Abstract
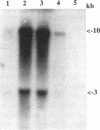
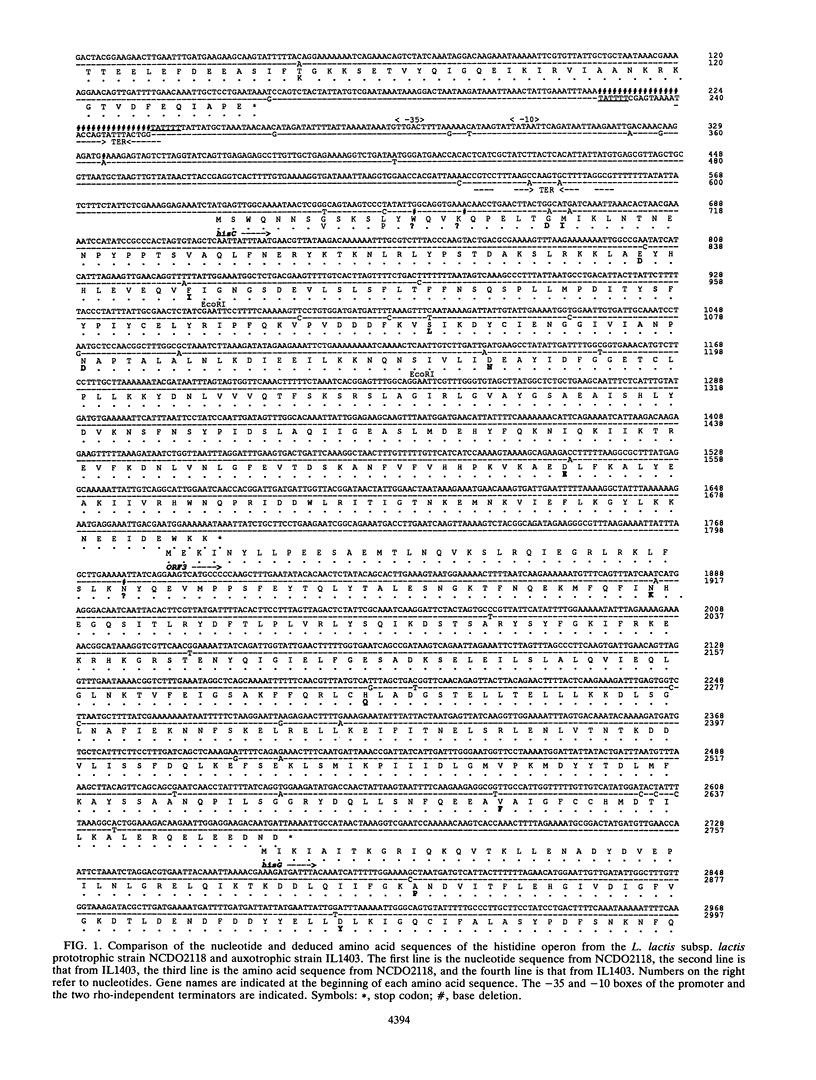
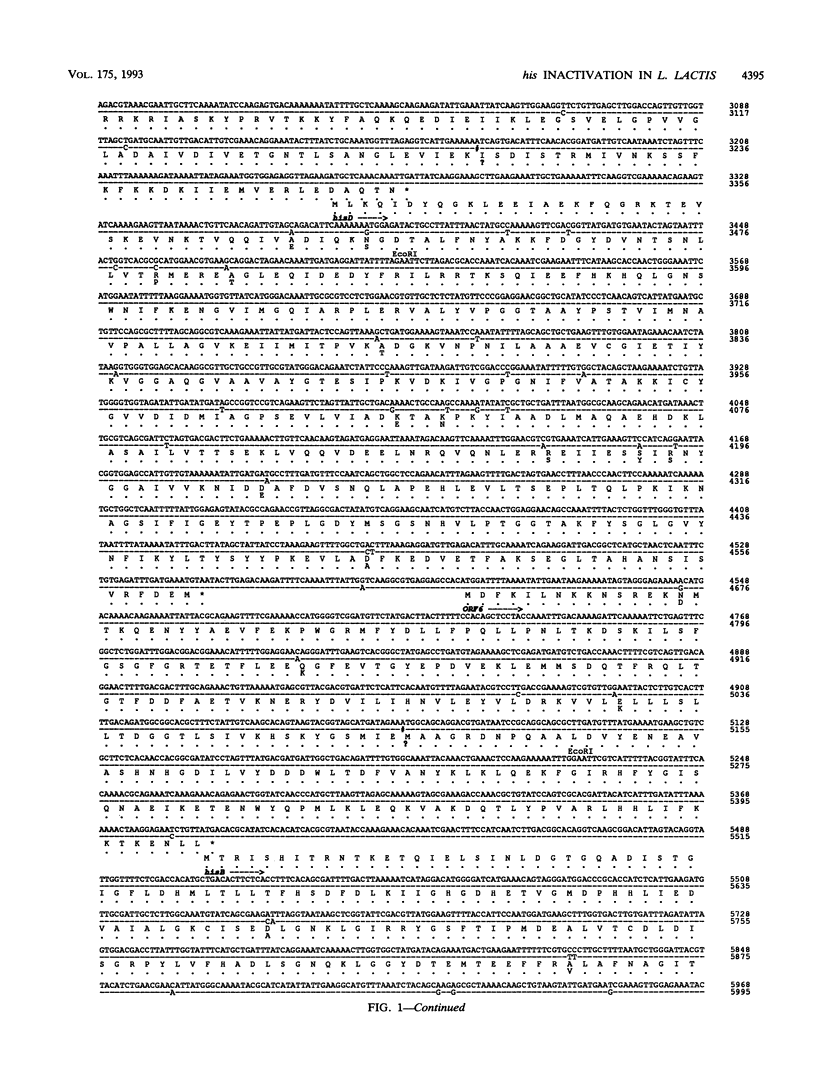
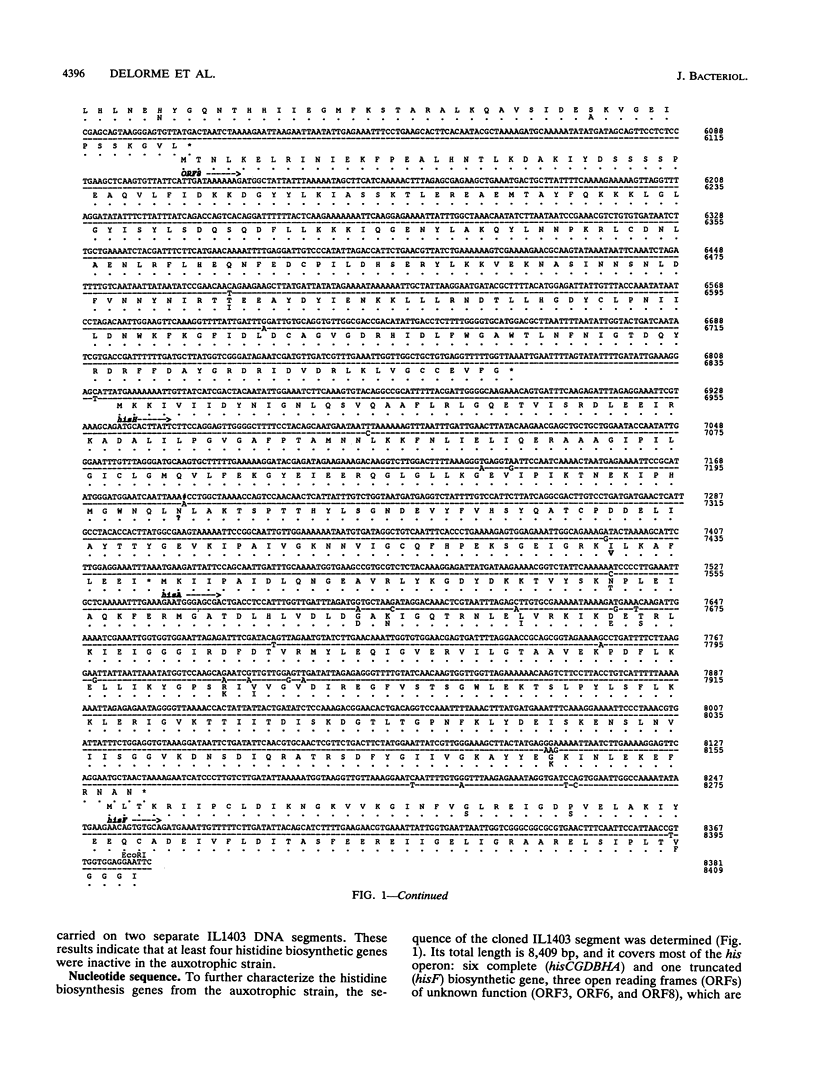
Lactococcus lactis strains from dairy and nondairy sources were tested for the ability to grow in the absence of histidine. Among 60 dairy strains tested, 56 required histidine, whereas only 1 of 11 nondairy strains had this requirement. Moreover, 10 of the 56 auxotrophic strains were able to grow in the presence of histidinol (Hol+), the immediate histidine precursor. This indicates that adaptation to milk often results in histidine auxotrophy. The histidine operon was detected by Southern hybridization in eight dairy auxotrophic strains tested. A large part of the histidine operon (8 kb, containing seven histidine biosynthetic genes and three unrelated open reading frames [ORFs]) was cloned from an auxotroph, which had an inactive hisD gene, as judged by its inability to grow on histidinol. Complementation analysis of three genes, hisA, hisB, and hisG, in Escherichia coli showed that they also were inactive. Sequence analysis of the cloned histidine region, which revealed 98.6% overall homology with that of the previously analyzed prototrophic strain, showed the presence of frameshift mutations in three his genes, hisC, hisG, and hisH, and two genes unrelated to histidine biosynthesis, ORF3 and ORF6. In addition, several mutations were detected in the promoter region of the operon. Northern (RNA) hybridization analysis showed a much lower amount of the his transcript in the auxotrophic strain than in the prototrophic strain. The mutations detected account for the histidine auxotrophy of the analyzed strain. Certain other dairy auxotrophic strains carry a lower number of mutations, since they were able to revert either to a Hol+ phenotype or to histidine prototrophy.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Delorme C., Ehrlich S. D., Renault P. Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6571–6579. doi: 10.1128/jb.174.20.6571-6579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick-Silversmith L., Hartman P. E. Histidine-requiring mutants of Escherichia coli K12. Genetics. 1970 Oct;66(2):231–244. doi: 10.1093/genetics/66.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Delorme C., Bardowski J., Chopin M. C., Ehrlich S. D., Renault P. Gene inactivation in Lactococcus lactis: branched-chain amino acid biosynthesis. J Bacteriol. 1993 Jul;175(14):4383–4390. doi: 10.1128/jb.175.14.4383-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Delorme C., Ehrlich S. D., Renault P. Divergence of Genomic Sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1992 Dec;58(12):4045–4047. doi: 10.1128/aem.58.12.4045-4047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Yokoyama S., Calhoun D. H. Role of cryptic genes in microbial evolution. Mol Biol Evol. 1983 Dec;1(1):109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- Kimura M. Recent development of the neutral theory viewed from the Wrightian tradition of theoretical population genetics. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):5969–5973. doi: 10.1073/pnas.88.14.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourgeois P., Lautier M., Mata M., Ritzenthaler P. Physical and genetic map of the chromosome of Lactococcus lactis subsp. lactis IL1403. J Bacteriol. 1992 Nov;174(21):6752–6762. doi: 10.1128/jb.174.21.6752-6762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matney T. S., Goldschmidt E. P., Erwin N. S., Scroggs R. A. A preliminary map of genomic sites for F-attachment in Escherichia coli K12. Biochem Biophys Res Commun. 1964 Oct 14;17(3):278–281. doi: 10.1016/0006-291x(64)90397-3. [DOI] [PubMed] [Google Scholar]
- McKay L. L. Functional properties of plasmids in lactic streptococci. Antonie Van Leeuwenhoek. 1983 Sep;49(3):259–274. doi: 10.1007/BF00399502. [DOI] [PubMed] [Google Scholar]
- Parker L. L., Hall B. G. Characterization and nucleotide sequence of the cryptic cel operon of Escherichia coli K12. Genetics. 1990 Mar;124(3):455–471. doi: 10.1093/genetics/124.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991 Jul;33(1):23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- Tulloch D. L., Finch L. R., Hillier A. J., Davidson B. E. Physical map of the chromosome of Lactococcus lactis subsp. lactis DL11 and localization of six putative rRNA operons. J Bacteriol. 1991 May;173(9):2768–2775. doi: 10.1128/jb.173.9.2768-2775.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]