Abstract
Each plant cell type expresses a unique transcriptome and proteome at different stages of differentiation dependent on its developmental fate. This study compared gene expression and protein accumulation in cell-cycle-competent primary root pericycle cells of maize (Zea mays) prior to their first division and lateral root initiation. These are the only root cells that maintain the competence to divide after they leave the meristematic zone. Pericycle cells of the inbred line B73 were isolated via laser capture microdissection. Microarray experiments identified 32 genes preferentially expressed in pericycle versus all other root cells that have left the apical meristem; selective subtractive hybridization identified seven genes preferentially expressed in pericycle versus central cylinder cells of the same root region. Transcription and protein synthesis represented the most abundant functional categories among these pericycle-specific genes. Moreover, 701 expressed sequence tags (ESTs) were generated from pericycle and central cylinder cells. Among those, transcripts related to protein synthesis and cell fate were significantly enriched in pericycle versus nonpericycle cells. In addition, 77 EST clusters not previously identified in maize ESTs or genomic databases were identified. Finally, among the most abundant soluble pericycle proteins separated via two-dimensional electrophoresis, 20 proteins were identified via electrospray ionization-tandem mass spectrometry, thus defining a reference dataset of the maize pericycle proteome. Among those, two proteins were preferentially expressed in the pericycle. In summary, these pericycle-specific gene expression experiments define the distinct molecular events during the specification of cell-cycle-competent pericycle cells prior to their first division and demonstrate that pericycle specification and lateral root initiation might be controlled by a different set of genes.
Maize (Zea mays) primary roots have a radial organization in the transverse orientation that is determined by the presence of various functionally diverse cell types (Sass, 1977). The central cylinder of the root contains vascular xylem and phloem elements necessary for water and nutrient transport. The outermost cell layer of the central cylinder is a single anatomically distinct layer of thin-walled pericycle cells (Feldman, 1994). Longitudinally, maize roots can be divided into a meristematic zone at the root tip, followed by an elongation zone and a differentiation zone characterized by root hairs (Ishikawa and Evans, 1995). After root cells have left the root apical meristem, they are specified into different cell types. A unique attribute of pericycle cells compared to other root cells that have left the meristematic zone is the competence of a subset of these genes to re-enter the cell cycle and become founder cells of lateral root meristems. The mitotic activation of pericycle cells can be triggered by both endogenous and exogenous signals (Dubrovsky et al., 2000). Whether pericycle cells in maize are already differentiated when they re-enter the cell cycle is still under debate (Dubrovsky et al., 2000; Casimiro et al., 2003). However, the observation that in some maize cultivars pericycle cells start initiating lateral root primordia about 8 h after they have left the meristematic zone of the root apex (Dubrovsky and Ivanov, 1984) supports the notion that these pericycle cells are not completely differentiated before they re-enter the cell cycle. In many species, it has been demonstrated that the pericycle maintains its competence to divide constitutively (Beeckman et al., 2001; Roudier et al., 2003). In Arabidopsis (Arabidopsis thaliana) and many other species, pericycle cells opposite the xylem pole become founder cells (De Smet et al., 2006). In contrast, in maize (Casero et al., 1995) and other grasses, including rice (Oryza sativa; Nishimura and Maeda, 1982) and wheat (Triticum vulgare; Foard et al., 1965), pericycle cells that become founder cells are located at the phloem poles. The positioning of these founder cells must have an important developmental function. It is assumed that the direct contact of the founder cells with the vascular transport system might be beneficial because the xylem is responsible for root-to-shoot transport of water and dissolved ions (De Smet et al., 2006). In this context, the positioning of the founder cells in the maize pericycle between two xylem strands might even enhance this process (Bell and McCully, 1970).
In maize, two mutants, lrt1 (Hochholdinger and Feix, 1998) and rum1 (Woll et al., 2005), have been identified that do not initiate lateral roots from pericycle cells of the embryonic primary and seminal roots. The observation that the mutants do not affect lateral root initiation in the postembryonically formed shoot-borne roots implies the existence of root-type-specific differences in pericycle cell specification or lateral root initiation (Hochholdinger et al., 2004c, 2004d). This, together with the distinct positioning of pericycle cells that will become root founder cells in maize versus Arabidopsis, makes it likely that the molecular events during the specification of the pericycle are in some points fundamentally different between monocots and dicots. A recent pericycle-specific transcriptome analysis of the maize rum1 mutant that does not initiate lateral roots has revealed a number of genes related to signal transduction, transcription, and cell cycle that were differentially expressed between wild-type and mutant pericycle cells (Woll et al., 2005). These genes might thus be related to the process of lateral root initiation (Woll et al., 2005).
A global gene expression map of the Arabidopsis primary root was created by separating different cell types via protoplasting of cell-type-specific promoter∷GFP marker lines in a fluorescence-activated cell sorter (Birnbaum et al., 2003). RNA of the different cell types was subsequently hybridized to Affymetrix 22K Arabidopsis microarray chips. Pericycle-specific gene expression was not addressed in this study. Laser capture microdissection (LCM) is an alternative technology that enables the analysis of cell-type-specific gene expression profiles in species like maize, where no cell-type-specific marker lines are available (Asano et al., 2002; Kerk et al., 2003; Nakazono et al., 2003; Woll et al., 2005). In this approach, frozen or fixed cells of interest are physically linked to a thermoplastic film with a low-power laser beam or catapulted into a collection tube with a defocused laser (Schnable et al., 2004). After isolation and amplification of RNA from these cells, their corresponding cDNA can be used for further analyses.
The identification of genes and proteins predominantly expressed in pericycle versus nonpericycle cells that have left the meristematic zone of the young maize primary root will help to define a set of genes that might be related to the unique attribute of pericycle cells to maintain competence for cell division and to dissect molecular differences between the developmental processes of pericycle specification and lateral root initiation. Finally, such data could be helpful for future identification of molecular differences between monocot and dicot pericycle cells.
RESULTS
B73 Primary Root Pericycle Cells Do Not Divide in the First 3 d after Germination
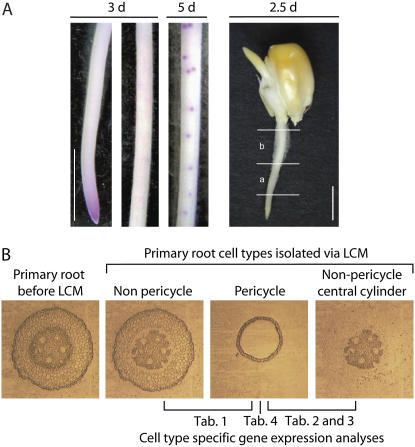
To detect the earliest cell divisions in the maize primary root pericycle of the inbred line B73, we analyzed the time course of lateral root initiation by whole-mount staining of young primary roots with Schiff's reagent (Fig. 1A). Schiff's reagent specifically stains nuclei and can thus be used to detect meristematic tissue, which contains a high concentration of dividing cells. No lateral root primordia were detectable in the differentiation zone of any wild-type (B73) primary root sample 3 d after germination (Fig. 1A, 3 d). These roots displayed meristematic activity only in the primary root tip (Fig. 1A, 3 d). The first lateral root primordia were detected as faint spots in 4-d-old primary roots. These primordia became more pronounced 5 d after germination (Fig. 1A, 5 d), shortly before they penetrated the primary root surface, and finally became visible as lateral roots from outside. The absence of cell divisions in pericycle cells outside the meristematic zone of 3-d-old primary roots was confirmed by microscopic analyses of serial cross sections of maize primary roots (data not shown).
Figure 1.
A, Feulgen staining of B73 roots shows lateral root initiation via emerging primordia. No lateral root primordia are visible 3 d after germination (3 d), whereas primordia were detected at 5 d. Sampling strategy for LCM experiments on 2.5-d-old root: (a) elongation zone; (b) differentiation zone. Approximately 3 mm of the root tip were discarded. Same number of sections of a and b make one biological replicate. Bars indicate 5 mm. B, B73 root cross sections 2.5 d: before and after capturing pericycle, nonpericycle, and nonpericycle central cylinder cells.
Isolation of Pericycle and Nonpericycle Cells via LCM
Pericycle cells represent the outermost cell layer of the central cylinder and are the only root cells that maintain competence for cell division outside the apical meristem. Pericycle-specific gene expression cannot be studied in whole-root extracts because its expression profile would be masked by the expression profiles of the other cell types that compose the majority of the root. Therefore, isolation of pericycle cells from the surrounding cell layers is required to study the transcriptome and proteome of this cell type. Because we were interested in the analysis of pericycle-specific gene expression before the first divisions of the founder cells (i.e., during the specification of this cell type), cells from cross sections of primary roots that were cultivated in paper rolls (Hoecker et al., 2006) were collected 64 h (2.5 d) postimbibition. This sampling strategy controlled the variance associated with the first cell divisions that occur between 72 to 96 h (3–4 d) after germination (Fig. 1A). In each experiment, similar numbers of cells were collected from the lower part of the root, which included the elongation zone (Fig. 1A, 2.5 d, part a) and the upper part of the root, which included the root hair or differentiation zone (Fig. 1A, 2.5 d, part b). Because elongation and differentiation zones are partly overlapping (Ishikawa and Evans, 1995), these zones could not be clearly separated. Only every fifth cross section that was obtained with the microtome along the primary root was collected to achieve equally distributed sampling throughout the complete primary root. The size of the meristematic zone of the primary root tip was determined via Feulgen staining (Fig. 1A, 3 d) and subsequent size determination via Image Pro Express software. On average, in 2.5-d primary roots, the meristematic zone has a size of 2.7 ± 0.03 mm (n = 48). Therefore, 3 mm of the root tip that contain the meristematic zone of the primary root were discarded. Hence, only nondividing pericycle cells outside the meristematic zone were included in our analyses.
Figure 2.
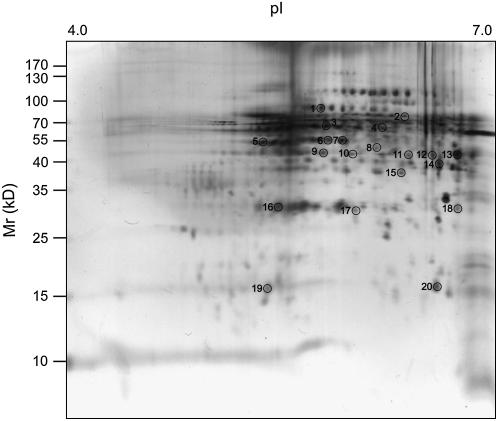
Representative proteomic 2-D map of soluble protein extracts from pericycle cells isolated via LCM from 2.5-d-old maize primary roots. Proteins were separated in the first dimension according to their pIs on IPG strips pH 4 to 7 and in the second dimension according to their molecular masses on a 12% SDS-polyacrylamide gel. Proteins were detected with silver staining. Proteins that were identified via ESI-MS/MS and the MS search engine MASCOT are numbered on the map.
We compared gene expression in the pericycle with two types of root cells (Fig. 1B). First, we analyzed gene expression within pericycle cells with all other transverse nonpericycle cells via 12K microarray hybridization experiments (Table I) of the Iowa State University (ISU) GenII vA chip (GenII vA: gene expression omnibus platform GPL 4876; http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GPL4876). Second, we performed gene expression analysis of the central cylinder by comparing the expression within pericycle cells that represent the outermost cell layer of the central cylinder with the functionally diverse nonpericycle cells of the central cylinder comprising xylem and phloem cells, via suppression subtractive hybridization (SSH) and EST sequencing (Tables II and III). Finally, we generated a reference proteome map of the pericycle (Table IV). All pericycle and nonpericycle cells subjected to the different gene expression experiments were isolated via LCM (“Materials and Methods”). Total RNA was extracted from approximately 13,000 captured root cells per biological replicate. This number of cells provided sufficient RNA (25–150 ng) for subsequent RNA amplifications. Two linear amplifications (Woll et al., 2005) yielded between 6 to 20 μg of amplified RNA (aRNA).
Table I.
Genes identified via microarray experiments to be preferentially expressed in pericycle versus nonpericycle cells
AC, Accession no.
| Gene No. | Gene IDa | Fc Arrayb | P Valuec | Fc Blotd | Function (BLASTx) [Species] ACe |
|---|---|---|---|---|---|
| Protein Synthesisf | |||||
| 1 | 486043A01.x3 | 1.96 | 3.85 × 10−03 | 4.09 | 60S ribosomal protein L38 [O. sativa] BAC45072.1 |
| 2 | MEST33-A07 | 1.82 | 9.33 × 10−03 | 1.97 | Translation initiation factor 3, subunit 11 [O. sativa] Q94HF1 |
| 3 | 618008D05.x1 | 1.78 | 7.25 × 10−03 | 2.07 | Ribosomal S29-like protein [A. thaliana] NP_189984.1| |
| 4 | MEST15-H09 | 1.60 | 6.71 × 10−03 | 1.64 | 60S ribosomal protein L22 [A. thaliana] NP_187207.1 |
| Transcription | |||||
| 5 | 606065E07.x1 | 2.05 | 1.65 × 10−03 | 1.63 | Transcription elongator protein 3 [O. sativa] CAD40910.2 |
| 6 | 618046D09.y1 | 2.03 | 3.44 × 10−03 | 1.98 | Zinc finger DHHC domain-containing protein 2 [O. sativa] NP_915461.1 |
| 7 | 606063C12.x1 | 1.89 | 5.69 × 10−03 | 3.71 | Putative homeodomain Leu zipper protein [O. sativa] Q6YWR4 |
| 8 | MEST125-D04 | 1.86 | 9.22 × 10−03 | 1.68 | RNase L inhibitor-like protein [O. sativa] AAM19067.1 |
| 9 | 486057C02.x2 | 1.73 | 8.64 × 10−03 | 1.58 | Ethylene-insensitive-3-like protein [O. sativa] BAB78462.1 |
| 10 | 606011B10.x1 | 1.62 | 8.39 × 10−03 | 2.77 | tRNA-pseudouridine synthase [A. thaliana] AAT06466 |
| Signal Transduction | |||||
| 11 | 614051F10.x2 | 2.18 | 9.13 × 10−03 | 1.52 | Ser/Thr protein kinase [O. sativa] CAD41330.2 |
| 12 | 606065H03.x1 | 1.63 | 2.20 × 10−03 | 4.90 | Calcium-dependent protein kinase [O. sativa] BAB92912.1 |
| Disease/Defense | |||||
| 13 | 486056D01.x1 | 1.98 | 9.98 × 10−03 | 2.04 | DnaJ-class molecular chaperone [A. thaliana] NP_191819.1 |
| 14 | 614016D12.y1 | 1.77 | 9.28 × 10−03 | 2.66 | Pathogen-related protein PR10 [O. sativa] AAL27005.1 |
| 15 | MEST39-B06 | 1.70 | 2.77 × 10−03 | 1.75 | Pollen-specific protein C13 [O. sativa] AAM08621.1 |
| 16 | 486086H01.x1 | 1.58 | 6.35 × 10−04 | 2.15 | Glutathione S-transferase GST 13 [Z. mays] Q9FQC6 |
| Energy | |||||
| 17 | 486045A09.x1 | 1.76 | 8.84 × 10−03 | 3.45 | Digalactosyldiacylglycerol synthase [Glycine max] AAT67420 |
| Metabolism | |||||
| 18 | MEST26-D05 | 1.68 | 2.26 × 10−03 | 2.20 | Polyamine oxidase [Z. mays] CAC04002.1 |
| Protein Fate | |||||
| 19 | 614083G03.x1 | 2.37 | 9.60 × 10−03 | 3.47 | Cys proteinase [O. sativa] AAK73137.1 |
| 20 | MEST34-E09 | 1.98 | 6.49 × 10−03 | 3.20 | Peptidyl-prolyl cis-trans isomerase [Z. mays] P21569 |
| Subcellular Localization | |||||
| 21 | 707031D05.x2 | 1.94 | 5.87 × 10−04 | 1.66 | Ankyrin [A. thaliana] NP_787123.1 |
| 22 | 945031C06.Y1 | 1.72 | 1.52 × 10−04 | 3.21 | Tonoplast membrane integral protein (ZmTIP4-1) [Z. mays] Q9ATL6 |
| Unknown | |||||
| 23 | 707041F03.x1 | 3.83 | 9.77 × 10−03 | 2.49 | No database hit |
| 24 | 606017E12.x2 | 2.88 | 9.22 × 10−03 | 2.84 | Unknown protein [O. sativa] CAC39068.1 |
| 25 | 614022F10.y1 | 2.65 | 1.95 × 10−03 | 1.70 | No database hit |
| 26 | 707041E07.x1 | 2.15 | 5.30 × 10−03 | 1.60 | No database hit |
| 27 | 707064C07.y1 | 2.02 | 2.21 × 10−03 | 4.89 | Unknown protein [O. sativa] CAE01522.1 |
| 28 | 603021F03.x1 | 1.85 | 5.22 × 10−03 | 1.71 | Unknown protein [A. thaliana] NP_564254.1 |
| 29 | 486032D01.x2 | 1.71 | 4.95 × 10−03 | 7.57 | No database hit |
| 30 | 486030G07.x1 | 1.66 | 5.83 × 10−03 | 2.03 | No database hit |
| 31 | 606026E02.x2 | 1.57 | 3.68 × 10−03 | 3.56 | Unknown protein [O. sativa] BAB32964.1 |
| 32 | 687023G11.x2 | 1.55 | 9.34 × 10−03 | 1.61 | No database hit |
EST sequence that was spotted on the microarray.
Fc obtained in microarray experiments (cutoff value: 1.5).
P value of microarray experiments (cutoff value: 0.01).
Fc obtained in reverse RNA gel-blot experiments (cutoff value: 1.5).
Obtained via BLASTx (cutoff value: 1e−10).
Classification of the proteins according to Arabidopsis MIPS database (Schoof et al., 2002).
Table II.
Genes identified via SSH and confirmed via qRT-PCR to be preferentially expressed in pericycle versus nonpericycle central cylinder cells
AC, Accession no.
| Gene IDa | Function (BLASTx) [Species] ACb | Fcc |
|---|---|---|
| Protein Synthesisd | ||
| BT016828 | Elongation factor 2 [O. sativa] XP_465992 | 5.69**e |
| CB380608 | 60S ribosomal protein L10A (RPL10aC) [O. sativa] XP_483755 | 6.88** |
| Cellular Transport | ||
| CK827793 | PDR-like ABC transporter [O. sativa] CAD59574 | 4.93** |
| Disease/Defense | ||
| BM079363 | Heat shock protein 90 [O. sativa] BAD04054 | 2.65** |
| Metabolism | ||
| CF637826 | Terpene synthase 7 [O. sativa] ABF95916 | 3.48* |
| Signal Transduction | ||
| BT016182 | Translationally controlled tumor protein-like protein [Z. mays] AAN40686 | 2.03** |
| Unknown Function | ||
| AI861099 | Unknown protein [O. sativa] XP_465991 | 7.93** |
EST that corresponds to sequence obtained via SSH.
Obtained via BLASTx (cutoff value: 1e−10).
Fc obtained in qRT-PCR experiments.
Classification of the proteins according to Arabidopsis MATDB (Schoof et al., 2002).
Significance level in t test. **, P < 0.01; *, P < 0.05.
Table III.
Anchoring, functional classification, and clustering of 701 pericycle and nonpericycle ESTs to PUT (cDNA) and MAGI (genomic DNA) databases
AC, Accession no.
| Pericyclea | Nonpericycle Central Cylinder | Totalb | |
|---|---|---|---|
| (a) EST Anchoring | |||
| Anchored to PUTsc | 307 (246 + 28) | 283 (204 + 31) | 590 (421 + 63) |
| Anchored to MAGIsd | 21 (19 + 1) | 10 (8 + 1) | 31 (27 + 2) |
| Novel genese | 49 (44 + 2) | 31 (31 + 0) | 80 (75 +2) |
| Total No. | 377 (309 + 31) | 324 (243 + 32) | 701 (523 + 67) |
| Pericycle | Nonpericycle Central Cylinder | Total | P Valuef | |
|---|---|---|---|---|
| (b) Functional Classification | ||||
| Unknown | 119 (32%) | 102 (31%) | 221 (32%) | 1.000 |
| Metabolism | 49 (13%) | 51 (16%) | 100 (14%) | 0.330 |
| Protein synthesis | 52 (14%) | 24 (7%) | 76 (11%) | 0.007 |
| Protein with binding function | 35 (9%) | 21 (6%) | 56 (8%) | 0.209 |
| Protein fate | 24 (6%) | 24 (7%) | 48 (7%) | 0.654 |
| Disease/defense | 19 (5%) | 25 (8%) | 44 (6%) | 0.161 |
| Energy | 19 (5%) | 17 (5%) | 36 (5%) | 1.000 |
| Signal transduction | 16 (4%) | 20 (6%) | 36 (5%) | 0.304 |
| Transport | 18 (5%) | 12 (4%) | 30 (4%) | 0.576 |
| Transcription | 16 (4%) | 10 (3%) | 26 (4%) | 0.548 |
| Cell fate | 4 (1%) | 15 (5%) | 19 (3%) | 0.004 |
| Cell cycle | 5 (1%) | 2 (1%) | 7 (1%) | 0.460 |
| Transposons | 1 (<1%) | 1 (0%) | 2 (0%) | 1.000 |
| Total No. | 377 | 324 | 701 |
| PlantGDB ACg | No. of ESTsh | Function (BLASTx) [Species] GenBank ACi |
|---|---|---|
| (c) EST Clustering | ||
| Pericycle (377 ESTs)j | ||
| PUT-155a-Zea_mays-109877741 | 5 (3) | HMGB1 [Z. mays] P27347 |
| PUT-155a-Zea_mays-126577737 | 3 (0) | No database hit |
| PUT-155a-Zea_mays-120277744 | 3 (4) | 60S ribosomal protein L17 [Z. mays] O48557 |
| Contig 2 | 3 (0) | No database hit |
| Nonpericycle Central Cylinder (324 ESTs) | ||
| PUT-155a-Zea_mays-40243 | 8 (1) | No database hit |
| PUT-155a-Zea_mays-082734 | 4 (1) | Photosynthetic reaction center [Medicago truncatula] ABE92785.1 |
| PUT-155a-Zea_mays-120277744 | 4 (3) | 60S ribosomal protein L17 [Z. mays] O48557 |
| PUT-155a-Zea_mays-38477738* | 4 (0) | Glutathione S-transferase I [Z. mays] P12653 |
| PUT-155a-Zea_mays-95577739 | 3 (1) | Low-temperature/salt-responsive protein [Pennisetum glaucum] AAV88601.1 |
| PUT-155a-Zea_mays-111477743 | 3 (2) | No database hit |
| PUT-155a-Zea_mays-109877741 | 3 (5) | HMGB1 [Z. mays] P27347 |
| PUT-155a-Zea_mays-018803 | 3 (0) | Physical impedance-induced protein [Z. mays] AAC31615.1 |
| PUT-155a-Zea_mays-018690 | 3 (0) | Dynamin GTPase effector [M. truncatula] ABD28549.1 |
Number in brackets indicates: (singletons + contigs).
Note that singleton and contig numbers of pericycle and nonpericycle central cylinder ESTs are not necessarily additive because some contigs have only one member in a library and become only a contig when both libraries are analyzed in common.
ESTs that were anchored to known EST contigs (PUTs) via sequence information retrieved from www.plantgdb.org.
ESTs that were anchored to known genomic sequences (MAGIs) via sequence information retrieved from http://magi.plantgenomics.iastate.edu.
ESTs that did not fit to any known EST or genomic sequence and can thus be considered novel sequence information.
P values for Fisher's exact test of difference between proportions.
PUTs that are represented by at least three ESTs in a library.
Number of ESTs retrieved per contig by randomly sequencing library clones. Number of the corresponding contig in the second library in brackets.
Obtained via BLASTx versus plantGDB “all plant proteins” db (cutoff value: 1e−10).
Library and number of sequenced clones.
Table IV.
Primary root pericycle proteins isolated from B73 seedlings identified after 2-D electrophoresis and ESI-MS/MS analysis of trypsin-digested proteins matched against NCBI nonredundant protein database entries
AC, Accession no.
| Spot No. | Protein [Species] ACa | Molecular Mass (kD) Predicted/Gelb | pI Predicted/Gelc | Sequence Coveraged | MASCOT Scoree | No. Matched Peptidesf |
|---|---|---|---|---|---|---|
| Signal Transductiong | ||||||
| 15 | Guanine nucleotide-binding protein β-subunit protein [O. sativa] NP_001056254 | 36.4/38 | 6.06/6.0 | 10% | 128 | 4 |
| Cellular Organization | ||||||
| 5 | Actin [Saccharum officinarum] AAU93346 | 41.7/53 | 5.24/5.3 | 12% | 171 | 4 |
| Disease/Defense | ||||||
| 18 | 1-Cys peroxiredoxin antioxidant [Z. mays] ABD24377 | 25.0/30 | 6.31/6.2 | 17% | 84 | 4 |
| 19 | Pathogenesis-related protein 10 [Z. mays] AAY29574h | 16.9/16 | 5.39/5.3 | 21% | 134 | 3 |
| Energy | ||||||
| 4 | ATPase subunit 1 [Z. mays] ABE98710 | 55.1/62 | 5.85/5.9 | 12% | 203 | 6 |
| 7 | 3-Phosphoglycerate kinase [Z. mays] AAO32644 | 31.6/53 | 5.01/5.7 | 26% | 465 | 8 |
| 11 | Glyceraldehyde-3-P dehydrogenase [Z. mays] Q09054 | 36.5/43 | 6.41/6.1 | 15% | 127 | 5 |
| 13 | Glyceraldehyde-3-P dehydrogenase [Z. mays] Q09054 | 36.5/43 | 6.41/6.3 | 16% | 204 | 5 |
| 12 | Glyceraldehyde-3-P dehydrogenase [Z. mays] AAA33466 | 26.4/42 | 6.25/6.2 | 23% | 156 | 5 |
| 14 | Glyceraldehyde-3-P dehydrogenase [Z. mays] AAA87579 | 36.4/39 | 6.61/6.2 | 18% | 255 | 6 |
| Metabolism | ||||||
| 1 | Met synthase [Z. mays] AAL33589 | 84.4/87 | 5.73/5.6 | 4% | 88 | 3 |
| 2 | Phe ammonia-lyase [Z. mays] AAL40137 | 74.9/75 | 6.52/6.0 | 4% | 77 | 3 |
| 3 | Wheat adenosylhomocysteinase-like protein [O. sativa] AAO72664 | 53.2/63 | 5.62/5.6 | 12% | 295 | 7 |
| 6 | S-adenosylmethionine synthetase [O. sativa] AAT94053i | 43.2/55 | 5.74/5.6 | 16% | 234 | 6 |
| 8 | Glu dehydrogenase [Z. mays] AAB51596 | 44.0/48 | 5.96/5.9 | 8% | 146 | 3 |
| 9 | α-1,4-Glucan-protein synthase [Z. mays] P80607 | 41.2/45 | 5.75/5.6 | 17% | 206 | 7 |
| 10 | α-1,4-Glucan-protein synthase [Z. mays] P80607 | 41.2/44 | 5.75/5.8 | 9% | 69 | 4 |
| 16 | Glutathione S-transferase I [Z. mays] P12653 | 23.8/31 | 5.44/5.4 | 15% | 250 | 6 |
| 17 | Glutathione S-transferase IV [Z. mays] P46420 | 24.6/30 | 5.77/5.8 | 13% | 172 | 4 |
| 20 | Nucleoside diphosphate kinase [S. officinarum] P93554 | 16.6/16 | 6.30/6.2 | 21% | 184 | 3 |
Identified proteins obtained via automated algorithm of the MASCOT software (www.matrixscience.com) from the NCBI nonredundant database.
Molecular mass of predicted protein/of protein on gel.
Isoelectric point of predicted protein/of protein on gel.
Percentage of predicted protein sequence covered by matched peptides (cutoff value: 4%).
−10*log(P), where P is the probability that the observed match is a random event. Scores >39 indicate identity or extensive homology (P < 0.05). Protein scores are derived from ion scores as a nonprobabilistic basis for ranking protein hits (cutoff value: >41).
Number of peptides that were identified of a particular protein via ESI-MS/MS (cutoff value: 3).
Classification of the proteins according to Arabidopsis MIPS database (Schoof et al., 2002).
This gene was differentially expressed in a microarray dataset comparing the expression of wild-type versus mutant rum1 pericycle cells (Woll et al., 2005).
Microarray Analysis of Pericycle versus Nonpericycle Primary Root Cells
Six biological replicates of aRNA from pericycle and nonpericycle cells (Fig. 1B; Table I) were generated and hybridized in pairwise fashion to the 12K spotted cDNA microarray slides, including a dye swap. Statistical analyses using specific variances for each gene revealed an initial dataset of 70 genes that displayed significantly higher expression in pericycle cells as compared to nonpericycle primary root cells of the corresponding transverse sections (fold change [Fc] > 1.5; P < 0.01; estimated false discovery rate [FDR] 35% by the method of Allison et al. [2002]). Due to the relatively low Fcs and high FDR, we subjected all 70 genes to independent confirmation experiments via reverse RNA gel blots. We were able to PCR amplify 65 of 70 differentially expressed genes from the corresponding clones of the maize unigene collection (www.maizegdb.org). These amplified gene sequences were blotted on two nylon membranes. aRNA of pericycle and nonpericycle cells was blotted as a normalization control on the nylon membrane. The membranes were hybridized in parallel with 32P-labeled pericycle and nonpericycle cDNA. Among the 65 genes, expression of 11 genes was below the detection limit of the reverse RNA dot-blot experiments and thus eliminated from subsequent analyses. Among the remaining 54 genes, 32 were confirmed to be differentially expressed with Fc of at least 1.5 and therefore accepted as preferentially expressed in the pericycle (Table I). Because ESTs typically cover only part of a full-length gene, we searched maize putative assembled unique transcripts (PUTs) at PlantGDB (www.plantgdb.org) to identify additional sequence information for all differentially expressed genes. Annotated protein homologs of those EST contigs were identified via the BLASTx algorithm with a cutoff value of 1e−10 (Altschul et al., 1997). For 22 of 32 genes preferentially expressed in pericycle cells, an annotated protein sequence was detected in the National Center for Biotechnology Information (NCBI) nonredundant database (as of December 12, 2006). These proteins were classified into functional categories according to the Arabidopsis Munich Information Center for Protein Sequences (MIPS) database, version 2.0 (http://mips.gsf.de/proj/thal/db). Among the 22 proteins with known function, six were classified in the transcription category, four in the protein synthesis category, and four in the disease/defense category. The remaining proteins fell in the signal transduction (n = 2), metabolism (n = 1), energy (n = 1), protein fate (n = 2), and subcellular localization (n = 2) categories. Among the remaining 10 genes, four genes displayed high similarity to proteins of unknown function, whereas no database hits were detected for the remaining six genes.
Identification of Genes That Are Preferentially Expressed in Pericycle versus Nonpericycle Central Cylinder Cells via SSH
SSH (Diatchenko et al., 1996) is a powerful technique to enrich genes that are differentially expressed between two tissues of interest. This technique is of particular interest when not all genes of a species are available on microarray chips. Thus far, SSH has been mainly applied to study differential gene expression in whole organs. We performed SSH with aRNA that enriched genes that are preferentially expressed in pericycle versus nonpericycle central cylinder cells (Fig. 1B; Table II). SSH products that were enriched for preferential expression in pericycle cells were cloned into the pGEM vector system and transformed into Escherichia coli JM109 cells. Confirmation of pericycle-specific gene expression was conducted via a two-step procedure. First, all candidates were prescreened via reverse RNA gel-blot hybridization. Second, expression of all genes that exhibited differential regulation in the RNA gel-blot experiment was tested via quantitative real-time PCR (qRT-PCR) experiments. For the reverse RNA gel-blot prescreening, 464 SSH products that were expected to be preferentially expressed in pericycle cells were randomly picked. Inserts of these clones were PCR amplified with SSH-specific primers forward and reverse (Supplemental Data S1). Each PCR product was blotted on two nylon membranes in parallel. One nylon membrane was hybridized with 32P-dCTP-labeled pericycle cDNA, whereas the second membrane was hybridized with 32P-dCTP-labeled cDNA from the nonpericycle central cylinder. The constitutively expressed GAPDH (GenBank accession no. X75326) and actin (GenBank accession no. AY104722) genes were used for normalization. In this first round of screening, 15 transcripts displayed preferential expression in the pericycle. These transcripts were sequenced and selected for qRT-PCR confirmation. Four independent biological replicates of both cell types were used to perform qRT-PCR experiments in which thioredoxin (AF435816) was used as a reference gene. Seven of the 15 genes were confirmed to be preferentially expressed in pericycle cells (Table II) via Student's t test (Fc > 2; P < 0.05). The seven differentially expressed genes belonged to the functional categories of protein synthesis (n = 2), cellular transport (n = 1), disease/defense (n = 1), signal transduction (n = 1), and metabolism (n = 1). One transcript encoded a protein of unknown function.
Analysis of Pericycle versus Nonpericycle Central Cylinder ESTs
Cell-type-specific high-throughput EST sequencing provides clues as to genes that are predominantly expressed in a particular cell type, but may also identify differentially expressed gene candidates between the analyzed tissues. Moreover, it allows for the identification of the most prevalent functional classes of expressed genes in a cell type and provides the opportunity to isolate genes not yet deposited in maize databases. We therefore generated cDNA (EST) libraries of pericycle and nonpericycle central cylinder cells (Fig. 1B; Table III). ESTs were cloned nondirectionally and sequenced from one direction, thus providing 5′ or 3′ sequences. After automated sequence cleanup, a total of 701 sequences were analyzed. All sequences were clustered into contigs by anchoring them to maize PUTs and maize assembled genomic islands (MAGIs; Fu et al., 2005; version 4.0 at http://magi.plantgenomics.iastate.edu) or among themselves if they were novel ESTs not yet available in any database. All ESTs were submitted to GenBank and to dbEST (for GenBank and dbEST accessions and functional annotation, see Supplemental Data S1). The 701 ESTs represented 523 singletons (74% of all ESTs) and 67 clusters consisting of at least two ESTs (Table IIIa). Among these 67 clusters, only 25 were present in both pericycle and nonpericycle central cylinder cells, suggesting considerable diversity in gene expression within these two tissues. The 377 ESTs of pericycle cells were grouped into 340 independent transcripts containing 309 singletons (82%), whereas 324 ESTs of nonpericycle central cylinder cells generated 275 independent transcripts containing 243 singletons (85%). Among the pericycle ESTs, 81% (307/377) were anchored to known maize PUTs, whereas 87% (283/324) of the nonpericycle central cylinder ESTs were anchored to known PUTs. Remarkably, 70 pericycle ESTs (19%) and 41 nonpericycle central cylinder ESTs (13%) did not match maize PUTs. Twenty-one of 70 novel pericycle ESTs and 10 of 41 nonpericycle ESTs were anchored to MAGIs. Hence, 49 ESTs obtained from pericycle cells (representing 46 different transcripts) and 31 ESTs retrieved from nonpericycle central cylinder cells represented potentially novel genes. Thus, in total, 77 of 590 unique EST contigs (13%) identified in this study are considered to be maize EST contigs not previously deposited in any public database. The consensus sequences of the 590 unique EST contigs were used to perform BLASTx searches against the NCBI nonredundant protein database as of December 12, 2006, using 1e−10 as the cutoff value. BLASTx results were subsequently categorized into transcripts encoding proteins of known function and transcripts encoding proteins of unknown function (Table IIIb). Classification of sequences derived from pericycle ESTs revealed that 258 of 377 ESTs (68%) represented genes of known function, whereas the remaining 118 ESTs (32%) were associated with genes of unknown function (Table IIIb). Similarly, 222 of 324 nonpericycle central cylinder ESTs (69%) represented genes of known function, whereas 102 ESTs (31%) were transcribed from genes of unknown function (Table IIIb). Sequences encoding functionally annotated proteins of the two analyzed cell types were grouped into functional categories according to the MIPS (Schoof et al., 2002) classification system (Table IIIb). In the pericycle dataset, protein synthesis (14%) and metabolism (13%) were the most abundant functional classes followed by proteins with binding function (9%), protein fate (6%), disease/defense (5%), energy (5%), transport (5%), signal transduction (4%), transcription (4%), cell fate (1%), cell cycle (1%), and transposons (<1%; Table IIIb). Notably, distribution of the functional classes was very similar between pericycle and nonpericycle central cylinder cells. However, Fisher's exact test (Fisher, 1935) revealed two functional classes that were represented by significantly different proportions of ESTs (Table IIIb). Whereas 14% of the ESTs retrieved from pericycle cells belonged to the protein synthesis category, only 7% of the ESTs identified from nonpericycle central cylinder cells fell in this functional category (P = 0.007). Similarly, whereas 5% of the ESTs retrieved from nonpericycle central cylinder cells were related to cell fate, only 1% of the pericycle ESTs were associated with this functional category (P = 0.004). EST clusters derived from pericycle and nonpericycle central cylinder cells were sorted according to the number of members (Table IIIc). For pericycle cells, four clusters containing at least three transcripts were detected, whereas for nonpericycle central cylinder cells nine clusters contained at least three ESTs. The clusters representing high mobility group protein B1 (P27347) and an unknown protein (O48557) were present in both libraries with at least three ESTs. The relatively low abundance of specific clusters in pericycle and nonpericycle central cylinder cells suggests that there is no prevalence for the expression of highly abundant genes in these cell types. The probability that our analysis missed EST clusters that make up at least 5% of the pericycle transcriptome is P = 0.00013 and P = 0.0175 for the nonpericycle transcriptome, assuming binomial distribution of the transcripts.
Proteome Analysis of the Most Abundant Soluble Proteins of the Maize Primary Root Pericycle
Proteomics can detect and identify the most abundant proteins of a particular root type at a certain developmental stage (Hochholdinger et al., 2006). To date, proteome analyses of maize roots have been limited to the analysis of whole roots due to the protein amount required for two-dimensional (2-D) electrophoresis and subsequent identification of proteins by mass spectrometry (MS; Hochholdinger et al., 2004a, 2004b, 2005; Sauer et al., 2006; Liu et al., 2006). Hence, up to this time, no cell-type-specific proteome dataset is available for roots. We have therefore generated a reference map of the most abundant soluble proteins of the maize primary root pericycle 2.5 d after germination. We isolated approximately 1,000 rings of pericycle cells from root cross sections that represent approximately 200,000 pericycle cells via LCM according to the sampling procedure described for the microarray experiments (Fig. 1B). These cells yielded approximately 30 μg of protein extract, which was subsequently separated via isoelectric focusing on a linear gradient of pH 4 to 7. After isoelectric focusing, proteins were separated according to their molecular masses in a second dimension on 12% SDS-polyacrylamide gels and silver stained according to a MS-compatible protocol (Blum et al., 1987; Fig. 2). The pericycle reference map was made in triplicate from independent pericycle root protein preparations and all identified proteins were detected in all replications. The 56 most abundant protein spots were picked from a representative 2-D gel, digested with trypsin, and the eluted peptides were subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses (Table IV). Automated MASCOT software (http://www.matrixscience.com) was used for searching the NCBI nonredundant protein databases on all available higher plant proteins (Streptophyta) because the maize genome has not yet been completely sequenced and many genes are highly conserved among higher plants. Twenty of the 56 proteins were identified by matching known plant proteins with a MASCOT score >41 and the conservative criteria that were defined for the mass spectrometric identification of proteins (“Materials and Methods”). Peptide sequences and MASCOT scores of the individual peptides are provided in Supplemental Data S1. The 20 identified proteins, which are encoded by 18 different genes, were classified according to the MIPS annotation system (http://mips.gsf.de). The most abundant functional class was metabolism (10/20), followed by energy (6/20) and disease and defense (2/20). The cellular organization and signal transduction categories were represented by one protein each.
Comparative Analysis of Pericycle-Specific Transcriptome and Proteome Datasets
This study provided data on the most abundantly expressed genes and proteins in the pericycle as well as of genes preferentially expressed in pericycle versus nonpericycle tissue and thus candidate genes involved in maize pericycle specification. Previously, we identified genes that were differentially expressed in the pericycle of the wild type versus the lateral root initiation mutant rum1, thus providing clues as to which genes might be related to lateral root initiation (Woll et al., 2005). To identify genes that might be related to pericycle specification as well as lateral root initiation, we compared these different datasets with each other as well as with the most abundant pericycle genes and proteins via BLAST searches (alignment score >90%, sequence length >100 bp). In summary, little overlap exists between the datasets related to lateral root initiation (Woll et al., 2005) and pericycle specification (Tables I and II). Among the 163 genes differentially expressed between wild-type and mutant rum1 pericycle cells, only one gene with similarity to a pollen-specific protein C13 precursor (BG842708) also displayed preferential expression in pericycle versus nonpericycle primary root cells. This might indicate significantly different regulation of these distinct developmental processes. Remarkably, three genes that were differentially expressed between wild-type and rum1 pericycle cells and might thus be related to lateral root initiation were also among the most abundant pericycle transcripts and proteins, including transcripts encoding a HMG1-like protein (PUT-155a-Zea_mays-109877741) and a 60S ribosomal protein (PUT-155a-Zea_mays-120277744), as well as a protein representing a S-adenosylmethionine synthetase (AAT94053.1). Similarly, a pericycle-specific gene that encodes for a pathogenesis-related protein 10 (AAY29574) that might be related to pericycle specification was also among the most abundant proteins.
DISCUSSION
LCM Isolation of the Maize Pericycle Allows the Dissection of Undifferentiated Root Cells during Specification
LCM (Schnable et al., 2004; Ohtsu et al., 2007) of defined internal cell types of a plant organ in combination with downstream molecular analyses of transcripts or proteins isolated from these cell can reveal insight about how genes function and how their gene products interact during development (Kerk et al., 2003; Woll et al., 2005). To date, only a few such studies are available that analyzed cell-type-specific gene expression in plants via microarray analyses (e.g. Nakazono et al., 2003; Casson et al., 2005; Klink et al., 2005; Woll et al., 2005; Jiang et al., 2006). Among those, two cell-type-specific microarray studies have analyzed different aspects of maize root formation. Jiang et al. (2006) compared gene expression profiles of the apical meristem, the quiescent center, and the root cap of maize primary root tips and revealed the up-regulation of gene clusters in the root cap that were linked to major metabolic processes. Woll et al. (2005) compared gene expression in the pericycle of wild-type seedlings prior to lateral root initiation versus gene expression in the mutant rum1 that does not initiate lateral roots and identified a subset of genes related to signal transduction, cell cycle, transcription, and translation that might be related to the process of lateral root initiation. In this study, gene expression profiles were compared between maize primary root pericycle versus nonpericycle cells that left the meristematic zone before the first cell divisions, hence during the specification of these cells. These cells are characterized by their competence for cell division, which all other nonpericycle root cells that have left the meristematic zone do not have. Some pericycle cells, typically those next to the phloem poles (Casero et al., 1995), become pericycle founder cells that later divide and develop into lateral roots.
Enhanced Transcription and Protein Synthesis-Related Gene Expression Suggests Distinct Metabolic Activity of Cell-Cycle-Competent Pericycle Cells
Pericycle and surrounding nonpericycle cells that have left the meristematic zone differ in their capacity to divide and thus in their differentiation status. This study demonstrated that the competence of pericycle cells to re-enter the cell cycle is correlated with the preferential expression of a subset of genes related to protein synthesis, transcription, and signal transduction. Remarkably, these functional classes comprise 29% of all genes preferentially expressed in the pericycle (Tables I and II) and are thus significantly more abundant than one would expect from their distribution in the completely sequenced rice genome (Goff et al., 2002). Because the cells of the young primary roots analyzed in this study have left the meristematic tissue only shortly before analysis, this altered metabolic activity in the pericycle versus nonpericycle might also support the notion by Dubrovsky and Ivanov (1984) that pericycle cells do not differentiate after they leave the meristematic zone and dedifferentiate before the initiation of lateral roots, but are rather undifferentiated until they initiate lateral roots.
Lateral Root Initiation and Pericycle Determination Might Be Controlled by a Different Set of Genes
This study identified genes that are preferentially expressed in pericycle versus nonpericycle cells of the primary root that might therefore be related to the process of pericycle specification. A previous cell-type-specific microarray study from our laboratory compared the pericycle transcriptomes of the wild type and mutant rum1. Because the mutant rum1 does not initiate lateral roots, differentially expressed genes might be related to lateral root formation (Woll et al., 2005). Among the 167 genes that were differentially expressed between wild-type and rum1 pericycle cells, only one gene was also differentially expressed between pericycle and nonpericycle cells. This is even more striking because in both analyses the same microarray chips and the same developmental stage of primary roots were analyzed. Interestingly, the functional classes of transcription, translation, and signal transduction that were prevalent in pericycle versus nonpericycle cells were also represented by genes differentially expressed between wild-type and rum1 pericycle cells. However, these functional classes were represented in the different datasets by different genes. This might imply that there are significant differences in the molecular networks that determine the developmental processes of pericycle specification and lateral root initiation.
Pericycle-Specific EST Sequencing Reveals Putative Novel Genes and High Diversity of Gene Expression in the Pericycle
Generation of cell-type-specific ESTs and high-throughput EST sequencing provides several types of information. First, EST sequencing detects the most abundant transcripts within a tissue that has been subjected to LCM, which is of particular interest in species like maize, where not all genes are yet available on microarray chips. In wheat egg cells and two-celled proembryo cells, the most abundant EST clusters representing an ECA-1-like gene and a histone H4 comprised approximately 7% (49/735) and 8% (39/462) of all sequenced ESTs, respectively (Sprunck et al., 2005). In contrast, the most abundant root pericycle EST cluster representing the high-mobility group B1 (HMGB1) gene made up only about 1% of all sequenced ESTs (5/377), whereas the most abundant nonpericycle central cylinder cluster representing a protein of unknown function made up 2% of all sequenced ESTs of that library (8/324). This indicates a higher degree of gene expression diversity in maize root cells than in wheat embryo cells. Notably, the HMGB1 gene was expressed at relatively high levels in pericycle and nonpericycle central cylinder cells. It has been demonstrated that ectopic expression of this maize gene in tobacco (Nicotiana tabacum) seedlings specifically affects root development, leading to reduced primary root elongation and cortical cell size in primary roots, whereas aboveground development of these plants was not altered (Lichota et al., 2004). Remarkably, this HMGB1 gene was also preferentially expressed in wild-type pericycle cells versus mutant rum1 pericycle cells in a previous dataset from our laboratory at the same developmental stage investigated in this study (Woll et al., 2005). Second, high-throughput EST sequencing allows for quantification of the relative abundance of functional classes of transcripts of a particular cell type. The relative abundance of most functional categories was similar in pericycle and nonpericycle central cylinder cells. However, the functional class of genes encoding proteins involved in protein synthesis was significantly higher in pericycle cells (14%) compared to nonpericycle central cylinder cells (7%) and also exceeded the proportion of this gene class in the Arabidopsis (Arabidopsis Genome Initiative, 2000) and rice genomes (Goff et al., 2002; Yu et al., 2002), where approximately 4% of genes are related to protein synthesis. This observation is in line with the microarray and SSH experiments in this survey supporting the notion of increased transcriptional and translational activity in this cell type during pericycle specification. Finally, EST sequencing allows for the identification of transcripts that might be underrepresented in whole-organ EST sequencing efforts and have therefore not yet been identified. Among 340 pericycle gene clusters identified in this study, 46 (14%) represented genes that have not been previously identified in EST or genomic databases. Similarly, 31 (11%) of the nonpericycle central cylinder clusters were neither available in EST nor in genomic databases. Moreover, for 19% of the pericycle EST clusters, expression has not been previously demonstrated, whereas for 15% of the nonpericycle central cylinder EST clusters, expression was shown in this study. These numbers are comparable to the figures obtained by Sprunck et al. (2005), who isolated 735 ESTs from egg cells and 462 ESTs from two-celled proembryos from wheat and identified 18% gene clusters in egg cells and 11% gene clusters in two-celled proembryos that have not been previously deposited in public databases. Interestingly, maize, wheat, and rice are the plant species with the best representation by ESTs. As of May 25, 2007, 1.2 million maize EST sequences and 1.1 million wheat EST sequences were available in public databases. Thus, identification of such a considerable percentage of genes not present in public databases supports the feasibility of cell-type-specific gene expression analyses for the discovery of putatively novel expressed genes.
Two Genes Differentially Expressed in Pericycle-Specific Datasets Are among the Most Abundant Proteins of the Pericycle
Proteins are the primary effectors of biological function in living organisms. It is therefore desirable to extend high-throughput gene expression analyses to the protein level, especially because it has been demonstrated that RNA and protein levels do not always correlate (e.g. Liu et al., 2006). We have therefore initiated an effort to identify the most abundant proteins during pericycle specification by combining the isolation of pericycle cells via LCM of root cryosections with 2-D electrophoresis and silver staining that is compatible with electrospray ionization (ESI)-MS/MS (Blum et al., 1987). The identification of 20 proteins represents a first step toward a reference map of the maize pericycle. The number of pericycle cells (200,000) and the amount of isolated protein (30 μg) is comparable to the 250,000 vascular bundle cells representing 25 μg of protein that have been isolated by Schad et al. (2005) from Arabidopsis. Whereas Schad et al. (2005) used a silver-staining technique that was not compatible with MS and therefore identified 33 proteins in a non-gel-based LC-MS/MS system, we identified 20 of 56 picked proteins directly from silver-stained gels. None of the 20 proteins was also identified in the EST sequencing projects, which is not surprising because none of the transcripts accumulated to particularly high expression levels and, in any case, RNA and protein levels do not necessarily correlate with each other (e.g. Liu et al., 2006). However, it was surprising that, among the 20 proteins identified from the most abundant soluble proteins of the pericycle, two genes were also differentially expressed either between wild-type and rum1 pericycle cells (GenBank accession no. AAT94053.1; Woll et al., 2005) or between pericycle and nonpericycle cells (GenBank accession no. AAY29574.1; Table I). This might imply that even some of the most abundant soluble proteins might play important roles in pericycle specification and subsequent lateral root initiation and support the value of protein profiling.
In summary, this study provides specific analysis of gene expression of the maize pericycle cells during specification by combining the isolation of pericycle cells and their mRNA from primary root tissue via LCM with the downstream analysis of gene expression via microarray analyses, SSH, qRT-PCR, EST sequencing, and proteome profiling. The rationale behind the application of various transcriptome profiling techniques was that only a fraction of all maize genes are currently available on maize microarray chips. SSH therefore facilitated the identification of additional differentially accumulated genes, whereas EST sequencing discovered a considerable proportion of genes in the pericycle that were not yet present in maize sequence databases. With the completion of the maize genome sequence and hence the availability of almost all maize genes on microarray chips, future cell-type-specific expression analyses in maize are expected to focus on microarray analyses in combination with confirmatory high-throughput qRT-PCR experiments.
The results of the analyses provided here are an initial step toward the identification of genes that are involved in pericycle specification and give a glimpse on molecular differences between root cells that have the competence to divide and cells that do not have this competence. Because genetic and anatomical data imply differences in pericycle founder cell positioning between monocots and dicots, these differences might already be manifested during pericycle specification. It will therefore be interesting in the future to compare the data of this study with pericycle-specific gene expression profiles of the dicot model organism Arabidopsis upon availability of such datasets.
MATERIALS AND METHODS
Microarray Experiments
Staining of Root Meristems with the Feulgen Technique
The length of the meristematic zone of 2.5-d maize (Zea mays) primary roots was determined from scanned primary roots (hp scanjet 7400C; Hewlett-Packard) with Image Pro Express software (Media Cybernetics) after Feulgen staining according to Dubrovsky and Ivanov (1984) as described by Woll et al. (2005). Three-, 4-, and 5-d-old primary roots were fixed and stained with Schiff's solution as described by Hoecker et al. (2006). The staining pattern of the roots was documented with an Olympus SC35 type 12 digital camera under a binocular (Stemi SV8; Zeiss).
Plant Material, Growth Conditions, and Fixation of Primary Root Samples for LCM
Roots were grown at 28°C in the dark in paper rolls (Hoecker et al., 2006). To avoid circadian effects on gene expression, roots were always germinated at 6 pm and harvested after 64 h (2.5 d) at 10 am. Roots with a length between 1.5 and 2 cm were harvested after the apical 0.3 cm of the root apex, including the meristematic zone and the distal elongation zone, were discarded. The remaining differentiation and elongation zones of the roots were collected in 0.5-cm samples, fixed, and embedded in Tissue-Tek O.C.T. medium (Sakura Finetek) according to the protocol described by Nakazono et al. (2003). Separate pools of three (microarray experiments, RT-PCR) to eight (SSH experiments) primary roots represented one biological replicate.
Cryosectioning and LCM
Preparation of 10-μm primary root cross sections was performed at −20°C using a cryostat (Leica CM1850) and mounted on adhesive slides using the CryoJane tape-transfer system (Instrumedics). At most, only every fifth section of a series was collected on an adhesive tape window that was brought into contact with the specimen before sectioning. The tape window containing the cross section was transferred and firmly cross-linked to an adhesive-coated slide with a flash of 360-nm UV light. After removal of the tape window, cross sections were dehydrated in an ethanol series as described by Woll et al. (2005). Sections were kept in fresh xylene until they were used for LCM.
In the PixCellII LCM system (Arcturus Bioscience), pericycle cells were isolated as described by Woll et al. (2005). Circles of pericycle cells each representing approximately 200 cells were captured using the following parameters: laser spot size of 7.5 μm, laser power of 50 mW, and laser pulse duration of 550 to 650 μs.
RNA Extraction and Amplification
Total RNA of approximately 13,000 captured and pooled pericycle cells was extracted using the RNaqueous-micro kit (Ambion) and treated with the RNase-free DNaseI set (Qiagen). Approximately 50 ng total RNA from captured pericycle and nonpericycle central cylinder cells were transcribed into cDNA and amplified via the BD SMART PCR cDNA synthesis kit (BD Biosciences) according to the manufacturer's protocol for the SSH, qRT-PCR, and EST sequencing experiments, whereas the method described by Woll et al. (2005) was applied for the microarray experiments. The efficiency of amplification was quantified using the RiboGreen RNA quantification reagent (Molecular Probes) by measuring RNA yield after the first and second round of amplification.
Microarray Hybridization, Scanning, Spot Quantification Data Analysis, and Reverse RNA Gel-Blot Confirmation
Six independent biological replications from each cell type were profiled on six microarrays. Microarray probe synthesis and hybridization of spotted 12K maize cDNA microarray slides (GenII vA: gene expression omnibus platform GPL 4876; http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GPL4876) were conducted as described by Woll et al. (2005). Samples from the two cell types were paired on each array. Dyes were assigned to samples in a way that each genotype was measured an equal number of times with both dyes (dye swap). After removal of empty spots and spots that yielded more than one band during PCR amplification, 11,767 of the 13,076 spots on the microarray chip were analyzed. Dried slides were scanned three times at different scan settings with a ScanArray 5000 scanner (Packard) for each channel (Cy3 and Cy5) with laser power and PMT gain settings adjusted so that the signal intensity for both channels was equal for one slide. ImaGene software (Biodiscovery) was used to quantify the spot intensities on the slides. The lowess normalization method originally described by Dudoit et al. (2002) was applied as described by Woll et al. (2005). For each of 11,767 sequences, a mixed linear model analysis of the normalized log-scale signal intensities was conducted to identify transcripts whose expression differed significantly between pericycle and nonpericycle cells. The mixed linear model included genotype and dye terms as fixed effects as well as slide terms, and general error terms as random effects. A t test for cell type differences was conducted as part of a mixed linear model analysis for each gene (Wolfinger et al., 2001), yielding 11,767 P values. As described by Allison et al. (2002), a mixture of uniform and β-distribution was fit to the observed distribution of the 11,767 P values obtained from the mixed linear model analysis. The estimated parameters from the fit of the mixture model were used to estimate the posterior probability of differential expression for each gene and to estimate the FDR among all genes with P values ≤0.01 and estimated Fc > 1.5.
For confirmatory reverse RNA gel-blot experiments, PCR products of the differential genes were generated with the general vector-specific oligonucleotide primers T3, T7, GAD10-F, GAD10-R, and Gal4-R, and DNA of the corresponding clones from the maize unigene collection (www.maizegdb.org) as a template. PCR products were separated on 1% agarose gels and 0.75 μg of amplified pericycle and nonpericycle RNA was run in each gel as a loading control for normalization. Gels were incubated in denaturing solution (1.5 m NaCl, 0.5 m NaOH) and neutralization solution (0.5 m Tris-Cl, pH 7.2, 1 m NaCl) for 1 h each. Blotting on Hybond NX membranes (Amersham Biosciences), cross-linking, generation of radioactively labeled cDNA probes generated from pericycle and nonpericycle RNA of 2.5-d-old primary root of maize, hybridization, washing, and x-ray film exposure (Agfa Cronex 5) was preformed as described by Woll et al. (2005). Signals were quantified with Quantity One software (Bio-Rad).
SSH
SSH is a technique that allows for the comparison of genes that are preferentially expressed in a particular tissue (Diatchenko et al., 1996). Two SSH aRNA (cDNA) libraries were generated from total RNA isolated from LCM-captured pericycle and nonpericycle central cylinder cells via the BD SMART PCR cDNA synthesis kit (BD Biosciences) according to the manufacturer's instructions. SSH was performed with pericycle aRNA as tester and nonpericycle central cylinder aRNA as driver via the BD CLONTECH PCR-select cDNA subtraction kit (BD Biosciences) according to the manufacturer's protocol. The tester cDNA population contains preferentially expressed genes of interest, whereas the driver population contains the reference cDNA. Putative pericycle-specific transcripts were cloned into the pGEM T-Easy vector (Promega) and transformed into Escherichia coli JM109 cells. Subsequently, putative pericycle-specific transcripts were amplified via colony PCR of overnight bacterial cultures with the SSH-specific oligonucleotide primers SSH forward and reverse and screened for pericycle specificity via reverse gel-blot analyses. Blotting and cross-linking of the PCR products, radioactive labeling of pericycle and nonpericycle RNA samples of 2.5-d-old primary roots, hybridization, washing of the membrane, and visualization of the signals via x-ray films were performed as described by Woll et al. (2005).
qRT-PCR, Data Analysis, and Statistics
SSH clones that displayed pericycle-specific expression in the reverse RNA gel-blot screen were subjected to qRT-PCR validation. Sequences of differential clones were BLASTed (Altschul, 1991) against the maize EST database (www.maizegdb.org) and maize genomic sequence database MAGI 4.0 (Fu et al., 2005) to recover additional sequence information. Primers were designed with Primer3 software (Rozen and Skaletsky, 2000) according to the criteria set up by Swanson-Wagner et al. (2006). All primers were BLASTed to the MAGI database to confirm their gene specificity. The specificity of the primers was verified by gel electrophoresis and melting-curve analyses of the iCycler (Bio-Rad). Only primers that yielded a single peak in both analyses were used in the validation experiments. The specific primer sequences of the target and control genes are listed in Supplemental Data S1. RNA samples from four biological replications of pericycle and nonpericycle central cylinder cells were isolated and amplified as described in the section on RNA isolation and amplification. Amplification of a GAPDH (X07156; for primer sequences, see Supplemental Data S1) fragment with oligonucleotide primers flanking an intron excluded the possibility of genomic DNA contamination of the aRNA samples. The template amount for qRT-PCR was 5 ng of amplified cDNA in each PCR reaction. PCR reactions were performed in a thermocycler (iCycler iQTM multi-color real-time PCR detection system; Bio-Rad) using a commercial fluorescence detection kit (QuantiTect SYBR Green PCR kit; Qiagen). Primer annealing was performed at 55°C for 30 s and elongation at 72°C for 60 s. Fluorescence was measured in each cycle at 72°C. As a reference gene, we used the housekeeping gene thioredoxin (AF435816; for primer sequences, see Supplemental Data S1) as previously reported by Casati and Walbot (2004). Experiments for each gene in each tissue and biological replicate were repeated four times.
Statistical data analysis was based on the threshold cycles (CT) of the PCR products and performed as described in Buck et al. (2004). The CT value is defined as the PCR cycle at which the fluorescence intensity of a transcript crosses a threshold line in the exponential amplification phase. The CT provides information about the amount of starting material. The efficiency (E) of the PCR reaction for each primer pair was determined by a dilution series ranging from 8 to 0.125 ng per well and calculated by the equation E = 10(−1/slope). This formula yields values between 1 (0% E) and 2 (100% E). The slope (S) was calculated by the iCycler program by correlating the mean CT value of each dilution sample versus the logarithm of the sample concentration. The mean CT values of measurements of each primer combination in the three biological replications of a cell type were used for further statistical analysis. The log-transformed mean-normalized expression values were calculated to compare relative expression levels between the different tissues of pericycle and central cylinder as previously described (Simon, 2003). Fcs were tested for significance (P < 0.05) against the null hypothesis that there is no expression difference between the two cell types in an unpaired bidirectional Student's t test.
EST Analysis
EST Library Construction and Sequencing
EST libraries of pericycle and nonpericycle central cylinder cells were generated from LCM isolated cells via the SMART PCR cDNA synthesis kit (BD Biosciences) as described above. Fifty nanograms of amplified cDNA from each cell type were cloned into the pGEM vector system in a nondirectional manner and transformed into JM109 cells. Randomly picked colonies were sequenced with the vector-specific standard M13 reverse primer. The quality value files were generated by ABI KB-base caller and then imported into the Lucy program for trimming vector and low-quality regions (Lucy parameters used: size 9, error 0.01 0.01, bracket 30 0.01). PolyA tails in the Lucy-trimmed sequences were trimmed using The Institute for Genomic Research (TIGR) SeqClean.
EST Anchoring
Maize PUTs (version 155a; November 11, 2006) were downloaded from MaizeGDB (http://www.maizegdb.org) and BLASTed against 701 ESTs (parameters used: -W 24, -F F, -e 1e-20). Only the alignments with ≥97% similarity and overall PUT coverage >0.5 or ≥100 bp were included for further analyses. The EST versus PUT alignment with the highest bit score was used to unambiguously anchor each EST to a maize PUT. When multiple best EST/PUT alignments existed, the corresponding EST could not be anchored and was therefore not included for abundance analysis. The remaining ESTs that were not anchored to maize PUTs were BLASTed against the MAGI database (Fu et al., 2005) according to the same alignment parameters defined above for the PUT anchoring. This anchored previously unknown ESTs to genomic sequences. The potential novel transcripts were screened against a repeat database containing publicly available transposable elements provided by Dr. Jeff Bennetzen at the University of Georgia using Repeatmasker (http://www.repeatmasker.org).
Proteomics Experiments
Protein Isolation
Pericycle cells were isolated via LCM as described above. LCM caps containing pericycle cells were used as lids for 0.5-mL tubes containing 30 μL extraction solution (7 m urea, 2 m thiourea, 2% [w/v] CHAPS, 1.25% [v/v] Bio-Lytes 3/10 [Bio-Rad], 50 mm dithiothreitol, traces of bromphenol blue, and 1 tablet per 10 mL of solution protease inhibitor complete [Roche]). Proteins were dissolved by shaking the tube, thus emerging the pericycle cells on the cap in the solution. Samples were first incubated on ice for 15 min. For efficient protein extraction, four alternating cycles of 2 min of ultrasonication followed by 2 min of incubation on ice followed by 15 min of incubation at room temperature were performed. This procedure was repeated twice with each cap. All steps were repeated after placing a new cap with pericycle cells on the tube. Protein extracts of five tubes were pooled and yielded a total of 150 μL of protein extract. Samples were then treated with 150 units of endonuclease (Sigma) before being exposed to an additional 2 min of ultrasonication and 2 min of incubation on ice four times. The insoluble fraction was removed via centrifugation at 14,000g for 40 min. The supernatant containing the soluble protein fraction was immediately subjected to 2-D electrophoresis. Approximately 30 μg of protein were isolated from 1,000 rings of pericycle cells representing approximately 200,000 cells.
2-D Separation of Pericycle Proteins
Isoelectric focusing of proteins was performed with 30 μg of protein extract using an IPG Phor isoelectric focusing unit (Amersham Biosciences) and 7-cm immobilized, linear pH 4 to 7 gradients (Immobiline drystrips; Amersham Biosciences). Rehydration was performed at 50 V overnight. The voltage settings of isoelectric focusing were 0- to 100-V gradient for 1 min, 100 V for 2 h, 100- to 4,000-V gradient for 90 min, 4,000 V for 5 h, 4,000- to 100-V gradient to a total of 26,900 Vh. Equilibration of strips was performed as previously described (Sauer et al., 2006). Proteins in the equilibrated strips were then separated on the basis of their Mrs in 12% SDS-PAGE 7-cm × 8-cm minigels (Bio-Rad). After electrophoresis, proteins were stained with a silver-staining procedure that is compatible with MS (Blum et al., 1987).
Nano-HPLC-ESI-MS/MS
The most abundant pericycle proteins were excised from a representative gel and digested in-gel using trypsin (sequencing grade; Promega). The eluted, trypsin-generated peptides were subsequently processed with a Dionex LC Packings HPLC system (Dionex LC Packings) containing the components Famos (autosampler), Switchos (loading pump and switching valves), and Ultimate (separation pump and UV detector). Subsequently, ESI-MS/MS mass spectra were recorded using the high-performance quadrupole time-of-flight mass spectrometer QStar Pulsar i (Applied Biosystems) equipped with a nano-ESI source (column adapter [ADPC-PRO] and distal-coated SilicaTips [FS360–20–10–D–20]; both from New Objective). The same composition and gradient of mobile phase A was used as described by Liu et al. (2006).
Analysis of Spectrometric Data
Measured peptides were searched in the NCBI nonredundant protein sequence database Viridiplantae (green plants) as of July 18, 2007, using the MOWSE algorithm as implemented in the MS search engine MASCOT (Matrix Science). All experimental data, achieved by 2-D electrophoresis and MS, and corresponding search results were stored in a LIMS database (Proteinscape 1.3; Bruker Daltonics). Database searches were performed on all available higher plant proteins because the maize genome has not been completely sequenced and many proteins are well conserved among higher plants.
Only proteins that met the following criteria were accepted as unambiguously identified: (1) number of matched peptides >2; (2) MASCOT score >41 [probability-based MOWSE score: −10*log(P)], where P is the probability that the observed match is a random event (scores >41 indicate identity or extensive homology; P < 0.05); (3) sequence coverage ≥4%; (4) allowed missed cleavage: 1; (5) deviation of predicted molecular mass and molecular mass of a protein on the gel: ±20%; (6) allowed modifications: carbamidomethylation (C), oxidation (M); and (7) maximum allowed molecular mass deviation: 0.5 D. Additionally, every peptide used for protein identification was checked for (1) y-ion series: ≥80% of the y-ions should be available; (2) presence of the b2-ion; (3) peptide score >20; and (4) e value <1e−10 (probability that the observed match is a random peptide). Identified proteins were functionally annotated via the MIPS database (Schoof et al., 2002).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EH210606 to EH211306.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data S1. Oligonucleotide primers for reverse RNA gel blot, annotated list of pericycle and nonpericycle ESTs, and peptide sequences and peptide MASCOT scores of proteins from Table IV.
Supplementary Material
Acknowledgments
We thank Marianne B. Smith (Iowa State University) for technical support and advice on cryosectioning; Margie Carter (ISU Image Analysis Facility) and Hailing Jin and David Skibbe (both of the Schnable laboratory) for helpful discussion on microarray experiments; Huaiyu Yang (University of Tuebingen) for help with organizing the EST data; and Christine Brand (University of Tuebingen) for advice on qRT-PCR.
This work was supported in part by SFB446 “cell behavior in eukaryotes,” Wilhelm-Schuler-Stiftung, and Rainer-und-Maria-Teufel-Stiftung. M.S. was supported by a German Academic Exchange Service fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Frank Hochholdinger (frank.hochholdinger@zmbp.uni-tuebingen.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allison DB, Gadbury GL, Heo M, Fernández JR, Lee CK, Prolla TA, Weindruch R (2002) A mixture model approach for the analysis of microarray gene expression data. Comput Stat Data Anal 39 1–20 [Google Scholar]
- Altschul SF (1991) Amino acid substitution matrices from an information theoretic perspective. J Mol Biol 219 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 3 401–408 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inze D (2001) The peri-cell-cycle in Arabidopsis. J Exp Bot 52 403–411 [DOI] [PubMed] [Google Scholar]
- Bell JK, McCully ME (1970) A histological study of lateral root initiation and development in Zea mays. Protoplasma 70 179–205 [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blum WF, Ranke MB, Lechner B, Bierich JR (1987) The polymorphic pattern of somatomedins during human development. Acta Endocrinol (Copenh) 116 445–451 [DOI] [PubMed] [Google Scholar]
- Buck C, Schaeffel F, Simon P, Feldkaemper M (2004) Effects of positive and negative lens treatment on retinal and choroidal glucagon and glucagon receptor mRNA levels in the chicken. Invest Ophthalmol Vis Sci 45 402–409 [DOI] [PubMed] [Google Scholar]
- Casati P, Walbot V (2004) Rapid transcriptome responses of maize (Zea mays) to UV_B in irradiated and shielded tissues. Genome Biol 5 R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero PJ, Casimiro I, Lloret PG (1995) Lateral root initiation by asymmetrical transverse divisions of pericycle cells in four plant species: Raphanus sativus, Helianthus annuus, Zea mays and Daucus carota. Protoplasma 188 49–58 [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 4 165–171 [DOI] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42 111–123 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inze D, Beeckman T (2006) Lateral root initiation or the birth of a new meristem. Plant Mol Biol 60 871–887 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campell AP, Crenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al (1996) Suppressive subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Ivanov VB (1984) Certain mechanisms of lateral root initiation in germinating maize roots. Physiol Biochem Cult Plants 16 279–284 [Google Scholar]
- Dubrovsky JG, Rost TL, Colon-Carmona A, Doerner P (2000) Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol 124 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP (2002) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statist Sinica 12 111–140 [Google Scholar]
- Feldman L (1994) The maize root. In M Freeling, V Walbot, eds, The Maize Handbook. Springer, New York, pp 29–37
- Fisher RA (1935). The Design of Experiments, Ed 8. Oliver and Boyd, Edinburgh
- Foard DE, Haber AH, Fishman TN (1965) Initiation of lateral root primordia without completion of mitosis and without cytokinesis in uniseriate pericycle cells. Am J Bot 52 580–590 [Google Scholar]
- Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, Aluru S, Schnable PS (2005) Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc Natl Acad Sci USA 102 12282–12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Feix G (1998) Early post-embryonic root formation is specifically affected in the maize mutant lrt1. Plant J 16 247–255 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Guo L, Schnable PS (2004. a) Cytoplasmic regulation of the accumulation of nuclear-encoded proteins in the mitochondrial proteome of maize. Plant J 37 199–208 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Guo L, Schnable PS (2004. b) Lateral roots affect the proteome of the primary root of maize (Zea mays L.). Plant Mol Biol 56 397–412 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K (2004. c) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Sauer M, Dembinsky D, Hoecker N, Muthreich N, Saleem M, Liu Y (2006) Proteomic dissection of plant development. Proteomics 6 4076–4083 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Guo L, Schnable PS (2005) The accumulation of abundant soluble proteins changes early in the development of the primary roots of maize (Zea mays L.). Proteomics 5 4885–4893 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D (2004. d) Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programs. Ann Bot (Lond) 93 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker N, Keller B, Piepho HP, Hochholdinger F (2006) Manifestation of heterosis during early maize (Zea mays L.) root development. Theor Appl Genet 112 421–429 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML (1995) Specialized zones of development in roots. Plant Physiol 109 725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Zhang S, Lee S, Tsai G, Kim K, Huang H, Chilcott C, Zhu T, Feldman LJ (2006) Transcription profile analyses identify genes and pathways central to root cap functions in maize. Plant Mol Biol 60 343–363 [DOI] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Alkharouf N, MacDonald M, Matthews B (2005) Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode). Plant Mol Biol 59 965–979 [DOI] [PubMed] [Google Scholar]
- Lichota J, Ritt C, Grasser KD (2004) Ectopic expression of maize chromosomal HMGB1 protein causes defects in root development of tobacco seedlings. Biochem Biophys Res Commun 318 317–322 [DOI] [PubMed] [Google Scholar]
- Liu Y, Lamkemeyer T, Jakob A, Mi G, Zhang F, Nordheim A, Hochholdinger F (2006) Comparative proteome analyses of maize (Zea mays L.) primary roots prior to lateral root initiation reveal differential protein expression in the lateral root initiation mutant rum1. Proteomics 6 4300–4308 [DOI] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 3 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Maeda E (1982) Cytological studies on differentiation and dedifferentiation in pericycle cells of excised rice roots. Jpn J Crop Sci 51 553–560 [Google Scholar]
- Ohtsu K, Takahashi H, Schnable PS, Nakazono M (2007) Cell type-specific gene expression profiling in plants by using a combination of laser microdissection and high-throughput technologies. Plant Cell Physiol 48 3–7 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fedorova E, Lebris M, Lecomte P, Gyorgyey J, Vaubert D, Horvath G, Abad P, Kondorosi A, Kondorosi E (2003) The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol 131 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132 365–386 [DOI] [PubMed] [Google Scholar]
- Sass JE (1977) Morphology. In GF Sprague, ed, Corn and Corn Improvement. American Society of Agronomy Publishers, Madison, WI, pp 89–110
- Sauer M, Jakob A, Nordheim A, Hochholdinger F (2006) Proteomic analysis of shoot-borne root initiation in maize (Zea mays L.). Proteomics 6 2530–2541 [DOI] [PubMed] [Google Scholar]
- Schad M, Lipton MS, Giavalisco P, Smith RD, Kehr J (2005) Evaluation of two-dimensional electrophoresis and liquid chromatography—tandem mass spectrometry for tissue-specific protein profiling of laser-microdissected plant samples. Electrophoresis 26: 2729-2738; erratum Schad M, Lipton MS, Giavalisco P, Smith RD, Kehr J (2005) Electrophoresis 26: 3406 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Hochholdinger F, Nakazono M (2004) Global expression profiling applied to plant development. Curr Opin Plant Biol 7 50–56 [DOI] [PubMed] [Google Scholar]
- Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes HW, Mayer KF (2002) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res 30 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P (2003) Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19 1439–1440 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T (2005) The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J 41 660–672 [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1-hybrid and its inbred parents. Proc Natl Acad Sci USA 103 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8 625–637 [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk L, Stransky H, Nettleton D, Schnable PS, Hochholdinger F (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.