Abstract
The plastid genome of higher plants is transcribed by two different types of RNA polymerases named nucleus encoded RNA polymerase (NEP) and plastid encoded RNA polymerase. Plastid encoded RNA polymerase is a multimeric enzyme comparable to eubacterial RNA polymerases. NEP enzymes represent a small family of monomeric phage-type RNA polymerases. Dicotyledonous plants harbor three different phage-type enzymes, named RPOTm, RPOTp, and RPOTmp. RPOTm is exclusively targeted to mitochondria, RPOTp is exclusively targeted to plastids, and RPOTmp is targeted to plastids as well as to mitochondria. In this article, we have made use of RPOTp and RPOTmp T-DNA insertion mutants to answer the question of whether both plastid-located phage-type RNA polymerases have overlapping or specific functions in plastid transcription. To this aim, we have analyzed accD and rpoB messenger RNAs (mRNA; transcribed from type I NEP promoters), clpP mRNA (transcribed from the −59 type II NEP promoter), and the 16S rRNA (transcribed from the exceptional PC NEP promoter) by primer extension. Results suggest that RPOTp represents the principal RNA polymerase for transcribing NEP-controlled mRNA genes during early plant development, while RPOTmp transcribes specifically the rrn operon from the PC promoter during seed imbibition.
Regulation of transcription in plant plastids is surprisingly complex (for review, see Shiina et al., 2005; Liere and Börner, 2006). Most of the genes that have been preserved during evolution from the cyanobacterial ancestor of plastids have been (Martin, 2003) and are still (Timmis et al., 2004) transferred to the nucleus, leaving only a limited number of genes (about 120) on the present day plastome. These genes encode a whole set of tRNAs and proteins functioning in photosynthesis, transcription, translation, RNA splicing, protein degradation, and lipid biosynthesis (for review, see Sugita and Sugiura, 1996). This small genome is transcribed by several RNA polymerases that are either nucleus encoded (nucleus encoded RNA polymerases [NEP]) or plastid encoded (plastid encoded RNA polymerase [PEP]). PEP is of eubacterial type, the core enzyme being composed of several subunits (α, β, β′, β″). The subunit composition is most comparable to that of cyanobacterial RNA polymerases where the β-subunit is subdivided into two independent proteins when compared to RNA polymerases of other bacteria (Bergsland and Haselkorn, 1991; for review, see Igloi and Kössel, 1992). Promoter specificity and transcription initiation by PEP needs nucleus-encoded σ-like factors (for review, see Allison, 2000; Lysenko, 2007).
NEP enzymes are single subunit phage-type RNA polymerases (Lerbs-Mache, 1993; Chang et al., 1999). Three different genes encoding NEP enzymes have been identified in Arabidopsis (Arabidopsis thaliana) coding for proteins that are localized in mitochondria (RPOTm, At1g68990), plastids (RPOTp, At2g24120), or in both organelles (RPOTmp, At5g15700; Hedtke et al., 1997, 2000). However, only two genes (RPOTp and RPOTm) have been detected in monocotyledons (Chang et al., 1999; Ikeda and Gray, 1999; Emanuel et al., 2004; Kusumi et al., 2004), thus raising the question of the function of RPOTmp in dicotyledons. Evidence for the importance of RPOTmp comes from RPOTp/RPOTmp double mutants that show developmental arrest early after germination (Hricova et al., 2006). A first analysis of part of the plastid transcriptome of RPOTmp knockout mutants showed a reduction of light-induced accumulation of several plastid mRNAs when compared with wild-type plants (Baba et al., 2004). However, this reduction concerned PEP as well as NEP transcripts and promoter usage has not yet been analyzed in these mutants. Thus, no really specific function can actually be attributed to RPOTmp. Recent in vitro transcription studies have shown that only RPOTm and RPOTp are able to correctly initiate transcription from a subset of consensus (YRTA) type promoters without help of auxiliary factors (Kühn et al., 2007). For RPOTmp, such factors are probably obligatory for correct transcription initiation, suggesting a specific, different function of RPOTmp in plastid transcription. Transcription initiation factors that interact with NEPs are not yet characterized on the molecular level. One good candidate for such a factor is CDF2, a sequence-specific DNA-binding factor that is implicated in rDNA transcription regulation in spinach (Spinacia oleracea; Baeza et al., 1991; Iratni et al., 1994; Bligny et al., 2000).
Promoters that are recognized by NEPs have been characterized by using plant material lacking PEP activity (Allison et al., 1996; Hajdukiewicz et al., 1997) or having very low levels of PEP activity (Vera and Sugiura, 1995; Vera et al., 1996; Kapoor et al., 1997; Hübschmann and Börner, 1998; Miyagi et al., 1998; Silhavy and Maliga, 1998a, 1998b). From these experiments, NEP promoters can be principally classified into two groups. Consensus-type or type I promoters (e.g. accD and rpoB) are characterized by a YRTA motif located closely upstream of the transcription start site. Nonconsensus-type or type II promoters lack this motif, and critical promoter sequences are located downstream of the transcription start site as shown for the −59 clpP promoter (Sriraman et al., 1998a) and some tRNA promoters (Gruissem et al., 1986; Cheng et al., 1997; Wu et al., 1997).
An exceptional promoter represents the rrn PC promoter. In spinach, this promoter is recognized by NEP2 with the help of CDF2 that binds immediately upstream of the transcription start site (Baeza et al., 1991; Iratni et al., 1994; Bligny et al., 2000). The PC promoter is used in spinach, Arabidopsis, and mustard (Sinapis alba; Baeza et al., 1991; Pfannschmidt and Link, 1997; Sriraman et al., 1998b). In other dicotyledons like tobacco (Nicotiana tabacum), carrot (Daucus carota), pea (Pisum sativum), and in all thus-far-analyzed monocotyledons, transcript initiation at PC could not be demonstrated, and transcription of the rrn operon starts at the eubacterial type P2 promoter (for review, see Lerbs-Mache, 2000). From our own results obtained with spinach, we have concluded that PC is recognized by a second NEP enzyme (NEP2) that is different from the principal plastid phage-type polymerase (NEP1; Bligny et al., 2000). On the other hand, in vitro transcription studies using the peak A and peak B enzymes from mustard plastids led to the conclusion that the PC promoter is transcribed by PEP (Pfannschmidt and Link, 1997).
In this article, we have made use of Arabidopsis RPOTp and RPOTmp T-DNA insertion mutants to analyze which NEP recognizes in vivo the consensus-type promoters like accD and rpoB, the nonconsensus-type clpP −59 promoter, and the exceptional rrn PC promoter. To answer the question of whether the rrn PC promoter might be recognized by PEP, we have also grown wild-type plants in the presence of Tagetin, a specific inhibitor of PEP activity (Mathews and Durbin, 1990, 1994). Total RNA obtained from these mutants and from wild-type plants without and after Tagetin treatment has been analyzed by primer extension. Many plastid genes harbor multiple promoters and primer extension allows us to map the 5′ ends of the RNAs and thus to determine which of the promoters is used. Experiments have been carried out with seeds and very young plantlets 2 or 3 d after germination because de novo rRNA synthesis is highest during early developmental periods (Baumgartner et al., 1993; for review, see Lerbs-Mache, 2000), and all three plastid RNA polymerases are expressed at that developmental stage (Demarsy et al., 2006).
RESULTS
Isolation of RPOTmp T-DNA Insertion Lines
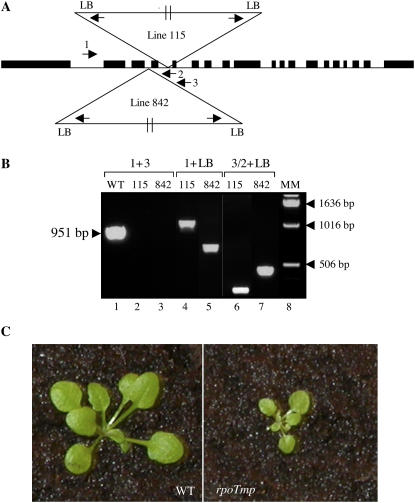
Two different Arabidopsis RPOTmp T-DNA insertion lines have been obtained from the Salk collection (SALK_132842 and SALK_086115). They are named 842 and 115 in the following. The T-DNA is inserted in the third and fourth intron, respectively, of the RPOTmp gene, as indicated in Figure 1A. Homozygous plants were selected by PCR screening using primer pairs 1 and 2 to reveal wild-type DNA and primer pairs 1 and left border (LB) to reveal the T-DNA insertion. After two successive backcrosses, one homozygous plant for each insertion mutant has been selected and the insertion has been further characterized by PCR using LB and 1, 2, 3 primers (Fig. 1B). Results indicate that, in both lines, two T-DNAs are inserted in head-to-tail orientation. Three-week-old wild-type and 115 plants, grown for 1 week on agar plates and subsequently transferred to soil, are shown in Figure 1C. The mutant line 115 shows a general reduction in growth as already reported for mutant lines 833 and 286E07 (Baba et al., 2004).
Figure 1.
Characterization of two different RPOTmp T-DNA insertion mutants. A, Schematic presentation of the T-DNA insertions and the locations of the primers that have been used for PCR analyses. B, Selection of two different homozygous lines, 842 and 115, after the second backcross. The presence or absence of wild-type DNA was verified by PCR using primers 1 and 3 (lanes 1–3) and the presence and orientation of the T-DNA insertion was analyzed by PCR using primers LB and 1 (lanes 4 and 5) or LB and either 2 or 3 (lanes 6 and 7). C, Wild-type plants and the homozygous RPOTmp T-DNA insertion mutant (line 115) grown under a 16-h-light/8-h-dark cycle on agar plates for 1 week and then transferred to soil for 2 weeks. [See online article for color version of this figure.]
Isolation of a RPOTp T-DNA Insertion Line
As RPOTp T-DNA insertion mutant, we have used the same mutant that has been recently described under the name sca3-3 (Hricova et al., 2006). The mutant SALK_067191 was acquired at the Nottingham Stock Center and a homozygous line has been selected by PCR screening as indicated in Figure 2 using primers 4 and 5 to verify the presence of wild-type DNA and primer pairs 4 and LB or 5 and LB to reveal the T-DNA (see Fig. 2A for primer location and Fig. 2B for PCR amplification products). The T-DNA is again inserted as dimer in a head-to-tail orientation (Fig. 2A). Reverse transcription (RT)-PCR analysis of the two different mutants, rpoTp and rpoTmp, shows the absence of transcripts downstream of the T-DNA insertion (Fig. 2C). The rpoTp mutant is much retarded in growth and development and displays pale-green cotyledons and leaves (Fig. 2D). The rpoTp mutant has been cleaned by three successive backcrosses before usage in further experiments.
Figure 2.
Selection and characterization of a homozygous line for rpoTp. A, Schematic presentation of the T-DNA insertions and the location of the primers that have been used for PCR analyses. B, Characterization of a homozygous line after the third backcross. The absence of wild-type DNA was verified by PCR using primers 4 and 5 (lanes 1 and 2) and the presence and orientation of the T-DNA insertion was analyzed by PCR using primers LB and 5 (lines 3 and 4) or primers LB and 4 (lines 5 and 6). C, The absence of RPOTp (line 2) or RPOTmp (line 3) transcripts in the two T-DNA insertion mutants has been verified by RT-PCR. Transcripts for RPOTm and APT have been analyzed as controls. D, Wild-type and homozygous RPOTp T-DNA insertion mutants grown in soil under a 16-h-light/8-h-dark cycle for 3 weeks. Scale bar = 1 cm. [See online article for color version of this figure.]
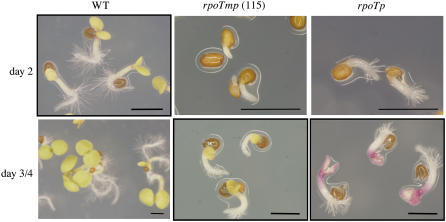
Comparison of PEP- and NEP-Initiated Precursor RNAs in Wild-Type Plants and rpoTp and rpoTmp Mutants
As shown in Figures 1 and 2, both mutants are retarded in growth when compared to wild-type plants. To ensure that differences in precursor RNAs do not simply reflect changes in the developmental stage of the plants, we used plantlets of visually comparable developmental stages for further experiments, i.e. 2-d-old plantlets for wild type and 3- to 4-d-old plantlets for mutant plant material (Fig. 3, stages that have been used for RNA extraction are boxed). We analyzed precursor RNA of two genes that are transcribed from consensus-type NEP promoters (accD and rpoB), two genes that are transcribed only from PEP promoters (rbcL and psbA), one gene that is transcribed from a nonconsensus-type NEP promoter (clpP), and finally the rrn operon that is transcribed from a PEP (P2, nomenclature of the rrn operon as in Lerbs-Mache, 2000) and the exceptional NEP PC promoter in Arabidopsis (Pfannschmidt and Link, 1997).
Figure 3.
Phenotypes of wild-type and mutant plants 2 to 4 d after germination. Physiological stages of plantlets that have been used for microarray and primer extension experiments are boxed. The line indicates 5 mm.
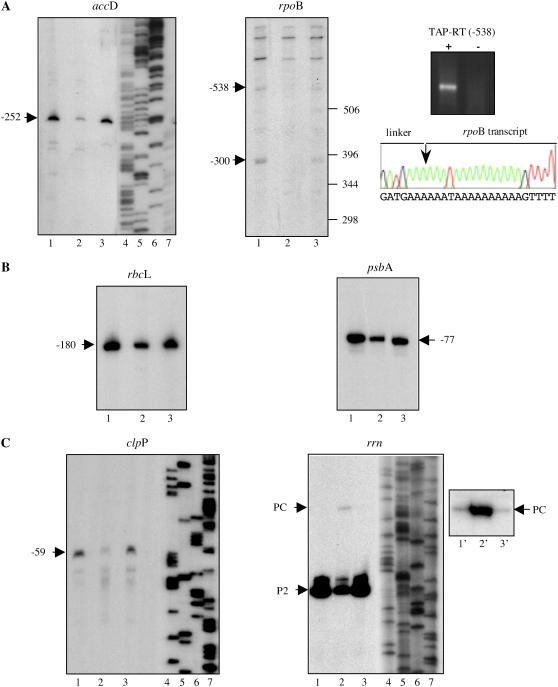
In the case of accD, we could only detect the −252 precursor RNA (Fig. 4A, left). The promoter that produces the −174 precursor RNA, recently described by Swiatecka-Hagenbruch et al. (2007) using 3-week-old plant material, is probably not active in 2- to 4-d-old plantlets. The −252 accD transcript is considerably diminished in rpoTp plants but not in rpoTmp plants, suggesting that the promoter is specifically recognized by RPOTp. When analyzing rpoB mRNAs, we detected several RNAs (Fig. 4A, middle). One of them corresponds to the recently described primary transcription product starting at position −300 from a type I NEP promoter (Swiatecka-Hagenbruch et al., 2007). The −538 transcript has not been described previously. Determination of the 5′ end of this transcript by 5′-RACE using tobacco acid pyrophosphatase (TAP)-treated and nontreated RNA reveals this RNA as primary transcript (Fig. 4A, right top) and locates the transcription start site at position −538 (Fig. 4A, right bottom). The larger RNAs could not be detected by 5′-RACE and might therefore correspond to artefacts. Both rpoB mRNAs, −300 and −538, are strongly diminished in the rpoTp mutant, indicating that these two NEP promoters are also specifically recognized by RPOTp. The two mRNAs, rbcL and psbA, which are exclusively produced from PEP promoters, are also diminished in the rpoTp mutant (Fig. 4B). This might be explained as a secondary effect due to the general diminution of transcription of the rpo operon (Hricova et al., 2006), i.e. to a diminution of PEP enzyme. The −59 type II NEP promoter of the clpP gene is also specifically recognized by RPOTp, because transcripts starting at this position are only diminished in the rpoTp mutant but not in the rpoTmp mutant (Fig. 4C, left). In contrast to the type I and type II NEP promoters, the −136 rrn PC promoter seems to be activated in the rpoTp mutant (Fig. 4C, right, lanes 1–3). In general, in wild-type plants and the rpoTmp mutant, the amount of PC-initiated rrn transcripts is very low when compared to P2-initiated transcripts. A repetition of the experiment with longer exposure shows a slight diminution of PC transcripts in rpoTmp mutants when compared to wild type (Fig. 4C, right, lanes 1′–3′). Altogether, these results indicate that the exceptional NEP PC promoter is not under specific control of RPOTp. The PC promoter represents a likely candidate to be recognized specifically by RPOTmp, but RPOTp might recognize the promoter to some low extent in rpoTmp mutants.
Figure 4.
Characterization of different NEP- or PEP-specific precursor RNAs. Total RNA was prepared from 2-d-old wild-type (lane 1) and 3- to 4-d-old rpoTp (lane 2) or rpoTmp (lane 3) Arabidopsis plantlets and analyzed by primer extension. A, Analyses of RNAs of two exclusively NEP-transcribed genes, e.g. accD and rpoB. The insert on the right shows 5′-RACE of the −538 rpoB transcript with (+) or without (−) TAP treatment of mRNAs. The sequence of the 5′ end and the beginning of the linker is shown below. B, Analyses of RNAs of two exclusively PEP-transcribed genes, rbcL and psbA. C, Analyses of RNAs of two genes, clpP and rrn16S, that are transcribed from nonconsensus-type NEP promoters. The insert on the right shows the PC-initiated transcript after longer exposure. [See online article for color version of this figure.]
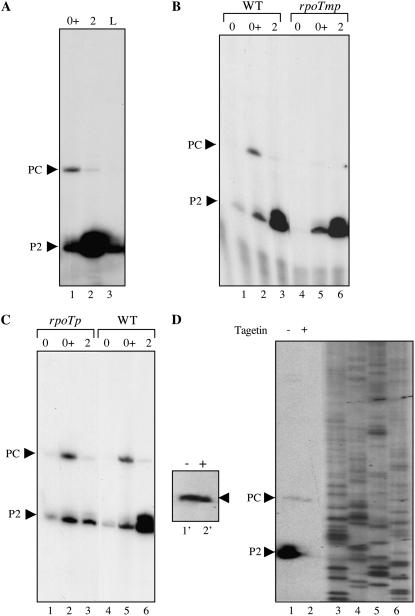
We have recently shown that in Arabidopsis there is a tremendous rise in RNAs coding NEP, i.e. NEP enzymes should be highly active during the period of imbibition (Demarsy et al., 2006), and it has been known for a long time that 16S rRNA is highly expressed in the very early stages of plastid development (Baumgartner et al., 1993). A comparison of rrn precursor RNAs in very young seedlings and leaves of mature plants confirms these former observations (Fig. 5A). To get a clearer picture on rrn transcription, we therefore reanalyzed PC-initiated rrn precursor RNAs by primer extension using dry seeds (stage 0), seeds after imbibition (stage 0+), and seedlings 2 d after germination (stage 2). The result shows an intermittent activation of the rrn PC promoter during the period of imbibition in wild-type plants. In the rpoTmp mutant, the PC-initiated transcript is absent in the two insertion lines, 115 and 842. Figure 5B (lanes 1–6) shows the result obtained with line 115. If we compare rrn transcripts of the same developmental stages from the rpoTp mutant with wild-type plants, we do not detect considerable differences in PC-initiated transcripts at stage 0+ (Fig. 5C, lanes 1–6). These experiments have been repeated several times showing the same result, i.e. we can conclude that RPOTmp specifically transcribes the rrn operon from the PC promoter during imbibition.
Figure 5.
Characterization of the 16S PC promoter. 16S precursor RNAs have been analyzed by primer extension using total RNA prepared from different plant materials. A, Dry seeds (lane 1), seeds after imbibition (lane 2), and leaves from mature Arabidopsis plants (lane 3). B, Dry seeds (lanes 1 and 4), seeds after vernalization (lanes 2 and 5), and plantlets 2 d after germination (lanes 3 and 6) of wild-type (lanes 1–3) and rpoTmp plantlets (lanes 4–6). C, Dry seeds (lanes 1 and 4), seeds after vernalization (lanes 2 and 5), and plantlets 2 d after germination (lanes 3 and 6) of rpoTp (lanes 1–3) and wild-type plantlets (lanes 4–6). D, Seeds of the tt2-1 mutant have been germinated and grown in the absence (−) or presence (+) of Tagetin and total RNA has been prepared 1 d after germination. 16S precursor RNAs have been analyzed by primer extension. The insert on the left shows a longer exposure of the PC-initiated rrn transcript.
Finally, we wanted to assure that PC is indeed recognized by a NEP enzyme and not by PEP. For this objective, imbibition was performed using the transparent testa mutant having a highly permeable seed coat (tt2-1; Debeaujon et al., 2000; Rajjou et al., 2004) in the presence of Tagetin, a specific inhibitor of PEP (Mathews and Durbin, 1990, 1994) under the same experimental conditions as already described (Demarsy et al., 2006), and RNA was analyzed by primer extension 42 h after cold release of seeds (Fig. 5D). The PEP-initiated P2 transcript disappears after Tagetin treatment, while the PC transcript remains present at the same amount as in the untreated control (Fig. 5D, lanes 1 and 2 and 1′ and 2′).
DISCUSSION
Transcription of the plastid rrn operon is species specific. The rrn operon upstream sequence harbors multiple promoter elements that are differentially used in different plant species (for review, see Lerbs-Mache, 2000). In most of the already-analyzed plant species, transcription starts at the eubacterial type P2 promoter (for nomenclature of rrn promoters, see Lerbs-Mache, 2000) that can be considered as the principal rrn PEP promoter in most plant species. Exceptions are spinach and mustard in which the PC promoter represents the principal rrn operon promoter (Baeza et al., 1991; Pfannschmidt and Link, 1997). PC is located between P1 and P2, two initiation sites that are used by Escherichia coli RNA polymerase under in vitro transcription conditions (Iratni et al., 1994; Pfannschmidt and Link, 1997). More precisely, PC is located between the −10 and −35 consensus elements of the eubacterial type P2 promoter. This location excludes a priori that PC is used by an eubacterial type RNA polymerase because the P1 promoter is located too far upstream to allow initiation at PC. Nevertheless, the usage of the PC promoter is still ambiguous. In spinach, experiments using spectinomycin to induce ribosome depletion in chloroplasts and thus prevent the formation of PEP show a strong diminution of the PEP-transcribed rbcL transcript and a remarkable augmentation of NEP-initiated transcripts like clpP-59 and rrn 16S PC (Bligny et al., 2000). In mustard, rrn transcription starting at the PC promoter has been shown by in vitro transcription using two different RNA polymerase fractions, one of them being inhibited by rifampicin, a specific inhibitor of eubacterial-type multimeric RNA polymerases (Pfannschmidt and Link, 1997). By using biochemical fractionation of spinach chloroplast protein extracts, our group has obtained two different NEP activities; only one of them (NEP2) was able to correctly initiate at the PC promoter. This fraction did not react with antibodies that had been made against the C-terminal part of maize (Zea mays) RPOTp. From these results, it was not clear which RNA polymerase (PEP, RPOTp, or RPOTmp) initiates at the PC promoter. In addition, the existence of a fourth RNA polymerase of eukaryotic type in plastids could not be excluded (Bligny et al., 2000). The latter hypothesis was supported by several facts, such as the existence of internal promoter regions (Gruissem et al., 1986; Wu et al., 1997; Sriraman et al., 1998a), the importance of TATA-like sequence elements in plastid transcription (Link, 1984; Eisermann et al., 1990), plastid-associated location of eukaryotic-type transcription factors like TFIIS (da Costa e Silva, 2004) and pBrp (Lagrange et al., 2003), and bioinformatic prediction of plastid localization of at least 48 eukaryotic-type transcription factors (Wagner and Pfannschmidt, 2006; Schwanke et al., 2007). The recently discovered plant-specific RNA polymerase IV (RNAPIV; Pontier et al., 2005) would have been a likely candidate for a plastid-specific function, e.g. for PC transcription.
To answer the question of whether PC is transcribed by one of the two plastid localized phage-type RNA polymerases or by RNAPIV, we decided to analyze rrn precursor RNAs in wild-type and rpoTp and rpoTmp plants and also in the nrpd1 double mutant in which the two largest subunits of RNAPIV (NRPD1a and NRPD1b) are mutated (Pontier et al., 2005). The rpoTp and rpoTmp T-DNA insertion lines have been obtained either from the Salk collection or from the Nottingham Stock Center. The two rpoTmp mutants are different from those that have been published previously by Baba et al. (2004). The rpoTp mutant corresponds to the same mutant that has been described by Hricova et al. (2006). So far, plastid transcripts of rpoTp and rpoTmp mutants have been partially characterized by RT-PCR or dot blot hybridization, but none of the transcripts has been analyzed by primer extension. However, due to the fact that many of the plastid transcription units are transcribed from multiple promoter regions, dot blot or array hybridization studies are not sufficient to characterize changes in plastid transcription.
Figure 3 shows that rpoTp and rpoTmp mutants are retarded in germination and early seedling outgrowth. To avoid that observed changes in promoter usage are simply due to the slower growth of the rpoT mutants, we have chosen wild-type and mutant plants having the same visible phenotype, i.e. 2-d-old wild-type plants and 3- to 4-d-old mutants (see Fig. 3) for primer extension analyses of some selected mRNAs. The abundance of rbcL and psbA mRNAs, two RNAs that are exclusively transcribed by PEP, is diminished in the rpoTp mutant (Fig. 4B). This diminution of PEP transcripts is probably due to a general diminution of PEP enzyme in rpoTp mutants because the rpoB/C1/C2 operon is transcribed by RpoTp (see Fig. 3A, rpoB). Type I and type II NEP promoters as shown by the accD −252 promoter, the rpoB −300 promoter, and the clpP −59 promoter, are all recognized mainly by RPOTp. By revealing the −538 rpoB transcript and confirming it as primary transcript by TAP RT-PCR, we detected an additional, as-yet-unknown rpoB promoter that is also under the control of RPOTp (Fig. 4A, middle and right).
The only one of the analyzed RNAs that is specifically transcribed by RPOTmp is the PC-initiated rrn transcript (Fig. 5, B and C). This result already demonstrates that the PC promoter is recognized by RPOTmp. Nevertheless, as a supplementary control, we also analyzed rrn precursor RNAs in the nrpd1 double mutant in which the two largest subunits of RNAIV are inactivated by primer extension. The PC-initiated precursor RNA is not affected in this mutant, thus confirming specific recognition of PC by RPOTmp (data not shown).
Taken together, our results suggest that RPOTp is the principal NEP enzyme in Arabidopsis chloroplasts and RPOTmp seems to be highly active in transcribing the rrn operon at the PC promoter during seed imbibition. Both RNA polymerases, RPOTp and RPOTmp, have specific functions and cannot completely replace each other. However, RPOTmp seems to be able to transcribe from type I and type II NEP promoters with low efficiency. This can be supposed from the presence of low transcript levels in the RPOTp T-DNA insertion mutant (see Fig. 4) and from quantification of the precursor RNAs using ImageJ software that reveals a slight diminution of the transcripts also in RPOTmp mutants (Supplemental Table S1). Thus, RPOTmp probably contributes to a low extent also to the transcription of type I and type II NEP promoters. This low gene expression by RPOTmp seems to be sufficient to assure plant survival of RPOTp mutants (RPOTp/RPOTmp double mutants are lethal; see Hricova et al., 2006), but it is not sufficient to restore a normal green phenotype of these mutants.
These results suggest that RPOTmp is also present in chloroplasts in later developmental stages and not only in proplasts/amyloplasts of seeds. However, different reports show the absence or only very low quantities of PC-initiated transcripts in leaves and cotyledons (Sriraman et al., 1998b; Bisanz et al., 2003; Zoschke et al., 2007) and enhancement of PC initiation in a mutant that is deficient in rRNA processing (Bisanz et al., 2003). These results indicate that RPOTmp should be present and active in mature chloroplasts only at a very low level and an enhancement of activity and/or quantity occurs in the processing-defective mutant. Several experiments to detect RPOTmp in purified chloroplasts by western-blot analysis were negative (data not shown). The presence of only a very low quantity of RPOTmp in chloroplasts of Arabidopsis might be the reason for the controversial results that have been obtained using GFP fusion constructs to analyze the localization of the enzyme (Hedtke et al., 2000; Kabeya and Sato, 2005).
From our results we can conclude that RPOTmp should be present and active in proplastid/amyloplasts of germinating seeds. In chloroplasts of mature plants, the quantity of the enzyme should be very low. Although it is still unclear whether transcription from the rrn PC promoter is the only really specific function of RPOTmp in dicotyledonous plants, such function would explain why monocotyledons exist quite well without RPOTmp. Up to now, the PC promoter has not been found in monocotyledons like rice (Oryza sativa; Silhavy and Maliga, 1998a, 1998b), barley (Hordeum vulgare; Hübschmann and Börner, 1998), or maize (Strittmatter et al., 1985), and in dicotyledons, the usage of PC varies strongly between species. For example, in spinach and mustard, the PC promoter is the principal rrn promoter (Iratni et al., 1997; Pfannschmidt and Link, 1997), but in Arabidopsis, PC is strongly used during imbibition and represents a minor promoter in later developmental stages (see Fig. 5; Sriraman et al., 1998b; Zoschke et al., 2007). In tobacco, usage of the PC promoter has not been observed (Vera and Sugiura, 1995; Sriraman et al., 1998b; Suzuki et al., 2003). All these results suggest a species-specific function of RPOTmp and organ- and/or development-specific activity of RPOTmp in plastids of Arabidopsis. Organ- and/or development-specific activity of RPOTmp in Arabidopsis plastids could be regulated by organ-/development-specific supply of transcription factors, organ-/development-specific expression of the RPOTmp gene, and/or organ-/development-specific regulated import of RPOTmp into plastids. To distinguish between these three possibilities of regulation of RPOTmp activity represents a challenge for future experiments.
CONCLUSION
We have made use of rpoTp and rpoTmp mutants to analyze promoter usage of the two different NEP enzymes. While RPOTp seems to be the main mRNA transcribing NEP in Arabidopsis plastids during early seedling development, RPOTmp seems to have a specific function in rrn transcription during seed imbibition. Type I NEP promoters and the −59 type II clpP NEP promoter are principally recognized by RPOTp during early seedling development, while RPOTmp specifically recognizes the exceptional PC promoter during seed imbibition. From our results, we can conclude that the two NEP enzymes have different, development-related functions in plastids of Arabidopsis.
MATERIALS AND METHODS
Plant Material and RNA Isolation
Surface-sterilized Arabidopsis (Arabidopsis thaliana) seeds (0) were spread on Murashige and Skoog agar plates, kept for 72 h at 4°C in darkness (0+), and then transferred into a growth chamber and grown for up to 3 d at 23°C under a 16-/8-h light/dark cycle at 70 μmol of photons m−2 s−1. tt2.1 seeds were spread on Murashige and Skoog agar plates containing 100 μm tagetin, kept for 24 h at 4°C in darkness, and then transferred into a growth chamber and grown for 42 h at 23°C as described previously (Demarsy et al., 2006). Total RNA was prepared from stage 2/3 seedlings as described by Privat et al. (2003). For germination studies (stages 0, 0+, and 2), RNA was prepared as described by Demarsy et al. (2006).
Isolation of RpoTmp T-DNA Insertion Lines
For all three mutants (RPOTmp: SALK_132842 and SALK_086115; RPOTp: SALK_067191) screening for the insertion and for homozygous plants has been done by PCR using the following primers (nos. correspond to those used in Figs. 1 and 2): (1) 5′-GCCTTTAGGGTTCCTTGATTGTC-3′; (2) 5′-GCGACATTCACATTTCCAACAA-3′; (3) 5′-ATGGCATCACTGCATATCTCCC-3′; (LB) 5′-GCGTGGACCGCTTGCTGCAACT-3′; (4) 5′-GAATCATACCCGAATCTCGTG-3′; and (5) 5′-CTGCAGCGAGAGGGTAACACG-3′.
Primer Extension
Using isolated total DNA from Arabidopsis as template, the clpP, rpoB, accD, rbcL, psbA, and rrn promoter regions have been PCR amplified and cloned into pCRR2.1-TOPOR (Invitrogen) with the following primers: 5′-ATGTAACTTTATTGCATTGG-3′ and 5′-TCATAGTTGCATTACT-3′ (clpP), 5′-GGTATGCAATCGAATTGG-3′ and 5′-CTTCTATTAAACCCTGATC-3′ (rpoB), 5′-CTTTCGTGTCAGGGCTTG-3′ and 5′-GAACGCTCATCCCAACC-3′ (accD), 5′-GCATATCCGGTTATGCG-3′ and 5′-GCCAAGATATCAGTATCC-3′ (rbcL), and 5′-CCGTATCATCTTGACTTGG-3′ and 5′-GGGCAGGTTCTTACGCG-3′ (rrn). Primer extension experiments have been performed as described (Favory et al., 2005) using 10 μg of total RNA, except for the analyses of the rrn and the rpoB transcripts, where 1 μg or 20 μg was used, respectively. The following primers have been used for primer extension and to establish the accompanying sequence ladders: 5′-GATGTATCTCCTTCTCC-3′ (clpP), 5′-CCCTGATCAATAAACCG-3′ (rpoB), 5′-GCTTTACTTAGCTCACC-3′ (accD), 5′-CCCAACACTTGCTTTAG-3′ (rbcL), 5′-TGCGATAATAAAAACAGAAGTTGCG-3′ (psbA), and 5′-TTCATAGTTGCATTACT-3′ (rrn).
5′-RACE
The discrimination between transcription start sites and processing sites of precursor RNAs was done by RNA ligase-mediated (RLM) RACE (RLM-RACE kit, Ambion) without and with previous TAP treatment of RNAs. Reactions were performed according to the suppliers' protocol but without removal of free 5′ phosphates by calf intestine alkaline phosphatase. PCR products were analyzed on agarose gels after two successive PCR amplifications, the first using two outer primers and the second using two inner primers. Primers are as follows: rpoB as outer: 5′-GAAATACCGCTGGAACTTACG-3′, rpoB as inner: 5′-CCGCTGGAACTTACGGAG-3′. The inner and outer adapter primers are those of the RLM-RACE kit.
Semiquantitative RT-PCR
Two micrograms of DNase I-treated RNA was reverse transcribed using 400 units of Superscript II (Invitrogen SARL) according to the manufacturer's protocol. The reaction was performed in the presence of 1 μg random hexamers in a total volume of 60 μL at 42°C for 50 min. Aliquots of 3 μL of this reaction were afterward used as template for semiquantitative PCR in a 25-μL reaction mix containing 1 unit of BioTaq (Bioline). To ensure that amplification is in the linear range, the optimal number of cycles have been determined for each couple of primers separately.
PCR was carried out under the following conditions: 5 min denaturation at 94°C followed by n cycles of amplification (30 s at 94°C; 30 s at 55°C; 1 min at 72°C) and a final 10-min elongation step at 72°C. ADENINE PHOPHORIBOSYLTRANSFERASE (APT) mRNA amplification was used as an internal standard (Moffatt et al., 1994). Primers and number n of cycles are as follows: rpoTp, 5′-CTTGCTCCCTTCTTCAG-3′ and 5′-CCTTGAAGATTTGCTCC-3′, n = 37; rpoTmp, 5′-GATTTTGGTGATGAAAAAGAAG-3′ and 5′-CTCCCAAACCGGGATTTC-3′, n = 33; rpoTm, 5′-CAGATGACTGCTTTTGCACC-3′ and 5′-GGAATGTTGATGGTTAACCTCAA-3′, n = 33; and APT, 5′-TCCCAGAATCGCTAAGATTGCC-3′ and 5′-CCTTTCCCTTAAGCTCTG-3′, n = 24.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers RPOTp (At2g24120) and RPOTmp (At5g15700).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Quantification of plastid precursor RNAs from wild-type plants and RPOTp and RPOTmp mutants.
Supplementary Material
Acknowledgments
We are grateful to A. Cottet and E. Lambert for excellent technical assistance and to T. Lagrange and I. Debeaujon for kindly providing nrpd1 and tt2-1 seeds, respectively.
This work was supported by the European Community (grant no. FP6–2002–LifeSciHealth, PLASTOMICS, proposal no. 503238, L.M. was employed under this contract).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Silva Lerbs-Mache (silva.lerbs-mache@ujf-grenoble.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Allison LA (2000) The role of sigma factors in plastid transcription. Biochimie 82 537–548 [DOI] [PubMed] [Google Scholar]
- Allison LA, Simon LC, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Baba K, Schmidt J, Espinosa-Ruiz A, Villarejo A, Gardeström P, Sane AP, Bhalerao P (2004) Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J 38 38–48 [DOI] [PubMed] [Google Scholar]
- Baeza L, Bertrand A, Mache R, Lerbs-Mache S (1991) Characterization of a protein binding sequence in the promoter region of the 16S rRNA gene of the spinach chloroplast genome. Nucleic Acids Res 19 3577–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE (1993) Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development. Plant Physiol 101 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland K, Haselkorn R (1991) Evolutionary relationships among eubacteria, cyanobacteria, and chloroplasts: evidence from the rpoC1 gene of Anabaena sp. strain PCC 7120. J Bacteriol 173 3446–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisanz C, Bégot L, Carol P, Perez P, Bligny M, Pesey H, Lerbs-Mache S, Mache R (2003) The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol Biol 51 651–663 [DOI] [PubMed] [Google Scholar]
- Bligny M, Courtois F, Thaminy S, Chang CC, Lagrange T, Baruah-Wolff J, Stern D, Lerbs-Mache S (2000) Regulation of plastid rDNA transcription by interaction of CDF2 with two different RNA polymerases. EMBO J 19 1851–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Sheen J, Niwa Y, Lerbs-Mache S, Stern DB (1999) Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell 11 911–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YS, Lin CH, Chen LJ (1997) Transcription and processing of the gene for spinach chloroplast threonine tRNA in a homologous in vitro system. Biochem Biophys Res Commun 233 380–385 [DOI] [PubMed] [Google Scholar]
- da Costa e Silva O, Lorbiecke R, Garg P, Müller L, Wassmann M, Lauert P, Scanlon M, Hsia AP, Schnable PS, Krupinska K, et al (2004) The Etched1 gene of Zea mays (L) encodes a zinc ribbon protein that belongs to the transcriptionally active chromosome (TAC) of plastids and is similar to the transcription factor TFIIS. Plant J 38 923–939 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S (2006) Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol 142 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisermann A, Tiller K, Link G (1990) In vitro transcription and DNA binding characteristics of chloroplast and etioplast extracts from mustard (Sinapis alba) indicate differential usage of the psbA promoter. EMBO J 9 3981–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel C, Weihe A, Graner A, Hess WR, Börner T (2004) Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J 38 460–472 [DOI] [PubMed] [Google Scholar]
- Favory JJ, Kobayshi M, Tanaka K, Peltier G, Kreis M, Valay JG, Lerbs-Mache S (2005) Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Res 33 5991–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W, Elsner-Menzel C, Latshaw S, Narita JO, Schaffer MA, Zurawski G (1986) A subpopulation of spinach chloroplast tRNA genes does not require upstream promoter elements for transcription. Nucleic Acids Res 19 7541–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PTJ, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277 809–811 [DOI] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (2000) One RNA polymerase serving two genomes. EMBO Rep 5 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricova A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschmann T, Börner T (1998) Characterisation of transcript initiation sites in ribosome-deficient barley plastids. Plant Mol Biol 36 493–496 [DOI] [PubMed] [Google Scholar]
- Igloi GL, Kössel H (1992) The transcriptional apparatus of chloroplasts. Crit Rev Plant Sci 10 525–558 [Google Scholar]
- Ikeda TM, Gray MW (1999) Identification and characterization of T3/T7 bacteriophag-like RNA polymerase sequences in wheat. Plant Mol Biol 40 567–578 [DOI] [PubMed] [Google Scholar]
- Iratni R, Baeza L, Andreeva A, Mache R, Lerbs-Mache S (1994) Regulation of rDNA transcription in chloroplasts: promoter exclusion by constitutive repression. Genes Dev 8 2928–2938 [DOI] [PubMed] [Google Scholar]
- Iratni R, Diederich L, Harrak H, Bligny M, Lerbs-Mache S (1997) Organ-specific transcription of the rrn operon in spinach plastids. J Biol Chem 272 13676–13682 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Sato N (2005) Unique translation initiation at the second AUG codon determines mitochondrial localization of the phage-type RNA polymerases in the moss Physcomitrella patens. Plant Physiol 138 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor JY, Suzuki JY, Sugiura M (1997) Identification and functional significance of a new class of non-consensus-type plastid promoters. Plant J 11 327–337 [DOI] [PubMed] [Google Scholar]
- Kühn K, Bohne AV, Liere K, Weihe A, Börner T (2007) Arabidopsis phage-type RNA polymerase: accurate transcription of organellar genes. Plant Cell 19 959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Yara A, Mitsui N, Tozawa Y, Iba K (2004) Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol 45 1194–1201 [DOI] [PubMed] [Google Scholar]
- Lagrange T, Hakimi M-A, Pontier D, Courtois F, Alcaraz JP, Grunwald D, Lam E, Lerbs-Mache S (2003) Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Mol Cell Biol 23 3274–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S (1993) The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc Natl Acad Sci USA 90 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S (2000) Regulation of rDNA transcription in plastids of higher plants. Biochimie 82 525–535 [DOI] [PubMed] [Google Scholar]
- Liere K, Börner T (2006) Transcription of plastid genes. In KQ Grasser, ed, Regulation of Transcription in Plants. Blackwell Publishing Ltd., Oxford, pp 184–224
- Link G (1984) DNA sequence requirements for the accurate transcription of a protein-coding plastid gene in a plastid in vitro system from mustard (Sinapis alba L.). EMBO J 3 1697–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko AL (2007) Plant sigma factors and their role in plastid transcription. Plant Cell Rep 26 845–859 [DOI] [PubMed] [Google Scholar]
- Martin W (2003) Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci USA 100 8612–8614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DE, Durbin RD (1990) Tagetitoxin inhibits RNA synthesis directed by RNA polymerases from chloroplasts and Escherichia coli. J Biol Chem 265 493–498 [PubMed] [Google Scholar]
- Mathews DE, Durbin RD (1994) Mechanistic aspects of tagetitoxin inhibition of RNA polymerase from Escherichia coli. Biochemistry 33 11987–11992 [DOI] [PubMed] [Google Scholar]
- Miyagi T, Kapoor S, Sugita M, Sugiura M (1998) Transcript analysis of the tobacco plastid operon rps2/atpI/H/F/A reveals the existence of a non-consensus type II (NCII) promoter upstream of the atpI coding sequence. Mol Gen Genet 257 299–307 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, McWhinnie EA, Agarwal SK, Schaff DA (1994) The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143 211–216 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G (1997) The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol Gen Genet 257 35–44 [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privat I, Hakimi MA, Buhot L, Favory JJ, Lerbs-Mache S (2003) Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol Biol 55 385–399 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of a-Amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanke R, Fischer K, Ketelsen B, Krupinska K, Krause K (2007) Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol Genet Genomics 277 631–646 [DOI] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Yand Khan MS (2005) Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol 244 1–68 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Maliga P (1998. a) Mapping of the promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr Genet 33 340–344 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Maliga P (1998. b) Plastid promoter utilization in a rice embryonic culture. Curr Genet 34 67–70 [DOI] [PubMed] [Google Scholar]
- Sriraman P, Silhavy D, Maliga P (1998. a) The phage-type PclpP-53 plastid promoter comprises sequences downstream of the transcription initiation site. Nucleic Acids Res 26 4874–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman P, Silhavy D, Maliga P (1998. b) Transcription from heterologous rRNA operon promoters in chloroplasts reveals requirement for specific activation factors. Plant Physiol 117 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter G, Gozdzicka-Jozefiak A, Kössel H (1985) Identification of an rRNA operon promoter from Zea mays chloroplasts which excludes the proximal tRNAVal from the primary transcript. EMBO J 4 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Sugiura M (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol 32 315–326 [DOI] [PubMed] [Google Scholar]
- Suzuki JY, Sriraman P, Svab Z, Maliga P (2003) Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell 15 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M, Liere K, Börner T (2007) High diversity of plastid promoters in Arabidopsis thaliana. Mol Genet Genomics 277 725–734 [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5 123–135 [DOI] [PubMed] [Google Scholar]
- Vera A, Hirose T, Sugiura M (1996) A ribosomal protein gene (rpl32) from tobacco chloroplast DNA is transcribed from alternative promoters—similarities in promoter region organization in plastid housekeeping genes. Mol Gen Genet 27 280–284 [DOI] [PubMed] [Google Scholar]
- Vera A, Sugiura M (1995) Chloroplast rRNA transcription from structurally different tandem promoters: an additional novel-type promoter. Curr Genet 27 280–284 [DOI] [PubMed] [Google Scholar]
- Wagner R, Pfannschmidt T (2006) Eukaryotic transcription factors in plastids: bioinformatic assessment and implications for the evolution of gene expression machineries in plants. Gene 381 62–70 [DOI] [PubMed] [Google Scholar]
- Wu CY, Lin CH, Chen LJ (1997) Identification of the transcription start site for the spinach chloroplast serine tRNA gene. FEBS Lett 418 157–161 [DOI] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differently regulated during leaf development. Plant J 50 710–722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.