Abstract
Environmental chemicals are known to induce a high degree of hydroxyl radical-mediated damage in DNA. Accordingly, we tested the hypothesis that this exposure leads to new forms of DNA using principal components analysis of Fourier transform infrared spectra. The hepatic DNA of English sole (controls) from an essentially clean environment was compared with that of sole inhabiting a chemically contaminated environment. All livers studied were cancer-free; however, a high incidence of liver cancer has been found in the exposed population. The exposed sole were sampled twice, 2 years apart, while the sediments in which they live were under remediation. After obtaining infrared spectra, the first three principal components (PC1, PC2, and PC3) were calculated and found to represent 97% of the total spectral variance. When the principal component scores were plotted in 3-dimensional space, clusters of points were obtained that represented the DNA from the control and exposed groups. Each of the points was derived from ≈106 wavenumber–absorbance correlations. The spatial location of a point was a highly discriminating measure of DNA structure. The clusters of points were completely separated, demonstrating that the three groups could be 100% correctly classified. The points from the control group were tightly clustered whereas those from the exposed groups were highly diverse. The findings demonstrate that exposure to environmental chemicals results in new, structurally diverse forms of DNA that likely play an important role in carcinogenesis.
Keywords: free radicals, hydroxyl radical, cancer etiology, Fourier transform infrared spectroscopy, principal components analysis
English sole exposed to environmental chemicals in their natural habitat (Duwamish River, Seattle, WA) have been shown to have a remarkably high degree of base damage resulting from the attack of the hydroxyl radical (·OH) on DNA (1). The base damage included substantial increases in mutagenic hydroxy derivatives [8-hydroxyadenine (8-OH-Ade) and 8-hydroxyguanine (8-OH-Gua)] (2, 3) and ring-opened products [e.g., 2,6-diamino-4-hydroxy-5-formamidopyrimidine] compared with controls. In another study of English sole from the Duwamish River (4), positive correlations were found between 8-OH-Ade and 8-OH-Gua and incidences of a variety of nonneoplastic hepatic lesions. These included basophilic foci (a putatively preneoplastic lesion) and megalocytic hepatosis (a putatively degenerative or cytotoxic lesion) (5, 6). The ·OH was likely a common etiological factor in the formation of the base modifications and the hepatic lesions, which is consistent with its proposed role in tumor formation (7–11). The sole population from the Duwamish River was shown to have a high incidence of liver cancer; however, this condition rarely occurs in fish from uncontaminated environments (5, 6).
The ·OH is believed to arise from H2O2 via the Fe2+-mediated Fenton reaction (12). H2O2, which readily crosses the nuclear membrane, is likely produced via redox cycling of sediment contaminants, such as polynuclear aromatic hydrocarbons and chlorinated pesticides, to which the fish are exposed in their contaminated habitat (13).
Substantial modifications of the phosphodiester–deoxyribose structure accompany the formation of the mutagenic base changes in the liver of fish from the Duwamish River, as demonstrated by Fourier transform infrared (FT-IR) spectral models of DNA (1). This is consistent with the fact that the ·OH is capable of abstracting hydrogens from the furanose moiety and introducing hydroxyl groups at various positions on the ring structure. Ultimately, the radical attack inflicts substantial damage to the phosphodiester–deoxyribose structure, which leads to the loss of phosphoric acid and strand breaks (14).
Recently, principal components analysis (PCA) of FT-IR spectra provided a unique perspective of structural changes in DNA associated with human breast (9, 15) and prostate (16) cancer. Wavenumber–absorbance relationships were expressed as points in 2- or 3-dimensional PC plots, each point representing a highly discriminating measure of DNA structure. Structural alterations, such as those introduced by the ·OH, can alter vibrational and rotational motion, thus changing the spatial location of the points. The breast (9, 15) and prostate (16) studies showed that clusters of points representing DNA from normal and cancer tissues could be separated based on PC scores with a sensitivity and specificity of 83% and 100%, respectively. This implies that substantial and specific structural alterations lead to the development of new forms of DNA in the progression of normal cells to cancer cells.
PCA/FT-IR technology has now been applied to the analysis of DNA from English sole inhabiting the Duwamish River and from sole inhabiting an essentially clean control environment (Quartermaster Harbor, WA). The purpose was to test the hypothesis that new forms of DNA are created as a consequence of exposure to environmental chemicals. FT-IR spectral models were used to identify structural changes in DNA reflected in the height, width, and location of absorbance bands representing various DNA functional groups, such as NH2, PO2−, and CO (17, 18). PCA of individual spectra yielded plots in which ≈106 wavenumber–absorbance correlations were represented by a single point in 3-dimensional space. A plot of PC scores showed that clusters of points representing DNAs from the control and exposed fish were separated without any overlap. Also, there was a significant increase in structural diversity between the DNAs of the control and exposed groups.
MATERIALS AND METHODS
English sole were obtained from a relatively clean rural environment, Quartermaster Harbor, WA (19), and a chemically contaminated urban environment, Duwamish River, Seattle, WA (13). All livers were examined histologically (5, 6) and were found to be cancer-free; however, those from the Duwamish River contained various nonneoplastic lesions (5) characteristic of fish from contaminated environments (6, 13).
The Duwamish River flows into Puget Sound through a heavily industrialized area. The sediments contain a variety of carcinogens and other xenobiotics, such as polynuclear aromatic hydrocarbons and chlorinated pesticide residues (13); however, a restoration program (initiated in 1991) is in progress to reduce the sediment contamination (20).
Two groups of sole were obtained from the Duwamish River, one in October, 1993 (DUW93; n = 8) and the other in October, 1995 (DUW95; n = 10). Because of the restoration program, the DUW95 samples were expected to reflect significantly less sediment contamination than the DUW93 samples but greater contamination than the Quartermaster Harbor control samples (QMH; n = 7). These were acquired in October, 1995. The lengths ± SD of the QMH, DUW95, and DUW93 fish were 29.5 ± 4.2 cm, 23.6 ± 1.6 cm, and 24.1 ± 0.8 cm, respectively. The weights were 254.3 ± 115.0 g, 125.6 ± 16.2 g, and 125.0 ± 22.5 g. Positive correlations between 8-OH-Ade and 8-OH-Gua and the incidences of five nonneoplastic hepatic lesions were reported previously for the three groups of fish used in this study (4).
Isolation of DNA from hepatic tissue and PCA analyses of FT-IR spectra were undertaken as described (8, 9). Each FT-IR spectrum was normalized over the range 1750–700 cm−1. PCA was used to identify a few variables (components) that capture most of the information in the original, long list of variables (the spectral absorbancies at each of the 1051 wavenumbers from 1750–700 cm−1). PC scores were calculated with the grand mean of all spectra subtracted from each spectrum. Thus, the PC scores represent variations in spectral (structural) features as they differ from the grand mean spectrum. The Kruskal–Wallis and Mann–Whitney tests were used to calculate the statistical significance of differences in PC scores between groups. The same procedures were used to test for differences in spectral diversity, which was defined for a group as the mean distance of spectra to the group centroid. The unequal variance t test was used to compare the mean normalized absorbance between groups. The t test was carried out at each of the 1051 wavenumbers from 1750–700 cm−1. Fish age, reflected in length and mass, was a potentially confounding variable, and this possibility was addressed in the analysis.
RESULTS
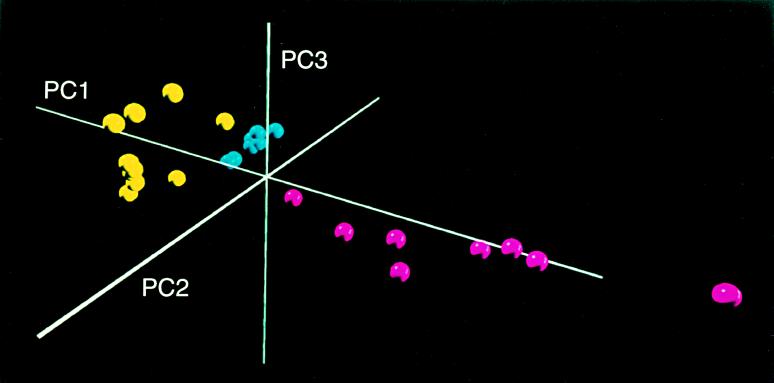
The clusters of points derived from the first three PC scores, which summarized spectral features of the DNA from the QMH and DUW groups, are shown in a 3-dimensional projection (Fig. 1). The hypothesis that all groups have the same mean values of PC scores (thus, similar spectra) is rejected (Kruskal–Wallis P value < 0.001), and the hypothesis that any two of the groups have the same mean values of PC scores also is rejected (Mann–Whitney P value 0.04 to <0.001). The three groups are distinct without any overlap (Fig. 1). PC1 and PC2, combined, account for 94% of the spectral variation and thus provide a good means for representing the variety of spectra encountered. PC3 is used for display purposes (Fig. 1) although it explains only 3% of the spectral variation.
Figure 1.
Three-dimensional PC plot representing completely separated clusters of DNA structures. The DNA forms of the QMH (blue), DUW95 (yellow), and DUW93 (maroon) groups are shown, illustrating substantial differences in cluster diversity between groups.
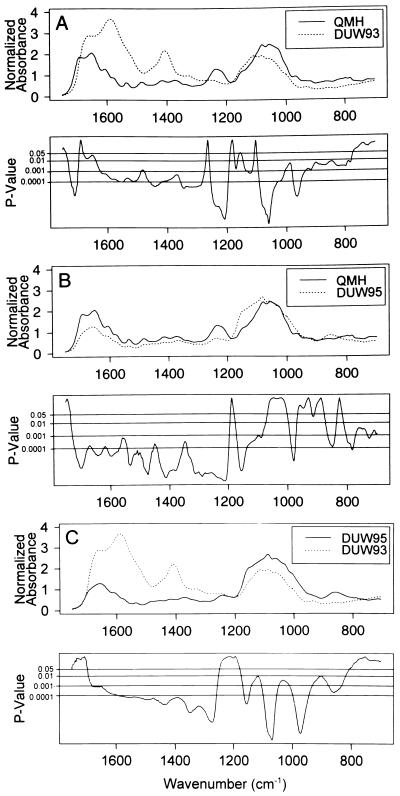
The differences between groups occur at many frequencies. The upper part of each panel in Fig. 2 shows the mean spectrum for each of two groups (QMH/DUW93; QMH/DUW95; and DUW95/DUW93). The bottom part of each panel shows P values for each spectral comparison, one P value per wavenumber. The comparisons yield P < 0.05 at 78–87% of the 1051 wavenumbers, thus demonstrating that the structures of the DNAs from the DUW93 and DUW95 groups are markedly different from each other and the QMH group. Accordingly, the findings substantially invalidate the null hypothesis that the mean, normalized spectra are equal between groups. The spectral differences are notable with respect to the antisymmetric stretching vibrations of the PO2− structure (≈1240 cm−1) (17, 18). The band at this spectral region is present in the QMH group, as in the spectra of other vertebrates (17, 18), but is virtually lost in the spectra of the DUW93 and DUW95 groups. Other major differences are evident in spectral regions representing vibrations associated with the nucleic acids (≈1700–1300 cm−1) and deoxyribose (≈1150–950 cm−1).
Figure 2.
The mean, normalized spectra of DNA from the QMH, DUW95, and DUW93 groups (top panels). P values for spectral difference, occurring over wide regions of the spectra, are given for each wavenumber (lower panels).
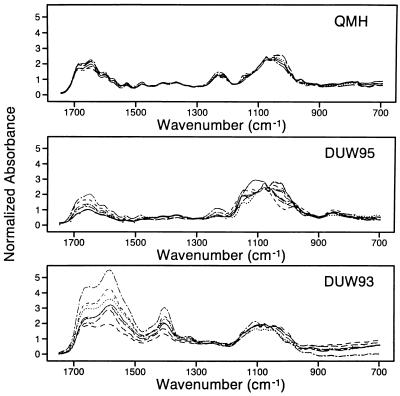
It is obvious (Fig. 1) that the samples can be 100% correctly classified into groups (separated) on the basis of the PC scores (Table 1). Fig. 1 shows that the diversity of spectra (note the spread of points) is substantially greater in the DUW93 and DUW95 groups compared with the QMH group. The varying diversity between the groups and the spectral differences that separate them are also evident in Fig. 3, in which the individual spectra of each group are overlaid. The tightness of the QMH spectra and the increasing spectral diversity from the QMH to the DUW groups is notable in the region ≈1700–1450 cm−1, which includes strong C-O stretching and NH2 bending vibrations of the nucleic acids. Also in the DUW93 group, compared with the other groups, there is a pronounced increase in absorbance and spectral diversity in the ≈1450–1300 cm−1 region assigned to weak NH vibrations and CH in-plane deformations of the nucleic acids (17, 18). The region ≈1150–950 cm−1, which includes strong stretching vibrations associated with deoxyribose, increases in spectral diversity from QMH to DUW95 but tightens in the DUW93 group. The differences between the spectral properties are consistent with the discrimination between groups shown in Table 1 and the increased diversity of the clusters illustrated in Fig. 1.
Table 1.
Principal component scores by group and statistical significance of differences between groups
| QMH (n = 7) | DUW95 (n = 10) | DUW93 (n = 8) | KW P value for overall differences | MW P value for QMH vs. DUW93 | MW P value for QMH vs. DUW95 | MW P value for DUW93 vs. DUW95 | |
|---|---|---|---|---|---|---|---|
| PC1 | −6.1 ± 1.4 | −12.8 ± 2.8 | 21.3 ± 12.3 | <0.001 | <0.001 | <0.001 | <0.001 |
| PC2 | 6.1 ± 1.3 | −3.3 ± 2.6 | −1.3 ± 1.4 | <0.001 | <0.001 | <0.001 | 0.04 |
Scores are mean ± SD. KW, Kruskal–Wallis; MW, Mann–Whitney.
Figure 3.
Individual spectra are overlaid showing pronounced differences in structural diversity between groups, such as in regions of the spectrum assigned to nucleic acid (≈1700–1300 cm−1) and deoxyribose (1150–950 cm−1) vibrations. Also, note the substantial reduction in absorbance at ≈1240 cm−1 in the DUW95 group assigned to antisymmetric stretching vibrations of the PO2− structure and the virtual disappearance of this band in the DUW93 group (17, 18).
A formal test for diversity differences (Kruskal–Wallis test for the null hypothesis that all groups have the same mean distance to the group centroid) yields P = 0.002, strongly suggesting unequal diversity among groups. These mean distances to the centroid provide a scale for measuring diversity. A larger mean distance indicates that a group is more spread out (Fig. 1); that is, the spectra are more diverse. The DUW95 group has a mean distance that is four times that of the QMH group, representing a 4-fold greater diversity (Table 2). Two of the three pairwise comparisons of diversity were significant (P < 0.05); however, the comparison between the DUW95 and DUW93 groups was not significant (Mann–Whitney P value = 0.2) although the DUW93 group (representing DNA with the most altered base structure) (1, 4) is more diverse than the DUW95 group.
Table 2.
Spectral diversity for three groups
| Group | Distance to group centroid (diversity), mean ± SD | n |
|---|---|---|
| QMH | 2.5 ± 1.1 | 7 |
| DUW95 | 5.8 ± 2.1 | 10 |
| DUW93 | 10.2 ± 7.2 | 8 |
P values for null hypotheses: (i) all three groups have the same mean diversity, Kruskal–Wallis P value = 0.002; (ii) mean QMH = mean DUW95, Mann–Whitney P value = 0.003; (iii) mean QMH = mean DUW93, Mann–Whitney P value = 0.001; and (iv) mean DUW93 = mean DUW95, Mann–Whitney P value = 0.2.
It is unlikely that the varying diversities of the groups are due to age variables. The QMH group is the most diverse in length and mass, yet it shows the least spectral diversity. The QMH group shows a length SD that is two to five times larger than that of the DUW95 and DUW93 groups and a mass SD that is five to seven times larger. However, the mean distance of the QMH spectra to their centroid is 2- to 4-fold smaller than that of the DUW groups. These results would be highly inconsistent if age were a significant factor in spectral diversity. Length and mass also appear to have little effect in creating the spectral differences by location (Fig. 1). In regression analysis, length and mass combined explained only 7% of the variation in PC1 and 40% of the variation in PC2. PC1 is by far the more important component in explaining spectral diversity. Length and mass explain only ≈9% of the overall spectral variation whereas location explains 77%.
DISCUSSION
The DNA structures isolated from the QMH, DUW95, and DUW93 fish were each unique in that the PC plot revealed a complete separation of clusters (Fig. 1). In addition, the DNAs from the exposed groups were substantially more diverse than those of the control group, and the DUW93 group was more diverse than the DUW95 group (Table 2; Fig. 1). These distinctions, which were not significantly age-related, likely arose from structural features induced in DNA by different environmental factors. Among the environmental factors likely contributing to the cluster separations and the differences in diversity are the type, degree, and duration of exposure to toxic chemicals in the sediments. Striking differences occurred between the three groups in regions of the spectra assigned to the nucleic acids and the phosphodiester–deoxyribose structure (Figs. 2 and 3), suggesting that alterations in these structures contributed substantially to the separation of clusters and the differences in diversity among groups.
In a previous study (1), a remarkably high degree of ·OH-induced base modification occurred in the DNA from the DUW93 group compared with that from a control group. GC–MS analyses of several modified bases (8-OH-Ade, 8-OH-Gua, 2,6-diamino-4-hydroxy-5-formamidopyrimidine, and 4,6-diamino-5-formamidopyrimidine) (1) showed that the mean values for the control group were <0.20 nmol/mg DNA whereas comparable values for the exposed group ranged from 0.77 nmol/mg for 8-OH-Ade to 39.2 nmol/mg for 2,6-diamino-4-hydroxy-5-formamidopyrimidine. Moreover, ≈40% of the wavenumbers in the infrared region studied (1500–700 cm−1) were significantly different at P ≤ 0.05, further indicating that substantial damage to DNA had occurred.
A comparison also has been made (4) between 8-OH-Ade and 8-OH-Gua concentrations in the DNA of the three fish groups used in the present study. The values for the QMH, DUW95, and DUW93 groups increased progressively and were statistically different using the GC-MS rank model that combined both base modifications (P = 0.004).
Additional evidence that supports the role of the ·OH in altering DNA is the statistical difference obtained in spectral regions assigned to the phosphodiester–deoxyribose moiety (Fig. 2). For example, the abstraction of hydrogens from the furanose ring by the ·OH (14) would be expected to significantly alter wavenumber–absorbance relationships in the 1150- to 950-cm−1 region, as found in the present work, leading to point shifts in PC plots (Figs. 1 and 2). Moreover, FT-IR spectral analysis of calf thymus and normal breast DNA exposed for various periods to ·OH-generating systems (Fe2+/H2O2) showed pronounced alterations in regions of the spectrum assigned to vibrations of the deoxyribose moiety (D.C.M, S.J.G., and J. Cramer, unpublished results).
A number of factors may contribute to alterations in the DNA structure. For example, exposure of normal breast cells to polynuclear aromatic hydrocarbons results in depurinating adducts and mutagenic changes (21). In addition, altered patterns of cytosine methylation are associated with free radical injury to DNA, notably in the formation of 8-OH-Gua (22).
There was a statistically significant increase in the diversity of clusters representing the two DUW groups compared with the tight cluster of the reference group (Fig. 1; Table 2). In addition, the DUW93 group that had the greatest diversity also had the highest degree of base modification (1), thus supporting the concept that increased DNA damage is associated with increased diversity. Increased diversity may be especially important in carcinogenesis in that it sets the stage for the selection of DNA forms that give rise to malignant cellular phenotypes, as suggested (9, 15). The high degree of diversity in the exposed fish groups may serve the same function.
Cluster separation in PC plots was found in studies of breast (9, 15) and prostate (16) cancer. With the prostate, for example, virtually perfect discrimination was achieved between DNA from normal and adenomacarcinoma tissues. Perfect discrimination was obtained between clusters in the present work, thus demonstrating that the DNA structures had unique properties representing new forms of DNA. Considering that fish in the Duwamish River are prone to liver tumors (5), the distinctly different forms of DNA found in the DUW95 and DUW93 groups likely constitute critical stages in the progression to cancer.
The PCA/FT-IR spectral data support our previous findings (4) showing that the DNA from the three groups can be readily distinguished on the basis of mutagenic ·OH-induced changes in adenine and guanine; that is, concentrations of 8-OH-Ade and 8-OH-Gua increased among the QMH, DUW95, and DUW93 groups. This is consistent with the concept that the ·OH, which likely arises from H2O2 (12) derived via redox cycling of sediment chemicals (23), plays a major role in altering base structures and contributes to cluster discrimination and diversity.
In conclusion, this study has shown that damage to the DNA of English sole exposed to environmental chemicals (1) leads to new, diverse forms of DNA. These new forms may play a pivotal role in carcinogenesis and ultimately contribute to the development of liver cancer in the fish population. In addition, the findings raise the question whether environmental chemicals play a role in generating the new forms of DNA recently found in breast (9, 15) and prostate (16) cancers.
Acknowledgments
We thank Derek Stanford for computing, David Fraser for computing and graphics, and Heather McSpadden for technical support. Helpful comments on the manuscript were provided by Drs. David L. Eaton, Mortimer M. Elkind, Daniel L. Gustafson, Anil Singhal, and Charles Waldren. This work was supported by grant ES04696 from the National Institutes of Health Sciences as a subcontract from the University of Washington Superfund Basic Research Program.
ABBREVIATIONS
- PC
principal component
- PCA
principal components analysis
- FT-IR
Fourier transform infrared
- ·OH
hydroxyl radical
- 8-OH-Ade
8-hydroxyadenine
- 8-OH-Gua
8-hydroxyguanine
- DUW
Duwamish River
- QMH
Quartermaster Harbor
References
- 1.Malins D C, Gunselman S J. Proc Natl Acad Sci USA. 1994;91:13038–13041. doi: 10.1073/pnas.91.26.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamiya H, Miura H, Murata-Kamiya N, Ishikawa H, Sakaguchi T, Inoue H, Sasaki T, Masutini C, Hanaoka F, Nishimura S, Ohtsuka E. Nucleic Acids Res. 1995;23:2893–2899. doi: 10.1093/nar/23.15.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Nature (London) 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 4.Malins D C, Polissar N L, Garner M M, Gunselman S J. Cancer Res. 1996;56:5563–5565. [PubMed] [Google Scholar]
- 5.Myers M S, Rhodes L D, McCain B B. J Natl Cancer Inst. 1987;78:333–363. [PubMed] [Google Scholar]
- 6.Moore M J, Myers M S. In: Aquatic Toxicology: Molecular, Biochemical and Cellular Perspectives. Malins D C, Ostrander G K, editors. Boca Raton, FL: Lewis; 1994. pp. 327–386. [Google Scholar]
- 7.Floyd R A. Carcinogenesis. 1990;11:1447–1450. doi: 10.1093/carcin/11.9.1447. [DOI] [PubMed] [Google Scholar]
- 8.Malins D C, Holmes E H, Polissar N L, Gunselman S J. Cancer. 1993;71:3036–3043. doi: 10.1002/1097-0142(19930515)71:10<3036::aid-cncr2820711025>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 9.Malins D C, Polissar N L, Gunselman S J. Proc Natl Acad Sci USA. 1996;93:2557–2563. doi: 10.1073/pnas.93.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng K C, Cahill D S, Kasai H, Nishimura S, Loeb L A. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 11.Musarrat J, Arezina-Wilson J, Wani A A. Eur J Cancer. 1996;32A:1209–1214. doi: 10.1016/0959-8049(96)00031-7. [DOI] [PubMed] [Google Scholar]
- 12.Imlay J A, Chin S M, Linn S. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 13.Malins D C, McCain B B, Brown D W, Chan S-L, Myers M S, Landahl J T, Prohaska P G, Friedman A J, Rhodes L D, Burrows D G, Gronlund W D, Hodgins H O. Environ Sci Technol. 1984;18:705–713. [Google Scholar]
- 14.Von Sonntag C, Hagen U, Schön-Bopp A, Schulte-Frohlinde D. Adv Radiat Biol. 1981;9:110–142. [Google Scholar]
- 15.Malins D C, Polissar N L, Gunselman S J. Proc Natl Acad Sci USA. 1996;93:14047–14052. doi: 10.1073/pnas.93.24.14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malins D C, Polissar N L, Gunselman S J. Proc Natl Acad Sci USA. 1997;94:259–264. doi: 10.1073/pnas.94.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutherland G B B M, Tsuboi M. Proc R Soc Ser A. 1957;239:446–463. [Google Scholar]
- 18.Parker F S. Applications of Infrared, Raman, and Resonance Raman Spectroscopy in Biochemistry. New York: Plenum; 1983. pp. 349–398. [Google Scholar]
- 19.Washington State Department of Ecology. Sediment Analysis Report. Olympia, WA: Washington State Department of Ecology; 1994. [Google Scholar]
- 20.King County Department of Natural Resources. Duwamish/Diagonal Phase II: Sampling and Analysis Plan. WA: King County Department of Natural Resources; 1996. [Google Scholar]
- 21.Chakravarti D, Pelling J C, Cavalieri E L, Rogan E G. Proc Natl Acad Sci USA. 1995;92:10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weitzman S A, Turk P W, Milkowski D H, Kozlowski K. Proc Natl Acad Sci USA. 1994;91:1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malins D C, Haimanot R. Aquat Toxicol. 1991;20:123–130. [Google Scholar]