Abstract
PsbP, an extrinsic subunit of photosystem II (PSII), is a nuclear-encoded protein that optimizes the water-splitting reaction in vivo. In addition to PsbP, higher plants have two nuclear-encoded genes for PsbP homologs (PsbP-like proteins [PPLs]) that show significant sequence similarity to a cyanobacterial PsbP homolog (cyanoP); however, the function of PPLs in higher plants has not yet been elucidated. In this study, we characterized Arabidopsis (Arabidopsis thaliana) mutants lacking either of two PPLs, PPL1 and PPL2. Phylogenetic analysis suggests that PPL1 would be an ortholog of cyanoP, and PPL2 and PsbP may have a paralogous relationship with PPL1. Analysis on mRNA expression profiles showed that PPL1 expressed under stress conditions and PPL2 coexpressed with the subunits of chloroplast NAD(P)H dehydrogenase (NDH) complex. Consistent with these suggestions, PSII activity in a ppl1 mutant was more sensitive to high-intensity light than wild type, and the recovery of photoinhibited PSII activity was delayed in ppl1 plants. Therefore, PPL1 is required for efficient repair of photodamaged PSII. Furthermore, the stoichiometric level and activity of the chloroplast NDH complex in thylakoids were severely decreased in a ppl2 mutant, demonstrating that PPL2 is a novel thylakoid lumenal factor required for accumulation of the chloroplast NDH complex. These results suggest that during endosymbiosis and subsequent gene transfer to the host nucleus, cyanoP from ancient cyanobacteria evolved into PPL1, PPL2, and PsbP, and each of them has a distinct role in photosynthetic electron transfer in Arabidopsis.
Current evidence suggests that chloroplasts arose more than 1.2 billion years ago (Butterfield, 2000) when a free-living cyanobacterium became an endosymbiont in a eukaryotic host. The subsequent evolutionary process of endosymbiotic gene transfer (Brown, 2003), which entailed the transfer of many genes to the eukaryotic host nucleus, dramatically reduced the size of the chloroplast genome. Endosymbiotic gene transfer appears to be a very widespread and continual process (Huang et al., 2005); about 18% of the nuclear genes in Arabidopsis (Arabidopsis thaliana) seem to have come from cyanobacteria, and obvious remnants of chloroplast DNA have been found in higher plant nuclear chromosomes. It is thought that the transfer of genes from the chloroplast genome to the nuclear genome results in greater diversification of gene structure and function; some of these nuclear-encoded genes gained a new function after undergoing internal gene duplication and they are involved in regulation of chloroplast functions (Leister, 2003). For example, the cyanobacterial high-light-inducible protein gene is thought to have evolved into nuclear genes encoding early-light-inducible proteins, stress-enhanced proteins, and light-harvesting complex proteins in higher plants and to have various functions, such as photoprotection or capturing light energy (Montané and Kloppstech, 2000). This diversification of gene function from ancient cyanobacteria enables chloroplasts to adapt their function to the living environment.
The characteristic common to chloroplasts and cyanobacteria is oxygenic photosynthesis, which generates molecular oxygen from the water-splitting reaction. This oxygen-evolving reaction is catalyzed by the protein complex, PSII, composed of intrinsic and extrinsic membrane protein subunits. Interestingly, many intrinsic subunits are still encoded by chloroplast genomes, whereas most extrinsic subunits of eukaryotic oxyphototrophs are nuclear-encoded proteins. This suggests that extrinsic subunits should be susceptible to diversification. In fact, the intrinsic subunits are highly conserved in all known oxygenic photosynthetic organisms, including higher plants, green and nongreen algae, and cyanobacteria, whereas the repertoire of extrinsic subunits that stabilize the function and activity of the oxygen-evolving center differs significantly among those organisms (for review, see Bricker and Burnap, 2005). Eukaryotic higher plants and green algae have a set of three extrinsic proteins (PsbO [33 kD; also referred to as OEC33], PsbP [23 kD; also referred to as OEC23], and PsbQ [17 kD; also referred to as OEC16]), whereas prokaryotic oxygenic photosynthetic bacteria have a different set of proteins (PsbO; [12 kD; PsbU], and PsbV [cytochrome c550]). PsbO, which is present in all of the known genomes of oxyphototrophs, plays an important role in stabilizing the manganese cluster that catalyzes the water-splitting reaction (Debus, 1992). The other extrinsic proteins, PsbU and PsbV in cyanobacteria and PsbP and PsbQ in higher plants, have been reported to play a role in optimizing the availability of Ca2+ and Cl− cofactors to maintain the Mn-Ca2+-Cl− cluster within PSII (Seidler, 1996). These facts suggest that functional replacement of PsbU and PsbV by PsbP and PsbQ occurred during the evolution of PSII (Enami et al., 2005).
Recently, genomic and proteomic studies have demonstrated the existence of the PsbP and PsbQ homologs, cyanobacterial PsbP (cyanoP) and PsbQ (cyanoQ), respectively, in cyanobacteria (De Las Rivas et al., 2004). In Synechocystis sp. PCC6803, cyanoQ is tightly associated with PSII and required for optimal O2 evolution (Roose et al., 2007a), whereas only a small fraction of cyanoP is associated with PSII (Thornton et al., 2004) and its contribution to PSII function is rather small (Ishikawa et al., 2005; Summerfield et al., 2005). These results do not correspond to observations from higher plants, where PsbP is much more important than PsbQ in regulating and stabilizing PSII (Ifuku et al., 2005). This suggests that cyanoP and PsbP have quite different functions and that PsbP has newly developed as an indispensable regulator of PSII during the evolution of oxyphototrophs. Consistent with this suggestion, it has been reported that higher plants have additional PsbP homologs that have higher similarity to cyanoP than to PsbP (De Las Rivas and Roman, 2005). In addition, Arabidopsis has many PsbP homologs, which are likely the result of gene duplication (Roose et al., 2007b). They have limited sequence identities with higher plant PsbP (approximately 25%) and their protein accumulation levels are 20 to 300 times lower than that of PsbP as estimated by proteomic analysis (Peltier et al., 2002; Schubert et al., 2002). Despite this interesting evolutionary feature, molecular function of PsbP homologs in higher plants has been completely unknown.
In this study, we characterized two PsbP homologs, PsbP-LIKE PROTEIN1 (PPL1 [At3g55330]) and PPL2 [At2g39470]), in Arabidopsis. To investigate their functions, we characterized mutants lacking PPL1 and PPL2. Analysis of a ppl1 mutant suggested that PPL1 is required for efficient repair of photodamaged PSII under high-intensity light. On the other hand, the stoichiometric level of a subunit of the chloroplast NAD(P)H dehydrogenase (NDH) complex in thylakoid membranes and activity of the complex were severely decreased in a ppl2 mutant, suggesting that PPL2 is a novel thylakoid lumenal factor required for the accumulation of the NDH complex. These results demonstrate the differential functions of PsbP and PPL proteins in the thylakoid lumen of Arabidopsis.
RESULTS
PsbP Protein Family in Arabidopsis
A position-specific iterated-BLAST search of the National Center for Biotechnology Information databases with the sequence of cyanoP (sll1418) hits eight PsbP homologs in addition to two authentic PsbPs (PsbP1 [At1g06680] and PsbP2 [At2g30790]) in Arabidopsis. Based on the amino acid sequence similarity with PsbP1, we classified them into three groups: two PsbP proteins, two PPL proteins, and six PsbP domain (PPD) proteins (Table I). Two PPL proteins show significantly higher similarity with the PsbP1 sequence (near 40% similarity) than the other six PPD proteins, and a BLASTP search of The Arabidopsis Information Resource database (http://www.arabidopsis.org) with the PsbP1 sequence also suggests a close relationship with PsbP1 (PPL1 [At3g55330], E value 9 × 10−6; PPL2 [At2g39470], E value 5 × 10−4). All PsbP homologs in Arabidopsis, except for PPD2, are found in the rice (Oryza sativa) EST database (http://www.ncbi.nlm.nih.gov/dbEST), suggesting that they are conserved in both dicot and monocot plants (Supplemental Fig. S1). All PsbP homologs have an obvious thylakoid lumen-targeting signal, consistent with previous proteomic studies, which has proven their localization in the thylakoid lumen, except for PsbP2 (Peltier et al., 2002; Schubert et al., 2002). In this study, we focused on two PPL proteins, PPL1 and PPL2, to compare their molecular function with that of PsbP.
Table I.
PsbP family members in Arabidopsis
| Name | Locus | Accession No.a | cyanoP
|
PsbP
|
||
|---|---|---|---|---|---|---|
| Ib | +c | I | + | |||
| % | % | % | % | |||
| PsbP1 | At1g06680 | Q42029 | 16.6 | 35.3 | ||
| PsbP2d | At2g30790 | O49344 | 21.4 | 35.2 | 86.2 | 92.3 |
| PPL1 | At3g55330 | P82538 | 32.5 | 48.7 | 24.7 | 44.8 |
| PPL2 | At2g39470 | O80634 | 27.2 | 44.3 | 24.7 | 39.9 |
| PPD1 | At4g15510 | O23403 | 28.8 | 40.2 | 20.7 | 34.2 |
| PPD2 | At2g28605 | NP_850123 | 16.7 | 35.2 | 10.7 | 24.9 |
| PPD3 | At1g76450 | Q9S720 | 14.3 | 29.8 | 19.0 | 33.3 |
| PPD4 | At1g77090 | O49292 | 19.5 | 36.6 | 13.2 | 29.8 |
| PPD5 | At5g11450 | Q8VY65 | 16.9 | 32.0 | 13.3 | 34.2 |
| PPD6 | At3g56650 | NP_191224 | 19.2 | 34.7 | 19.9 | 35.6 |
Two PPL Proteins, PPL1 and PPL2, Are Well Conserved among Higher Plants
Searching genome or EST databases by the tBLASTN program identified orthologs of PPL1 and PPL2 in all plant species we checked with highly conserved amino acid sequences (Supplemental Fig. S2). Chlamydomonas reinhardtii, which contains PsbP, has no PPL protein orthologs, whereas Chlamydomonas incerta has a PPL1 homolog in spite of PsbP proteins. Cyanidioschyzon merolae has two PPL proteins, whose sequences are closer to cyanoP than to PPL proteins in higher plants. These results suggest that both PPL1 and PPL2 would be well-conserved proteins in higher plants, but not in chlorophytes or rhodophytes.
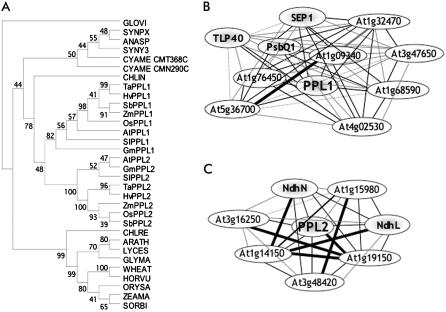
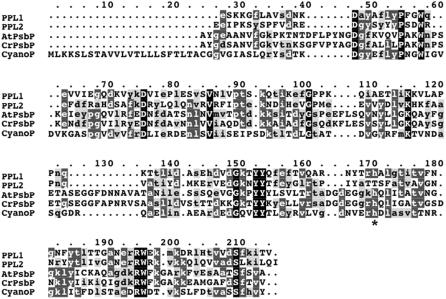
Based on the alignment of cyanoP, PPL, and PsbP sequences, a phylogenetic tree was constructed (Fig. 1A). As expected, cyanoP and PPL sequences branched into a different clade from PsbP sequences. Alignment of the amino acid sequences suggests that both PPL1 and PPL2 are much more similar to cyanoP than to PsbP in higher plants (Fig. 2). In particular, PPL1 is more similar to cyanoP than PPL2 and also contains a His residue conserved in all cyanoPs and PsbPs (Fig. 2, asterisk). These facts suggest that PPL1 is closely related to cyanoP, whereas PsbP and PPL2 may have a paralogous relationship with PPL1. High conservation of PsbP and PPLs indicates their important function in higher plants.
Figure 1.
Bioinfomatic analysis of PPL1 and PPL2 proteins. A, Protein phylogenetic trees of the PsbP and PPL protein family. A rooted tree of 32 protein sequences was produced using the neighbor-joining method with Gloeobacter as an outgroup. The sequences are named for each organism that corresponds to the cyanoP or PsbP protein sequence used (the protein ID no. from UniProt or GenBank is given together with the species-scientific name): GLOVI, Q7NKN7, Gloeobacter violaceus PCC 7421; ANASP, Q8YSK9, Anabaena sp. PCC 7120; SYNPX, Q7U7Q4, Synechococcus sp. WH8102; SYNY3, P73952, Synechocystis sp. PCC 6803; CMN290C and CMT368C, C. merolae annotated coding sequences (http://merolae.biol.s.u-tokyo.ac.jp/blast/blast.html); CHLRE, P11471, C. reinhardtii; CHLIN, ABA01138, C. incerta; ARATH, Q42029, Arabidopsis; LYCES, P29795, Lycopersicon esculentum (Solanum lycoperin); GLYMA, EST reconstructions, Glycine max (http://www.ncbi.nlm.nih.gov/dbEST); ZEAMA, EST reconstructions, Z. mays; SORBI, EST reconstructions, Sorghum bicolor; ORYSA, Q943W1, rice; WHEAT, Q00434, Triticum aestivum; HORVU, EST reconstructions, Hordeum vulgare. The sequences of PPL1 and PPL2 orthologs are as follows: TaPPL1 and TaPPL2, EST reconstructions, T. aestivum; HvPPL1 and HvPPL2, EST reconstructions, H. vulgare; SbPPL1 and SbPPL2, EST reconstructions, S. bicolor; ZmPPL1 and ZmPPL2, EST reconstructions, Z. mays; OsPPL1 and OsPPL2, Q6ZDL1 and EST reconstructions, rice; AtPPL1 and AtPPL2, P82538 and O80634, Arabidopsis; SlPPL1 and SlPPL2, EST reconstructions, S. lycoperin; GmPPL1 and GmPPL2, EST reconstructions, G. max. Bootstrap values from 1,000 replications are shown at the tree nodes. B, Network diagram of the genes that showed a high r value with each other and PPL1. C, Network diagram of the genes that showed a high r value with each other and PPL2. Thick black lines, r ≥ 0.925; thin black lines, 0.925 > r ≥ 0.900; gray lines, 0.900 > r ≥ 0.875; black dotted lines, r < 0.875.
Figure 2.
Comparison of amino acid sequences of PPL1, PPL2, PsbP, and cyanoP. The sequences of AtPPL1 (P82538, Arabidopsis), AtPPL2 (O80634, Arabidopsis), AtPsbP (Q42029, Arabidopsis), CrPsbP (P11471, C. reinhardtii), and cyanoP (P73952, Synechocystis sp. PCC 6803) were aligned with ClustalW. Black, dark gray-, and light gray-shaded boxes show all-identical, identical, and similar amino acids, respectively. *, His residue conserved in all cyanoPs and PsbPs.
mRNA Expression Profiles Differ Significantly between the Two PPLs
To estimate functions of PPL1 or PPL2, we searched for genes that are coexpressed with these genes using a program in ATTED-II using Pearson's correlation coefficient (r) for neighboring pairs of genes (see “Materials and Methods”). Nine genes were found to be coexpressed with PPL1 with a high r value (r ≥ 0.900; Supplemental Table S1), then a second search of ATTED-II was conducted after changing the query to the nine genes with high r values plus PPL1. As a result, eight genes with a high average r value (r ≥ 0.900) were selected. These eight genes, shown in a network diagram (Fig. 1B), include TLP40 (a thylakoid lumen peptidyl-prolyl cis-trans isomerase; At3g01480), PsbQ1 (At4g21280), and SEP1 (a stress-enhanced protein; At4g34190). TLP40 triggers dephosphorylation of the PSII reaction center proteins in response to heat shock in spinach (Spinacia oleracea; Rokka et al., 2000), PsbQ is required for PSII stability in Arabidopsis (Yi et al., 2006), and SEP1 is a high-light stress-enhanced protein related to the chlorophyll (chl) a/b-binding gene family (Heddad and Adamska, 2000).
In the case of PPL2, six genes were coexpressed with PPL2 with a high r value (r ≥ 0.900; Supplemental Table S2), and all of those six genes and PPL2 showed a high average r value (r ≥ 0.900) in a second search of ATTED-II after changing the query to the six genes with high r values plus PPL2. These six genes, also shown in a network diagram (Fig. 1C), include the genes encoding NdhL (At1g70760) and NdhN (At5g58260) proteins. NdhL and NdhN are stromal subunits of the NDH complex that catalyzes electron donation to plastoquinone (PQ) from the stromal electron pool. NDH in chloroplasts is thought to protect against photooxidative stress (Endo et al., 1999).
The above results indicate that both PPL1 and PPL2 would have a function related to photosynthesis, presumably under certain stress conditions, such as high-intensity light. However, based on microarray data, the spectrum of genes coexpressed with PPL1 is significantly different from that coexpressed with PPL2, suggesting that the function of PPL1 and PPL2 genes differs in Arabidopsis.
Photosynthetic Properties of ppl1 and ppl2 Mutants in Arabidopsis
To elucidate the functions of PPL1 and PPL2 proteins, we used a ppl1 mutant (salk_014843) containing a single T-DNA insertion located in intron 4 and a ppl2 mutant (salk_020674) containing two T-DNA insertions located in exon 5 (Supplemental Fig. S3A). The PPL1 or PPL2 mRNA was undetectable by reverse transcription (RT)-PCR in the respective mutants (Supplemental Fig. S3B), indicating that the T-DNA insertions had disrupted expression. Under normal growth conditions (10/14-h photoperiod at 100–200 μmol quanta m−2 s−1, 21°C), both ppl1 and ppl2 plants showed nearly wild-type growth (Supplemental Fig. S3C).
The parameters for the photosynthetic properties showed that the maximal quantum yield of PSII (Fv/Fm), the relative amount of far-red oxidizable P700, and the chl a/b ratio were not significantly different between mutants and wild-type plants (Table II). Therefore, the steady-state properties of PSII and PSI in dark-adapted leaves were not affected in both ppl1 and ppl2 plants. However, under actinic light (AL) illumination (144 μmol photons m−2 s−1), ppl2 plants showed significantly lower ΦPSII values (0.613 ± 0.015) and ppl1 plants showed slightly lower ΦPSII values (0.631 ± 0.025) compared to wild type (0.676 ± 0.023). These lower ΦPSII values were fully restored in mutant plants complemented with the wild-type gene, ppl1 + PPL1 and ppl2 + PPL2 (0.696 ± 0.039 and 0.690 ± 0.031, respectively), indicating the involvement of PPL1 and PPL2 in photosynthetic electron transfer in Arabidopsis.
Table II.
Photosynthetic measurements of wild-type, ppl1, and ppl2 plants
Data presented are means ± sd of three to six independent measurements.
| Photosynthetic Parameters | Wild Type | ppl1 | ppl2 |
|---|---|---|---|
| Fv/Fma | 0.785 ± 0.015 | 0.781 ± 0.014 | 0.786 ± 0.013 |
| ΦPSIIb | 0.671 ± 0.023 | 0.641 ± 0.023 | 0.613 ± 0.015* |
| Far-red oxidizable P700c | 1.00 ± 0.04 | 0.99 ± 0.03 | 0.97 ± 0.01 |
| Chl a/b ratio | 2.97 ± 0.14 | 2.99 ± 0.11 | 2.98 ± 0.12 |
| O2 evolutiond | |||
| (Maximum)e | 246 ± 14 | 213 ± 2* | 261 ± 18 |
| (Average)f | 168 ± 8 (68)g | 143 ± 6 (67)* | 182 ± 7 (70) |
Fv/Fm value was measured after incubation in the dark for 1 h.
ΦPSII was measured under illumination at 144 μmol quanta m−2 s−1.
Relative value to wild-type plants set as 1.00.
O2 evolution activity was indicated by μmol O2 [mg chl]−1 h−1.
Maximal activity of the O2 evolution from isolated thylakiod membranes.
Average activity of the O2 evolution during the first 5 min.
Figures in parentheses are relative % of the average to the maximal activity. Asterisks indicate the means that were statistically different from that of wild type (P < 0.05; Student's t test).
The oxygen-evolving activities of isolated thylakoid membranes were then examined on the chl basis with p-phenylbenzoquinone as an electron acceptor (Table II). The maximal activity from ppl1 thylakoids was significantly lower than those from wild-type and ppl2, and a similar observation was obtained in the further presence of 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinonine, which blocks the electron transfer between PSII and PSI (data not shown). Because the ratio of the average activity during first 5 min to the maximal (initial) activity after light illumination was almost the same among the samples (approximately 70%), the stability of active PSII was not significantly different, which was consistent with the normal value of Fv/Fm in ppl1 mutants.
Accumulation of Proteins in Isolated Thylakoids from ppl1 and ppl2 Mutants
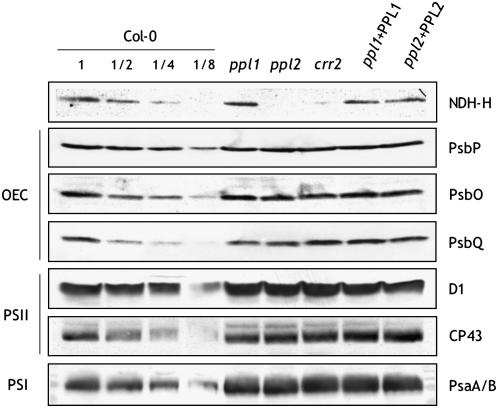
To investigate whether a deficiency of PPL1 or PPL2 affects the normal accumulation of thylakoid membrane proteins, immunoblot analysis was conducted on isolated thylakoid membranes of ppl1 and ppl2 mutants. In addition to the protein subunits of the major protein complexes, such as PSII and PSI located in thylakoids, a protein blot was also probed with antibodies against the NdhH subunit of the NDH complex because our bioinformatics analysis suggested that PPL2 was coexpressed with the subunits of the chloroplastic NDH complex (Fig. 1C; Supplemental Table S2). Previous study showed that the NdhH subunit is unstable without other NDH subunits (Munekage et al., 2004; Rumeau et al., 2005) and thus can be used to monitor the accumulation of complete NDH complexes.
Despite the lower activity of oxygen evolution, deletion of PPL1 did not substantially affect the accumulation of thylakoid membrane proteins (Fig. 3). On the other hand, interestingly, the amount of NdhH was markedly reduced in the ppl2 mutant (≤12.5% of wild-type NdhH levels) and comparable to that seen for the NDH mutant crr2 (chlororespiratory reduction2), which lacks the pentatrico peptide repeat protein needed for the intergenic RNA processing essential for the translation of NdhB (Hashimoto et al., 2003; Fig. 3). This ppl2 phenotype was fully recovered in ppl2 + PPL2 plants, suggesting that the lack of the NDH complex was attributable to loss of PPL2 expression. The amounts of other subunits were not noticeably different between ppl2 and the wild type.
Figure 3.
Immunodetection of various proteins in thylakoid membranes isolated from mutant and wild-type plants. Isolated thylakoid proteins were subjected to immunoblotting with antisera against the indicated proteins. Protein complexes found in the thylakoid membranes are shown at the left. OEC, Oxygen-evolving complex proteins. Samples containing 5 μg (Col-0 1, ppl1, ppl2, crr2, ppl1 + PPL1, ppl2 + PPL2), 2.5 μg (Col-0 1/2), 1.25 μg (Col-0 1/4), and 0.625 μg (Col-0 1/8) of chl were loaded in each well. crr2, A known mutant lacking the activity and accumulation of NDH complex; ppl1 + PPL1, ppl1 transformed with 35S-driven PPL1 cDNA; ppl2 + PPL2, ppl2 transformed with 35S-driven PPL2 cDNA. Equal sample loading was confirmed by Coomassie Brilliant Blue R-250 staining in replicate gels (data not shown).
Arabidopsis ppl1 Mutant Was Sensitive to High-Intensity Light-Induced PSII Photoinhibition
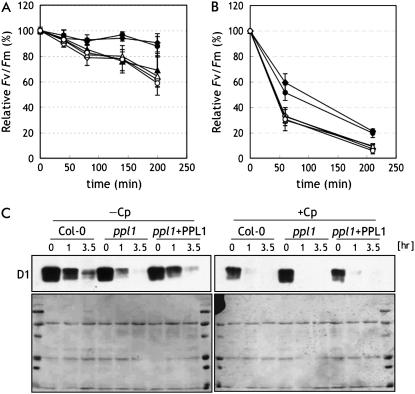
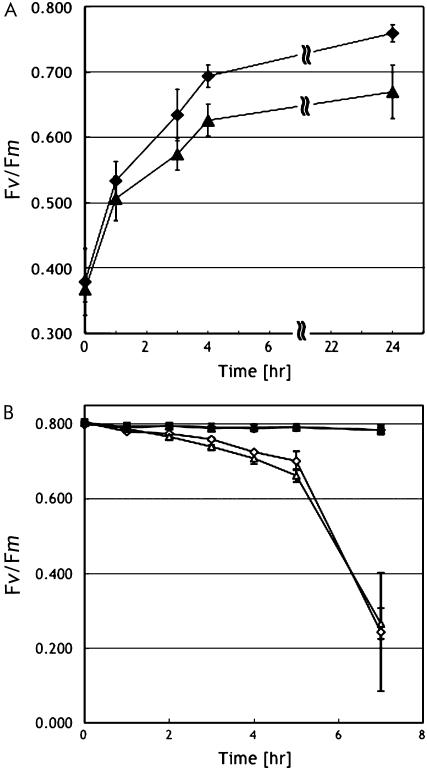
The bioinformatics analysis shows that PPL1 is coexpressed with TLP40 and SEP1, suggesting that its expression is induced in response to stress (Fig. 1B; Supplemental Table S1). Then, we examined the performance of PSII in wild type, ppl1, and ppl1 + PPL1 plants under high-intensity light. Detached leaves were irradiated with high-intensity light, and Fv/Fm was monitored. Under high-intensity light (550 μmol photons m−2 s−1), wild-type leaves showed little decline in Fv/Fm (Fig. 4A). However, in the ppl1 mutant, Fv/Fm decreased to about 70% of the initial value within 200 min of illumination, clearly showing increased photoinhibition in the ppl1 mutant compared to wild-type plants. In the presence of chloramphenicol, which inhibits plastid protein synthesis, Fv/Fm decreased to about 60% within 200 min of illumination in both ppl1 and wild-type leaves. Increased photoinhibition in ppl1 leaves compared to wild-type leaves was also observed under further higher intensity light (1,200 μmol photons m−2 s−1; Fig. 4B). In addition, the photobleaching phenotype was observed in the ppl1 leaves after high-intensity light treatment (data not shown). These results demonstrate that ppl1 leaves are more sensitive to high-intensity light-induced photoinhibition than wild-type leaves, whereas wild-type and ppl1 leaves have potentially similar rates of PSII photodamage.
Figure 4.
Photoinhibition of PSII activity and degradation of PSII protein in ppl1 under high-intensity light. Maximal photochemical efficiency of PSII (Fv/Fm) was measured in detached leaves from wild-type (diamonds), ppl1 mutant (triangles), and ppl1 + PPL1 (circles) plants during exposure to irradiance of 550 (A) or 1,200 (B) μmol quanta m−2 s−1 in the presence (white symbols) or absence (black symbols) of 200 μg mL−1 chloramphenicol. Values were means ± sd (n = 3). C, Immunodetection of PSII reaction center protein, D1. Proteins were extracted from detached leaves (n = 3) from ppl1 mutant and wild-type plants during exposure to irradiance of 1,200 μmol quanta m−2 s−1 in the presence (+Cp) or absence (−Cp) of 200 μg mL−1 chloramphenicol. Sample corresponding to 5.2 μg of solubilized protein was loaded in each well. The lower images show the Ponceau S-stained membranes onto which proteins separated by SDS-PAGE were electroblotted.
To examine whether high-intensity light-induced PSII inactivation led to a decrease in the steady-state level of PSII complexes, we analyzed the accumulation of D1 proteins in leaves from ppl1 mutants and wild-type plants under high-intensity light. As shown in Figure 4, the amount of D1 proteins declined gradually in leaves from ppl1 mutants and wild-type plants, and the extent of the decrease was much larger in the ppl1 mutant (Fig. 4C). In the presence of chloramphenicol, the amount of D1 proteins declined rapidly in both ppl1 mutants and wild-type plants and no apparent difference was observed. These phenotypes in the ppl1 mutant were fully recoverable in ppl1 + PPL1 leaves.
Arabidopsis ppl1 Mutant Showed Delayed Recovery of PSII Activity from Photoinhibition
To investigate further the reason for high sensitivity of ppl1 leaves to high light illumination, the capability for recovery from photoinhibition and the susceptibility to photodamage were separately compared in wild-type and ppl1 leaves under moderate light conditions. To analyze the recovery process, light intensity was lowered until 150 μmol photons m−2 s−1 after photoinhibitory treatment and recovery of the Fv/Fm values were monitored. As shown in Figure 5A, recovery of Fv/Fm values in ppl1 leaves was significantly slower than that in wild-type leaves: Fv/Fm values in the ppl1 leaves was still approximately 90% of the initial Fv/Fm values even after the 24-h recovery period. On the other hand, no noticeable difference of PSII damage was observed between wild-type and ppl1 leaves at this condition as observed under higher illumination (Figs. 4, A and B, and 5B). These results suggest that PPL1 is required for efficient repair of photodamaged PSII.
Figure 5.
Recovery of maximal photochemical efficiency of PSII after photoinhibition. A, Maximal photochemical efficiency of PSII (Fv/Fm) of detached leaves during recovery period at irradiance of 150 μmol quanta m−2 s−1, 30°C. The Fv/Fm was measured by following the photoinhibition treatment to 50% of the initial Fv/Fm values. B, The rate of PSII photodamage at irradiance of 150 μmol photons m−2 s−1, 30°C, in the presence (white symbols) or absence (black symbols) of 200 μg mL−1 chloramphenicol. Wild-type, Diamonds; ppl1 mutant, triangles. Values were means ± sd (n = 3).
Arabidopsis ppl2 Mutant Is Defective in NDH Activity in Vivo
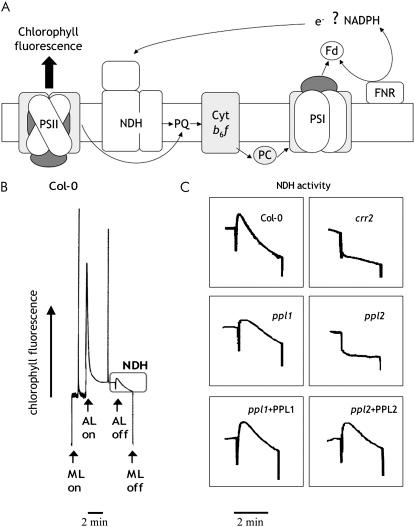
Because the immunoblot analysis suggested the absence of NDH complexes in ppl2 mutants, NDH activity of these plants was examined. The NDH complex catalyzes electron donation to PQ from the stromal electron pool (Fig. 6A). The activity of NDH can be monitored as a transient increase in chl fluorescence after turning off AL illumination (Fig. 6B; Burrows et al., 1998; Kofer et al., 1998; Shikanai et al., 1998). The increase in chl fluorescence is due to reduction of PQ by the stromal electron pool that accumulates during AL illumination. As a result, wild-type and ppl1 plants showed an increase in chl fluorescence, indicating NDH activity, whereas ppl2 mutants showed no increase in chl fluorescence (Fig. 6C), consistent with the immunoblot analysis results. The loss of NDH activity in ppl2 plants was also fully recovered in ppl2 + PPL2 plants. Therefore, we conclude that PPL2 is required for the accumulation and activity of the NDH complex in Arabidopsis thylakoid membranes.
Figure 6.
NDH activity monitored by chl fluorescence analysis. A, Schematic model of NDH function. The NDH complex functions in electron transport from an unidentified electron donor, possibly NAD(P)H or ferredoxin (Fd) to the PQ. Reduction of PQ in the dark depends on NDH activity and can be monitored as a transient increase of chl fluorescence after AL illumination. cyt b6f, Cytochrome b6f complex; PC, plastocyanin; FNR, ferredoxin-NADP+ oxidoreductase. B, Typical trace of chl fluorescence in wild type (Col-0). C, Curves in the boxes show NDH activity. Leaves were exposed to AL (144 μmol quanta m−2 s−1) for 4 min, then AL was turned off and the subsequent change in the level of chl fluorescence was monitored. ML, Measuring light.
PPL1 Proteins Are Not Tightly Associated with PSII
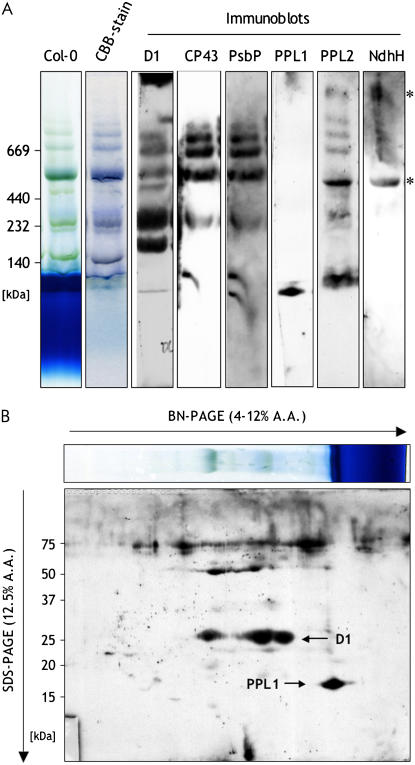
Our results described above suggested that PPL1 had a function in PSII, although its function was different from that of PsbP. We then examined whether PPL1 was associated with PSII by blue native (BN)-PAGE. Thylakoid membranes isolated from wild-type plants were solubilized with n-dodecyl-β-d-maltoside, and the protein complexes were separated in a BN gel. After BN-PAGE, the separated protein complexes were blotted on the membrane and further subjected to immunoblot analysis using specific antibodies against PPL1 and PPL2 (Supplemental Fig. S4). The immunoblots using anti D1 and CP43 antibodies showed signal bands from PSII supercomplexes, dimeric PSII, monomeric PSII, and CP43-less PSII at the positions of approximately 669, 440, 240, and 200 kD, respectively (Fig. 7A). PsbP was mainly comigrated with the PSII supercomplexes, dimeric PSII, and monomeric PSII. In contrast, the immunoblots using anti-PPL1 antibody showed that PPL1 existed as a free form in thylakoids. Molecular size of PPL1 signal was confirmed by two-dimensional (2D) BN-SDS-PAGE (Fig. 7B). These results suggest that PPL1 is not tightly associated with any type of PSII complexes. On the other hand, the immunoblots using anti-PPL2 antibody showed two specific bands at positions corresponding to approximately 1,000 and 500 kD (Fig. 7A, asterisks). Bands of the same size were also detected with anti-Ndh-H antibody, which might correspond to dimeric or monomeric NDH complexes (Darie et al., 2005), respectively. This result indicates that PPL2 would be a subunit of the chloroplastic NDH complex.
Figure 7.
Immunoblot analysis of thylakoid proteins separated by BN-PAGE or 2D BN-SDS-PAGE. Thylakoid membranes isolated from wild-type plants. Thylakoids containing 7.5 μg of chl were separated in a BN gel (A), or in the first dimension in a BN gel followed by SDS-PAGE in the second dimension (B). The resolved proteins were immunodetected with anti-D1, anti-CP43, anti-PsbP, anti-PPL1, anti-PPL2, and anti-NdhH antibodies. A.A., Acrylamide.
DISCUSSION
In this study, we demonstrate that two PPL proteins, PPL1 and PPL2, have distinct functions in the thylakoids in Arabidopsis: PPL1 is required for the efficient repair of photodamaged PSII and PPL2 is crucial for the accumulation of the chloroplastic NDH complex. PPL proteins are highly conserved among higher plants, suggesting their general importance in plant photosynthesis.
Phylogenic analysis suggests that the PPL1 protein is a putative ortholog of cyanoP in higher plants (Fig. 1A). Knockout of PPL1 in Arabidopsis made the plant sensitive to high-intensity light (Fig. 4). This change was similar to, but much milder than, the effects observed in PsbP-RNAi plants (Ifuku et al., 2005), suggesting that PPL1 and PsbP might have analogous function, but that PsbP has a more central role. In fact, protein abundance of PPL1 was much smaller than that of PsbP (Peltier et al., 2002). Furthermore, our BN-PAGE analysis could not identify PPL1 bound to PSII (Fig. 7), which would be due to the lower population and/or instability of the PSII-PPL1 complex. Because PPL1 was involved in the repair of the photodamaged PSII (Figs. 4 and 5), PPL1 would function in the dynamic life cycle of PSII and might associate with a PSII intermediate only transiently. Similar function has also been suggested for cyanoP because only a small fraction of cyanoP is associated with PSII (Thornton et al., 2004) and cyanoP has not been identified in x-ray structures of cyanobacterial PSII (Kamiya and Shen, 2003; Ferreira et al., 2004; Loll et al., 2005). However, expression of cyanoP has not been reported to be enhanced by high-intensity light (Hihara et al., 2001). Therefore, it is possible that PPL1 might have developed an additional function that cyanoP does not have.
Recently, it has been reported that another lumenal protein, Psb27, is also required for efficient repair of photodamaged PSII in Arabidopsis (Chen et al., 2006), suggesting that Psb27 and PPL1 could be functionally related. Cyanobacteria also have a PsbP27 ortholog, which is reported to stabilize an inactive PSII monomer without oxygen-evolving complex proteins in the PSII repair cycle (Nowaczyka et al., 2006). Cyanobacterial Psb27 has a sequence motif called a lipobox with the invariant Cys residue in the N-terminal leader sequence, which is unique to bacterial lipoproteins, whereas Arabidopsis Psb27 does not have the lipobox sequence motif (Nowaczyka et al., 2006). Similarly, cyanoP has a lipobox residue in its N terminus, whereas PPL1 does not, suggesting that Psb27 and PPL1 have similarity in their evolutionary and functional features. However, psb27 (At1g03600) was not included in the top 300 genes coexpressed with PPL1 (r < 0.770) in the gene search of ATTED-II. This result suggests that PPL1 is required for a different step or at a different time in the PSII repair cycle from Psb27.
Although PsbP, PPL1, and cyanoP seem to be functionally related, PPL2 has quite a different role related to the chloroplastic NDH complex. Whereas many subunits of the chloroplastic NDH complex are similar to those of the prokaryotic respiratory complex-I (NDH-I), the chloroplastic NDH complex has extra subunits that are unique to cyanobacteria and higher plants. Recently, a biochemical and genetic approach identified many candidate NDH subunits (Munshi et al., 2005); however, no thylakoid lumenal factor required for the accumulation of the chloroplastic NDH complex has been identified. It is worthwhile to note that coexpression analysis strongly indicates that a PsbQ homolog might have a function related to PPL2. Proteome analysis showed much lower stoichiometric levels of PPL2 and a PsbQ homolog in the thylakoid lumen than PsbP (Peltier et al., 2002), consistent with the less stoichiometric levels of the NDH complex than PSII in thylakoid membranes. This suggests that PPL2, and possibly a PsbQ homolog, would be a subunit of the NDH complex, and this suggestion was also supported by our BN-PAGE analysis (Fig. 7). Because PPL2 and the PsbQ homologs do not exist in cyanobacteria, a novel regulatory mechanism specific to higher plants is expected to control the accumulation of the NDH complex in the thylakoid membranes. Further biochemical characterization is obviously needed to address this issue.
CONCLUSION
We have demonstrated divergent and important functions of PPL proteins in higher plants. PsbP is a representative of the PsbP protein family (Pfam [PF01789]), and the protein folding of PsbP is classified in the Mog1p/PsbP-like superfamily in the Structural Classification of Proteins database (http://scop.mrc-lmb.cam.ac.uk/scop). In addition to PsbP and two PPL proteins, Arabidopsis has six PPD proteins, although nothing is known about their physiological functions (Table I). Despite the high-resolution structure of PsbP (Ifuku et al., 2004), the molecular mechanism of PsbP function (i.e. the binding mode to PSII and the manner of ion retention) has not been elucidated, and thus the importance of PsbP domain remains unclear. Further studies on the molecular function of PsbP, PPL, and other PPD proteins should reveal the nature of this diverged protein family and such information will be important for studies of the evolution of oxygenic photosynthetic organisms.
MATERIALS AND METHODS
Bioinformatics Analysis
PsbP homologs were searched for nonredundant protein sequence databases in Arabidopsis (Arabidopsis thaliana) by the position-specific iterated-BLAST program (http://www.ncbi.nlm.nih.gov) using cyanoP as a query (GenBank accession no. P73952). Amino acid sequences of PsbP family proteins were aligned by the ClustalW program and the percentage of amino acid sequence identity (similarity) was calculated for the regions corresponding to Met-1-Tyr-188 or Ala-78-Ala-263 of cyanoP or Arabidopsis PsbP1, respectively. PPL homologs (E value of the best match ≥ 10−5) in the Chlamydomonas reinhardtii genome were searched using the BLASTP program in the Joint Genome Institute C. reinhardtii database (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). Cyanidioschyzon merolae PPL proteins were searched by the BLASTP program using Arabidopsis PPL1 as a query for the annotated CDS databases in the C. merolae genome project (http://merolae.biol.s.u-tokyo.ac.jp/blast/blast.html; CMN290C, E value < 10−11; CMT368C, E value < 10−5). Phylogenetic analyses were conducted by the neighbor-joining algorithm using MEGA version 3.1 (Kumar et al., 2004). Transit peptide sequences were ignored when constructing the phylogenetic tree.
To search for genes coexpressed with PPL1 or PPL2, we used Pearson's correlation coefficient (r) for each pair of genes, which was calculated using the 771 Affymetrix ATH1 array data from AtGenExpress (Schmid et al., 2005) and is available to the public in the Arabidopsis trans-factor and cis-element prediction database (ATTED-II; http://www.atted.bio.titech.ac.jp; Obayashi et al., 2007).
Plant Materials and Growth Conditions
Arabidopsis wild-type (ecotype Columbia-0 [Col-0]), ppl1, ppl2, and crr2 were grown in soil under growth chamber conditions (10/14-h photoperiod at 100–200 μmol photons m−2 s−1, 21°C) for 3 to 4 weeks. Seeds for the T-DNA insertional mutants of PPL1 (At3g55330) and PPL2 (At2g39470) were purchased from a collection developed at the Salk Institute Genomic Analysis Laboratory (Alonso et al., 2003). For confirmation of the homozygosity of the T-DNA insertion lines, gene-specific and T-DNA-specific primers were used as follows: PPL1-LP (5′-AAGGTTGGTGTGATTCCTTCC-3′), PPL1-RP (5′-ATGTGAGAATTCATGGGGATG-3′), PPL2-LP (5′-GGTGGAGAAGTGTGTAATGGC-3′), PPL2-RP (5′-GGATCGAGAAGACGGGTATTC-3′), and T-DNA LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′). Confirmation of null expression in both mutants was carried out by RT-PCR with the same primer sets used to obtain PPL cDNAs as described below.
Complementation of ppl1 and ppl2
The following primer sets were used to amplify PPL1 and PPL2 cDNA: PPL1 (Fw 5′-CACCATGGCTTCTCTGAAGCTTTCAC-3′ and Rv 5′-TCAAACAGTGATCTTGAAGGAATCT-3′) and PPL2 (Fw 5′-CACCATGGCAGTCTCCTCACTCTCA-3′ and Rv 5′-TCAAATCTGAAGGATCTTCAAAGAGT-3′). The PCR products were cloned into the pENTR TOPO vector (Invitrogen) and transferred from the entry constructs into the pGWB2 vector by a LR recombination reaction (Gateway; Invitrogen). The plasmids pGWB2-PPL1 and pGWB2-PPL2, in which the expression of each cDNA is driven by the 35S promoter, were used to transform homozygous ppl1 and ppl2 mutants via Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998). The seeds were collected and transformants were selected on medium containing Murashige and Skoog salt mix, 40 μg mL−1 hygromycin, and 0.8% agar.
Chl Fluorescence Analysis
Chl fluorescence was measured with a PAM2000 (pulse-amplitude modulation) chl fluorometer (Walz). Minimal fluorescence at open PSII centers in the dark-adapted state (F0) was excited by a weak measuring light (650 nm) at a light intensity of 0.05 to 0.1 μmol photons m−2 s−1. A saturating pulse of white light (800 ms, 3,000 μmol photons m−2 s−1) was applied to determine the maximal fluorescence at closed PSII centers in the dark-adapted state (Fm) and during AL illumination (Fm′). The quantum yield of PSII (ΦPSII) was calculated as (Fm′ − Fs)/Fm′ (Genty et al., 1989) under AL illumination (144 μmol photons m−2 s−1). After illumination with AL for 4 min, the AL was turned off, and the transient increase in chl fluorescence under room light was monitored as an indicator of in vivo NDH activity (Shikanai et al., 1998; Rumeau et al., 2005).
The change in the absorbance of P700 at 810 nm was measured with a PAM101 chl fluorometer with an ED-P700DW-E emitter-detector unit (Walz). The change in absorbance induced by saturating far-red light represents the relative amount of photooxidizable P700.
Oxygen Evolution Measurements
Oxygen evolution from thylakoid membranes was measured at 25°C with a Clark-type oxygen electrode (Hanzatech) under saturating red light with an R-60 red long-pass filter (Kenko). The buffer for measurement was 50 mm HEPES-NaOH, pH 7.6, containing 0.4 m Suc, 10 mm NaCl, 5 mm MgCl2, with an uncoupler (5 mm NH4Cl) and an electron acceptor for PSII (0.5 mm p-phenylbenzoquinone).
SDS-PAGE and Immunoblotting
To isolate thylakoid membranes, leaves were chopped in a blender with ice-cold buffer (50 mm HEPES-NaOH, pH 7.6, containing 0.4 m Suc, 10 mm NaCl, 5 mm MgCl2, and 5 mm sodium ascorbate). The mixture was then filtered and centrifuged and the pellet was washed and resuspended in the same buffer without the sodium ascorbate. The chl content of thylakoid membranes was determined as described in Arnon and Whatley (1949). Proteins corresponding to 5 μg chl were solubilized and separated on 12.5% or 15% SDS-polyacrylamide gels. To analyze D1, CP43, and PsaA/B proteins, 6 m urea was included in the gels. Separated proteins were transferred to a nitrocellulose membrane using a semidry blotting system (Bio-Rad). Detection was performed using ECL (Amersham Biosciences). ECL plus (Amersham Biosciences) was used for NdhH and PPL2 detection.
Specific Antibody Production against PPL1 and PPL2 Proteins
The nucleotide sequences encoding the amino acid residues from 75 to 230 of PPL1 and from 84 to 240 of PPL2 were amplified by RT-PCR using the following primer sets: PPL1 (5′-CATATGGCAGAAAGCAAAAAAGGATTC-3′ and 5′-CTCGAGAACAGTGATCTTGAAGGAATCTAC-3′) and PPL2 (5′-CATATGGAAGAGATACCAAAAAGCTACTCG-3′ and 5′-CTCGAGAATCTGAAGGATCTTCAAAGAGTC-3′). Amplified cDNAs were inserted in frame into the expression vector pET22b (Novagen) such that the His tag was fused at the C-terminal end. The resulting plasmids of PPL1 and PPL2 were transformed into Escherichia coli strain Rosettagami (DE3; Novagen) or BL21 (DE3), respectively. Expression of the PPL1-His or PPL2-His fusion protein was induced by adding 1 mm isopropyl β-d-thiogalactoside for 12 or 4 h, respectively. Cells were harvested and resuspended in 10 mm NaH2PO4, 1.5 mm KH2PO4, pH 8.0, 137 mm NaCl, and 2.7 mm KCl. The bacterial lysate was sonicated 20 times for 1 s. The overexpressed proteins in inclusion bodies were pelleted at 8,000g for 10 min and the pellet was solubilized in 100 mm NaH2PO4, 8 m urea, and 10 mm Tris-HCl, pH 8.0. The PPL1-His or PPL2-His fusion protein was purified on TALON Metal Affinity Resins (CLONTECH) or a nickel nitrilotriacetic acid agarose resin matrix, respectively. Polyclonal antibodies were raised in rabbit with purified antigens. Specificity of anti-PPL1 or anti-PPL2 antibodies was checked by using thylakoids isolated from wild-type and ppl1 or ppl2 plants (Supplemental Fig. S4).
Antisera
Rabbit antibodies against spinach (Spinacia oleracea) PsbP, Arabidopsis PPL1, Arabidopsis PPL2, and tobacco (Nicotiana tabacum) NdhH were produced by the authors. Rabbit antibodies against D1 peptides were produced by Dr. Y. Yamamoto (Okayama University). Rabbit antibodies against spinach PsbO and PsbQ were provided by the late Dr. A. Watanabe (Tokyo University). Rabbit antibody against Chlamydomonas PsaA/B (CP1) was provided by Dr. A. Tanaka (Hokkaido University). Rabbit antibody against spinach CP43 was a gift from Dr. Y. Kashino (Hyogo Prefectural University).
Photoinhibition Treatment
Detached leaves, placing adaxial side up on the filter papers steeped with a sodium phosphate buffer (NaOH/pH7.0), were illuminated at 550 or 1,200 μmol photons m−2 s−1. To examine the role of chloroplast-encoded protein synthesis in the susceptibility of leaves to photoinhibition, detached leaves were incubated with the buffer including 200 μg mL−1 chloramphenicol under reduced pressure for 5 min prior to photoinhibitory light treatment. To analyze the accumulation of D1 proteins under high-intensity light, proteins were extracted from leaves ground in liquid nitrogen and used for SDS-PAGE and subsequent immunoblotting against D1 antibodies.
Recovery Treatment
Saturating white light (3 min, 3,000 μmol photons m−2 s−1) was illuminated five to eight times at 3-min intervals on detached leaves and resulted in about 50% of the initial Fv/Fm values in wild-type and ppl1 leaves. Following that, the changes in Fv/Fm values from wild-type and ppl1 leaves were seen during the recovery period at irradiance of 150 μmol photons m−2 s−1, 30°C.
BN-PAGE
BN-PAGE was performed as described (Schägger et al., 1994; Asakura et al., 2004). The thylakoid samples containing 7.5 μg of chl were solubilized with 1% (w/v) n-dodecyl-β-d-maltoside, separated on a 4% to 14% acrylamide gradient gel, and transferred to a polyvinylidene difluoride membrane using a semidry blotting system (Bio-Rad). For 2D BN-SDS-PAGE analysis, a lane of the BN polyacrylamide gel (4%–12% acrylamide gradient) was excised, denatured, and run in the second dimension in SDS-PAGE with 12.5% acrylamide and 6 m urea. Subsequently, the gel was used for immunoblotting.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_115391 (PPL1) and NM_129505 (PPL2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein phylogenetic trees of the PsbP family in higher plants.
Supplemental Figure S2. Alignment of PPL1 (A) and PPL2 (B) sequences in higher plants.
Supplemental Figure S3. Genetic characterization and phenotype of ppl1 and ppl2 mutant plants.
Supplemental Figure S4. Immunodetection of PPL1 (A) or PPL2 (B) protein in thylakoid membranes isolated from mutant and wild-type plants.
Supplemental Table S1. Genes that coexpressed with PPL1 with a high r value (r ≥ 0.900).
Supplemental Table S2. Genes that coexpressed with PPL2 with a high r value (r ≥ 0.900).
Supplementary Material
Acknowledgments
We thank all of the investigators who kindly provided specific antibodies as listed in “Materials and Methods.” We also thank Dr. T. Nakagawa (Research Institute for Molecular Genetics, Shimane University), who kindly provided us with pGWB2 vectors, and Dr. T. Shikanai (Kyushu University), who kindly provided us with the crr2 mutant.
This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan, for Scientific Research on Priority Areas (grant no. 17051016 to K.I. and F.S.) and for Young Scientists (B; grant no. 18770032 to K.I.), and a Research Grant from Nissan Science Foundation (to K.I.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kentaro Ifuku (ifuku@kais.kyoto-u.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Arnon DI, Whatley FR (1949) Is chloride a coenzyme of photosynthesis? Science 110 554–556 [DOI] [PubMed] [Google Scholar]
- Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, Yano S, Terashima I, Barkan A, Nakai M (2004) Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker TM, Burnap RL (2005) The extrinsic proteins of photosystem II. In T Wydzynsky, K Satoh, eds, Photosystem II: The Light-Driven Water:Plastoquinone Oxidoreductase. Springer, Dordrecht, The Netherlands, pp 95–120
- Brown JR (2003) Ancient horizontal gene transfer. Nat Rev Genet 4 121–132 [DOI] [PubMed] [Google Scholar]
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J 17 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield NJ (2000) Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 263 386–404 [Google Scholar]
- Chen H, Zhang D, Guo D, Wu H, Jin M, Lu Q, Lu C, Zhang L (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61 567–575 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Darie CC, Biniossek ML, Winter V, Mutschler B, Haehnel W (2005) Isolation and structural characterization of the Ndh complex from mesophyll and bundle sheath chloroplasts of Zea mays. FEBS J 272 2705–2716 [DOI] [PubMed] [Google Scholar]
- De Las Rivas J, Balsera M, Barber J (2004) Evolution of oxygenic photosynthesis: genome-wide analysis of the OEC extrinsic proteins. Trends Plant Sci 9 18–25 [DOI] [PubMed] [Google Scholar]
- De Las Rivas J, Roman A (2005) Structure and evolution of the extrinsic proteins that stabilize the oxygen-evolving engine. Photochem Photobiol Sci 4 1003–1010 [DOI] [PubMed] [Google Scholar]
- Debus RJ (1992) The manganese and calcium ions of photosynthetic oxygen evolution. Biochim Biophys Acta 1102 269–352 [DOI] [PubMed] [Google Scholar]
- Enami I, Suzuki T, Tada O, Nakada Y, Nakamura K, Tohri A, Ohta H, Shen J-R (2005) Distribution of the extrinsic proteins as a potential maker for the evolution of photosynthetic oxygen-evolving photosystem II. FEBS J 272 5020–5030 [DOI] [PubMed] [Google Scholar]
- Endo T, Shikanai T, Takabayashi A, Asada K, Sato F (1999) The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett 457 5–8 [DOI] [PubMed] [Google Scholar]
- Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303 1831–1838 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990 87–92 [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36 541–549 [DOI] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc Natl Acad Sci USA 97 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Grunheit N, Ahmadinejad N, Timmis JN, Martin W (2005) Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol 138 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Nakatsu T, Kato H, Sato F (2004) Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep 5 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Yamamoto Y, Ono TA, Ishihara S, Sato F (2005) PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol 139 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Schröder WP, Funk C (2005) Functional analysis of the PsbP-like protein (sll1418) in Synechocystis sp. PCC 6803. Photosynth Res 84 257–262 [DOI] [PubMed] [Google Scholar]
- Kamiya N, Shen JR (2003) Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc Natl Acad Sci USA 100 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofer W, Koop HU, Wanner G, Steinmüller K (1998) Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol Gen Genet 258 166–173 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- Leister D (2003) Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet 19 47–56 [DOI] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438 1040–1044 [DOI] [PubMed] [Google Scholar]
- Montané MH, Kloppstech K (2000) The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene 258 1–8 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429 579–582 [DOI] [PubMed] [Google Scholar]
- Munshi MK, Kobayashi Y, Shikanai T (2005) Chlororespiratory reduction 6 is a novel factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant J 44 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyka MM, Hebeler R, Schlodderc E, Meyerb HE, Warscheidb B, Rögner M (2006) Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35 D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka A, Aro EM, Herrmann RG, Andersson B, Vener AV (2000) Dephosphorylation of photosystem II reaction center proteins in plant photosynthetic membranes as an immediate response to abrupt elevation of temperature. Plant Physiol 123 1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Kashino Y, Pakrasi HB (2007. a) The PsbQ protein defines cyanobacterial photosystem II complexes with highest activity and stability. Proc Natl Acad Sci USA 104 2548–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Wegener KM, Pakrasi HB (2007. b) The extrinsic proteins of photosystem II. Photosynth Res 92 369–387 [DOI] [PubMed] [Google Scholar]
- Rumeau D, Becuwe-Linka N, Beyly A, Louwagie M, Garin J, Peltier G (2005) New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 17 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217 220–230 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schröder WP, Kieselbach T (2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem 277 8354–8365 [DOI] [PubMed] [Google Scholar]
- Seidler A (1996) The extrinsic polypeptides of photosystem II. Biochim Biophys Acta 1277 35–60 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield TC, Winter RT, Eaton-Rye JJ (2005) Investigation of a requirement for the PsbP-like protein in Synechocystis sp. PCC 6803. Photosynth Res 84 263–268 [DOI] [PubMed] [Google Scholar]
- Thornton LE, Ohkawa H, Roose JL, Kashino Y, Keren N, Pakrasi HB (2004) Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. Plant Cell 16 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Hargett SR, Frankel LK, Bricker TM (2006) The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J Biol Chem 281 26260–26267 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.