective in each gene were screened for altered sensitivity to abiotic stresses. A preliminary screen of mutants homozygous for the T-DNA insertion identified two mutants exhibiting increased tolerance to salt stress that contained a T- DNA insertion in genes encoding different DEAD-box RNA helicases (Fig. 1, A and B). Analysis of microarray data in our database indicated that these genes are down-regulated by salt, osmotic, and heat stress, suggesting that the encoded proteins may function to suppress Arabidopsis stress responses. We therefore designated the proteins as STRS1 and STRS2.
Figure 1.
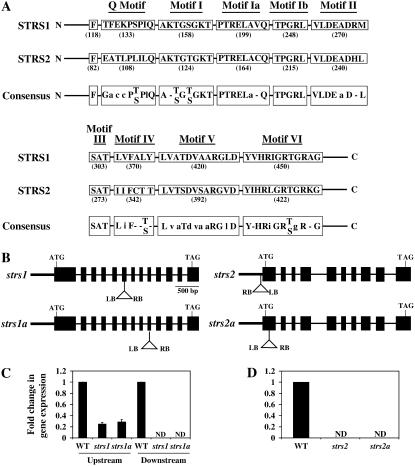
Arabidopsis STRS1 and STRS2 are DEAD-box RNA helicases whose expression is disrupted in T-DNA insertion mutants. A, Alignment of conserved motifs specific to DEAD-box helicases that are present in STRS1 and STRS2 with the consensus sequences of the DEAD-box family. Numbers in parentheses represent the amino acid position of the first residue in each motif. For the consensus sequences, capital letters denote amino acids that are conserved at least 80%, while lowercase letters denote amino acids that are conserved 50% to 79% (Tanner and Linder, 2001; Rocak and Linder, 2004). B, Scheme of the STRS1 and STRS2 genes. Black boxes represent exons and lines symbolize introns. The position and orientation of the T-DNA insertion is depicted (not to scale). LB, Left border sequence; RB, right border sequence. C and D, Real-time PCR analysis of STRS1 and STRS2 expression, respectively, in wild type and two independent T-DNA insertion mutants for each gene. Relative transcript levels were determined by real-time PCR according to the 2−ΔΔCT method using UBQ10 as an internal control (Livak and Schmittgen, 2001). Gene expression was normalized to the wild-type expression level, which was assigned a value of 1. Data represent the average of three independent experiments ± sd. Upstream, RT-PCR carried out with primers complementary to sequences upstream of the T-DNA insertion; Downstream, RT-PCR carried out with primers complementary to sequences downstream of the T-DNA insertion; ND, not detectable.
STRS1 (At1g31970) is predicted to encode a protein of 537 amino acid residues with an estimated molecular mass of 59.5 kD, while STRS2 (At5g08620) is predicted to be a protein of 563 amino acids with an estimated molecular mass of 62.5 kD. Database searches revealed that both proteins possess all nine conserved motifs that are characteristic of the DEAD-box protein family as well as an upstream conserved Phe (Fig. 1A; de la Cruz et al., 1999; Rocak and Linder, 2004). It has been estimated that the Arabidopsis genome encodes over 50 DEAD-box RNA helicases (Aubourg et al., 1999; Boudet et al., 2001), 32 of which were classified by Aubourg et al. (1999) as AtRH1 to AtRH32. Genomic and cDNA sequence analysis indicated that STRS1 is identical to AtRH5, while STRS2 is identical to AtRH25. The N- and C-terminal extensions of DEAD-box RNA helicases are of variable length, and it is thought that they confer substrate specificity (Aubourg et al., 1999; de la Cruz et al., 1999). Counting upstream from the conserved Phe and downstream from motif VI for N termini and C termini, respectively, STRS1 and STRS2 have N-terminal regions of 117 and 81 amino acids and C-terminal regions of 76 and 130 amino acids, respectively. Alignment of STRS1 and STRS2 protein sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) showed that the two proteins exhibit 32% sequence identity and 49% sequence similarity. However, all the sequence identity/similarity resides in the core helicase region of the protein (between the N- and C-terminal regions defined above) containing the conserved motifs, whereas there is no significant sequence similarity between the N-terminal and C-terminal regions.
The strs1 and strs2 Mutants Exhibit Enhanced Tolerance to Abiotic Stresses
To explore whether the STRS genes are involved in regulating a variety of abiotic stresses, the stress-responsive phenotypes of two independent T-DNA insertion lines for each gene were analyzed. The two strs1 mutant lines designated strs1 and strs1a contain a T-DNA insertion in exon 6 and exon 9, respectively (Fig. 1B). Real-time PCR analysis of STRS1 gene expression in unstressed wild-type and mutant plants using primers complementary to DNA downstream of the insertion showed that STRS1 transcript was undetectable in both mutant lines (Fig. 1C). However, when primers complementary to DNA upstream of the T-DNA insertion were employed, STRS1 transcripts were detected in strs1 and strs1a plants, albeit at 25% to 30% of wild-type STRS1 transcript levels. This suggests that truncated STRS1 transcripts are produced in both strs1 mutant lines but that they are less stable than wild-type transcripts. Furthermore, while it is unknown whether the truncated transcripts are translated, any truncated protein that may be produced in the strs1 mutants is unlikely to be functional due to the absence of essential protein motifs such as motif V and motif VI (Fig. 1A; de la Cruz et al., 1999; Rocak and Linder, 2004). The T-DNA insertion in strs2 is located in the STRS2 promoter region close to the transcription start site, whereas the insertion in strs2a is located in the first exon (Fig. 1B). No STRS2 transcript could be detected by real-time PCR in either line (Fig. 1D).
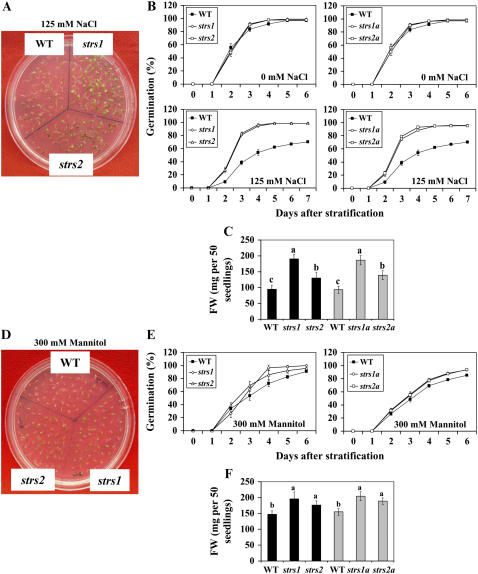
When grown under unstressed conditions, both seedling and adult strs mutants showed no morphological or developmental differences compared to wild type except for a very weak early flowering phenotype (data not shown). Percentage of seed germination of all four mutant lines on Murashige and Skoog (MS) plates in the absence of stress was virtually identical to wild type. However, germination of the strs mutants on MS plates supplemented with NaCl showed substantial tolerance to salt stress (Fig. 2, A and B). At only 2 d after stratification, the strs lines already showed 2- to 3-fold greater percentage of germination than wild-type seeds. By 5 d after stratification, strs seeds exhibited 95% to 99% germination, whereas wild type showed approximately 60% germination. In fact, under salt stress conditions, the final percentage of germination of wild-type seeds never reached more than 70%. In addition, all the strs lines grew faster than wild type under salt stress. Quantification of fresh weight (FW) at 7 d after germination demonstrated that strs1 and strs1a seedlings exhibited 100% greater FW than wild type, while strs2 and strs2a seedlings showed 37% and 49% greater FW, respectively, than wild type (Fig. 2C).
Figure 2.
Altered salt and osmotic stress tolerance of strs1 and strs2 mutants. A, Increased tolerance to salt stress. Seeds were germinated and grown on MS plates with and without 125 mm NaCl. Photographs were taken on the tenth day after stratification. WT, Wild type. B, Percentage of germination of wild type and two independent alleles of strs1 and strs2 on MS plates with and without 125 mm NaCl. Data are mean ± sd (n = 4). Fisher's protected lsd test showed no significant difference in germination percentage of wild type and mutants without NaCl. However, with NaCl, strs1, strs2, strs1a, and strs2a exhibited significantly higher germination percentage than wild type (P ≤ 0.05). C, FW of wild type and two independent alleles of strs1 and strs2 10 d after stratification on MS plates with and without 125 mm NaCl. Data are mean ± sd (n = 4). Bars with different letters indicate significant difference at P ≤ 0.05 (Fisher's protected lsd test). D, Increased tolerance to osmotic stress. Seeds were germinated and grown on MS plates with and without 300 mm mannitol. Photographs were taken on the tenth day after stratification. E, Percentage of germination of wild type and two independent alleles of strs1 and strs2 on MS plates with and without 300 mm mannitol. Data are mean ± sd (n = 4). Fisher's protected lsd test showed no significant difference in germination percentage between wild type and mutants without mannitol. However, with mannitol, strs1 and strs2 exhibited significantly higher germination than wild type at 4 and 5 d after stratification, while strs1a and strs2a exhibited significantly higher germination percentage than wild type at 4, 5, and 6 d after stratification (P ≤ 0.05). F, FW of wild type and two independent alleles of strs1 and strs2 10 d after stratification on MS plates with and without 300 mm mannitol. Data are mean ± sd (n = 4). Bars with different letters indicate significant difference at P ≤ 0.05 (Fisher's protected lsd test).
The strs mutants also showed tolerance to osmotic stress, albeit to a lesser extent than their tolerance to salt stress. When seedlings were germinated and grown on MS plates supplemented with mannitol, all four mutant lines showed between 11% and 33% greater germination than wild-type seedlings by 4 to 5 d after stratification (Fig. 2, D and E). Furthermore, strs1 and strs1a mutants exhibited over 30% greater FW than wild type, while strs2 and strs2a showed approximately 20% greater FW (Fig. 2F).
The strs mutants were next tested for altered basal and acquired thermotolerance. For basal thermotolerance, seeds were sown on MS plates, stratified for 4 d at 4°C, and then exposed to 1 to 4 h of 45°C (Hong and Vierling, 2000). Basal thermotolerance was quantified by two methods: (1) seeds were allowed to germinate and grow at 22°C with a 16-h photoperiod, and percentage of germination was recorded; and (2) seeds were allowed to germinate and grow for 6 d at 22°C in the dark, and hypocotyl elongation was measured. Figure 3, A and B, shows germination results of seeds given 3 h of heat stress. This duration of heat stress killed seeds of hot1-3, a mutant of HEAT SHOCK PROTEIN101 (HSP101) that is defective in basal and acquired thermotolerance (Hong and Vierling, 2000). Although a proportion of wild-type seeds was able to germinate, all the strs lines displayed greater germination at each time point after transfer to 22°C. Mutant seedlings also grew better than wild type after germination. While 3 h of heat stress led to a reduction in hypocotyl elongation in both wild type and strs lines compared to the control treatment (Fig. 3C), strs mutants still showed 2.5- to 3-fold greater hypocotyl elongation than wild type. Moreover, whereas 4 h of heat stress killed wild-type seeds altogether, the strs mutants survived and hypocotyl growth continued.
Figure 3.
Altered basal and acquired thermotolerance of strs1 and strs2 mutants. A, Basal thermotolerance. Stratified seeds sown on MS plates were exposed to 45°C for 3 h and then allowed to germinate and grow at 22°C. A representative plate is shown 6 d after transfer to 22°C. WT, Wild type; hot1-3, a mutant of HSP101 that is defective in basal and acquired thermotolerance (Hong and Vierling, 2000). B, Quantification of basal thermotolerance by percentage of germination of seeds treated at 45°C for 3 h. The results from two independent alleles of strs1 and strs2 are shown. Data are mean ± sd (n = 3). Fisher's protected lsd test showed that all strs mutant lines exhibited a significantly higher germination percentage than wild type and hot1-3 (P ≤ 0.05). C, Quantification of basal thermotolerance by hypocotyl elongation assay using two independent alleles of strs1 and strs2. Seeds were treated at 45°C for the indicated time periods and allowed to germinate in the dark on vertical plates. Hypocotyl length was measured 6 d after transfer to 22°C. Data are mean ± sd (n = 4). Each replicate consisted of approximately 20 seedlings. Bars with different letters indicate significant difference at P ≤ 0.05 (Fisher's protected lsd test). D, Acquired thermotolerance. Seedlings were grown on vertical plates in the dark for 3 d. Con, Control seedlings maintained at 22°C; PT, pretreatment of 38°C for 90 min; HS, heat stress of 45°C for 2 h; PT + 2, pretreatment followed by 2 h at 22°C and then 45°C for 2 h; PT + 3, pretreatment followed by 2 h at 22°C and then 45°C for 3 h. After heat treatment, seedlings were grown for a further 3 d before measurement of the post-stress increase in hypocotyl length. Data are mean ± sd (n = 4). Each replicate consisted of approximately 15 to 20 seedlings. Bars with different letters indicate significant difference at P ≤ 0.05 (Fisher's protected lsd test).
Acquired thermotolerance results from prior exposure to a pretreatment such as a sublethal high temperature (Lindquist, 1986). To assess whether the strs mutants possessed enhanced acquired thermotolerance, a quantitative hypocotyl assay was performed (Hong and Vierling, 2000). In brief, seedlings were grown on vertical MS plates in the dark for 2 to 3 d before application of heat stress treatments. The increase in hypocotyl length was recorded 3 d after the heat stress. Figure 3D shows that a heat stress of 45°C for 2 h killed both wild-type and mutant seedlings, as evidenced by lack of hypocotyl elongation. However, a pretreatment of 38°C followed by 45°C for 2 h or 3 h allowed seedlings to survive. The hot1-3 mutant was most severely affected by 2 h of heat stress, exhibiting an 80% reduction in hypocotyl elongation compared to the control (no heat treatment). Hypocotyl elongation of wild-type seedlings was reduced by 42%, whereas strs1 and strs2 exhibited a drop of only 4% and 13%, respectively. A heat stress of 3 h killed the hot1-3 mutant, while wild-type and strs seedlings showed a further reduction in hypocotyl elongation with the mutants displaying approximately one-half the reduction observed in wild type. A similar effect was observed for the strs1a and strs2a mutants. Taken together, these results suggest that the strs mutants exhibit enhanced basal and acquired thermotolerance. Plants were also tested for freezing tolerance, but no difference could be observed between wild type and mutants (data not shown).
The strs Mutants Exhibit Enhanced Expression of Stress-Responsive Genes and Their Upstream Regulators
To gain insight into the molecular basis of the stress-tolerant strs mutant phenotypes, we next investigated the expression of the well-characterized stress-responsive marker gene, RD29A (Yamaguchi-Shinozaki and Shinozaki, 1994). RD29A expression was analyzed by real-time PCR using UBIQUITIN10 (UBQ10) expression as an internal control. The two independent T-DNA insertion lines for each of the STRS genes exhibited identical expression phenotypes, and, therefore, only the results for the strs1 and strs2 mutants are shown. In wild-type plants subjected to salt, drought, or cold stress, RD29A expression was induced by each stress (Fig. 4, A–C). RD29A expression peaked at 6, 12, or 24 h after the onset of salt, drought, or cold stress, respectively, and then progressively declined in agreement with a previous report (Albrecht et al., 2003). RD29A expression was also induced by stress in the strs mutants with similar kinetics to that observed in wild-type plants. However, fold-induction of RD29A expression in the mutants was consistently higher than in wild-type plants, particularly at, and after, peak expression, suggesting that the STRS proteins function as negative regulators of stress-responsive gene expression.
Figure 4.
Expression of stress-responsive genes in wild-type and strs mutant plants subjected to salt, drought, and cold treatments. Two-week-old soil-grown plants were exposed to various stress treatments. Relative transcript levels were determined by real-time PCR according to the 2−ΔΔCT method using UBQ10 as an internal control (Livak and Schmittgen, 2001). Gene expression was normalized to the wild-type unstressed expression level, which was assigned a value of 1. Data represent the average of three independent experiments ± sd. A, D, G, J, L, N, and P, Salt treatment; 200 mm NaCl. B, E, H, K, M, O, and Q, Drought treatment; plants were removed from the soil and allowed to dry under 60% humidity. C, F, I, and R, Cold treatment; 4°C.
In unstressed plants, no differences in RD29A expression were observed between wild-type and mutant plants (Fig. 4, A–C), indicating that loss of STRS function alone is not sufficient for enhanced RD29A expression. We therefore surmised that derepressed expression of upstream transcription factors in the strs mutants might account for the enhanced RD29A expression. Consequently, we analyzed the expression of two members of the DREB transcription factor family that mediate stress-responsive RD29A expression via DRE elements in the RD29A promoter (Stockinger et al., 1997; Liu et al., 1998). In wild-type plants, DREB1A/CBF3 expression was induced by salt, drought, and cold stress with peak expression at 6 h after onset of salt stress and 3 h after onset of drought or cold stress (Fig. 4, D–F). Under salt and drought stress, expression dropped sharply by 6 h after stress and thereafter slowly declined, whereas under cold stress a more gradual decrease in DREB1A/CBF3 expression was observed. Furthermore, peak expression of DREB1A/CBF3 was an order of magnitude higher under cold stress than in salt and drought stress, reflecting the primary role of DREB1A/CBF3 in cold-responsive gene expression (Liu et al., 1998; Shinwari et al., 1998). In the strs mutants, expression kinetics of DREB1A/CBF3 was comparable to wild type, but fold-induction of DREB1A/CBF3 expression was higher in the mutant lines compared to wild type. The DREB2 proteins play a major role in drought and salt stress signaling networks (Liu et al., 1998; Nakashima et al., 2000). DREB2A expression was induced in wild-type plants by salt and drought stress and by cold stress, although cold-induced expression levels were two orders of magnitude lower than drought-induced expression (Fig. 4, G–I). Peak expression of DREB2A occurred at identical time points to RD29A expression. Once again, enhanced stress-induced expression of DREB2A was observed in the strs mutants with expression displaying similar kinetics to that observed in wild-type plants. No difference was observed in either DREB1A/CBF3 or DREB2A expression between unstressed wild-type and strs mutant plants. This suggests that the STRS proteins are not acting to repress stress-responsive gene expression in unstressed plants. Rather, the STRS proteins attenuate gene expression once it has been induced by stress.
We next tested whether the STRS genes also regulate stress-responsive genes that are not controlled by the DREB signaling subnetwork by analyzing the salt- and drought-induced expression of two non-DRE element genes, RD19 and RD22 (Yamaguchi-Shinozaki et al., 1992; Abe et al., 1997). Salt and drought led to induction of RD19 and RD22 expression with similar kinetics in both wild-type and mutant plants (Fig. 4, J–M). However, expression of both genes was enhanced in the strs1 and strs2 mutants, particularly at, and after, peak expression. Furthermore, salt- and drought-induced expression of AtMYC2, one of the transcription factors that regulates RD22 expression, also exhibited increased expression in the strs mutants (Fig. 4, N and O). These results suggest that STRS1 and STRS2 also function in DREB-independent signaling networks.
To ensure that the effects of the strs mutations were specific, we analyzed expression of the housekeeping gene ACTIN2 (ACT2). Figure 4, P to R, demonstrates that ACT2 expression was unaffected either by stress treatments or by absence of STRS1 or STRS2, thereby suggesting a specific role for the STRS proteins in Arabidopsis abiotic stress responses.
The molecular basis for the increased tolerance to heat stress of the strs mutants was examined by analyzing expression of the gene encoding HSP101. This protein has been shown to be essential for both basal and acquired thermotolerance (Hong and Vierling, 2000; Queitsch et al., 2000). In wild-type plants exposed to 40°C heat stress, induction of HSP101 expression was detected at 5 min after application of stress, reaching a peak at 2 h and declining by 3 h of stress (Fig. 5A). In both strs mutants, kinetics of stress-induced HSP101 expression followed closely that observed in wild type, but fold-induction was higher in the mutants than in wild type, particularly at, and after, peak HSP101 expression. No difference in HSP101 expression was observed between unstressed wild-type and mutant plants, similar to the other stress-response genes analyzed.
Figure 5.
Expression of heat stress-responsive genes in wild type and strs1 and strs2 mutants. A to D, Two-week-old plants were exposed to 40°C for the indicated time periods. Relative transcript levels were determined by real-time PCR according to the 2−ΔΔCT method using UBQ10 as an internal control (Livak and Schmittgen, 2001). Gene expression was normalized to the wild-type unstressed expression level, which was assigned a value of 1. Data represent the average of three independent experiments ± sd.
Heat shock proteins are primarily regulated at the transcriptional level by heat shock transcription factors (HSFs; Wu, 1995). In Arabidopsis, there are 21 putative HSFs (Nover et al., 2001), and of those that have been studied, some are constitutively expressed, while the expression of others is induced by heat stress (Lee et al., 1995; Prandl et al., 1998; Miller and Mittler, 2006). HSF4 and HSF7 are two genes whose expression is induced by heat stress (Prandl et al., 1998; AtGenExpress database, http://www.arabidopsis.org/info/expression/ATGenExpress.jsp), and we examined whether expression of HSF4 and HSF7 is enhanced in the strs mutants. Figure 5, B and C, shows that in wild type and strs mutants, the expression of both HSF4 and HSF7 was induced by heat stress, reaching peak expression at 30 min after onset of stress. Moreover, their expression was enhanced in the strs mutants predominantly at, and after, peak expression. As a control, the expression of TUBULIN5 (TUB5) in response to the heat treatment was examined. ACT2 expression was not used as a control, because its expression is affected by heat stress (data not shown). TUB5 expression was unaffected by heat stress in wild type and strs mutant seedlings (Fig. 5D), demonstrating that HSP101, HSF4, and HSF7 expression was specifically affected in the strs mutants. Thus, although there is, as yet, no evidence that HSF4 and/or HSF7 directly regulate HSP101 expression, our results suggest that the enhanced HSP101 expression observed in the strs mutants is due to derepression of upstream HSFs.
Taken together, our results suggest that the STRS proteins act as general attenuators of Arabidopsis stress responses and that increased salt-, drought-, cold-, and heat-induced gene expression in the strs mutants is due, at least in part, to derepressed, stress-mediated expression of upstream transcriptional activators.
STSR1 and STRS2 Regulate Both ABA-Dependent and ABA-Independent Stress Signaling Subnetworks
Drought-induced RD22 and AtMYC2 expression is mediated by ABA (Abe et al., 1997, 2003), and the finding that STRS1 and STRS2 negatively regulate RD22 and AtMYC2 expression (Fig. 4) suggests that the STRS proteins can function in ABA-dependent stress signaling. To further explore this notion, we analyzed the ABA-induced expression of the RD26 gene in wild-type and strs mutant seedlings exposed to ABA. RD26 is a dehydration-induced NAC protein that functions as a transcriptional activator in ABA-dependent stress signaling (Fujita et al., 2004). Figure 6A shows that in wild-type seedlings, RD26 exhibited a continual rise in expression up to at least 2 h after induction by ABA, consistent with previously reported findings (Fujita et al., 2004). On the other hand, fold-induction of ABA-induced RD26 expression was enhanced in the strs mutants, with maximum induction occurring at 1 h after transfer to ABA-containing plates. In wild-type and mutant seedlings transferred to MS plates without ABA, no induction of RD26 expression occurred (Fig. 6B), thereby demonstrating that the rise in RD26 expression in seedlings exposed to ABA was due to the action of ABA and not due to any stress caused by the transfer procedure itself.
Figure 6.
ABA-responsive gene expression and ABA sensitivity in wild type and strs1 and strs2 mutants. Seedlings were grown on vertical MS plates for 4 d after germination and then transferred to fresh treatment plates. Relative transcript levels were determined by real-time PCR according to the 2−ΔΔCT method using UBQ10 as an internal control (Livak and Schmittgen, 2001). Gene expression was normalized to the wild-type control expression level, which was assigned a value of 1. Data represent the average of four independent experiments ± sd (n = 4). A, Expression of RD26 in wild-type and strs mutant seedlings transferred to MS plates with 100 μm ABA. B, Expression of RD26 in wild-type and strs mutant seedlings transferred to MS plates without ABA. C, Expression of STRS1 and STRS2 in wild-type seedlings transferred to MS plates with 100 μm ABA. D, Expression of STRS1 and STRS2 in wild-type seedlings transferred to MS plates with 300 mm NaCl. E, Expression of STRS1 and STRS2 in aba2-1 (ABA-deficient) mutant seedlings transferred to MS plates with 300 mm NaCl. F, Percentage of germination of wild-type and strs1 and strs2 mutant seedlings after 6 d incubation on MS media containing different concentrations of ABA. Data are mean ± sd (n = 3). Fisher's protected lsd test showed that all strs mutant lines exhibited a significantly higher germination percentage than wild type upon exposure to ABA (P ≤ 0.01).
The STRS proteins could attenuate ABA-dependent stress-responsive gene expression either by modulating ABA signaling to its target genes or by acting directly in the ABA signaling pathways. In the latter case, it would be expected that STRS expression would respond to ABA. Figure 6C shows that exposure of wild-type seedlings to ABA led to rapid suppression of STRS expression, indicating that STRS1 and STRS2 can function as components of the ABA-dependent signaling subnetwork. However, the fact that the STRS proteins regulate stress-mediated DREB expression as well (Fig. 4) suggests that STRS1 and STRS2 also function in the ABA-independent stress signaling subnetwork. To test this hypothesis, we examined stress-mediated expression of STRS1 and STRS2 in wild type and the ABA-deficient aba2-1 mutant (Leon-Kloosterziel et al., 1996). Figure 6, D and E, shows that salt stress led to a reduction in STRS expression in both wild-type and aba2-1 plants, thus demonstrating that STRS1 and STRS2 can respond to stress signals in the absence of ABA. This finding supports the contention that STRS1 and STRS2 regulate both the ABA-independent and ABA-dependent stress signaling subnetworks.
Because the absence of STRS1 and STRS2 led to enhanced expression of ABA-responsive genes (Figs. 4, L–O, and 6A), we surmised that the strs1 and strs2 mutants might exhibit an ABA-hypersensitive phenotype. We therefore examined the effect of the strs mutations on ABA inhibition of seedling germination. Wild-type and mutant seed (both independent strs1 and strs2 T-DNA insertion lines) were germinated on MS plates containing 0, 0.5, 1, or 2 μm ABA. On control plates, no difference in germination could be observed between wild-type and mutant plants (Fig. 6F). Surprisingly, however, the strs mutant lines exhibited an ABA-insensitive phenotype, whereby ABA progressively inhibited wild-type germination more severely at each concentration than it did the strs mutants. This finding is similar to that found in the hos5 and fry1 mutants that also exhibit up-regulated stress gene expression but display an ABA-insensitive germination phenotype (Xiong et al., 1999, 2001b).
Expression of STRS1 and STRS2 in Wild-Type Plants Is Down-Regulated by Salt, Drought, and Heat Stress, But Not by Cold Stress
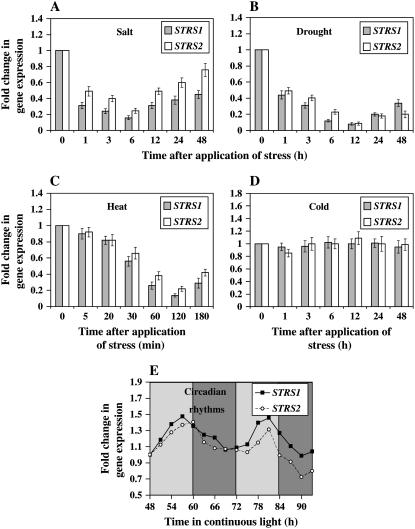
Inspection of our “stress gene” database along with results from testing STRS expression in wild-type and aba2-1 plants (Fig. 6, D and E) suggested that STRS1 and STRS2 expression is down-regulated by salt, osmotic, and heat stress. To confirm this observation and to examine the detailed temporal expression of STRS1 and STRS2, we analyzed STRS gene expression in wild-type plants in response to salt, drought, heat, and cold stresses. Salt and drought stresses led to an over 50% reduction in STRS1 and STRS2 expression by 1 h after the onset of stress (Fig. 7, A and B). Expression continued to decline to about 20% and 10% of control levels under salt and drought stress, respectively, with STRS expression progressively rising thereafter. However, the later rise in STRS expression levels was considerably less under drought stress than under salt stress. Furthermore, under salt stress, STRS1 expression exhibited greater stress-mediated repression than STRS2. Down-regulation of STRS expression was even more rapid after onset of heat stress (Fig. 7C). Expression levels reached their nadir by 2 h of heat stress and began rising again by 3 h of heat stress.
Figure 7.
Stress-responsive and circadian clock-controlled expression of STRS1 and STRS2. Two-week-old wild-type soil-grown plants were exposed to various stress treatments. Relative transcript levels were determined by real-time PCR according to the 2−ΔΔCT method using UBQ10 as an internal control (Livak and Schmittgen, 2001). Expression was normalized to the unstressed expression level of the respective gene, which was assigned a value of 1. Data represent the average of three independent experiments ± sd. A, Salt treatment; 200 mm NaCl. B, Drought treatment; plants were removed from the soil and allowed to dry under 60% humidity. C, Heat treatment; 40°C. D, Cold treatment; 4°C. E, Circadian clock control; 7-d-old wild-type seedlings were entrained in a 12-h-light:12-h-dark photoperiod for 4 d and then released into continuous light. Data are representative of similar results from two independent experiments. Light and dark shaded bars represent subjective day and subjective night, respectively.
We also noticed that trough STRS expression coincided with peak expression of RD29A and DREB2A at 6 and 12 h after the onset of salt or drought stress, respectively (compare Fig. 7A with Fig. 4, A and G; and Fig. 7B with Fig. 4, B and H). Furthermore, the fast post-peak recovery of STRS expression under salt and the slow recovery of expression under drought were also reflected in the kinetics of the post-peak decline of RD29A and DREB2A expression. Similarly, trough STRS expression coincided with peak HSP101 expression at 2 h of heat stress (compare Fig. 7C with Fig. 5A). These results suggest a remarkable temporal correlation between trough expression of the STRS genes and the peak expression of downstream stress-responsive genes within a particular stress treatment.
In contrast to salt, drought, and heat stress, cold treatment had no effect on the expression of the STRS genes (Fig. 7D). This result is in agreement with the finding that the strs mutants did not exhibit enhanced freezing tolerance (data not shown) and confirmed the notion that STRS1 and STRS2 are not involved in attenuation of cold stress-regulated gene expression. However, the strs mutants did display enhanced expression of the DREB genes and a downstream target gene in response to cold stress (Fig. 4, C, F, and I). This finding suggests that in the absence of STRS1 and STRS2, stress-induced expression of genes normally attenuated by the STRS proteins will show enhanced expression in response to any signal that triggers stress-responsive gene expression. This will occur even if that signal does not normally down-regulate STRS expression.
One limitation of using relative real-time PCR quantification is that, unlike northern analysis, a visualization of overall expression levels between various genes is not possible. Therefore, to determine expression levels of STRS1 and STRS2 compared to other stress-responsive genes, we quantified STRS transcript copy number in unstressed plants and compared them with transcript copy numbers of DREB1A/CBF3, RD29A, and HSP101 (Table I). STRS1 and STRS2 exhibited comparable amounts of unstressed transcript levels to each other, but these were an order of magnitude higher than RD29A and HSP101 and two orders of magnitude higher than DREB1A/CBF3 transcript levels. STRS1 and STRS2 expression was detected in all organs from unstressed plants that were examined (Table II). However, transcript copy numbers differed according to the organ analyzed. Highest STRS expression was observed in inflorescences and lowest levels in green siliques and roots.
Table I.
Comparison of STRS transcript levels with transcripts of other stress-responsive genes in unstressed wild-type plants
Quantification of transcript copy number was performed by relating the real-time PCR signal for each gene to a standard curve. The target gene transcript copy number was then adjusted for loading differences by dividing by normalized UBQ10 level. The table represents the average results from three independent experiments ± sd.
| Gene | Transcript Copy No. |
|---|---|
| STRS1 | 305 ± 42 |
| STRS2 | 308 ± 59 |
| DREB1A/CBF3 | 9 ± 0.9 |
| RD29A | 91 ± 9 |
| HSP101 | 86 ± 6 |
Table II.
Organ-specific STRS transcript copy number in unstressed wild-type plants
Quantification of transcript copy number was performed by relating the real-time PCR signal for each gene to a standard curve. The target gene transcript copy number was then adjusted for loading differences by dividing by normalized UBQ10 level. The table represents the average results from three independent experiments ± sd.
| Organ | STRS1 | STRS2 |
|---|---|---|
| Rosette leaves | 421 ± 45 | 614 ± 53 |
| Cauline leaves | 728 ± 68 | 1,020 ± 128 |
| Bolts | 1,073 ± 114 | 1,386 ± 127 |
| Inflorescence | 1,233 ± 96 | 1,894 ± 248 |
| Green siliques | 32 ± 4 | 48 ± 5 |
| Roots | 15 ± 1.5 | 17 ± 1.8 |
STRS1 and STRS2 Expression Is under the Control of the Circadian Clock
Transcript profiling has shown that the expression of many stress-responsive genes is under the control of the circadian clock (Harmer et al., 2000). Regulation by the clock may be an important means of coordinating plant stress responses to ensure optimum expression at periods when stresses are most likely to occur, thereby allowing anticipation of stress even in its absence. We therefore tested whether STRS1 and STRS2 expression is under the control of the circadian clock by entraining wild-type seedlings under a 12-h-light:12-h-dark photoperiod, releasing them into continuous light and taking samples every 3 h. Figure 7E shows that both STRS1 and STRS2 exhibited circadian rhythms in transcript accumulation with peak expression at mid- to late subjective afternoon. Because the STRS proteins attenuate stress-responsive gene expression, we expected that peak STRS expression would be close to peak expression of downstream stress-responsive genes. Indeed, peak STRS1 and STRS2 expression coincided with peak expression of DREB1A/CBF3 and other downstream stress genes (Harmer et al., 2000).
DISCUSSION
We have identified STRS1 and STRS2 as negative regulators of multiple abiotic stress responses in Arabidopsis. Disruption of the STRS1 and STRS2 genes by T-DNA insertions increases the tolerance of strs mutant seedlings to salt and osmotic stresses and enhances basal and acquired thermotolerance (Figs. 2 and 3). Consistent with their stress-tolerant phenotypes, the strs mutants exhibit enhanced expression of stress-responsive genes and their upstream transcriptional activators (Figs. 4 and 5). However, stress-responsive gene expression is not up-regulated in unstressed strs mutants. These findings, coupled with results showing stress-mediated down-regulation of STRS gene expression in wild-type plants (Fig. 7), suggest that STRS1 and STRS2 are required to attenuate expression of upstream stress signaling components.
STRS1 and STRS2 Are DEAD-Box RNA Helicases That Attenuate Arabidopsis Responses to Abiotic Stresses
The STRS1 and STRS2 genes encode proteins that are members of the large family of approximately 50 Arabidopsis DEAD-box RNA helicases, a larger number than the sequenced genomes of other organisms, including the fly and worm (Boudet et al., 2001). In nonplant systems, these proteins are involved in many aspects of RNA metabolism, particularly within supramolecular complexes. These processes include ribosome biogenesis, transcription, pre-mRNA splicing, mRNA export, RNA degradation, translation initiation, and organellar gene expression (Rocak and Linder, 2004; Linder, 2006). They are thought to function either as RNA chaperones that promote the formation of optimal RNA structure by local unwinding activity or by mediating RNA-protein association/dissociation (Schwer, 2001; Lorsch, 2002). However, very little is known about the function of the superfamily of RNA helicases in plants. The two exceptions are the DExH-box RNA helicase CARPEL FACTORY/DICER-LIKE1 (DCL1) and the DEAD-box RNA helicase LOS4. DCL1 is involved in processing microRNAs (Park et al., 2002; Reinhart et al., 2002) and has been shown to be involved in at least two stress-related mechanisms. DCL1 can form a complex with the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1), which functions to assist DCL1 in efficient and precise cleavage of primary microRNAs (Kurihara et al., 2006). HYL1 itself is also involved in ABA signaling, and the Arabidopsis hyl1 mutant is hypersensitive to ABA and exhibits enhanced ABA induction of downstream stress-responsive genes (Lu and Fedoroff, 2000). DCL1 is further involved in processing of the natural antisense siRNAs derived from Δ1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE (P5CDH) and SRO5 transcripts (Borsani et al., 2005). Under salt stress, this system acts to degrade the P5CDH transcript to allow accumulation of the compatible osmolyte Pro, while SRO5 acts to counteract the increased reactive oxygen species production caused by decreased P5CDH activity.
The los4-1 mutant exhibits severely reduced cold induction of DREB/CBF expression and its target genes and is more sensitive to cold stress (Gong et al., 2002). In contrast, the los4-2 mutation causes enhanced cold-induced DREB1C/CBF2 expression and its target genes and leads to plants that are more tolerant to freezing stress but more sensitive to heat stress (Gong et al., 2005). The LOS4 DEAD-box RNA helicase protein is enriched in the nuclear rim, and mRNA export is blocked at low and warm temperatures in the los4-1 mutant but only at warm temperatures in the los4-2 mutant.
Although our results suggest that STRS1 and STRS2 attenuate transcript accumulation of upstream stress transcription factors, their mode of action is unclear at present. They may directly affect transcription, pre-mRNA processing, mRNA stability, or other aspects of stress transcription factor RNA metabolism. Alternatively, they may function by regulating transcripts of enhancers or repressors of stress transcription factors. It is tempting to speculate, however, that STRS1 and STRS2 are involved in degrading the stress-induced mRNAs because the abundance of the STRS transcripts decreases as the abundance of the stress marker transcripts increases. If the STRS proteins themselves are short lived and their abundance parallels that of their transcripts, then a decline in STRS protein would allow accumulation of the stress-induced transcripts.
The virtually identical phenotypes of the strs1 and strs2 mutants plus the close pattern of expression of the two genes in response to the various stresses suggest that STRS1 and STRS2 may function together in a complex. It should also be noted that although the strs mutant phenotypes did not result from a perturbation of general gene expression, the STRS proteins may have additional functions to those in abiotic stress responses. This premise is supported by the observation that the strs mutants have a slightly early flowering phenotype, at least under long-day (16 h light:8 h dark) conditions (data not shown) and that the highest expression of both genes was detected in flowers (Table II).
STRS1 and STRS2 are two of several negative regulators of stress responses that have been identified in recent years. These include HOS1 (Lee et al., 2001), HOS5 (Xiong et al., 1999), FIERY1 (FRY1; Xiong et al., 2001b), and FRY2 (Xiong et al., 2002a). The hos1 mutation leads to enhanced cold induction of DREB/CBF expression and downstream genes but has no effect on ABA, salt, or osmotic stress-mediated gene expression. On the other hand, the hos5-1 mutation enhances osmotic, salt, and ABA stress-induced gene expression but not cold-induced gene expression, similar to the strs1 and strs2 mutations. These findings emphasize the distinct subnetworks for cold signaling and osmotic/salt signaling, Indeed, HOS1, which encodes a RING E3 ligase, mediates degradation of the ICE1 protein, a transcriptional activator of DREB1A/CBF3 expression specific to cold stress (Dong et al., 2006). In contrast, the fry1 and fry2 mutations lead to superinduction of stress gene expression by cold, osmotic, salt, and ABA, illustrating the links between the cold signaling and osmotic/salt signaling subnetworks.
Attenuation of the stress signaling networks by negative regulators is thus clearly important for proper regulation of the response to abiotic stresses. Constitutive activation of the stress response by ectopic expression of DREB1A/CBF3 leads to severe growth retardation in unstressed plants (Kasuga et al., 1999). Only when DREB1A/CBF3 expression is driven by the stress-inducible RD29A promoter is growth retardation greatly reduced. This suggests that attenuators are necessary to prevent overactivation of the stress response. This idea is further reinforced by our finding that peak circadian expression of STRS1 and STRS2 coincides with peak expression of DREB1A/CBF3 and other downstream stress genes (Fig. 7E; Harmer et al., 2000). Coexpression of genes encoding stress-transcriptional regulators, such as the DREB/CBFs, with genes encoding proteins, such as the STRSs that attenuate expression or activity of those transcription factors, might also ensure that transient stress conditions such as occur in the field do not lead to full activation of the stress response machinery. Indeed, the stress-mediated change in stress gene transcript abundance is itself transient (Fig. 4). The fact that the stress-mediated transient decline in STRS1 and STRS2 expression closely parallels that of the transient activation of stress transcriptional activators and their downstream targets (e.g. compare Fig. 7A with Fig. 4, A and G) suggests that STRS1 and STRS2 may be part of the basic machinery that is responsible for the transience of stress responses.
STRS1 and STRS2 Are Regulatory Nodes in the Heat, Osmotic, Salt, and ABA Signaling Subnetworks, But Not Cold Signaling Subnetworks
Our results further demonstrate the distinct regulation of the drought and salt signaling subnetworks and the cold signaling subnetwork. First, while strs mutants exhibit tolerance to salt and osmotic stresses (Fig. 2), they do not appear to be tolerant to freezing stress (data not shown). However, this finding must be further explored because freezing tests were carried out on detached leaves, whereas all other tolerance assays were performed on seedlings. Moreover, enhanced cold-induced gene expression was observed in the strs mutants (Fig. 4, C, F, and I), suggesting that mutant plants might indeed show tolerance to cold stress under certain conditions. Nevertheless, the freezing tolerance results are supported by the discovery that STRS1 and STRS2 expression is unaffected by cold stress, whereas it is down-regulated by salt and drought stresses (Fig. 7). In addition to the phenotypes of various stress repressor mutants such as hos1 and hos5-1 described above, there are several other lines of evidence pointing toward distinct signaling subnetworks for salt and drought stresses on the one hand and cold stress on the other hand. For instance, the DREB1 proteins and ICE1, the upstream regulator of DREB1A/CBF3 expression, are mainly involved in regulating cold-induced gene expression, while DREB2A and DREB2B control salt- and drought-induced gene expression (Liu et al., 1998; Shinwari et al., 1998; Nakashima et al., 2000). Furthermore, overexpression of constitutively active DREB2A leads to increased tolerance of Arabidopsis to drought stresses but only slight tolerance to freezing (Sakuma et al., 2006). However, there are clear regulatory links between the drought and salt signaling subnetworks and the cold signaling subnetwork. For instance, inducible expression or overexpression of DREB1A/CBF3 leads to increased tolerance to cold, salt, and drought stresses (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999).
The strs mutant phenotype showing increased basal and acquired thermotolerance (Fig. 3) and enhanced HSP101, HSF4, and HSF7 expression (Fig. 5) reveals interactions between the heat, salt, and osmotic signaling subnetworks. Analysis of the expression of all 21 Arabidopsis HSF genes has demonstrated that many HSFs are induced by multiple abiotic stresses, again illustrating the links between the signaling subnetworks (Miller and Mittler, 2006). However, few signaling components that may function as nodes linking the heat signaling subnetwork and other abiotic stress signaling subnetworks have been identified. Our results showing that STRS1 and STRS2 attenuate expression of upstream transcription factors involved in regulation of drought, salt, and heat stress-responsive gene expression and the exquisite temporal regulation of STRS1 and STRS2 expression in response to each stress suggest that STRS1 and STRS2 are nodes linking the salt, drought, and heat stress signaling subnetworks. Another recently discovered potential node linking heat stress signaling with other abiotic stress signaling subnetworks is the transcriptional activator MULTIPROTEIN BRIDGING FACTOR1c (MBF1c). Constitutive expression of MBF1c enhances the tolerance of transgenic Arabidopsis plants to heat, salinity, and osmotic stresses, and causes enhanced accumulation of stress-related transcripts (Suzuki et al., 2005). MBF1c does not appear to be involved in cold responses. It would be of interest, therefore, to investigate whether the STRS proteins and MBF1c can affect each other's expression.
The finding that STRS1 and STRS2 regulate the ABA-dependent stress signaling subnetwork (as well as the ABA-independent subnetwork) and that their expression is down-regulated by ABA (Fig. 6) provides another connection between heat stress responses and responses to other abiotic stresses. It has been found that ABA biosynthesis and signaling mutants are defective in acquired thermotolerance, while addition of exogenous ABA protects Arabidopsis plants from heat-induced oxidative damage (Larkindale and Knight, 2002; Larkindale et al., 2005). Because ABA is also involved in regulating stress responses to osmotic and salinity stress, ABA signaling might link STRS control of heat, salt, and osmotic stress responses. However, ABA does not appear to be involved in the induction of HSP expression (Larkindale et al., 2005). Thus, heat stress-mediated down-regulation of the STRS genes via ABA would, alone, not be sufficient to induce expression of HSPs. This is supported by the finding that absence of functional STRSs is, in itself, insufficient to enhance expression of heat stress genes in unstressed strs mutant plants. Positive stress-mediated signals are also required. Figure 8 presents a model of how STRS1 and STRS2 may function in the heat, salt, and osmotic stress signaling networks.
Figure 8.
A model of the regulation of abiotic stress signaling by STRS1 and STRS2. The STRS proteins attenuate stress-induced expression of upstream transcriptional activators operating in the ABA-independent and ABA-dependent stress signaling subnetworks. The STRS proteins act as regulatory nodes linking the salt/osmotic and heat stress signaling subnetworks, which repress STRS gene expression. ABA signaling might link STRS control of heat, salt, and osmotic stress responses.
In summary, this study has identified two negative regulators of ABA-dependent and ABA-independent upstream abiotic stress transcriptional activators. Furthermore, STRS1 and STRS2 are regulatory nodes linking salt, osmotic, heat, and ABA signaling subnetworks. Because STRS1 and STRS2 are members of the large family of DEAD-box RNA helicases, additional studies may identify other DEAD-box RNA helicases involved in abiotic stress signal transduction. Indeed, we have recently identified several other putative STRS genes in both our functional genomics screen and by inspection of the AtGenExpress database (http://www.arabidopsis.org/info/expression/ATGenExpress.jsp) that may be down-regulated by multiple abiotic stresses. Studies of their T-DNA insertion mutants are apace to determine whether they have similar phenotypes to the strs1 and strs2 mutants. Our findings also illustrate the growing importance of RNA metabolism in the regulation of Arabidopsis abiotic stress signal transduction (e.g. Lu and Fedoroff, 2000; Hugouvieux et al., 2001; Xiong et al., 2001a, 2002a; Borsani et al., 2005; Gong et al., 2005; Cao et al., 2006).
MATERIALS AND METHODS
Plant Materials and Growth
All Salk T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center (ABRC). The Salk ID for each mutant is as follows: strs1, Salk_062509; strs1a, Salk_147039; strs2, Salk_028850; and strs2a, Salk_005131 (Alonso et al., 2003). Lines homozygous for the T-DNA insert were isolated by PCR using gene-specific and T-DNA-specific primers (http://signal.salk.edu/tdnaprimers.2.html). The sequences of the gene-specific primers are as follows: Salk_062509 (F), 5′-ATAGTCGTTGCATCGTTTCTTGCT-3′, Salk_062509 (R), 5′-CCAAAACGCTTGGTATTAGTATGT-3′; Salk_147039 (F), 5′-ATGCAGGTCTTGGATGAACGTG-3′, Salk_147039 (R), 5′-TGTTCAGTGGCGTGAAGAAGGT-3′; Salk_028850 (F), 5′-TTTTCCTAGTGATCTGCTTTGGTT-3′, Salk_028850 (R), 5′-GCTCTGAAGCTATCTCCGAAAGAA-3′; and Salk_005131 (F), 5′-TTGGTAGTTGCAGAGCCTTACA-3′, Salk_005131 (R), 5′-TACCTGGAAAACCAAGGAAGAA-3′.
The F2 generation of homozygous mutants was used for all experiments. The wild-type control Arabidopsis (Arabidopsis thaliana) accession was Columbia. For plate experiments, seeds were surface sterilized by soaking in a solution of 50% bleach for 10 min and then rinsing five times with sterile water. Seeds were sown on nutrient agar plates containing MS salts (Murashige and Skoog, 1962), pH 5.8, 2% (w/v) Suc, 0.5 g/L MES, and 0.8% (w/v) agar. Seeds were stratified at 4°C for 4 d in the dark before being placed in a growth room at 22°C, 50% relative humidity, and a photoperiod of 16 h light (150 μmol photons m−2 s−1)/8 h dark. For soil experiments, seeds were suspended in 0.12% agarose and stratified at 4°C for 4 d in the dark before being sown in a 1:1 mixture of perlite (three mesh) and Arabidopsis growth medium (Weizmann Institute of Science) in pots. This soil mixture allowed leaching of nutrient solution to prevent build-up of salts and also permitted easy harvesting of whole plants for drought assays. Plants were irrigated with one-third Hoagland nutrient solution (Hoagland and Arnon, 1950). For analysis of organ-specific gene expression, plants were grown in soil in the growth room for approximately 4 weeks after germination. Plants were harvested after bolting when both inflorescences and siliques were visible. Only green siliques were taken for analysis. Roots were harvested from soil by gently washing in sterile water.
Abiotic Stress Assays
For salt and osmotic stress assays, 50 to 100 surface-sterilized wild-type and mutant seeds were sown on plates containing MS media with or without NaCl (salt stress) or mannitol (osmotic stress). Four replicate plates were used per treatment, and germination (emergence of radicals) was scored daily for 6 to 7 d until no further germination was observed. FW of seedlings was measured 10 d after stratification. Thermotolerance assays were performed essentially according to Hong and Vierling (2000). Seeds were surface sterilized, sown on plates containing MS media, and stratified at 4°C for 4 d. For basal thermotolerance, 50 to 100 seeds were heat treated at 45°C and then allowed to germinate in the growth room. Germination was recorded daily until no further germination was observed. Alternatively, heat-treated plates were placed vertically in the dark at 22°C to facilitate hypocotyl elongation along the plane of the agar. Hypocotyl elongation was measured after 6 d. For acquired thermotolerance, seedlings were grown for 3 d on vertical plates in the dark at 22°C and then heat stressed by subjecting seedlings to 45°C of various durations with and without a prior pretreatment. The pretreatment consisted of 38°C for 90 min followed by 2 h at 22°C. The increase in hypocotyl elongation following heat stress was measured after 3 d further growth in the dark at 22°C. Both basal (hypocotyl measurement) and acquired thermotolerance experiments were performed with four replicate plates per treatment with each plate containing approximately 12 to 15 seedlings. ABA sensitivity was measured by germinating seeds on four replicate MS plates per treatment containing 0, 0.5, 1, or 2 μm ABA (Sigma-Aldrich). All stress assay experiments were repeated at least twice. For analysis of stress-responsive gene expression, wild-type and mutant plants were grown in soil for 2 weeks and then exposed to various stresses. Salt stress was applied by irrigating with one-third Hoagland solution supplemented with 200 mm NaCl. Drought stress was induced by removal of whole plants from the soil, gently washing the roots, blotting dry, and then placing the plants in the growth room under 60% humidity (Lu et al., 2007). Cold stress was applied by placing plants at 4°C in the light. Heat stress was applied by exposing plants to 40°C. Each experiment consisted of three replicate pots with 10 to 12 plants per pot. ABA-responsive gene expression was measured by transferring 4-d-old seedlings grown on vertical MS plates to MS plates with or without 100 μm ABA. Four replicate plates were used per treatment. Three independent stress or ABA expression experiments were performed.
Analysis of Circadian Clock-Controlled STRS Expression
Fifty wild-type seedlings were germinated and grown on MS plates in the growth room for 7 d. After this period, seedlings were transferred to a tabletop growth chamber (LE509, MRC) at 22°C and entrained with a photoperiod of 12 h light (100 μmol photons m−2 s−1)/12 h dark. After 4-d entrainment, the photoperiod was changed to continuous light, and seedlings were harvested every 3 h over the third and fourth circadian cycles (Green and Tobin, 1999).
RNA Isolation, cDNA Preparation, Primer Design, and Quantitative Real-Time PCR
Total RNA was isolated, and cDNA was prepared according to Kant et al. (2006). Primers for amplification of PCR products of between 50 and 120 bp from Arabidopsis cDNA were designed using Arabidopsis sequences from GenBank and the Primer Express 2.0 software (Applied Biosystems). Primer sequences for each gene are shown in Table III. Real-time PCR was performed according to Kant et al. (2006). The relative quantification values for each target gene were calculated by the 2−ΔΔCT method using UBQ10 as an internal reference gene for comparing data from different PCR runs or cDNA samples (Livak and Schmittgen, 2001). To ensure the validity of the 2−ΔΔCT method, 2-fold serial dilutions of cDNA from unstressed Arabidopsis were used to create standard curves, and the amplification efficiencies of the target and reference genes were shown to be approximately equal (Livak and Schmittgen, 2001). For quantification of unstressed transcript levels, real-time PCR products for each gene were gel purified (QIAEX II Gel Extraction kit; Qiagen) and quantified with a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies). The 10-fold serial dilutions of each PCR product were used to create standard curves. At least three values corresponding to the absolute transcript copy number were produced for each sample in three independent experiments. As a loading control, the absolute transcript copy number of UBQ10 was also calculated and normalized to the highest UBQ10 level, which was assigned a value of 1. The target gene transcript copy number was then adjusted for loading differences by dividing by normalized UBQ10 level.
Table III.
Primers used for real-time PCR analysis of gene expression
F, Forward primer; R, reverse primer.
| Gene Name | Sequence (5′ to 3′) |
|---|---|
| ACT2 | F: TGGCCGATGGTGAGGATATT |
| R: AACCAGCCTTCACCATTCCA | |
| AtMYC2 | F: GGAATGACTGATTACCGGCTACA |
| R: TCCATCATAGAAGCGTTGTCGT | |
| DREB1A/CBF3 | F: TGGCTCCGATTACGAGTCTTC |
| R: GACCCGCCGGTTTCTTG | |
| DREB2A | F: TCTGGGAAGGAGATGGCAGT |
| R: AGCCACAGTAGTACCGTCACCTC | |
| HSF4 | F: CTTCATTCGTCAGCTCAACACTTAC |
| R: TCCCATTTATCCGGTACAGTTTTAC | |
| HSF7 | F: TTCGTCAGCTCAATACTTACGGATT |
| R: TTAGAAAACTCCCAACGATCTGG | |
| HSP101 | F: GCTAGCTGTGAATGCAGGACAT |
| R: ACCGGCACTAGAGATTGCTTG | |
| RD19 | F: CTCCAAAGGGAAAAAGACGTGT |
| R: GACGATCCATTTGATTTGCAATTA | |
| RD22 | F: CCAACTCCCAAAAATGGCG |
| R: TCCAATAACGCTCCGGTGTT | |
| RD26 | F: GAAAGCAACGGGTACTGACAAAA |
| R: AGCGATACTCGTGCATAATCCA | |
| RD29A | F: CCAGCAGCACCCAGAAGAA |
| R: TCATGCTCATTGCTTTGTCCAT | |
| STRS1 | F: CAGTCACATACGTGGCCGTT |
| R: CCAAAAGCCAATGTCTTACCTGA | |
| STRS2 | F: AGGCATACGCTTCACCGTTC |
| R: TCATGCCAACGAGTGGAGAA | |
| TUB5 | F: CCTTCACATTCAAGGTGGTCAA |
| R: TCGCAGATGACTTCCCAGAAC | |
| UBQ10 | F: CTCTCTACCGTGATCAAGATGCA |
| R: TGATTGTCTTTCCGGTGAGAGTC |
Acknowledgments
We express our appreciation to the ABRC for Arabidopsis T-DNA insertion mutant seeds and to Professors Elizabeth Vierling and Gadi Galili for their kind gift of the hot1-3 and aba2-1 mutants, respectively. We are much obliged to Dr. Matthew Hannah for carrying out the freezing tolerance assays. We are grateful to The Sol Leshin Fund for funding this research and to the Israel Science Foundation for funding the real-time PCR system. Many thanks to all members of the Barak laboratory for their support. We are also indebted to Prof. Yair Heimer, Dr. Gidon Grafi, Dr. Rachel Green, and the reviewers for critical reading of the manuscript. Finally, special thanks to Nick Poore of Bancroft's School for starting the journey.
This work was supported by The Sol Leshin Fund and by the Israel Science Foundation (to S.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Simon Barak (simon@bgu.ac.il).
Open Access articles can be viewed online without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Weinl S, Blazevic D, D'Angelo C, Batistic O, Kolukisaoglu U, Bock R, Schulz B, Harter K, Kudla J (2003) The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J 36 457–470 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Kreis M, Lecharny A (1999) The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res 27 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. CRC Crit Rev Plant Sci 24 23–58 [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A (2001) Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res 11 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E (2000) Responses to abiotic stresses. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 1158–1203
- Cao S, Jiang L, Song S, Jing R, Xu G (2006) AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell Mol Biol Lett 11 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing. Genes Dev 17 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HI, Hong JH, Ha JO, Kang JY, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275 1723–1730 [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD box proteins and related families. Trends Biochem Sci 24 192–198 [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Phan Tran LS, Yamaguchi-Shinozaki K, Kazuo Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39 863–876 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) ABA-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hoagland DF, Arnon DI (1950) The water culture for growing plants without soil. Berkeley California Agriculture Experimental Station Circular 347 39
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MG, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29 1220–1234 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K (1999) Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17 287–291 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Knight MR (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Hubel A, Schoffl F (1995) Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J 8 603–612 [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10 655–661 [DOI] [PubMed] [Google Scholar]
- Linder P (2006) Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res 34 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S (1986) The heat shock response. Annu Rev Biochem 55 1151–1191 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402– 408 [DOI] [PubMed] [Google Scholar]
- Lorsch JL (2002) RNA chaperones exist and DEAD box proteins get a life. Cell 109 797–800 [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PI, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63 289–305 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida H, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcription factor using two microarray systems. Plant J 38 982–983 [DOI] [PubMed] [Google Scholar]
- Miller G, Mittler R (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot (Lond) 98 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15 473–497 [Google Scholar]
- Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol 42 657–665 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandl R, Hinderhofer K, Eggers-Schumacher G, Schoffl F (1998) HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen Genet 258 269–278 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Linder P (2004) DEAD-box proteins: the driving force behind RNA metabolism. Nat Rev Mol Cell Biol 5 232–241 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B (2001) A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat Struct Biol 8 113–116 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high salinity stresses using a full-length cDNA microarray. Plant J 31 279–292 [DOI] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21 81–85 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6 410–417 [DOI] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250 161–170 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator Multiprotein Bridging Factor 1c. Plant Physiol 139 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8 251–262 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218 1–14 [DOI] [PubMed] [Google Scholar]
- Wu C (1995) Heat stress transcription factors. Annu Rev Cell Dev Biol 11 441–469 [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001. a) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1 771–781 [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (1999) HOS5: a negative regulator of osmotic stress-induced gene expression in Arabidopsis thaliana. Plant J 19 569–578 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Tanaka Y, Stevenson B, Koiwa H, Bressan RA, Hasegawa PM, Zhu JK (2002. a) Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc Natl Acad Sci USA 99 10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee HH, Ishitani M, Lee H, Zhang C, Zhu JK (2001. b) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Xhu JK (2002. b) Cell signaling during cold, drought and salt stress. Plant Cell (Suppl) 14 S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K (1992) Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol 33 217–224 [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A nove1 cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 6 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57 781–803 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]