Abstract
P1B-type heavy-metal ATPases (HMAs) are transmembrane metal-transporting proteins that play a key role in metal homeostasis. Despite their importance, very little is known about their functions in monocot species. We report the characterization of rice (Oryza sativa) OsHMA9, a member of the P1B-type ATPase family. Semiquantitative reverse transcription-polymerase chain reaction analyses of seedlings showed that OsHMA9 expression was induced by a high concentration of copper (Cu), zinc (Zn), and cadmium. We also determined, through promoter∷β-glucuronidase analysis, that the main expression was in the vascular bundles and anthers. The OsHMA9:green fluorescence protein fusion was localized to the plasma membrane. Heterologous expression of OsHMA9 partially rescued the Cu sensitivity of the Escherichia coli copA mutant, which is defective in Cu-transporting ATPases. It did not rescue the Zn sensitivity of the zntA mutant, which is defective in Zn-transporting ATPase. To further elucidate the functional roles of OsHMA9, we isolated two independent null alleles, oshma9-1 and oshma9-2, from the T-DNA insertion population. Mutant plants exhibited the phenotype of increased sensitivity to elevated levels of Cu, Zn, and lead. These results support a role for OsHMA9 in Cu, Zn, and lead efflux from the cells. This article is the first report on the functional characterization of a P1B-type metal efflux transporter in monocots.
A number of heavy metals, including copper (Cu), zinc (Zn), manganese, and iron, are essential micronutrients for a wide variety of physical processes. These micronutrients can serve structural roles in proteins, act as enzyme cofactors, and function in cellular redox reactions (Williams and Mills, 2005). However, when present in excess, they can have deleterious effects because of their reactive nature (Schutzendubel and Polle, 2002). For example, Cu is an essential element for plant growth and is important in various biochemical reactions but, at toxic levels, it interferes with numerous physiological processes (Fernandes and Henriques, 1991). Likewise, Zn acts as a nutrient, but can be very toxic at higher concentrations (Rout and Das, 2003). Furthermore, some metals, such as cadmium (Cd), mercury, silver, and lead (Pb), are generally considered nonessential to plants and are potentially highly toxic because of their reactivity with sulfur and nitrogen in amino acid side chains (Clemens, 2001). Accumulations of these heavy metals in plants can occur following their uptake from contaminated soil, which can then lead to toxic levels in animals feeding on them (Mills et al., 2005). To maintain the concentration of essential metals within physiological limits and to minimize the detrimental effects of nonessential metals, plants, like other organisms, trigger a complex network of homeostatic mechanisms to control their uptake, accumulation, trafficking, and detoxification (Clemens, 2001). Specialized transport proteins, in the form of channels, carriers, or pumps, mediate the movement of heavy metals through membranes (Williams et al., 2000). Several types of heavy-metal transporters have now been cloned from plants (Williams et al., 2000; Clemens, 2001).
The ion pumps in the P-type ATPase superfamily share a common enzymatic mechanism in which ATP hydrolysis aids in transporting ions across the membrane (Pedersen and Carafoli, 1987). P-type ATPases, found in all types of living organisms, are used to translocate a diverse set of ions, including H+, Na+/K+, H+/K+, and Ca2+, plus heavy metals and possibly lipids (Axelsen and Palmgren, 1998; Kuhlbrandt, 2004). This superfamily is divided into five major branches and 10 subfamilies, according to the substrate being transported. All the heavy-metal pumps from bacteria, plants, and humans share significant sequence similarities and are clustered together as the P1B subfamily (Axelsen and Palmgren, 1998). P1B-type heavy-metal ATPases (HMAs) have been implicated in the transport of a range of essential as well as potentially toxic metals across cell membranes. This HMA group can be subdivided into two distinct clusters through phylogenetic analyses (Rensing et al., 1999). The Cu cluster has members with a role in Cu and silver transport, whereas the Zn cluster proteins transport Zn and other heavy metals (e.g. cobalt, Cd, and Pb; Axelsen and Palmgren, 2001).
Of the eight P1B-ATPases in Arabidopsis (Arabidopsis thaliana), four belong to the Cu cluster and four to the Zn cluster (Baxter et al., 2003). AtHMA6/PAA1, the first HMA cloned from Arabidopsis (Tabata et al., 1997), plays a critical role in the Cu transport system in chloroplasts, being responsible for cofactor delivery to stomatal Cu/Zn superoxide dismutase (Shikanai et al., 2003). AtHMA8/PAA2, closely related to AtHAM6, transports Cu into the thylakoid lumen to supply plastocyanin (Abdel-Ghany et al., 2005). AtHMA5 is strongly and specifically induced by Cu in whole plants and T-DNA insertion alleles of AtHMA5 are hypersensitive to Cu, suggesting that AtHMA5 plays a key role in transmembrane transport while also interacting with plant metallochaperones (Andres-Colas et al., 2006). AtHMA7/RAN1 may be important in the delivery of Cu ions to ethylene receptors (Hirayama et al., 1999). Complementation of the yeast (Saccharomyces cerevisiae) ccc2 mutant by AtHMA7 confirms its function as a Cu transporter. Although AtHMA1 phylogenetically falls in the Zn cluster, yeast expression experiments demonstrated that AtHMA1 is involved in Cu homeostasis (Seigneurin-Berny et al., 2006). Proteomic analyses of the Arabidopsis chloroplast envelope identified AtHMA1 as one of the candidates for metal transporters. Characterization of Arabidopsis hma1 mutants revealed lower Cu content in chloroplasts and diminution of the total chloroplast superoxide dismutase activity. ATPase activity of AtHMA1 in purified chloroplast envelope membranes was specifically stimulated by Cu, demonstrating that the protein is an envelope ATPase, delivering Cu ions to the stroma (Seigneurin-Berny et al., 2006).
The Zn cluster P1B-ATPases play a role in metal detoxification. The increase in Zn2+ and Cd2+ levels in hma2 plants indicates that AtHMA2 drives the efflux of Zn2+ from the plant cells and also controls the levels of nonphysiological heavy metals, such as Cd2+ (Eren and Arguello, 2004). Disruption of AtHMA4 function resulted in increased sensitivity to elevated levels of Cd and Zn (Verret et al., 2004; Mills et al., 2005). An hma2 hma4 double mutant showed a chlorotic, stunted phenotype that can be rescued by exogenous Zn application, indicating that the primary role of these transporters is likely to be in the translocation of Zn (Hussain et al., 2004). AtHMA3 functions as a Cd/Pb transporter in yeast, whereas the AtHMA3∷GFP is localized to the vacuole, suggesting a role in the influx of Cd into the vacuolar compartment (Gravot et al., 2004). Unfortunately, most of the knowledge concerning plant P1B-type ATPases has been acquired from dicot species (Colangelo and Guerinot, 2006; Grotz and Guerinot, 2006; Broadley et al., 2007; Krämer et al., 2007). Therefore, the functional characterization of P1B-type HMA from monocot species is important to determine whether those same crucial roles are found in dicot species.
Rice (Oryza sativa) is an important food source for half the world's population. It is the first crop species to be sequenced and, therefore, has great impact in agricultural research (Goff et al., 2002; Yu et al., 2002; International Rice Genome Sequencing Project, 2005). Analysis of its genome sequences have resulted in the identification of nine P1B-type HMA genes (Williams and Mills, 2005). By contrast, 10 members of P1B-type HMAs have been reported in barley (Hordeum vulgare; Williams and Mills, 2005). In this study, we characterized a rice P1B-type ATPase, OsHMA9, through molecular and genetic approaches. We have previously reported the establishment of T-DNA insertional mutant lines in japonica rice (Jeon et al., 2000; Jeong et al., 2002; Ryu et al., 2004). To facilitate the use of those lines, we have also determined the sites where T-DNA is inserted and have established a database of the insertion sites (An et al., 2003; Jeong et al., 2006). From that mutant population, we have now obtained two independent T-DNA insertions in the OsHMA9 gene. Using knockout plants, we have investigated here the role of OsHMA9 for heavy-metal transport in rice.
RESULTS
Expression Analysis of Rice P1B-Type ATPase Genes
A database search for rice protein sequences belonging to the P1B-type ATPases identified a family of nine proteins (Baxter et al., 2003; Williams and Mills, 2005). Phylogenetic analysis showed that OsHMA1 through OsHMA3 belong to the Zn cluster, whereas OsHMA4 through OsHMA9 are part of the Cu cluster. Several of their ESTs were present in cDNA libraries prepared from panicles, seedling roots, or green shoots, indicating that these genes are functional in various organs. We investigated the expression patterns of the Cu subgroup (OsHMA4–OsHMA9) using semiquantitative reverse transcription (RT)-PCR with gene-specific primers located at the 3′ end of each gene, which does not cover any conserved sequences (Fig. 1). Because introns occurred between the primers, genomic DNA contamination could be distinguishable from the cDNA (i.e. PCR bands longer than the expected cDNA size were likely from the genomic DNA). We used short PCR cycles to avoid saturation and the products were hybridized with each OsHMA probe.
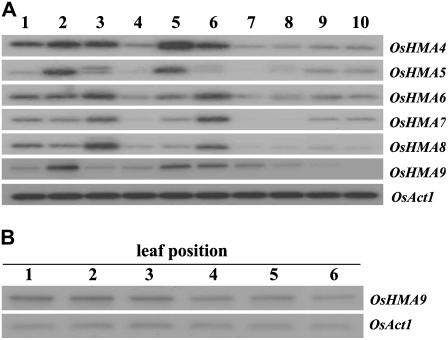
Figure 1.
Expression analysis of OsHMA4-9. A, Spatial expression patterns of genes OsHMA4 through OsHMA9. Semiquantitative RT-PCR was performed with cDNA prepared from various vegetative and reproductive organs: 1 and 2, Shoots and roots of 7-d-old seedlings; 3, 4, and 5, leaves, stems, and roots of 30-d-old plants; 6, fully expanded mature leaves at flowering stage; 7 and 8, panicles about 5 and 15 cm long; 9 and 10, immature seeds 3 and 9 d after pollination. OsAct1 is shown as internal control. B, Expression of OsHMA9 according to leaf position in 60-d-old plants. Positions are indicated by order of appearance. Experiments were performed at least twice or more when results were not consistent. The most representative results are shown.
During vegetative growth, OsHMA4, OsHMA5, and OsHMA9 were more strongly expressed in the roots of 30-d-old seedlings than in the leaves, whereas OsHMA6, OsHMA7, and OsHMA8 were more strongly expressed in the leaves (Fig. 1A). The OsHMA9 mRNA level was higher in fully expanded mature leaves at the flowering stage compared with young leaves, suggesting that expression is increased as leaves senesce (Fig. 1A). To further investigate this possibility, we examined 60-d-old plants producing six leaves from the main shoot. Our analysis showed that the OsHMA9 transcript level was higher in older leaves, implying that OsHMA9 might function in metal mobilization as those tissues mature. In Arabidopsis, AtHMA7/RAN1 also is induced during leaf senescence, contributing to nutrient mobilization (Himelblau and Amasino, 2001). All of the genes were weakly expressed in the stems and were also more weakly expressed in the reproductive organs than in the vegetative tissues (Fig. 1A). Expression of OsHMA4, OsHMA6, and OsHMA8 were relatively constant throughout the various stages of panicle and seed development. However, expression levels of OsHMA5 and OsHMA7 were stronger in the seeds, whereas that of OsHMA9 decreased in older panicles and seeds.
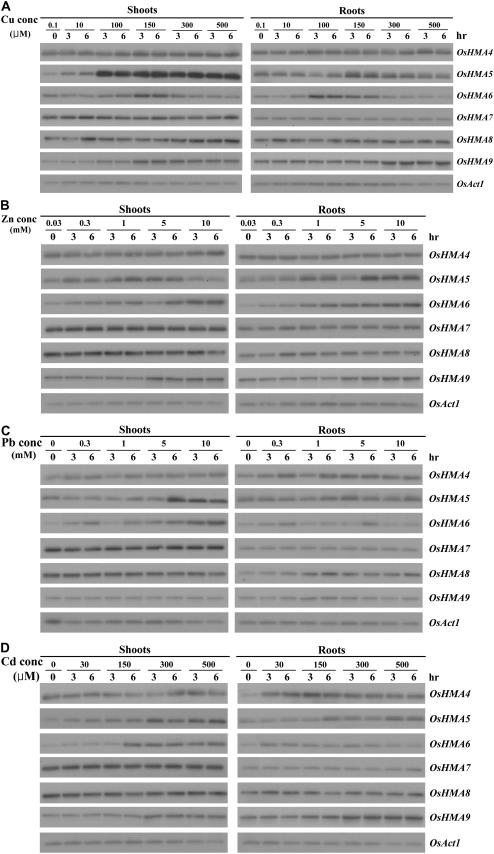
To evaluate the relationships between OsHMA genes and heavy metals, we performed dose-response experiments. Seven-day-old seedlings grown on Murashige and Skoog medium (containing 0.1 μm Cu and 30 μm Zn) were transferred to liquid medium supplemented with different concentrations of Cu, Zn, Pb, or Cd. Samples were harvested 3 or 6 h after this treatment (Fig. 2). Afterward, expression of OsHMA4, OsHMA7, and OsHMA8 was only slightly, but not significantly, enhanced under these conditions. In contrast, expression of OsHMA5 and OsHMA6 was induced by such treatment. Finally, in the case of OsHMA9, high concentrations of Cu, Zn, or Cd increased transcript levels, but Pb had no effect. Therefore, we might suggest that at least three OsHMAs (OsHMA5, OsHMA6, and OsHMA9) play roles in heavy-metal detoxification. Among those three, we selected OsHMA9 for further analysis.
Figure 2.
Dose-response analyses for expression of OsHMA4-9. Seven-day-old plants were treated for 3 or 6 h in liquid medium containing different concentrations of Cu, Zn, Pb, or Cd. Representative results from at least two replicates are shown for each section. In each experiment, three plants were used for RNA extraction of each indicated metal concentration and time.
Expression Analysis of OsHMA9 Using the GUS Reporter Gene
To study the spatial and temporal expression patterns of OsHMA9 in planta, we fused the 0.9-kb fragment carrying the OsHMA9 promoter to the GUS gene. Transgenic rice plants expressing that construct were generated by Agrobacterium-mediated cocultivation. Of the eight independent transformants examined here, seven manifested a consistent staining pattern. Histochemical GUS staining of 7-d-old seedlings showed that overall staining was much stronger after treatment for 6 h with 500 μm Cu (Fig. 3A), which coincided with RT-PCR results that demonstrated that the OsHMA9 gene was inducible by a high Cu level. When seedlings were treated for 6 h with 10 mm Zn or 500 μm Cd, GUS activity was increased, compared with the untreated plants (Supplemental Fig. S1). In cross sections of the seminal roots, strong GUS activity was observed in the vascular cylinder (Fig. 3B). In the differentiation zone, where lateral roots develop, expression expanded to the cortex region, but was not detectable in young lateral roots (Fig. 3C). In the leaves and sheaths, GUS activity was as strongly detected in the vascular bundles as in the roots and was also detected in the mesophyll cells, but only weakly in the epidermal cells (Fig. 3, D–F).
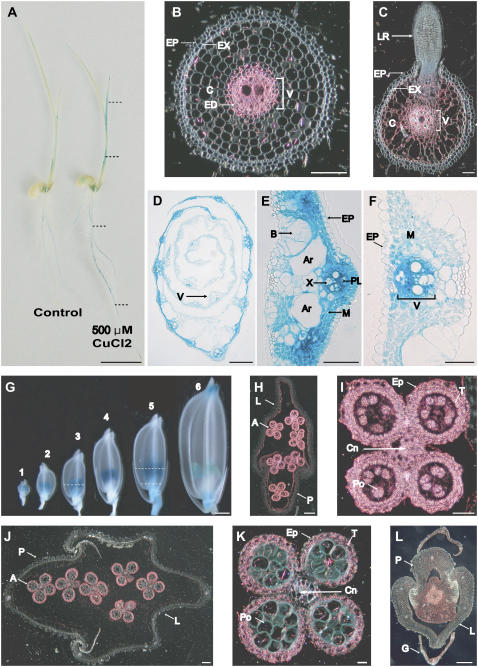
Figure 3.
OsHMA9 promoter-driven GUS expression pattern. A, Seven-day-old seedling grown in one-half-strength Murashige and Skoog medium (control; left) or treated with 500 μm CuCl2 for 6 h (right). B to F, Cross sections of 10-d-old seedlings at indicated position (dotted line) in A; seminal root (B), lateral root (C), shoot (D), midrib region (E), and leaf blade (F). G, Temporal and spatial expression patterns of OsHMA9-GUS fusion construct in spikelets at various stages, as defined by extent of anther development. Sample 1, Meiosis; 2, tetrad; 3, early young microspore; 4, late young microspore; 5, vacuolated pollen; and 6, late pollen mitosis. H, Cross section of sample 3 from G at indicated position. I, Close-up image of anther from H. J, Cross section of sample 5 from G at indicated position. K, Close-up image of anther from J. L, Cross section of sample 5 from G at indicated position. B, C, and H to L, Dark-field images where GUS activities are visualized as red signals. In other images, GUS staining is shown in blue. A, Anther; Ar, aerenchyma; B, bulliform cell; C, cortex; Cn, connective tissues; ED, endodermis; EP, epidermis; EX, exodermis; G, glume; L, lemma; LR, lateral root; M mesophyll; P, palea; PL, phloem; Po, pollen; T, tapetum; V, vascular bundle; X, xylem. Bars = 1 cm in A; 50 μm in B to F and L; 1 mm in G; 100 μm in H and J; and 25 μm in I and K.
In the developing spikelets, GUS activity was detectable mainly in the anthers (Fig. 3G). Cross sections at the young microspore stage showed activity in both the anther wall and the microspore (Fig. 3, H and I). At later stages, activity was mainly detectable in the anther wall (Fig. 3, J and K). In cross sections of the subordinate portions, GUS activity was present at low levels in the ovary (Fig. 3L). These results are, again, consistent with those from our RT-PCR analysis, demonstrating that OsHMA9 is strongly expressed in the stamens (Supplemental Fig. S2).
OsHMA9 Is a Membrane Protein
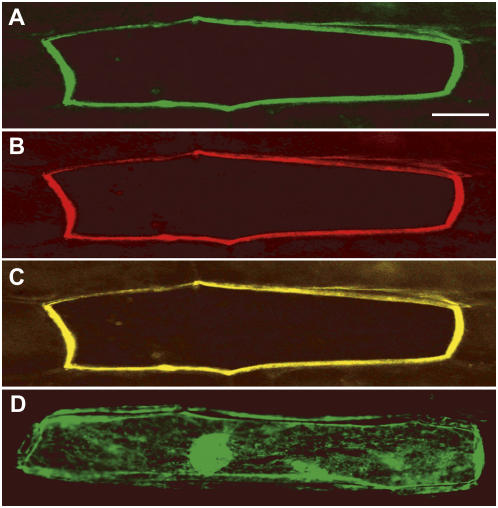
For further insight into the functioning of OsHMA9, we analyzed its subcellular distribution by expressing a fusion protein with GFP. The program ARAMEMNON (http://aramemnon.botanik.uni-koeln.de) predicted with high probability that OsHMA9 may be localized to the plasma membrane. To confirm this, DNA constructs encoding OsHMA9:GFP and AtAHA2:red fluorescent protein (RFP) under the control of the cauliflower mosaic virus 35S promoter were simultaneously introduced into onion (Allium cepa) epidermal cells by particle bombardment. AtAHA2 is an Arabidopsis H+-ATPase that localizes to the plasma membrane (Kim et al., 2001). After 16 h of incubation, expression of the introduced genes was examined under a fluorescent microscope with two different filters to visualize the GFP and RFP images (Fig. 4). The green fluorescent signal was detected from the plasma membrane (Fig. 4A) and coincided with an RFP signal driven by the AtAHA2 image (Fig. 4, B and C). In contrast, fluorescence of GFP alone was observed throughout the cytoplasm and nucleus (Fig. 4D). These results, together with its demonstrated inducibility by metals, suggest that OsHMA9 is a metal transporter located in the plasma membrane.
Figure 4.
Subcellular localization of OsHMA9:GFP fusion protein. Onion cells were cotransformed with OsHMA9:GFP and AHA2:RFP fusion constructs via particle bombardment; fluorescent signals were examined 16 h after transfection. A, Green fluorescence image. B, Red fluorescence image. C, Merged images of A and B. As a control, expression of GFP alone was analyzed (D). Bars = 25 μm.
Heterologous Expression of OsHMA9 in Escherichia coli
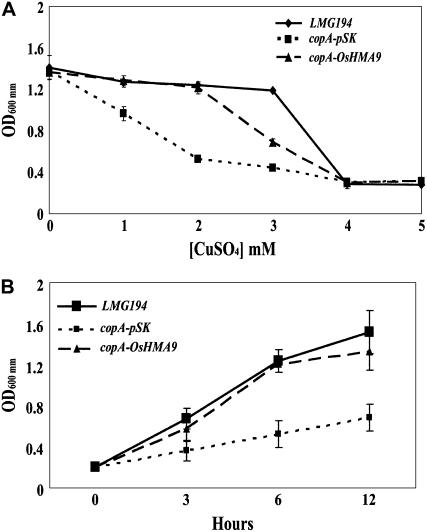
Because the OsHMA9 protein belongs to the Cu ATPase subclass phylogenetically, we investigated whether OsHMA9 could transport Cu in Escherichia coli. OsHMA9 cDNA was expressed in an E. coli mutant with a disruption in CopA, which encodes the Cu efflux P1B-type ATPase (Rensing et al., 2000). Expression of OsHMA9 partially rescued Cu sensitivity in the mutant (Fig. 5A), indicating that the protein is a Cu efflux transporter. Similar experiments using a range of Zn and Cd concentrations were also performed, but no differences in growth were seen between the copA mutant and the control, wild-type strains (data not shown). We also tested the growth rate of the copA mutant expressing OsHMA9 in Luria-Bertani medium containing 2 mm CuSO4 (Fig. 5B). Under such conditions, that rice gene was able to restore the mutant growth rate, suggesting a role in prompting Cu efflux from the cytoplasm.
Figure 5.
Heterologous expression of OsHMA9 in E. coli. A, Growth of wild-type (LMG194), copA mutant transformed with vector alone (copA-pSK) and copA mutant transformed with pBluescript/OsHMA9 (copA-OsHMA9) in Luria-Bertani medium with indicated concentration of CuSO4. B, Growth of wild-type, copA-pSK, and copA-OsHMA9 in Luria-Bertani medium supplemented with 2 mm CuSO4. Representative results from two replicates are shown for each image. Four samples were used in each replicate.
Because OsHMA9 is also induced by high Zn exposure, we examined whether OsHMA9 could transport Zn in E. coli. OsHMA9 cDNA was expressed in an E. coli mutant with a disruption in ZntA, which encodes the Zn efflux P1B-type ATPase (Rensing et al., 1997). Here, expression of OsHMA9 did not result in a significant increase in Zn resistance in the zntA mutant (Supplemental Fig. S3).
Isolation of T-DNA Insertion Mutant Alleles of OsHMA9
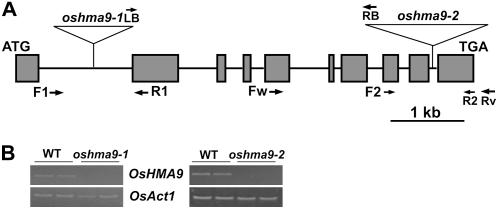
To further examine the functional roles of OsHMA9 in plants, we identified two independent T-DNA insertion alleles from the rice-flanking sequence tag database (An et al., 2003; Jeong et al., 2006). T-DNA was inserted into the first intron of OsHMA9 in oshma9-1 and into the ninth intron in oshma9-2 (Fig. 6A). T2 progeny were obtained and homozygous knockout plants were selected by PCR, using gene-specific primers, located on the 3′ end of OsHMA9, as well as T-DNA primers (Fig. 6A). When its transcript levels were examined in T-DNA homozygous plants, both alleles were found to be null (Fig. 6B). Because the T-DNA was inserted at the end of OsHMA9 in oshma9-2, we tested the presence of a truncated transcript using primers located before that T-DNA insertion (Supplemental Fig. S4). In fact, a truncated transcript was detected in oshma9-2 but not in oshma9-1. Although a truncated protein may have been produced in the former, it was unlikely to be functional because the phenotypes of oshma9-2 and oshma9-1 are identical. Therefore, we can conclude that, although the C-terminal region is not well conserved among the proteins in that family, it is necessary for protein functioning.
Figure 6.
Isolation of OsHMA9 knockout plants. A, Schematic diagram of OsHMA9 and insertion positions of T-DNA. Dark-gray boxes represent 10 exons; connecting black lines are nine introns. In line 1B03522, T-DNA was inserted into first intron (oshma9-1); in line 3A50792, into the ninth intron (oshma9-2) of OsHMA9. Horizontal arrows indicate primers (F1, R1, F2, and R2) used for genotyping T2 progeny. Fw, OsHMA9-specific forward primer; Rv, OsHMA9-specific reverse primer. B, RT-PCR analysis of OsHMA9 expression. Two replicates for wild type and oshma9 are shown. OsHMA9-specific primers (Fw and Rv) were used for RT-PCR analysis, with total RNA prepared from oshma9-1, oshma9-2, or wild-type (WT) plants at seedling stage. Rice actin1 (OsAct1) mRNA was amplified to show an equal amount of RNA in each sample.
OsHMA9 Knockout Plants Are Sensitive to Elevated Levels of Cu, Zn, Pb, and Cd
Because the expression of OsHMA9 was induced by heavy metals and its heterologous expression was restored in the E. coli copA mutant, we hypothesized that OsHMA9 might play a role in heavy-metal detoxification. The oshma9/oshma9 homozygous plants and their segregating wild-type siblings were indistinguishable when grown in soil or on solid medium containing one-half-strength Murashige and Skoog salts.
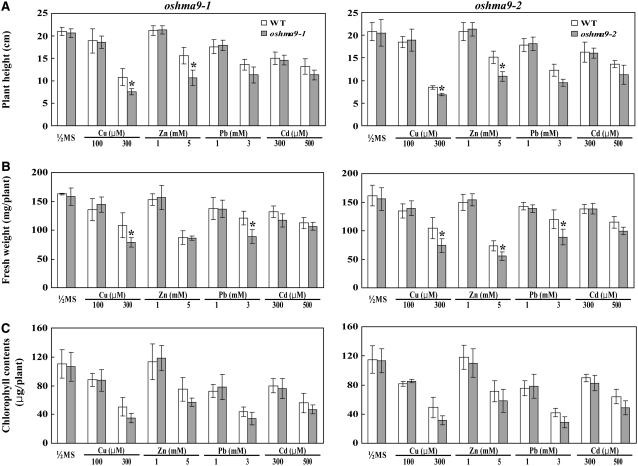
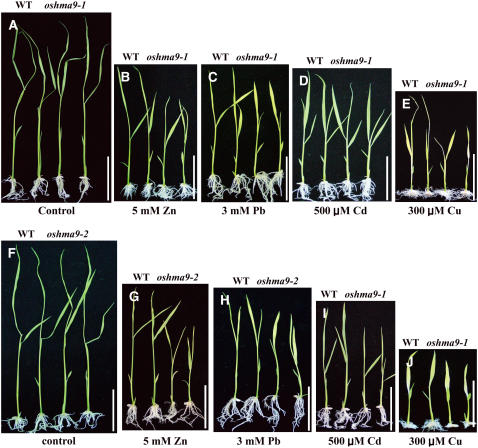
To test the effect of heavy metals on the growth of rice seedlings, we performed a dose-response experiment. Wild-type and knockout plants were grown for 12 d in solid medium supplemented with different concentrations of Cu, Zn, Pb, or Cd to determine at what levels the aberrant phenotypes would become apparent. Plant height, fresh weight, and chlorophyll content were measured to monitor sensitivity to heavy metals (Fig. 8; Supplemental Figs. S5–S8). When both genotypes were reared under normal conditions, all three growth parameters were similar between oshma9 and wild-type plants (Figs. 7A and 8). However, as concentrations of those four heavy metals increased, performance was inhibited in a dose-dependent manner (Supplemental Figs. S5–S8). Treatment with Cu at less than 100 μm did not lead to significant differences between genotypes, but, at 300 μm Cu, development by the knockout plants was diminished compared with the wild type (Fig. 7; Supplemental Fig. S5). Under such treatment, plant height for oshma9-1 and oshma9-2 was reduced to 70% and 81%, fresh weight to 73% and 71%, and chlorophyll content to 75% and 64%, respectively, compared with the wild type (Fig. 8; Supplemental Fig. S5). Likewise, growth of the mutants was retarded in Zn- or Pb-containing medium (Figs. 7 and 8; Supplemental Figs. S6 and S7). These results demonstrate that OsHMA9 is required for conferring tolerance to high levels of Cu, Zn, and Pb. The mutants also showed some sensitivity to Cd, but not as significant as the other metals (Supplemental Fig. S8).
Figure 8.
Phenotypic analysis of wild-type and oshma9 plants grown in medium containing indicated levels of heavy-metal ions. Seeds were germinated and grown for 12 d on one-half-strength Murashige and Skoog agar medium with or without the indicated concentration of CuCl2, ZnCl2, lead tartrate, or 500 μm CdCl2. Height (A) and fresh weight (B) were measured; data are from two independent experiments. In each experiment, eight plants were used for measuring height and fresh weight. Chlorophyll content (C) is average of four plants. Error bars represent se of 16 replicates for the height and fresh weight and eight replicates for chlorophyll content. Significant differences from wild type were determined by Student's t test. *, P < 0.05.
Figure 7.
Phenotype comparison of wild-type and oshma9 plants grown under elevated levels of heavy-metal ions. Wild-type, oshma9-1 (A–E), and oshma9-2 (F–J) plants were grown for 12 d on solid agar containing one-half-strength Murashige and Skoog medium (A and F), medium supplemented with 5 mm ZnCl2 (B and G), 3 mm lead tartrate (C and H), 500 μm CdCl2 (D and I), or 300 μm CuCl2 (E and J). For each, two plants at left are wild-type control and two plants at right are oshma9 grown under the same conditions. Bars = 5 cm.
oshma9 Mutants Accumulate More Zn, Cu, Pb, and Cd
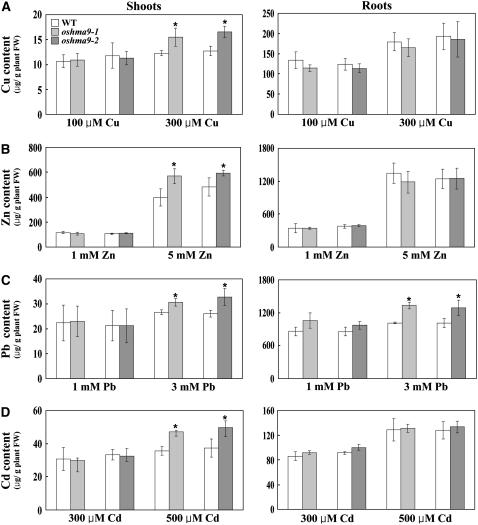
If OsHMA9 is indeed a metal efflux transporter, than metal content should be higher in the knockout plants than in wild types. We therefore measured those levels in 12-d-old knockout and wild-type seedlings that had been exposed to high concentrations of heavy metals. When plants were treated with 100 μm Cu, which did not cause any significant growth defects, metal content in shoots did not differ between the mutant and the wild type. However, when grown on medium containing 300 μm Cu, shoots of oshma9-1 and oshma9-2 plants contained 1.3-fold more Cu (Fig. 9). Similarly, higher amounts of Zn, Pb, and Cd were accumulated in the shoots when seedlings were grown with elevated concentrations of those metals. In contrast, none, except Pb, was accumulated in the roots (Fig. 9). Therefore, these results support our conclusion that OsHMA9 is a metal efflux transporter, especially in the shoots.
Figure 9.
Accumulations of Cu, Zn, Pb, and Cd in oshma9 plants in shoots and roots. Seeds were germinated and grown on for 12 d in one-half-strength Murashige and Skoog solid medium with the indicated concentration of CuCl2, ZnCl2, lead tartrate, or 500 μm CdCl2. Heavy-metal content was measured using atomic absorption spectroscopy. Average of two independent experiments is shown. Significant differences from wild type were determined by Student's t test. *, P < 0.05. Bars = se.
DISCUSSION
All living organisms must maintain an adequate supply of the metal ions required for vital cellular processes, but it is crucial that these be prevented from accumulating to toxic levels. Tolerance in plants grown on metal-polluted soil can be accomplished either by excluding the uptake mechanisms from the roots, or by metal efflux, compartmentation, and detoxification following that uptake. Interest is growing in research on the metal-transport mechanism to understand the behavior of heavy metals in the soil-plant system. In dicot species, P1B-type ATPases play a role in metal detoxification via efflux (Lee et al., 2003; Williams and Mills, 2005; Andres-Colas et al., 2006). Despite their importance, however, very little is known about the functioning of P1B-type ATPases in monocot species.
In this study, we determined the expression patterns for six members of the Cu cluster. Among them, OsHMA4, OsHMA5, and OHMA9 were more preferentially expressed in the roots, whereas the rest were detected almost equally throughout the seedlings. OsHMA9 and OsHMA5 are closely related to Arabidopsis AtHMA7 and AtHMA5, respectively. Expression of OsHMA4, OsHMA7, and OsHMA8 was not significantly enhanced under high concentration of Cu, Zn, Pb, or Cd. In contrast, expression of OsHMA5, OsHMA6, and OsHMA9 was induced by such treatment, suggesting that at least three OsHMAs (OsHMA5, OsHMA6, and OsHMA9) play roles in heavy-metal detoxification. Among those three, we selected OsHMA9 for further analysis because it was more specifically induced by Cu and Zn. The OsHMA9 gene was mainly expressed in vascular tissues, including the xylem and phloem, an observation consistent with a role in the loading and unloading of heavy metals in those tissues. In the leaves, OsHMA9 was also weakly expressed in the mesophyll tissues, a major storage site for heavy-metal ions, again indicating internal detoxification (Ma et al., 2004).
In developing spikelets, expression was strong in the anthers. At the early stages, OsHMA9 was expressed in all cell types within the anthers. However, in later development, its expression was prominent in the walls. The importance of metal transport in anthers has been previously reported. For example, promoter-GUS fusion analysis of AtHMA2 and AtHMA4 has revealed their strong expression in anthers, indicating these two transporters play a specific role in the delivery of Zn to male reproductive tissues (Hussain et al., 2004). COPT1, the Arabidopsis high-affinity Cu transporter, also is highly expressed in pollen; defective pollen development in COPT1 antisense plants is specifically reversed by Cu (Sancenon et al., 2004), supporting a role for its delivery during normal development. AtHMA5 is as strongly expressed as COPT1 in the pollen (Andres-Colas et al., 2006). As has been proposed with Arabidopsis, OsHMA9 probably plays a role in metal delivery to rice anthers. Weak expression in the ovary indicates that the protein might also deliver metals in the female organ. However, in our OsHMA9 knockout plants, we were unable to find any developmental defects in either anther or ovary. Therefore, we might infer that other Cu transporters adequately transport the Cu needed during the formation of those reproductive organs.
To elucidate the functional roles of OsHMA9 in planta, we assessed the growth of OsHMA9 knockout plants on medium containing elevated levels of Cu, Zn, Pb, and Cd. For these metals, growth was inhibited compared with the wild type, as manifested by their reduced height, fresh weight, and chlorophyll content. Moreover, the oshma9 mutant accumulated more Cu, Zn, Pb, and Cd than did the wild type in the shoots. These results indicate that OsHMA9 plays a role in metal detoxification when plants are placed in unfavorable environments, such as with elevated metal ion concentrations. Other evidence for OsHAM9 functioning as an efflux pump arose from our experiments with E. coli. Heterologous expression partially rescued the sensitivity to high Cu by the copA mutant, which is defective in its endogenous Cu efflux pump. However, OsHMA9 did not complement the zntA E. coli mutant, although disruption of OsHMA9 led to greater accumulation of Zn in the plants, indicating Zn transport activity. AtHMA4 confers Cd resistance when expressed in wild-type yeast, but not in the zntA E. coli mutant (Mills et al., 2003, 2005). Disruption of AtHMA4 also results in increased sensitivity to elevated levels of Cd and Zn. These suggest that AtHMA4 serves an important role in metal detoxification at higher metal concentrations (Mills et al., 2005). A range of yeast mutants could be tested to further examine the role of OsHMA9 in Zn transport.
Although the OsHMA9 gene is expressed in the roots and shows similar metal regulation to the shoots, oshma9 mutants accumulated metals (apart from Pb) only in the shoots. It is possible that heavy metals are more efficiently sequestered in roots.
OsHMA9 is in the same subclass phylogenetically with AtHMA7/RAN1, which functions in the delivery of Cu ions to the ethylene receptor (Hirayama et al., 1999). Further works are needed to elucidate the roles of OsHMA9 in ethylene-signaling pathways.
Rice contains nine P1B-type HMAs, a number high in comparison with other nonplant eukaryotic species, which have only one or two members (Baxter et al., 2003). None of these rice P1B ATPases has previously been characterized. Arabidopsis has eight members, which have been characterized to some extent (Williams and Mills, 2005; Andres-Colas et al., 2006). Among the rice transporters, OsHMA6 and OsHMA9 form a subclass with Arabidopsis AtHMA7/RAN1, which functions in the delivery of Cu ions (Hirayama et al., 1999). Although OsHMA6 and OsHMA9 shared high sequence similarity, their expression patterns differed, with the latter being strongly expressed in roots and the former being expressed mainly in the leaves. OsHMA9 was more strongly induced by Cu treatment than was OsHMA6. These results suggest a divergent role in Cu homeostasis, although both genes require more detailed analyses.
OsHMA9 contains conserved domains present in ATP-driven efflux pump proteins. These include an aspartyl kinase domain (DKTGT), a GDGXNDXP domain (GDGINDSP in OsHMA9), and an E1-E2 ATPase domain characterized by a region with PXD and TGE motifs. The most commonly discussed motif is the heavy-metal-associated domain (Jordan et al., 2001), which typically includes a GMXCXXC sequence. This is present in OsHMA9 in two copies—GMTCSAC and GMTCANC. Research on AtHMA5 has revealed two HMA domains present in the amino-terminal region, both of which interact with Arabidopsis ATX1-like chaperones in vivo, indicating a role for AtHMA5 in metal compartmentalization and detoxification (Andres-Colas et al., 2006). Further work is needed to determine the functional roles of these motifs. Nevertheless, taken together, our results strongly indicate that OsHMA9 is a heavy-metal efflux protein present in the plasma membrane.
MATERIALS AND METHODS
Plant Growth
Surface-sterilized seeds of oshma9-1, oshma9-2, and wild-type rice (Oryza sativa ‘Dongjin’) were germinated on one-half-strength Murashige and Skoog medium comprising 1.5% Suc, 0.2% phytagel, and 0.28 mm myoinositol (Sigma). Seedlings were grown for 7 d at 27°C under continuous light, then transferred to soil in the greenhouse and raised to maturity.
Semiquantitative RT-PCR Analysis
Total RNA was isolated from each tissue type with the RNA isolation kit (Tri Reagent; MRC). For cDNA synthesis, we used 2 μg of total RNA as template plus Moloney murine leukemia virus reverse transcriptase (Promega) in a 25-μL reaction mixture. RT-PCR was performed in a 50-μL solution containing a 1-μL aliquot of the cDNA reaction, 0.2 μm of gene-specific primers, 10 mm dNTPs, and 1 unit of rTaq DNA polymerase (TaKaRa Shuzo). PCR conditions included 23 cycles (OsAct1) or 25 cycles (OsHMA4–OsHMA9) of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, and extension at 72°C for 90 s. We performed a minimal number of cycles to ensure that the reaction was not saturated. PCR products were barely visible when the agarose gels were stained by ethidium bromide. These were separated by electrophoresis on a 1.2% agarose gel, blotted onto a nylon membrane, and hybridized with the 32P-labeled probe specific to each gene, generated from the nonconserved 3′ end regions. Primers are shown in Table I. To test the expression levels of OsHMA4 through OsHMA9 under heavy-metal stress, 7-d-old plants were floated on one-half-strength Murashige and Skoog medium with or without different concentrations of Cu, Zn, Pb, or Cd. After 3 and 6 h of treatment, leaf and root tissue samples were frozen in liquid nitrogen.
Table I.
Nucleotide sequences of the primers used for RT-PCR analysis
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| OsAct1 | GTATCCATGAGACTACATACAACT | TACTCAGCCTTGGCAATCCACA |
| OsHMA4 | CCAACCTTCGAGACTCTTCA | GCATGGGACCTCAGTAAATG |
| OsHMA5 | GGGGCTAATCATGGGGTACT | GAACTCCTGTCTTCGCGTTC |
| OsHMA6 | ATAATGGAGGAGGCCGAACT | CCTGCCTTTTCATGTGGAGT |
| OsHMA7 | TGCTGGGGAAATATCTGGAG | CTTCTGAACTGGAGCCCTTG |
| OsHMA8 | CGGTGAATGAGAATGGTGTG | TCTTGTCGAAAACGATGCAG |
| OsHMA9 | CAGTAATCGGAGGGACAATG | GTGATGAAACGGTGAAGAGG |
Isolation of OsHMA9 Knockout Plants
Two OsHMA9 knockout alleles were isolated from the rice-flanking sequence tag database (http:://www.postech.ac.kr/life/pfg). T2 progeny of the primary insertional mutants were grown to maturity to amplify the seeds. Genotypes of the progeny were determined by PCR, with three primers. For the oshma9-1 line, two OsHMA9-specific primers (F1, 5′-TGCAATTTTGGGTTTTAGCTG-3′ and R1, 5′-CTGGGATAATTTCTGCGTCAA-3′) and a T-DNA-specific primer (LB-2, 5′-ACGTCCGCAATGTGTTATTAA-3′) were used. For oshma9-2, the OsHMA9-specific primers (F2, 5′-ATCGGCCTTCCTTTTCTCAT-3′ and R2, 5′-TTTCTTTGGCAAGCTCCCTA-3′) and a T-DNA-specific primer (RB-2, 5′-CAAGTTAGTCATGTAATTAGCCAC-3′) were used.
Construction of the OsHMA9 Promoter-GUS Fusion Molecule and GUS Assay
Genomic sequences (−900 to −1 bp from the translation initiation codon) containing the promoter region of OsHMA9 were amplified by PCR, using two primers (pf, 5′-AAAAGCTTCACTGTCAGAGCGAAACAAAAATG-3′ and pr, 5′-AAAGGATCCGCCGAGAGCTGCAGGTGGGCCAT-3′). The fragment was connected to the GUS-NOSt cassette (derived from pBI 101.2) and ligated into pCAMBIA1302, resulting in pGA2880. This plasmid was transferred to Agrobacterium tumefaciens strain LBA4404 with the freeze-thaw method (An et al., 1988). Transgenic plants carrying the above construct were generated by Agrobacterium-mediated cocultivation (Lee et al., 1999). Histochemical GUS staining of transgenic plants was performed according to the method described by Jefferson et al. (1987), except for the addition of 20% methanol to the staining solution (Dai et al., 1996). Sections (10 μm) were prepared as previously described by Jung et al. (2005) and were observed with a microscope (Nikon) under bright- and dark-field illumination.
Subcellular Localization of the OsHMA9-GFP Fusion Protein in Onion Epidermal Cells
Full-length OsHMA9 cDNA was PCR amplified using the primer pair of gf (5′-AATCTAGACAGGCATGGCCCACCTGCAGCTC-3′) and gr (5′-AAAGATCTCTACAGTTATCTGCAACACGGTGG-3′). These primers contained XbaI or BglII sites (underlined sequences) to facilitate cloning of the amplified cDNA. After sequence analysis, the OsHMA9 cDNA was cloned into the XbaI and BamHI sites of the 326GFP vector (Lee et al., 2001). The AtAHA2:RFP fusion molecule, under the control of the cauliflower mosaic virus 35S promoter, was obtained from Inhwan Hwang (Pohang University of Science and Technology). Onion (Allium cepa) cells were put in Murashige and Skoog medium containing 0.4 mm mannitol during bombardment. The constructs were introduced into onion epidermal cells by particle bombardment with the Biolistic PDS-1000/He Particle Delivery system (Bio-Rad). Expression of GFP and RFP was monitored at 12 h after transformation using a fluorescent microscope (Axioplan2; Carl Zeiss).
Heterologous Expression of OsHMA9 in Escherichia coli
The cDNA fragment containing an entire open reading frame for OsHMA9 was amplified through RT-PCR with the primer pair F (5′-AAGGTACCCACCAAATATTCCCACCTCGCG-3′) and R (5′-AATCTAGAGTGATGAAACGGTGAAGAGG-3′). These primers contained a KpnI or XbaI site (underlined sequences) to facilitate cloning of the amplified cDNA. The fragment was first cloned into the pGEM-T vector (Promega). After sequence analysis, the OsHMA9 cDNA was subcloned into the KpnI and XbaI sites of pBluescript II SK− (Stratagene). The resulting plasmid was introduced into two E. coli strains: LMG194 or LMG194 with a copA mutation (Rensing et al., 2000). After these strains were grown for 12 h in Luria-Bertani liquid medium containing 0 to 5 mm CuSO4, their cell densities were spectrometrically measured at 600 nm.
Measurements of Plant Growth and Heavy-Metal Content
Wild-type and homozygote oshma9 seeds were germinated and their seedlings grown for 12 d on solid medium containing one-half-strength Murashige and Skoog salts and supplemented with or without different concentrations of Cu, Zn, Pb, and Cd. Plants were collected and their weights and shoot lengths were measured. To assess their chlorophyll content, leaves were harvested and chlorophyll was extracted with 1 mL 80% acetone from 0.1-g samples. An aliquot of the extracts was taken to measure A645 and A663 with a spectrophotometer. Specific chlorophyll content was determined according to the method of Arnon (1949). Heavy-metal content was measured as previously described by Kim et al. (2002). Shoots of the 12-d-old plants were harvested and dissolved in 11 N HNO3 in a 200°C oven. Total metal content was then analyzed by atomic absorption spectroscopy (Solaar 989; Unicam Atomic Absorption).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NP_001058305.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. OsHMA9 promoter-driven GUS expression pattern.
Supplemental Figure S2. Spatial expression pattern of OsHMA9 in wild-type spikelets.
Supplemental Figure S3. Heterologous expression of OsHMA9 in E. coli.
Supplemental Figure S4. RT-PCR analysis of the OsHMA9 transcript in oshma9 and wild-type plants.
Supplemental Figure S5. Height, fresh weight, and chlorophyll content of plants grown on medium containing different concentrations of Cu.
Supplemental Figure S6. Height, fresh weight, and chlorophyll content of plants grown on medium containing different concentrations of Zn.
Supplemental Figure S7. Height, fresh weight, and chlorophyll content of plants grown on medium containing different concentrations of Pb.
Supplemental Figure S8. Height, fresh weight, and chlorophyll content of plants grown on medium containing different concentrations of Cd.
Supplementary Material
Acknowledgments
We thank Priscilla Licht for critical reading of the manuscript. We also thank Professor Bharati Mitra for providing the E. coli mutant strains, Inhwan Hwang for the AtAHA2:RFP molecule, In-Soon Park and Kyungsook An for plant transformation, and Changduk Jung for growing our plants in the greenhouse.
This work was supported by grants from the Crop Functional Genomic Center, the 21st Century Frontier Program (grant no. CG1111); from the Biogreen 21 Program, Rural Development Administration; and from the National Research Laboratory Program. Y.-Y.K. was a recipient of the Brain Korea 21 fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gynheung An (genean@postech.ac.kr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abdel-Ghany SE, Muller-Moule P, Niyogi KK, Pilon M, Shikanai T (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–19
- An S, Park S, Jeong DH, Lee DY, Kang HG, Yu JH, Hur J, Kim SR, Kim YH, Lee M, et al (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133 2040–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Colas N, Sancenon V, Rodriguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Penarrubia L (2006) The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J 45 225–236 [DOI] [PubMed] [Google Scholar]
- Arnon DI (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46 84–101 [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley MR, White PJ, Hammond JP, Zelko I, Lux A (2007) Zinc in plants. New Phytol 173 677–702 [DOI] [PubMed] [Google Scholar]
- Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212 475–486 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2006) Put the metal to the petal: metal uptake and transport throughout plants. Curr Opin Plant Biol 9 322–330 [DOI] [PubMed] [Google Scholar]
- Dai Z, Gao J, An K, Lee JM, Edwards GE, An G (1996) Promoter elements controlling developmental and environmental regulation of a tobacco ribosomal protein L34. Plant Mol Biol 32 1055–1065 [DOI] [PubMed] [Google Scholar]
- Eren E, Arguello JM (2004) Arabidopsis HMA2, a divalent heavy metal-transporting P(IB)-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol 136 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes JC, Henriques FS (1991) Biochemical, physiological, and structural effects of excess copper in plants. Bot Rev 57 246–273 [Google Scholar]
- Goff SA, Ricke D, Lan TH, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100 [DOI] [PubMed] [Google Scholar]
- Gravot A, Lieutaud A, Verret F, Auroy P, Vavasseur A, Richaud P (2004) AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett 561 22–28 [DOI] [PubMed] [Google Scholar]
- Grotz N, Guerinot ML (2006) Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim Biophys Acta 1763 595–608 [DOI] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158 1317–1323 [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR (1999) RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transport, is required for ethylene signaling in Arabidopsis. Cell 97 383–393 [DOI] [PubMed] [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 11 793–800 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45 123–132 [DOI] [PubMed] [Google Scholar]
- Jordan IK, Natale DA, Koonin EV, Galperin MY (2001) Independent evolution of heavy metal-associated domains in the copper chaperones and copper-transporting ATPases. J Mol Evol 53 622–633 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YY, Yang YY, Lee Y (2002) Pb and Cd uptake in rice roots. Physiol Plant 116 368–372 [Google Scholar]
- Krämer U, Talke IN, Hanikenne M (2007) Transition metal transport. FEBS Lett 581 2263–2272 [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W (2004) Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol 5 282–295 [DOI] [PubMed] [Google Scholar]
- Lee J, Bae H, Jeong J, Lee JY, Yang YY, Hwang I, Martinoia E, Lee Y (2003) Functional expression of a bacterial heavy metal transporter in Arabidopsis enhances resistance to and decreases uptake of heavy metals. Plant Physiol 133 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Jeon JS, Jung KH, An G (1999) Binary vector for efficient transformation of rice. J Plant Biol 42 310–316 [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Ueno D, Zhao FJ, McGrath SP (2004) Subcellular localisation of Cd and Zn in the leaves of a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Planta 220 731–736 [DOI] [PubMed] [Google Scholar]
- Mills RF, Francini A, Ferreira da Rocha PS, Baccarini PJ, Aylett M, Krijger GC, Williams LE (2005) The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett 579 783–791 [DOI] [PubMed] [Google Scholar]
- Mills RF, Krijger GC, Baccarini PJ, Hall JL, Williams LE (2003) Functional expression of AtHMA4, a P1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J 35 164–176 [DOI] [PubMed] [Google Scholar]
- Pedersen P, Carafoli E (1987) Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem Sci 4 146–150 [Google Scholar]
- Rensing C, Fan B, Sharma R, Mitra B, Rosen BP (2000) CopA: an Escherichia coli Cu(II)-translocating P-type ATPase. Proc Natl Acad Sci USA 97 652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Ghosh M, Rosen BP (1999) Families of soft-metal-ion-transporting ATPases. J Bacteriol 181 5891–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Mitra B, Rosen BP (1997) The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA 94 14326–14331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout GR, Das P (2003) Effect of metal toxicity on plant growth and metabolism: I. Zinc. Agronomie 23 3–11 [Google Scholar]
- Ryu CH, You JH, Kang HG, Hur J, Kim YH, Han MJ, An K, Chung BC, Lee CH, An G (2004) Generation of T-DNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol Biol 54 489–502 [DOI] [PubMed] [Google Scholar]
- Sancenon V, Puig S, Mateu-Andres I, Dorcey E, Thiele DJ, Penarrubia L (2004) The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J Biol Chem 279 15348–15355 [DOI] [PubMed] [Google Scholar]
- Schutzendubel A, Polle A (2002) Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot 53 1351–1365 [PubMed] [Google Scholar]
- Seigneurin-Berny D, Gravot A, Auroy P, Mazard C, Kraut A, Finazzi G, Grunwald D, Rappaport F, Vavasseur A, Joyard J, et al (2006) HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J Biol Chem 281 2882–2892 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Muller-Moule P, Munekage Y, Niyogi KK, Pilon M (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K, Kashiwagi S, Mori H, Ueguchi C, Mizuno T (1997) Cloning of a cDNA encoding a putative metal-transporting P-type ATPase from Arabidopsis thaliana. Biochim Biophys Acta 1326 1–6 [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P (2004) Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett 576 306–312 [DOI] [PubMed] [Google Scholar]
- Williams LE, Mills RF (2005) P1B-ATPases—an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 10 491–502 [DOI] [PubMed] [Google Scholar]
- Williams LE, Pittman JK, Hall JL (2000) Emerging mechanisms for heavy metal transport in plants. Biochim Biophys Acta 1465 104–126 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.