Abstract
The circadian clock is an endogenous mechanism that generates rhythms with an approximately 24-h period and enables plants to predict and adapt to daily and seasonal changes in their environment. These rhythms are generated by molecular oscillators that in Arabidopsis (Arabidopsis thaliana) have been shown to consist of interlocking feedback loops involving a number of elements. An important characteristic of circadian oscillators is that they can be entrained by daily environmental changes in light and temperature. Previous work has shown that one possible entrainment point for the Arabidopsis oscillator is the light-mediated regulation of expression of one of the oscillator genes, CIRCADIAN CLOCK ASSOCIATED1 (CCA1). In this article, we have used transgenic plants with constitutive CCA1 expression to show that light also regulates CCA1 transcript stability. Our experiments show that CCA1 messenger RNA is relatively stable in the dark and in far-red light but has a short half-life in red and blue light. Furthermore, using transgenic plants expressing chimeric CCA1 constructs, we demonstrate that the instability determinants in CCA1 transcripts are probably located in the coding region. We suggest that the combination of light regulation of CCA1 transcription and CCA1 messenger RNA degradation is important for ensuring that the Arabidopsis circadian oscillator is accurately entrained by environmental changes.
Many organisms have circadian clocks, endogenous 24-h systems that allow them to anticipate the daily changes in their environmental conditions (Bell-Pedersen et al., 2005). In plants, the circadian system is important for the regulation of a wide variety of processes from leaf and petal movements to stomatal opening, protein phosphorylation (Yakir et al., 2007), and transcript accumulation from up to 16% of the genome (Harmer et al., 2000; Schaffer et al., 2001; Michael and McClung, 2003; Edwards et al., 2006).
In the model plant, Arabidopsis (Arabidopsis thaliana), the oscillator that generates circadian rhythms consists of interlocking positive/negative feedback loops of several genes including CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION1 (TOC1; Millar et al., 1995; Wang et al., 1997; Schaffer et al., 1998; Wang and Tobin, 1998). CCA1 and LHY expression rises before dawn and suppresses the expression of TOC1 by binding to its promoter. In the evening, when CCA1 and LHY levels decrease, TOC1 expression rises. TOC1 then activates CCA1 and LHY expression by an as-yet-unknown mechanism (Alabadi et al., 2001). Other genes such as GIGANTEA (GI), EARLY FLOWERING4, LUX ARRHYTHMO, TIME FOR COFFEE, and ARABIDOPSIS PSEUDORESPONSE REGULATOR3/5/7/9 (APRR3/5/7/9) might operate in the oscillator or close to it (Doyle et al., 2002; Hazen et al., 2005; Locke et al., 2005; Nakamichi et al., 2005; Edwards et al., 2006; Gould et al., 2006; Ding et al., 2007; McWatters et al., 2007).
An important adaptive feature of the circadian clock is its ability to be entrained by environmental signals such as changes in light or temperature. This can be achieved by affecting one or more of the stages of the oscillator feedback loop (e.g. changing transcription rates, posttranscriptional events, translation rates, or posttranslational events). Several putative entrainment points have been described for the Arabidopsis oscillator. One such entrainment point is the activation of CCA1 and LHY transcription by light (Wang and Tobin, 1998; Kim et al., 2003). Another well-characterized effect light has on the Arabidopsis oscillator is the regulation of proteasomal degradation of TOC1 and APRR5 proteins that is mediated by ZEITLUPE (ZTL), an F-box motif and Kelch domain protein (Mas et al., 2003; Kiba et al., 2007). ZTL itself is stabilized by a blue light-mediated interaction with GI (Kim et al., 2007). Finally, the translation rate of LHY may be light regulated; light causes an increase in the translation of LHY in transgenic plants expressing LHY from a constitutive promoter (Kim et al., 2003).
In other organisms, there is evidence for entrainment by posttranscriptional events and RNA modification. For example, in Neurospora crassa, there is a transcribed antisense sequence for the oscillator component frequency (frq), which is expressed with an opposite phase to the sense sequence of frq and appears to play a role in entrainment by light signals (Kramer et al., 2003). In addition, frq shows temperature-dependent alternative splicing, creating FRQ proteins with different lengths (Colot et al., 2005). The Drosophila melanogaster oscillator component PERIOD (PER) has an alternative splicing region in its 3′ untranslated region (UTR). The ratio of spliced and unspliced PER mRNA is controlled by temperature and light and can affect the rate of PER accumulation and activity rhythms (Majercak et al., 1999, 2004).
In general, RNA stability has an important role in the regulation of gene expression in eukaryotic cells (Ross, 1995; Meyer et al., 2004). One of the best characterized elements affecting RNA stability is the AUUUA sequence that appears, at differing frequencies and often in overlapping patterns, in the 3′ UTR of eukaryotic transcripts like c-fos and interleukin-3. This group of sequences (also known as AU-rich elements) can accelerate the decay of an mRNA containing them (Chen and Shyu, 1995). Another sequence affecting RNA stability is the so-called downstream element (DST), discovered first in the 3′ UTR of the auxin-inducible SMALL AUXIN-UP RNA gene family of soybean (Glycine max; McClure et al., 1989). The DST sequence was shown to destabilize mRNA in tobacco (Nicotiana tabacum) cells and probably plays a similar role in RNA stability in Arabidopsis (Newman et al., 1993; Gil and Green, 1996; Johnson et al., 2000). Furthermore, a DST element is present in some circadian-regulated genes, and, in some cases, destabilization of mRNA by DST is under circadian clock control (Lidder et al., 2005). mRNA stability is also frequently regulated by small complementary RNAs. Several such small RNA pathways have been characterized in plants. The best characterized is the miRNA pathway involving small RNA molecules derived from endogenous noncoding transcripts with an extensive stem and loop structure (Brodersen and Voinnet, 2006).
Some mRNAs in plants show altered half-lives in response to external signals. For example, the FERREDOXIN-1 transcript in pea (Pisum sativum) is destabilized by darkness. This process is controlled by multiple CAUU sequences in the 5′ UTR (Petracek et al., 1998; Bhat et al., 2004). In Arabidopsis and pea, some LIGHT-HARVESTING CHLOROPHYLL-BINDING transcripts are destabilized by blue light perceived by the phototropin-1 receptor (Anderson et al., 1999; Folta and Kaufman, 2003). Furthermore, in beans (Phaseolus vulgaris), PROLINE-RICH PROTEIN1 mRNA is destabilized by treatment with a fungal elicitor (Zhang et al., 1993).
In this article, we show that CCA1 transcript stability is regulated by light. We suggest that the combination of light regulation of CCA1 transcription and RNA degradation is important for ensuring that the circadian oscillator is accurately entrained by environmental changes.
RESULTS
Light Regulates CCA1 Posttranscriptionally
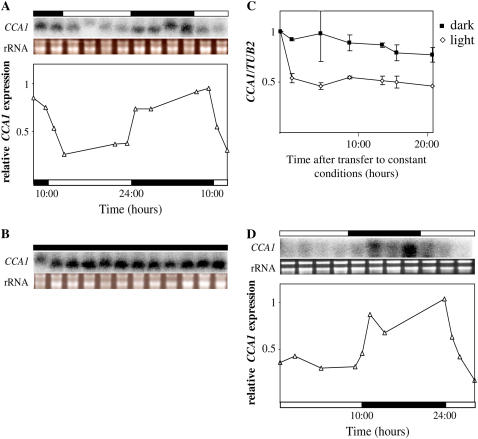
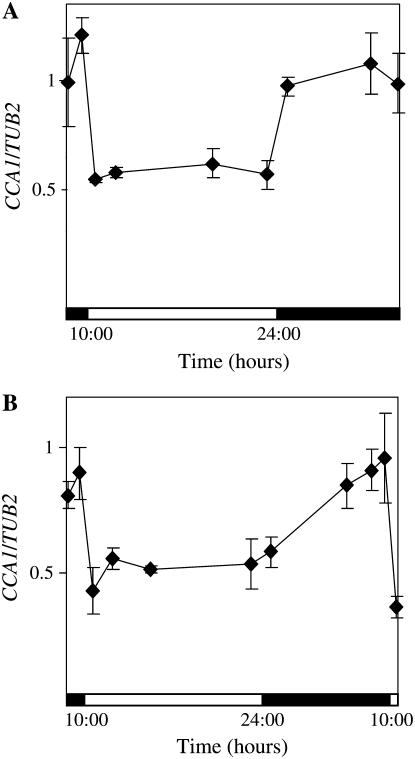
To determine whether light has posttranscriptional effects on CCA1 levels in addition to its known role in activating CCA1 expression (Wang et al., 1997; Wang and Tobin, 1998; Kim et al., 2003), we examined CCA1 transcript levels in transgenic plants expressing CCA1 driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter (CCA1-ox 034; described in Wang and Tobin, 1998). Although in these plants CCA1 should be constitutively expressed, we observed that CCA1 mRNA levels cycled according to the light/dark period. Figure 1A shows that after the lights came on, there was a sharp decrease in total CCA1 transcript levels and that the levels stayed low until the lights went off again. In CCA1-ox 034 plants maintained in constant dark, CCA1 mRNA levels were consistently high (Fig. 1, B and C). By contrast, the levels of CCA1 mRNA were significantly lower in plants grown in constant light conditions (Fig. 1C). Thus, it appears that the levels of CCA1 transcript are light dependent, with CCA1 mRNA being degraded significantly more quickly in the light than in the dark. Although the TUB2 control used for the quantitative real-time PCR has been reported to be circadian regulated (Edwards et al., 2006), the changes are very small and do not significantly affect our results (Supplemental Fig. S1, A and B).
Figure 1.
In plants expressing CCA1 transcript under the control of constitutive CaMV 35S promoter, CCA1 mRNA levels are higher in the dark than in the light. Three-week-old CCA1-ox 034 and CCA1-ox 038 plants were kept in light:dark cycles (14 h light:10 h dark). The levels of CCA1 mRNA were determined by northern analysis (A, B, and D; shown above the graph) or quantitative real-time PCR (C) and plotted on a graph relative to the maximum levels of expression. Aliquots of 1.5 μg of total RNA from each sample were run on an agarose gel to check for quality and to verify quantification (A, B, and D; shown above the graph). The white and black bars represent light and dark periods, respectively. The experiments were repeated at least twice. A, CCA1-ox 034 in light:dark. B, CCA1-ox 034 in constant dark. C, CCA1-ox 034 in constant light and constant dark. Dark, Black squares; light, white diamonds. D, CCA1-ox 038 in light/dark.
There might be, however, alternative explanations for the differences in CCA1 transcript levels that we observed. One possible explanation is that there is a light-dependent difference in transcription rates that is a result of the specific integration site of the transgene. To address this question, we examined CCA1 transcript levels in an independent transgenic line, CCA1-ox 038 (Wang and Tobin, 1998), by both quantitative real-time PCR and northern analysis. Our results (Fig. 1D; Supplemental Fig. S2) show that the CCA1 mRNA levels were low in the light and high in the dark, ruling out the possibility of light-dependent insertion site differences in transcription.
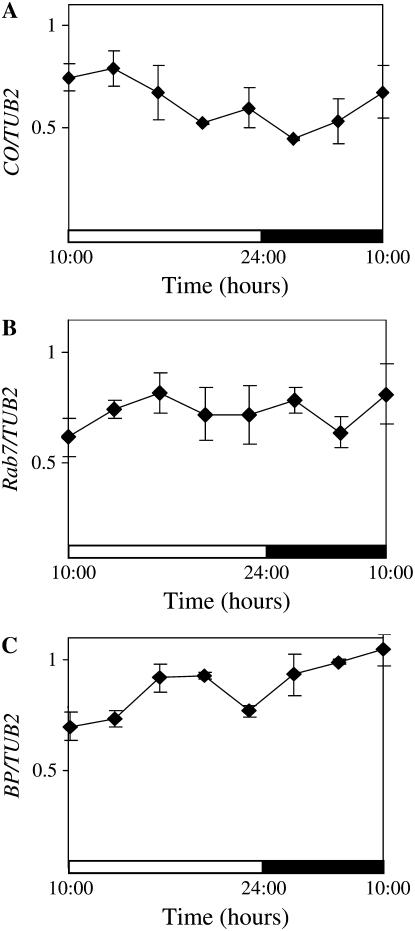
Another explanation for the changes in CCA1 transcript levels under different light conditions could be that light alters the transcription rate from the CaMV 35S promoter itself. Although, as far as we know, there is no evidence showing a light- or dark-dependent regulation of transcription from the CaMV 35S promoter, we wanted to rule out this possibility. We therefore examined mRNA levels of three different transcripts, CONSTANS (CO), AtRabG3e (Rab7), and BREVIPEDICELLUS (BP; Lincoln et al., 1994; Onouchi et al., 2000; Mazel et al., 2004), expressed from the CaMV 35S promoter in transgenic plants (see “Materials and Methods”). As shown in Figure 2A, CO transcript levels were not significantly altered in light and dark. This result is consistent with previous reports showing CO mRNA levels are not light dependent (Valverde et al., 2004). Rab7 and BP transcripts also remain high and did not show significant changes in light and dark (Fig. 2, B and C). Thus, expression from the CaMV 35S promoter does not appear to be regulated by light.
Figure 2.
Transcription from the constitutive CaMV 35S promoter is not light dependent. Three-week-old 35S∷CO, AtRab7-7, and 35S∷BP plants were grown in light:dark cycles (14 h light:10 h dark). The levels of CO, Rab7, and BP RNA were determined by quantitative real-time PCR relative to the levels of TUB2. The averages of three independent experiments are shown together with the ses. The white and black bars represent light and dark periods, respectively. A, CO levels in 35S∷CO. B, Rab7 levels in AtRab7-7. C, BP levels in 35S∷BP.
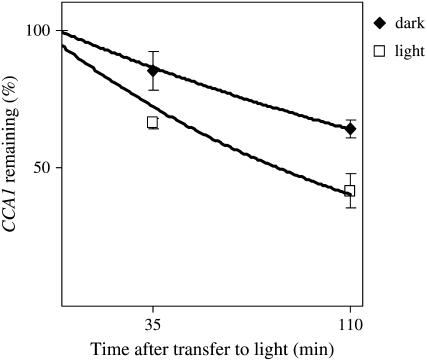
To further confirm that the differences we observe in CCA1 mRNA accumulation in dark and light are due to differences in the rate of mRNA degradation, we inhibited transcription using cordycepin (3′-deoxyadenosine). The cordycepin was applied to whole CCA1-ox 034 plants (as described in the “Materials and Methods”) after 10 h of growth in the dark. The plants were subsequently transferred to light or left in the dark and the levels of CCA1 transcripts examined. Figure 3 shows that in plants transferred to light, the levels of CCA1 mRNA decreased faster than in plants left in the dark.
Figure 3.
CCA1 mRNA decay is faster in light than in dark. Three-week-old CCA1-ox 034 plants were treated with cordycepin (3′-deoxyadenosine), as described in “Materials and Methods,” and then transferred to light or kept in the dark. The levels of CCA1 mRNA were determined by quantitative real-time PCR. Dark, Black diamonds; light, white squares. The averages of seven independent experiments are shown together with the ses. CCA1 half-life in light, 90 ± 13 (se) min. CCA1 half-life in dark, 173 ± 15 (se) min.
We also calculated the half-life of CCA1 mRNA in light and dark. In general, most sense mRNA degradation obeys first-order kinetics (Lam et al., 2001) and CCA1 transcript degradation can be fitted with first-order kinetics (R2 greater than 0.9 for both light and dark). Using the equation t1/2 = 0.693/kd (Petracek et al., 1998) to calculate the half-life of CCA1 mRNA, we found that in plants transferred to light, CCA1 mRNA half-life is 1.5 h. In the dark, however, CCA1 transcript degradation rate is doubled, and the half-life is about 3 h. Using a binomial test, we calculated that the levels of CCA1 mRNA in the light are significantly lower than CCA1 mRNA levels in the dark (P < 0.015).
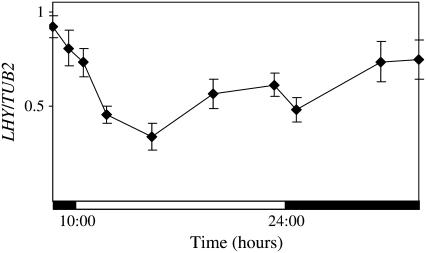
CCA1 is highly homologous to another oscillator gene, LHY (55% ClustalW score for the two cDNAs and more than 70% ClustalW score for the MYB domain). Previous work has shown that in plants expressing LHY from the constitutive CaMV 35S promoter (lhy-1TN104), LHY transcript levels did not cycle under conditions of alternating light and dark (Kim et al., 2003). However, under the conditions that we used in our experiments, we found that LHY transcript levels are lower in the light than in the dark although, unlike CCA1, do not show a clear square wave form of accumulation (Fig. 4 compared to Fig. 1, A and D). Thus, it seems that LHY transcripts may be light regulated but possibly in a different way from CCA1 transcripts.
Figure 4.
In plants expressing LHY transcript under the control of the constitutive CaMV 35S promoter, LHY levels are light regulated but not as much as CCA1 in CCA1-ox. Three-week-old lhy-1TN104 plants were grown in light:dark cycles (14 h light:10 h dark). The levels of LHY mRNA were determined by quantitative real-time PCR relative to the levels of TUB2. The averages of five independent experiments are shown together with the ses. The white and black bars represent light and dark periods, respectively.
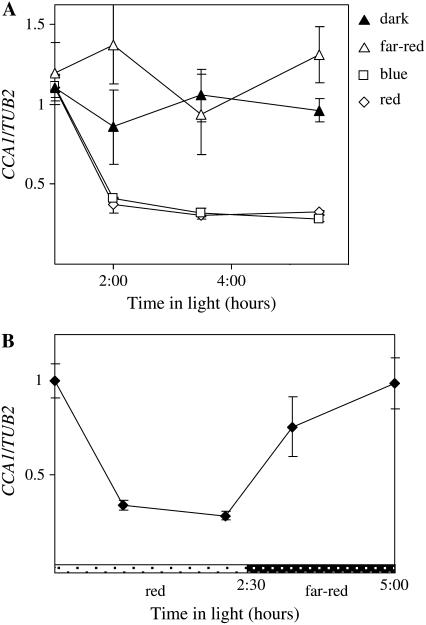
Red and Blue, But Not Far-Red, Light Promotes CCA1 Transcript Degradation
Plants have specific photoreceptors that are sensitive to different wavelengths of light. To determine which wavelengths and, thus, which photoreceptors are important for CCA1 mRNA degradation, the levels of CCA1 transcript were measured in CCA1-ox 034 plants transferred from dark to red, blue, or far-red light or kept in the dark. Figure 5A shows that in the dark the levels of CCA1 transcripts were constantly high. However, by 1 h after exposure to red or blue light there was a sharp decrease in CCA1 transcript to less than 40% of the levels observed in the dark. By contrast, CCA1 mRNA levels remained high in far-red light. When dark-adapted plants were first exposed to red light before being transferred to far red, CCA1 transcript levels decreased in red light and increased again in far red to the levels observed in the dark (Fig. 5B). Taken together, our results show that red and blue light induce CCA1 mRNA degradation and that this effect is reversed by far-red light.
Figure 5.
CCA1 mRNA is less stable in red and blue light than in far-red light or dark. A, After 10 h in the dark, 3-week-old CCA1-ox 034 plants were transferred to red, blue, or far-red light or kept in the dark. The levels of CCA1 mRNA were determined by quantitative real-time PCR relative to the levels of TUB2. The averages of three independent experiments are shown together with the ses. Dark, Black triangles; far-red, white triangles; red, white diamonds; blue, white squares. B, After 10 h in the dark, 3-week-old CCA1-ox 034 plants were transferred to red light for 2.5 h (white bars with black dots) and then to far-red light (black bars with white dots). The levels of CCA1 mRNA were determined by quantitative real-time PCR. For each treatment, the averages of two independent experiments were plotted together with the ses.
cis-Elements Determining CCA1 Transcript Stability
To start to determine the mechanism(s) for regulating CCA1 transcript stability, we looked for the location of the cis-acting stability elements. Elements affecting posttranscriptional regulation of transcripts are often located in the untranslated regions of genes, the 5′ and 3′ UTRs, and introns (Mignone et al., 2002). Therefore, it might be expected that the elements determining CCA1 mRNA stability are found in the untranslated regions of CCA1.
We made a novel construct (CCA1∷eIF4A-ox) to test the contribution of the untranslated regions of CCA1 mRNA to stability. CCA1∷eIF4A-ox contains the coding region of CCA1 and 19 nt (out of 237 nt) of 5′ UTR under the control of the CaMV 35S promoter. Furthermore, in the CCA1∷eIF4A-ox construct, the 3′ UTR of CCA1 has been replaced with the 3′ UTR from AteIF4A, a highly stable transcript (Gutierrez et al., 2002). Plants were transformed with the CCA1∷eIF4A-ox construct and homozygous lines recovered. Figure 6, A and B, shows that in two independent transgenic lines of CCA1∷eIF4A-ox, CCA1 transcripts were still unstable in the light. Our results indicate that the elements that control different stability are most likely to be at least partly localized in the coding region rather than in the 5′ and 3′ UTRs and introns.
Figure 6.
The cis-elements regulating the differences in CCA1 mRNA stability in light and dark are probably not located in the noncoding regions. Three-week-old CCA1∷eIF4A-ox 1 and CCA1∷eIF4A-ox 2 plants were grown in light:dark cycles. The levels of CCA1 mRNA were determined by quantitative real-time PCR and plotted on a graph relative to the levels of TUB2. The averages of three independent experiments are shown together with the ses. The white and black bars represent light and dark periods, respectively. A, CCA1∷eIF4A-ox 1. B, CCA1∷eIF4A-ox 2.
DISCUSSION
It has been known for several years that the plant circadian clock is entrained by light signals that affect the transcription of its components, such as CCA1 and LHY (Wang and Tobin, 1998; Kim et al., 2003). More recently, evidence suggests that there are also additional levels of light entrainment that affect posttranscriptional events (e.g. TOC1 and GI protein stability and LHY translation rate; Kim et al., 2003; Mas et al., 2003; David et al., 2006). Here, we present evidence that light has an additional role in posttranscriptional regulation by controlling the stability of CCA1 RNA.
The light-induced changes in CCA1 mRNA stability were followed using plants expressing CCA1 under the control of the constitutive CaMV 35S promoter. As we have demonstrated, expression of CO, Rab7, and BP transgenes from this promoter is not regulated by light. Other groups have shown similar results (Kim et al., 2003; Valverde et al., 2004; David et al., 2006). However, CCA1 transcript levels under the control of the CaMV 35S promoter cycle according to the light/dark conditions. In the dark, CCA1 mRNA levels are constantly high, while in the light, they rapidly decline to about 40% of the dark levels. We showed that the changes in CCA1 mRNA levels are not dependent on the site of transgene integration and hypothesize that it is the result of light-dependent changes in transcript stability. Our measurements do not distinguish between endogenous CCA1 mRNA and CCA1 mRNA from the transgene. However, because endogenous CCA1 expression is induced by light, albeit to lower levels in CCA1-ox 034 plants than wild type, endogenous CCA1 mRNA would, if anything, mask the reduction in CCA1 transcripts we observed in the light (Wang et al., 1997). Our results that CCA1 mRNA is less stable in light are consistent with an earlier report that showed CCA1 transcript levels increasing in CCA1-ox 034 plants transferred from light to constant dark (Wang and Tobin, 1998).
The use of the transcription inhibitor, cordycepin, enabled us to demonstrate that in vivo CCA1 transcripts are less stable in light than in dark. We also used the cordycepin results to estimate the CCA1 mRNA half-life. The half-life values are, however, only approximate due to the limited sample time frequency. Based on our cordycepin results, CCA1 transcript half-life is around 1.5 h in the light; thus, CCA1 represents one of the unstable to moderately unstable Arabidopsis genes, a group that comprises only about 4% of Arabidopsis genome (Gutierrez et al., 2002). In the dark, however, CCA1 mRNA is significantly more stable. Our cordycepin experiments also suggest the differences in CCA1 transcript levels in light and dark are not the result of differential transcription regulation controlled by cis-acting elements within in the CCA1 coding region. Finally, the cordycepin experiments show that light-dependent transcription is not necessary for CCA1 mRNA degradation. Thus, if small RNAs are involved in this process, they are not transcribed by light but may be light activated, as their effect can be seen in light even without transcription.
We also examined the factors regulating differences in CCA1 mRNA stability in light and dark. Plants have several known photoreceptors that are activated by different wavelengths of light. Our experiments showed that red but not far-red light caused CCA1 mRNA degradation, indicating that the active form of phytochrome (Pfr) might be involved. However, it is possible that blue-light photoreceptors are also involved, as blue-light caused a reduction in CCA1 mRNA levels. Using plants transformed with chimeric CCA1∷eIF4A constructs, we demonstrated that the cis elements affecting RNA degradation are probably part of the coding region, because even though the 3′ UTR was replaced and the 5′ UTR was almost abolished, CCA1 transcripts were still unstable in light.
Because CCA1 mRNA levels are clearly regulated by light at the level of transcription, why might there be a need for additional control at the level of transcript stability? It is possible that the differences in CCA1 mRNA decay rates we observed in light and dark are important for maintaining an accurately entrained circadian clock by allowing plants to synchronize CCA1 expression precisely to sunrise. Thus, in wild-type plants grown in light/dark cycles, CCA1 expression starts early in the morning, before sunrise (Wang and Tobin, 1998). Then at daybreak, a change in CCA1 transcript stability causes a rapid disappearance of CCA1 mRNA. The combination of these two events, induction of transcription and transcript degradation, will result in a sharper peak of CCA1 mRNA accumulation, allowing a more precise detection of early morning, irrespective of seasonal changes. Consistent with the idea that mRNA stability is important for the pattern of oscillator transcript accumulation, experiments carried out in mice have shown that increasing the stability of a chimeric construct based on Period3, an oscillator gene, significantly broadens the peak of expression (Kwak et al., 2006).
Light-mediated changes in transcription rate and transcript stability can also be important for modifying the effects of environmental noise in clock entrainment. Kramer et al. (2003) have shown that in Neurospora, frq antisense transcripts are activated by light and are probably important for limiting the response of the clock to extremes in the environment.
In summary, we have found that light controls CCA1 mRNA degradation. Our findings, together with recent results from other groups, suggest that there are multiple entrainment points in the Arabidopsis circadian system (e.g. light activation of CCA1 and LHY transcription and LHY translation and light repression of TOC1 and GI degradation; Wang and Tobin, 1998; Kim et al., 2003; Mas et al., 2003; David et al., 2006). There is thus a complex network of light regulation throughout the day to ensure tight control of oscillator function and entrainment.
MATERIALS AND METHODS
Plant Materials and Growth
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) was used for all experiments unless stated otherwise. CCA1-ox 034 and CCA1-ox 038 contain an insertion of CaMV35S∷CCA1 cDNA in Col-0 (Wang and Tobin, 1998). This insertion includes 23 nt of the 5′ UTR, the coding region, and all the 3′ UTR of CCA1. 35S∷CO contains an insertion of the genomic coding sequence under control of the 35S promoter in Col-0 (Onouchi et al., 2000). AtRab7-7 contains an insertion of 35S∷Rab7 cDNA in Wassilewskija (Mazel et al., 2004). 35S∷BP contains an insertion of BP cDNA under control of the 35S promoter in Nossen (Lincoln et al., 1994). lhy-1TN104 contains an insertion of the LHY genomic sequence under control of the 35S promoter in Landsberg erecta (Schaffer et al., 1998).
All seeds were imbibed and cold treated at 4°C for 4 d before germination. All plants, except those used for transformations, were grown in petri dishes on Murashige and Skoog (Weigel and Glazebrook, 2002) medium from Duchefa Biochemie supplemented with 1% Suc (w/v). Plants used for transformations were grown on soil. Plants used for examining mRNA levels in different light conditions were grown under 14:10 light:dark (125 μE m−2 s−1) cycles for 3 weeks before being transferred to constant dark, red light (50 μE m−2 s−1), blue light (50 μE m−2 s−1), far red (50 μE m−2 s−1), or left in light:dark. All experiments were performed at a constant 23°C. Philips fluorescent lights, TLD 18W/29 and TLD18W/33CW, and LED lights provided lighting for plant growth.
RNA Analysis
RNA extraction and northern analysis were carried out as previously described (Green and Tobin, 1999). As a control to ensure that we could directly compare results from northerns and quantitative real-time PCR, we analyzed a set of CCA1 mRNA by both techniques. Supplemental Figure S1 shows that in our experimental system, northerns and quantitative real-time PCR give very similar results. The primers used to make RNA probes for the northern analyses were: CCA1 forward: GTTGCAGCTGCTAGTGCTTG, CCA1 reverse: TGTAATACGACTCACTATAGGGAAGATCGAGCCTTTGATGC, TUB2 forward: CCTTAAATCTCCGGATTTCGATTC, and TUB2 reverse: TGTAATACGACTCACTATAGGGCCGGTTGGATCGATGCCGTG.
For quantitative real-time PCR, RNA samples were treated with DNase (DNA-free from Ambion) according to the manufacturer's instructions. DNA-free RNA samples were then used as a template to produce cDNA using Reverse-iT Max 1st Strand Synth kit from ABgene with random-hexamer primers according to the manufacturer's instructions. cDNA samples were diluted 5-fold and used as templates for the real-time PCR reaction by using ABsolute SYBR Green ROX mix from ABgene according to the manufacturer's instructions. Reactions were performed in a Rotagene real-time PCR machine. The primers for quantitative real-time PCR were: CCA1 forward, TCCAGATAAGAAGTCACGCTCA, CCA1 reverse, TCTAGCGCTTGACCCATAGC; CCA1∷eIF4A forward, AGAAAGATCCCAAACGGATG, CCA1∷eIF4A reverse, AGTACGGCAGAGCAAACACA; CO forward, ATATGGCTCCTCAGGGACTCACTA, CO reverse, ACTCCGGCACAACACCAGTTT; Rab7 forward, AGACGCGTTCCTGTGCATAACT, Rab7 reverse, TGTGGATTGCTGGTTCCGACAT; LHY forward, GCTAAGGCAAGAAAGCCATA, LHY reverse, TGCCAAGCTCTTCCATAAAG; TUB2 forward, GGTTGAGCCTTACAACGCTACTCT, TUB2 reverse, GTGGTTCAAATCACCAAAGCTGGG; eIF4A forward, GCACACAGTTTGATGCACGTCAGT, eIF4A reverse, GGTTCTCTTGAAGACCCATGGCA; BP forward, TCCCATTCACATCCTCAACA, and BP reverse, GTTTCCCCTCCGCTGTTATT.
RNA Stability Measurements
At the end of a dark period, 3-week-old plants were transferred to incubation buffer containing 1 mm PIPES, pH 6.25, 1 mm sodium citrate, 1 mm KCI, and 15 mm Suc. After 30 min, 3′-deoxyadenosine (cordycepin; Sigma) was added to a final concentration of 0.6 mm. A vacuum treatment was applied for 45 s. The manipulations were carried out in a darkroom under a green safe light. Plants were then kept in the dark for another 20 min before being transferred to light (125 μE m−2 s−1) or kept in the dark. Samples were taken at intervals as described in the text. As a control for cordycepin activity, the levels of an unstable transcript encoded by the expansin-like gene At3G45970 were measured (Gutierrez et al., 2002). As expected, At3G45970-encoded mRNA levels dropped rapidly in both light and dark following cordycepin treatment (data not shown).
Preparation of the CCA1∷eIF4A-ox Construct
The pBS-CCA1 plasmid, containing the CCA1 coding region cloned into the pBlueScript II KS+ (gi: 58061) plasmid between the PstI and BamHI sites, was obtained from Elaine Tobin, UCLA. The eIF4A 3′ UTR was amplified by reverse transcription-PCR and cloned into the pBlueScript between the EcoRI and BamHI sites to create pBS-eIF4A. pBS-CCA1 was digested with XbaI to isolate the CCA1 coding region, which was then cloned into the XbaI site on pBS-eIF4A. The direction of the insertion was confirmed by sequencing. The plasmid was then digested with BamHI and religated to minimize the space between the CCA1 coding region and the eIF4A 3′ UTR. The resulting plasmid was called pBS-CCA1∷eIF4A.
To generate additional cloning sites, pHY-Bar binary vector for plant transformation obtained from Elaine Tobin, UCLA, was digested with BamHI and XbaI and religated with the linker: forward, GATCCTCTAGAATCGATCTCGAGG, reverse, CTAGCCTCGAGATCGATTCTAGAG. pBS-CCA1∷eIF4A was digested with XbaI and XhoI and the CCA1∷eIF4A fragment was inserted between XbaI and XhoI to create pHY-Bar-CCA1-eIF4A, which contains the CCA1∷eIF4A-ox construct.
Plant Transformation
GV3101∷pMP90RK Agrobacteria containing the binary vector pHY-Bar-CCA1-eIF4A was cultured in Luria-Bertani medium at 28°C with agitation until OD600 nm = 1. Three-week-old flowering Arabidopsis plants were dipped in floral dip medium for 5 min (Weigel and Glazebrook, 2002). Plants were left horizontally in the dark for 24 h, then grown for three more weeks until the seeds were ready for harvesting. Transformed plants were identified by their resistant to 1% Basta (glufosinate ammonium).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantitative PCR analysis of CCA1 mRNA levels is not affected by minor oscillations in TUB2.
Supplemental Figure S2. Estimations of CCA1 mRNA levels by northern and by quantitative real-time PCR are comparable.
Supplementary Material
Acknowledgments
We thank Elaine Tobin, Pamela J. Green, Frédéric F. Souret, Alex Levine, Naomi Ori, and Alon Samach for their generous gifts of plasmids and seeds, David Greenberg for technical advice, and Simon Barak and Shai Yerushalmi for their critical reading of the manuscript.
This work was supported by the Israel Science Foundation (grant no. 0397232) and by the Enrico Berman Fund (grant no. 0347865).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rachel M. Green (rgreen@vms.huji.ac.il).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 [DOI] [PubMed] [Google Scholar]
- Anderson MB, Folta K, Warpeha KM, Gibbons J, Gao J, Kaufman LS (1999) Blue light-directed destabilization of the pea Lhcb1*4 transcript depends on sequences within the 5′ untranslated region. Plant Cell 11 1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S, Li T, Angela DK, Christopher LS, Sharon RF, Lynn FD, Marie EP (2004) The Fed-1 (CAUU)4 element is a 5′ UTR dark-responsive mRNA instability element that functions independently of dark-induced polyribosome dissociation. Plant Mol Biol 56 761–773 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 268–280 [DOI] [PubMed] [Google Scholar]
- Chen C-YA, Shyu A-B (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20 465–470 [DOI] [PubMed] [Google Scholar]
- Colot HV, Loros JJ, Dunlap JC (2005) Temperature-modulated alternative splicing and promoter use in the circadian clock gene frequency. Mol Biol Cell 16 5563–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KM, Armbruster U, Tama N, Putterill J (2006) Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett 580 1193–1197 [DOI] [PubMed] [Google Scholar]
- Ding Z, Millar AJ, Davis AM, Davis SJ (2007) TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Kaufman LS (2003) Phototropin 1 is required for high-fluence blue-light-mediated mRNA destabilization. Plant Mol Biol 51 609–618 [DOI] [PubMed] [Google Scholar]
- Gil P, Green PJ (1996) Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J 15 1678–1686 [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Locke JCW, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Perez-Amador MA, Lidder P, Green PJ (2000) Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc Natl Acad Sci USA 97 13991–13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by a SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Kim JY, Song HR, Taylor BL, Carré IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449 356–360 [DOI] [PubMed] [Google Scholar]
- Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK (2003) Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421 948–952 [DOI] [PubMed] [Google Scholar]
- Kwak E, Kim TD, Kim KT (2006) Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of Mouse Period3 mRNA. J Biol Chem 281 19100–19106 [DOI] [PubMed] [Google Scholar]
- Lam L, Pickeral O, Peng A, Rosenwald A, Hurt E, Giltnane J, Averett L, Zhao H, Davis RE, Sathyamoorthy M, et al (2001) Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biology 2 research0041.0041–research0041.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ (2005) Circadian control of messenger RNA stability: association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138 2374–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1 E1–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Chen WF, Edery I (2004) Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol 24 3359–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I (1999) How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24 219–230 [DOI] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570 [DOI] [PubMed] [Google Scholar]
- Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ (1989) Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E (2004) Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 39 197–216 [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2003) Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol 132 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biology 3 reviews0004.0001–reviews0004.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46 686–698 [DOI] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Nguyen TT, Gatz C, Sowinski DA, Allen GC, Thompson WF (1998) Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci USA 95 9009–9013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J (1995) mRNA stability in mammalian cells. Microbiol Rev 59 423–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Yakir E, Hilman D, Harir Y, Green RM (2007) Regulation of output from the plant circadian clock. FEBS J 274 335–345 [DOI] [PubMed] [Google Scholar]
- Zhang S, Sheng J, Liu Y, Mehdy MC (1993) Fungal elicitor-induced bean proline-rich protein mRNA down-regulation is due to destabilization that is transcription and translation dependent. Plant Cell 5 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.