Abstract
Oligomeric proanthocyanidins (PAs) composed primarily of epicatechin units accumulate in the seed coats of the model legume Medicago truncatula, reaching maximal levels at around 20 d after pollination. Genes encoding the single Medicago anthocyanidin synthase (ANS; EC 1.14.11.19) and leucoanthocyanidin reductase (LAR; EC 1.17.1.3) were cloned and the corresponding enzymes functionally identified. Recombinant MtANS converted leucocyanidin to cyanidin, and, more efficiently, dihydroquercetin to the flavonol quercetin. Levels of transcripts encoding dihydroflavonol reductase, ANS, and anthocyanidin reductase (ANR), the enzyme responsible for conversion of anthocyanidin to (−)-epicatechin, paralleled the accumulation of PAs in developing seeds, whereas LAR transcripts appeared to be more transiently expressed. LAR, ANS, and ANR proteins were localized to the cytosol in transfected tobacco (Nicotiana tabacum) leaves. Antisense down-regulation of ANS in M. truncatula resulted in reduced anthocyanin and PA levels, but had no impact on flavonol levels. Transgenic tobacco plants constitutively overexpressing MtLAR showed reduced anthocyanin content, but no catechin or increased levels of PAs were detected either in leaves or in flowers. Our results confirm previously ascribed in vivo functions for ANS and ANR. However, the apparent lack of catechin in M. truncatula PAs, the poor correlation between LAR expression and PA accumulation, and the lack of production of catechin monomers or oligomers in transgenic plants overexpressing MtLAR question the role of MtLAR in PA biosynthesis in Medicago.
Flavonoids comprise one of the largest groups of plant secondary metabolites and the biochemistry and genetics of their biosynthesis have been intensively studied over the past decades (Holton and Cornish, 1995; Winkel-Shirley, 2001). Several different classes of flavonoids, including anthocyanins, flavonols, isoflavones, and oligomeric proanthocyanidins (PAs; also called condensed tannins) contribute in many ways to the growth and survival of plants (Harborne and Williams, 2001; Dixon and Sumner, 2003). Furthermore, PAs affect taste in beverages, such as fruit juice, wine, or tea, and also benefit human health by protecting against free radical injury and cardiovascular disease (Bagchi et al., 2000; Cos et al., 2004). PAs in forage crops can reduce both proteolysis during ensiling and release of methane during rumen fermentation, which in turn protects ruminants against potentially lethal pasture bloat (Aerts et al., 1999; Douglas et al., 1999). PAs can also act as antinutrients, affecting both dietary protein and micronutrient availability for humans (Antony and Chandra, 1998).
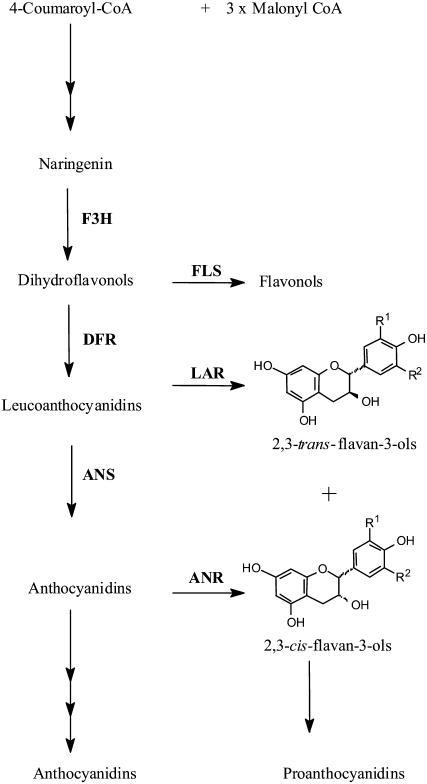
PAs are derived from the pathway leading to anthocyanins, a class of flavonoids well understood both biochemically and genetically (Winkel-Shirley, 2001). Three enzymes (leucoanthocyanidin reductase [LAR], anthocyanidin synthase [ANS], and anthocyanidin reductase [ANR]) function at branches between anthocyanin and PA biosynthesis (Fig. 1). LAR (a member of the reductase-epimerase-dehydrogenase family and closely related to isoflavone reductase from the isoflavonoid pathway [Tanner et al., 2003]) and ANS (a member of the 2-oxoglutarate-dependent dioxygenase [2-ODD] family) share the same substrate, flavan-3,4-diol (leucoanthocyanidin), which is converted to the PA unit 2,3-trans-flavan-3-ol (catechin) by LAR (Tanner et al., 2003), or to anthocyanidin by ANS (Saito et al., 1999). The latter compound can serve as substrate for ANR to produce another major PA unit, 2,3-cis-flavan-3-ol (epicatechin; Xie et al., 2003; Fig. 1), or be converted to anthocyanins by glycosylation and esterification. Thus, although catechin and epicatechin only differ by the stereochemical configuration at the C2 and C3 positions (trans- or cis-, respectively), they are synthesized by two quite distinct pathways (Fig. 1). The biosynthesis of PA oligomers is believed to proceed by addition of an extension unit (derived from leucoanthocyanidin, catechin, or epicatechin) to a starter unit (catechin or epicatechin) with sequential addition of further extension units, although the exact details of the PA polymerization process are still unclear (Dixon et al., 2005; Lepiniec et al., 2006).
Figure 1.
Schematic representation of the biosynthetic pathway for anthocyanins and PAs.
Most genetic studies on PA biosynthesis to date have been performed in Arabidopsis (Arabidopsis thaliana; Lepiniec et al., 2006) or barley (Hordeum vulgare; Jende-Strid, 1993). Because of the importance of PAs for the quality of forage legumes, it is important to develop a model system for studies on the molecular genetics of PA biosynthesis in legumes. Medicago truncatula has been selected as a model legume for genetic and genomic studies (Oldroyd and Geurts, 2001) and a wide range of genomic and genetic resources are now available for this species, including a whole-genome sequence that should be complete in 2008. We describe the distribution, nature, and deposition kinetics of PAs in M. truncatula, and the functional characterization and tissue-specific expression of MtLAR and MtANS in relation to PA biosynthesis. Our results provide the background for further development of Medicago as a model system for studying PA biosynthesis and deposition in forage legumes. Furthermore, they raise questions as to the role of LAR in the PA pathway in Medicago seeds.
RESULTS
Tissue-Specific Accumulation of PAs in M. truncatula
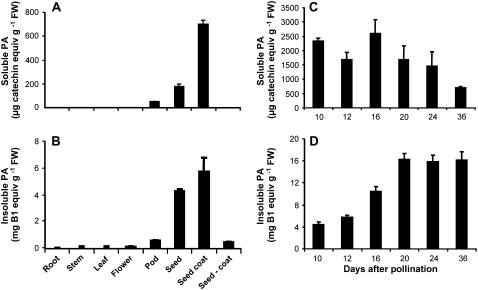
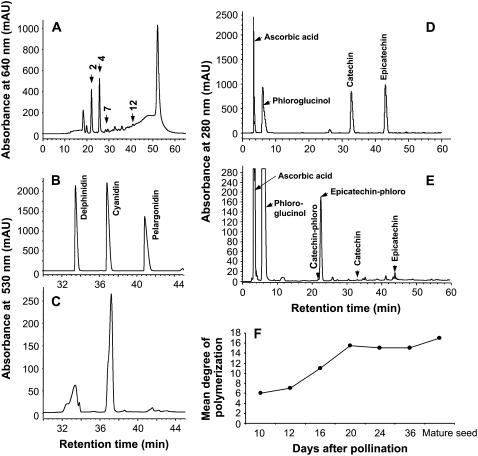
Tissues of M. truncatula ‘Jemalong A17’ were treated with 70% acetone:0.5% acetic acid to extract soluble PAs, which were then quantified by reaction with dimethylaminocinnamaldehyde (DMACA) reagent. Highest PA levels (about 0.7 mg catechin equivalents/g fresh weight [FW]) were found in the seed coat, with very low levels (1.9 μg catechin equivalents/g FW) in the remainder of the seed after the coat had been removed (Fig. 2A). Because of the high sensitivity of the detection method, very low levels of soluble PAs were detectable in flowers, leaves, roots, and stems. The size heterogeneity of the soluble PA oligomers from seed coats was subsequently determined by HPLC analysis with postcolumn derivatization with DMACA reagent (Peel and Dixon, 2007). This revealed the presence of low levels of free (−)-epicatechin monomers, along with a range of oligomers with an estimated highest degree of polymerization (DP) >12 (Fig. 3A).
Figure 2.
PA levels in M. truncatula ‘Jemalong A17’. A, Levels of soluble PAs in a range of tissues from mature plants. B, Levels of insoluble PAs in a range of tissues from mature plants. C, Levels of soluble PAs in seeds at various stages of development (dap). D, Levels of insoluble PAs in seeds at various stages of development. Soluble PAs were determined by reaction with DMACA reagent, insoluble PAs by butanol-HCl hydrolysis and estimation of resulting anthocyanidin.
Figure 3.
Composition of PAs in the seed coat of M. truncatula. A, Size heterogeneity of the soluble PA fraction from mature seed coats as determined by normal-phase HPLC and postcolumn derivatization with DMACA reagent. Numbers with arrows indicate the estimated number of units based on parallel analysis of size standards from D. uncinatum. B, Size standards of the anthocyanidin products dephinidin, cyanidin, and pelargonidin resolved by HPLC. C, HPLC analysis of anthocyanidins released by acid-butanol hydrolysis of soluble PAs from developing seed coats. D, Catechin and epicatechin standards analyzed by HPLC; these arise from starter units following phloroglucinolysis of intact PAs. E, Analysis of the products (starter and extension units) released by phloroglucinolysis of intact PAs from mature seed coats; the phloroglucinol adducts arise from extension units. F, Mean DP of PAs in seeds at various stages of development.
Insoluble (protein- or cell wall-bound) PAs were extracted from the residues left after extraction of soluble PAs by repeated sonication in butanol-HCl, followed by heating to generate colored anthocyanin from degradation of the PAs. Using this method, the highest levels of insoluble PAs were detected in the seed coat (about 5.8 mg procyanidin B1 equivalents/g FW; Fig. 2B) and in whole-seed and immature pods, in both cases presumably the result of the presence of the seed coats. Much lower levels were detected in flowers, leaves, stems, and roots. Due to the possible interference of cell wall components with the quantification of insoluble PAs (Marles et al., 2003), the insoluble PA content could be an underestimate. Furthermore, because different methods are used for quantification of soluble and insoluble PAs, it is difficult to compare the relative amounts of the two fractions within the different tissues; nevertheless, both fractions were clearly highest in seed coats, and insoluble PAs predominated.
PAs were extracted from developing seeds at various days after pollination (dap). Levels of soluble PAs were already close to maximal (around 2.5 mg catechin equivalents/g FW) at the earliest time point sampled (10 dap), whereas insoluble PAs associated with the cell wall fraction reached maximal levels (around 16 mg/g FW) at 20 dap (Fig. 2, C and D).
Nature of M. truncatula PAs
The butanol-HCl hydrolysate of the insoluble PA fraction from the seed coat was subjected to HPLC analysis and shown to contain primarily cyanidin, with lower amounts of delphinidin and pelargonidin (Fig. 3, B and C). By these criteria, M. truncatula PAs are composed primarily of epicatechin and/or catechin units (which yield cyanidin on hydrolysis), with much lower levels of epigallocatechin or gallocatechin (yielding delphinidin) and epiafzelechin or afzelechin (yielding pelargonidin) units (Fig. 3C).
To further determine the structure of M. truncatula seed coat PAs, the soluble PA fraction was isolated from seeds at various stages of development and subjected to cleavage with phloroglucinol. The released phloroglucinol adducts (extension units) and free starter units were then separated by HPLC and quantified (Fig. 3, D and E). By these criteria, soluble seed coat PAs were demonstrated to contain primarily epicatechin as a starter unit, and exclusively epicatechin extension units, consistent with the butanol-HCl hydrolysis data. Only very small peaks were present at retention times corresponding to catechin starter or extension (phloroglucinol adduct) units, and these may reflect (−)-catechin units that occur via nonenzymatic epimerization of (−)-epicatechin (Xie et al., 2004b). Phloroglucinolysis analysis revealed that the mean DP of the PAs increased from a value of 6 in seeds at 10 dap to a value of 17 in mature seeds (Fig. 3F), consistent with normal-phase HPLC analysis.
Identification and Properties of MtLAR
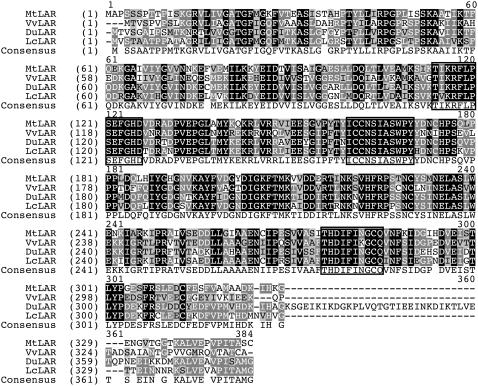
Informatic analysis revealed one candidate LAR homolog from M. truncatula (GenBank accession no. BN000703) and DNA gel-blot analysis confirmed the presence of a single LAR gene in the M. truncatula genome (Supplemental Fig. S1). The open reading frame (ORF) of this MtLAR gene was isolated by reverse transcription (RT)-PCR. The 1,050-bp sequence encodes a 349-amino acid polypeptide, with 66%, 62%, and 60% amino acid sequence identity to LARs from Lotus corniculatus (LcLAR), Desmodium uncinatum (DuLAR), and grape (Vitis vinefera; VvLAR), respectively (Fig. 4). A phylogenetic tree of plant reductase-epimerase-dehydrogenase enzymes, including the candidate MtLAR, has appeared elsewhere (Paolocci et al., 2007). The deduced MtLAR protein features conserved regions of 13, 11, and 10 amino acids containing, respectively, the LAR-specific amino acid motifs RFLP, ICCN, and THD as described previously (Tanner et al., 2003; Bogs et al., 2005). As shown in Figure 4, the conserved sequences including and immediately following these motifs were identical in MtLAR and DuLAR, with only two amino acid substitutions from the VvLAR and LcLAR sequences. The calculated pI and molecular mass of MtLAR were 5.88 and 38 kD, respectively.
Figure 4.
Alignment of deduced amino acid sequences of LAR genes. Sequences are from M. truncatula (ABE90657), grape (CAI56326), D. uncinatum (Q9SEV0), and L. corniculatus (ABC71327). Identical amino acids are indicated by white letters on a black background, conservative amino acids by white on a dark gray background, and similar amino acids by black on a light gray background. The RFLP, ICCN, and THD motifs are boxed.
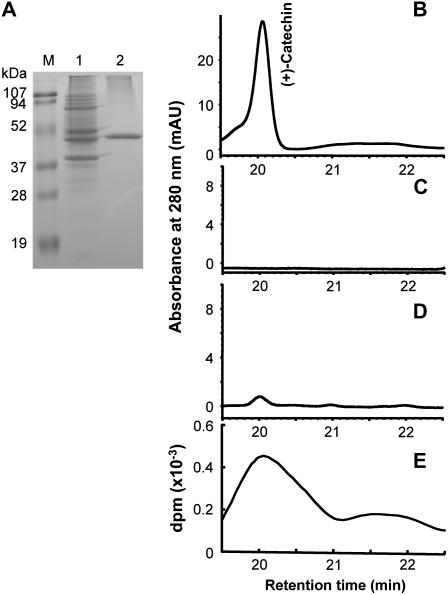
To determine the in vitro activity of recombinant MtLAR protein, the ORF was subcloned into the Escherichia coli expression vector pQE30 fused with a 6×-His tag at the N terminus. Recombinant MtLAR protein had a molecular mass of about 45 kD by analysis on a 12.5% SDS-PAGE protein gel (Fig. 5A). The discrepancy between the calculated and the apparent molecular masses of the MtLAR protein could be due to its particular amino acid composition, which can affect electrophoretic migration (Klenova et al., 1997). The protein was purified and assayed using 3H-labeled leucocyanidin as substrate in the presence of NADPH, followed by analysis of products by HPLC-UV and radioactivity detection. The vector control extract showed no product formation (Fig. 5C), whereas a peak with the same retention time as that of (+)-catechin was detected when MtLAR protein was incubated with 3H-labeled leucocyanidin (Fig. 5, D and E).
Figure 5.
Expression and assay of recombinant MtLAR. A, Analysis of MtLAR protein on a 12.5% SDS-PAGE gel. M, Protein molecular weight markers; 1, total protein from E. coli M15 containing pQE30-MtLAR induced by IPTG; 2, purified recombinant MtLAR protein used for enzyme activity assay. B, HPLC analysis of an authentic standard of (+)-catechin. C, HPLC analysis of product from incubation of leucocyanidin with protein extract from vector control. D, Product from incubation of purified recombinant MtLAR with 3H-leucocyanidin monitored by UV absorption. E, Product from incubation of purified recombinant MtLAR with 3H-leucocyanidin monitored by scintillation counting.
Identification and Properties of MtANS
A partial fragment of a putative MtANS sequence was selected from several M. truncatula ESTs by querying The Institute for Genomic Research EST database with Arabidopsis ANS (Wilmouth et al., 2002). The full-length cDNA sequence of MtANS was isolated by 5′-RACE (GenBank accession no. EF544389); the cDNA was 1,348 bp long with 39 bp of 5′-untranslated region and 238 bp of 3′-untranslated region, and a single putative polyadenylation signal sequence (AATAAA) 155 bp downstream from the stop codon. DNA gel-blot analysis revealed that ANS is encoded by a single gene in the M. truncatula genome (data not shown). The ORF of the MtANS gene is 1,071 bp, encoding a protein of 356 amino acids, and shows 78% identity to ANS from Malus × domestica, 72% identity to ANS from Perillia frutescens, and 76% identity to the Arabidopsis ANS for which the crystal structure has been elucidated (Wilmouth et al., 2002). Multiple sequence alignment (Supplemental Fig. S2) confirmed the presence of three conserved residues (His-232, His-288, and Asp-234 for MtANS) required to coordinate ferrous iron at the catalytic center of iron-containing soluble oxygenases, and an Arg residue (Arg-298 for MtANS) that is assumed to contribute to the specific binding of 2-oxoglutarate (Britsch et al., 1993; Prescott and John, 1996; Lukacin and Britsch, 1997). The theoretical pI and molecular mass of the deduced MtANS protein were predicted to be 5.9 and 40 kD, respectively.
MtANS shares around 40% identity at the amino acid level with other plant 2-ODDs (flavonol synthase [FLS] and flavanone 3-β-hydroxylase [F3H]) involved in flavonoid biosynthesis (Prescott and John, 1996). The deduced amino acid sequence of MtANS, together with those of FLS and F3H from several plant species, were aligned using ClustalW (Thompson et al., 1994) and subjected to phylogenetic analysis using the neighbor-joining method (Saitou and Nei, 1987). As viewed with Phylodraw (Choi et al., 2000), the three groups of functionally distinct plant 2-ODDs, FLS, F3H, and ANS, are clearly divided into three distinct clusters, and MtANS clustered within the ANS group (Supplemental Fig. S3).
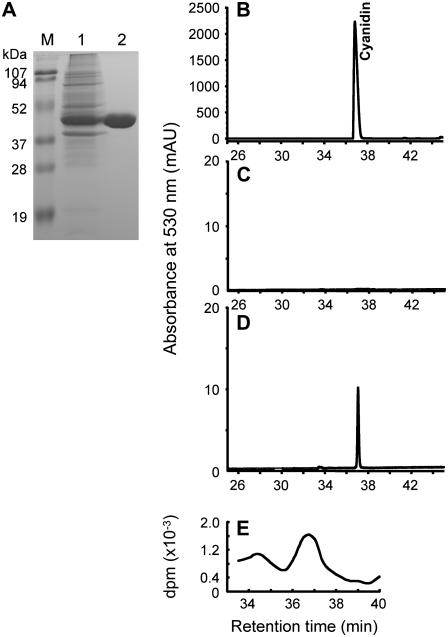
The MtANS ORF was expressed as a 6×-His-tagged N-terminal fusion in E. coli. Recombinant MtANS protein had a molecular mass of around 40 kD (Fig. 6A), as predicted by bioinformatics tools. The recombinant protein was purified and its activity assayed using radiolabeled 3H-leucocyanidin as substrate in the presence of ferrous iron and 2-oxoglutaric acid. The product was detected by HPLC monitored at 530 nm and its identity confirmed as cyanidin by comparison of retention time and UV/Vis spectrum with those of authentic standards (Fig. 6, B and D). The cyanidin peak was collected and confirmed to contain tritium label by scintillation counting (Fig. 6E), although overall enzymatic activity was low (50 pmol min−1 mg−1 protein), comparable to that of recombinant LAR. No 3H-leucocyanidin was converted to cyanidin in incubations with protein from the vector control (Fig. 6C).
Figure 6.
Expression and assay of recombinant MtANS. A, Analysis of MtANS protein on a 12.5% SDS-PAGE gel. M, Protein molecular weight markers; 1, total protein from E. coli M15 containing pQE30-MtANS induced by IPTG; 2, purified recombinant MtANS protein used for enzyme activity assay. B, HPLC analysis of an authentic standard of cyanidin. C, HPLC analysis of product from incubation of leucocyanidin with protein extract from vector control. D, Product from incubation of purified recombinant MtANS protein with 3H-leucocyanidin, monitored by UV absorption. E, Product from incubation of purified recombinant MtANS protein with 3H-leucocyanidin, monitored by scintillation counting.
Leucocyanidin is quite unstable and converts to its precursor dihydroquercetin during storage. Some 3H-quercetin was detected in incubations of MtANS with 3H-leucocyanidin, raising the question of whether it was derived from 3H-dihydroquercetin. To test this hypothesis, the same batch of purified recombinant MtANS used for the assays with labeled leucocyanidin was incubated with unlabeled dihydroquercetin in the presence of ferrous iron and 2-oxoglutaric acid. Significant production of quercetin was observed by HPLC analysis (Supplemental Fig. S4C), whereas no quercetin was observed when dihydroquercetin was incubated with protein from an empty-vector control (Supplemental Fig. S4B). Thus, MtANS is a bifunctional enzyme within the anthocyanidin/PA and flavonol biosynthetic pathways, at least in vitro. Similar observations have been made with ANS from Arabidopsis and rice (Oryza sativa; Turnbull et al., 2000; Tanner et al., 2003; Reddy et al., 2007). The rate of reaction with dihydroquercetin was almost 10-fold higher (460 pmol min−1 mg−1 protein) than with leucocyanidin.
Tissue-Specific Expression of PA Biosynthetic Genes in M. truncatula
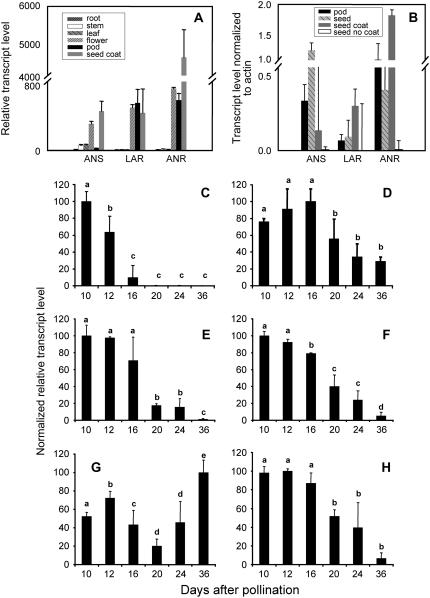
To assess the potential involvement of MtLAR and MtANS in PA biosynthesis, their transcript levels were first estimated by microarray analysis in a variety of tissue types, along with MtANR transcripts. Affymetrix microarray data of M. truncatula flavonoid and PA biosynthetic gene transcript levels were obtained from the recently compiled M. truncatula gene expression atlas assembled by our colleagues at the Noble Foundation (V.A. Benedito, I. Torres-Jerez, J. Murray, A. Andriankaja, S. Allen, K. Kakar, M. Wandrey, R. Thomson, T. Ott, S. Moreau, A. Niebel, J. He, X. Zhao, Y. Tang, and M.K. Udvardi, unpublished data). Tissues analyzed included roots, stems, leaves, flowers, and pods, as well as a time course profile for seed development. Seed coat RNA samples were generated and analyzed as part of the present work. Consistent with its role in PA biosynthesis, MtANR exhibited the highest expression level in seed coats, with lower expression in flowers and immature pods, and no expression in other tissues (Fig. 7A). In contrast to MtANR, MtLAR had similar low expression levels in flowers, pods, and seed coats. MtANS was expressed primarily in flowers and seed coats (Fig. 7A).
Figure 7.
Transcript levels of flavonoid/PA pathway genes in different tissues of M. truncatula. A, Affymetrix microarray analysis of RNA from the six tissues shown. B, Quantitative real-time PCR analysis of ANS, LAR, and ANR transcript levels in pods, whole seeds, isolated seed coats, and seeds without seed coats. C to H, Relative transcript levels of the indicated genes (C, LAR; D, ANS; E, ANR; F, DFR1; G, DFR2; H, F3′H) in seed coats at various stages of seed development (dap) as determined by Affymetrix microarray analysis. Letters reflect differences that were statistically significant at P = 0.05 by t test.
To further define the site of expression in seeds, RNA from pods, whole seeds, seed coats, and seeds without seed coats was analyzed by quantitative real-time PCR (Fig. 7B). In spite of high variation at the lowest transcript levels, the data suggest that the expression of ANS, ANR, and LAR in seeds is almost entirely the result of expression in the seed coat, thereby ruling out the possibility of significant contamination of seed coats from underlying tissue.
Microarray analysis indicated that ANS, dihydroflavonol reductase 1 (DFR1), and flavanone 3′-hydroxylase (F3′H) exhibited similar developmental expression patterns in seeds; transcripts corresponding to these three enzymes were already present at high levels at 10 dap, the first time practicable for dissection of developing seeds from the pods, and remained elevated through 16 dap (Fig. 7, D, F, and H). In contrast, LAR transcript levels had declined significantly by 16 dap (Fig. 7C), although total PA levels increased until 20 dap (Fig. 2D). ANR transcripts (Fig. 7E) appeared to follow the same pattern as ANS, DFR, and F3′H, although the 16-dap values showed quite high variation. Because of the low level of activity and the minute amounts of biological material available at early times postpollination, we did not determine extractable activities of the PA biosynthetic enzymes during seed development.
Subcellular Localization of MtANS, MtLAR, and MtANR
Because of the suggestion that the PA pathway may exist as a metabolic channel associated with cellular membranes (Winkel, 2004), it was of interest to determine the subcellular localization of LAR, ANS, and ANR. This was achieved by making gene constructs encoding C-terminal enhanced green fluorescent protein (EGFP)/yellow fluorescent protein (YFP) fusions of the respective ORFs for transient expression by particle bombardment and localization of fluorescence by confocal microscopy. As a control for endomembrane localization, we utilized a construct in which EGFP was fused to the membrane anchor of the endoplasmic reticulum (ER)-localized cytochrome P450 enzyme cinnamate 4-hydroxylase (C4H), as described previously (Achnine et al., 2004).
The ORFs of MtANS-YFP, MtLAR-EGFP, MtANR-EGFP, C4H-EGFP, and free EGFP were cloned into pRTL2 vector under the control of a double 35S promoter and transfected into tobacco (Nicotiana tabacum) leaves via gold particle bombardment. Confocal images of transfected tobacco leaf epidermal cells clearly indicated that MtANS, MtLAR, and MtANR proteins were each localized in the cytosol, with similar patterns to free EGFP protein (presence of diffuse fluorescence, visibility of cytoplasmic strands, and association of fluorescence with the outside of the nucleus), whereas the C4H-EGFP fusion gave the expected fine reticulate membrane localization (Supplemental Fig. S5). Patterns of cytoplasmic and membrane localization were as observed and confirmed in previous studies (e.g. Achnine et al., 2004).
Functional Characterization of MtLAR and MtANS through Modifying Their Expression in Transgenic Plants
To further investigate the function of MtLAR, the ORF was introduced into tobacco for constitutive ectopic expression under control of the cauliflower mosaic virus 35S promoter. Because expression of many foreign flavonoid pathway genes (including ANR) gives a clear metabolic phenotype in solanaceous plants (Muir et al., 2001; Tian and Dixon, 2006; Xie et al., 2006), we predicted formation of catechin in tobacco tissues that overexpresses LAR and also expresses the anthocyanin pathway (e.g. flowers), even if PAs themselves may not be made. From 38 independent kanamycin-resistant transgenic tobacco lines harboring the 35S:MtLAR construct, the five lines with the highest MtLAR transcript levels (lines 3, 5, 10, 12, and 38) were selected for further analysis. Leaf extracts from these plants showed no catechin or PA accumulation compared with control lines (data not shown), even though significant transcript levels for MtLAR were detected in this tissue (Supplemental Fig. S6A).
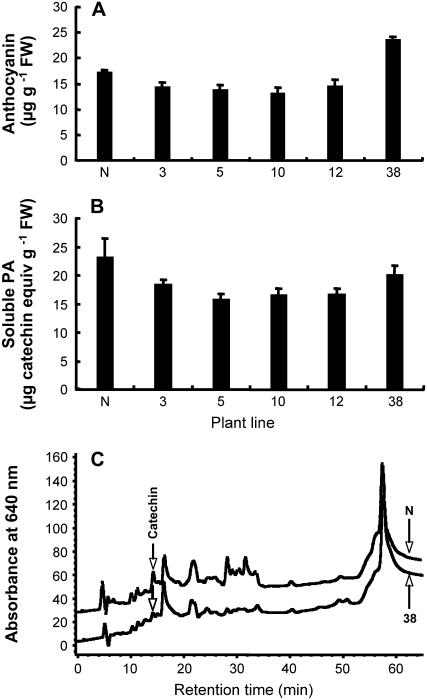
Lack of production of catechin or PAs in MtLAR-expressing leaves could result from lack of anthocyanidin substrate. Such substrate limitation does not occur in tobacco flowers, which produce increased levels of PAs at the expense of anthocyanins if expressing MtANR (Xie et al., 2003, 2006). Flower tissues from the five lines were first subjected to RT-PCR to confirm high MtLAR expression (Supplemental Fig. S6B), and then extracted and analyzed for anthocyanin and soluble and insoluble PA levels. Several transgenic lines exhibited a small reduction in anthocyanin levels (although in line 38 they were increased). However, PA levels were reduced rather than increased in all transgenic lines (Fig. 8, A and B), and no increase in material eluting from HPLC at the retention time of free catechin was observed (Fig. 8C). Similarly, no production of catechin or PAs was reported in tobacco or white clover (Trifolium repens) expressing Desmodium LAR, even though the recombinant enzyme was shown to be catalytically active after expression in planta (Tanner et al., 2003). We were unable to detect catechin in transgenic tobacco leaves or flowers expressing Desmodium LAR (data not shown).
Figure 8.
Anthocyanin and PA levels in flowers of transgenic tobacco constitutively expressing MtLAR. A, Anthocyanin levels as determined by extraction and UV absorption. B, Soluble PA levels as determined by extraction and reaction with DMACA reagent. C, Reverse-phase HPLC analysis (with postcolumn derivatization with DMACA) of the soluble PA fractions. Numbers refer to independent transgenic lines. N is empty-vector control line.
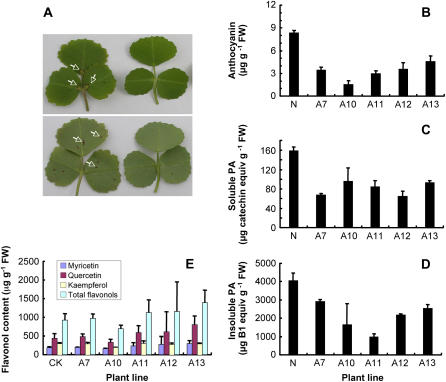
The potential in vivo functions of MtANS were determined by down-regulating its expression in M. truncatula genotype R108 by expression of a 35S-promoter-driven antisense construct. The presence of a red anthocyanin-rich circle at the base of the axial side of the leaflet and small red dots on the adaxial side (Fig. 9A) is a unique feature of anthocyanin deposition in this genotype. From 16 independent transgenic antisense MtANS lines, six plants lacked the unique red circle and spots, whereas the remainder had reduced levels of pigmentation (Fig. 9A). RT-PCR analysis confirmed reduced MtANS transcript levels in both leaves and flowers of the antisense lines when compared to the empty-vector control line (Supplemental Fig. S7). Reduction in MtANS expression did not affect expression of ANR or LAR in seeds (Supplemental Fig. S7).
Figure 9.
Anthocyanin and PA levels in transgenic M. truncatula ‘R108’ plants expressing an antisense MtANS transgene. A, Visible phenotypes of the adaxial side (top) and abaxial side (bottom) of leaves from plants transformed with an empty-vector control (left) or expressing the MtANS antisense construct (right). Note the loss of visible anthocyanin pigmentation (regions marked with white arrows) in the antisense line. B, Anthocyanin content of leaf tissue from five independent transgenic antisense (designated A) lines or empty-vector control (N). C, Soluble PA levels in seeds of the above lines. D, Insoluble PA levels in seeds of the above lines. E, Levels of flavonols in leaves of the above lines. Soluble PAs were determined by reaction with DMACA reagent, insoluble PAs by butanol-HCl hydrolysis, and estimation of resulting anthocyanidin. Flavonols were determined by HPLC analysis (see Supplemental Fig. S8).
Anthocyanin levels were strongly reduced in leaf tissues of all five antisense lines studied, in particular in line A10 with the lowest ANS transcript levels and complete absence of the red leaf spot (Fig. 9B). There also was a strong reduction in the levels of both soluble and insoluble PAs in seeds of the same lines (Fig. 9, C and D), consistent with involvement of ANS in PA biosynthesis, presumably to produce an anthocyanidin precursor for the ANR reaction leading to (−)-epicatechin.
To investigate the potential additional function for ANS in formation of flavonols from dihydroflavonols, as suggested from the in vitro activity of the enzyme, total flavonoid profiles of vector control and ANS antisense lines were compared by HPLC (Supplemental Fig. S8). We could not detect any significant difference in the levels of individual or total flavonols in lines in which ANS down-regulation led to a reduction in foliar anthocyanin levels (Fig. 9E).
DISCUSSION
M. truncatula as a Model for the Study of PA Biosynthesis
Arabidopsis has become the major model for studies on PA biosynthesis, largely as a result of the generation and analysis of a number of transparent testa (tt) and tannin-deficient seed (tds) mutants that lack PAs in the seed coat (Abrahams et al., 2003; Lepiniec et al., 2006). Most of the genetic loci associated with the tt phenotype have been characterized and encode enzymes of flavonoid and PA monomer formation, transporters, and transcription factors. Arabidopsis PAs appear to contain only (−)-epicatechin units (Routaboul et al., 2006) and, because Arabidopsis does not appear to possess a close ortholog of LAR, this has been presented as genetic evidence of LAR function in catechin formation (Tanner et al., 2003). Because of the impact of PAs on the quality of forage legumes, it is necessary to obtain functional orthologs of the Arabidopsis PA genes and other genes potentially specific for PA formation in legumes as a basis for genetic manipulation of the PA pathway in alfalfa (Medicago sativa) and clover, species that lack foliar PAs. The availability of extensive genomic (Cannon et al., 2006) and genetic (Tadege et al., 2005; Wang et al., 2006) resources makes M. truncatula the ideal model for such studies. However, apart from the biochemical characterization of ANR from M. truncatula (Xie et al., 2004b), little is known about PAs, the enzymes specific for their biosynthesis, or the transcriptional regulation of the PA pathway in this species.
Nature and Distribution of PAs in M. truncatula
PAs have been described in alfalfa, a species very closely related to M. truncatula. Essentially, PAs are either absent or present in very small amounts in the aerial organs of alfalfa. They are present at relatively high levels in seed coats, where they have been reported to exist as oligomers of DP from 4.4 to 6.5, with a mean DP of 5, containing epicatechin as the major extension unit and catechin as the major starter unit (Koupai-Abyazani et al., 1993).
Our results demonstrate major localization of PAs to the seed coat in M. truncatula, with very small amounts (close to detection limits) also present in flowers, leaves, roots, and stems. PAs start to accumulate in the M. truncatula seed coats before 10 dap at times when sampling is difficult because of the minute nature of the developing seeds. Levels reach a maximum at around 20 dap and the decrease in the soluble fraction as the insoluble fraction increases suggests that the PAs are initially soluble (potentially in the cell vacuole) and are subsequently insolubilized (presumably in the cell wall). The mean DP of the soluble fraction increases from 6 at 10 dap to around 17 in mature seeds. It is not easy to determine the DP of the insoluble fraction, but this is likely 17 or greater based on the size range of the soluble PAs.
Our most surprising finding was that Medicago PAs are composed almost entirely of (−)-epicatechin units. The hydroxylation pattern of the flavonoid B rings in the PA polymers was predominantly ortho-di-hydroxy, as revealed by HPLC analysis of the anthocyanidins released by butanol-HCl hydrolysis, consistent with catechin or epicatechin building blocks, but the phloroglucinolysis assays revealed insignificant levels of catechin as either starter or extension units. When PA preparations from D. uncinatum or barley were analyzed in parallel, catechin units were readily revealed (data not shown), confirming the validity of our experimental procedures.
Functions of ANS, ANR, and LAR in PA Biosynthesis in Medicago
Both genetic and biochemical evidence indicate clear roles for ANS and ANR in PA biosynthesis. Mutations in ANS and ANR result in a tt phenotype in Arabidopsis, with the ANR mutant (BANYULS) also accumulating anthocyanin pigment in the seed coat (Devic et al., 1999; Abrahams et al., 2003; Xie et al., 2003). Antisense down-regulation of ANS in M. truncatula resulted in reduction in the levels of anthocyanin pigments in leaves and of soluble and insoluble PAs in seeds. MtANR transcripts are most highly expressed in seed coats, which are the sites of maximal accumulation of PAs. ANS is expressed in the Medicago seed coat, but also in other tissues consistent with its role in anthocyanin as well as PA biosynthesis. Recombinant MtANS and MtANR, when expressed in E. coli, catalyze the formation of cyanidin from leucocyanidin (this work), and (−)-epicatechin from cyanidin (Xie et al., 2004b), respectively, consistent with their involvement in the pathway shown in Figure 1. Although MtANS can also catalyze the conversion of dihydroflavonol to flavonol in vitro, this is of questionable significance in vivo because flavonol levels were not noticeably reduced in transgenic Medicago lines in which antisense down-regulation of MtANS led to reduced anthocyanin and PA levels.
To date, the genetic evidence for LAR function is the observation that Arabidopsis does not possess an obvious LAR ortholog and has a PA composed of only epicatechin (Abrahams et al., 2003; Tanner et al., 2003; Routaboul et al., 2006). Furthermore, LAR expression is found in nonseed tissues of some plants, such as L. corniculatus (Paolocci et al., 2007) and grape (Bogs et al., 2005), which accumulate high levels of PAs in these tissues, and LAR is coregulated with ANR via MYB family transcription factors (Bogs et al., 2007; Paolocci et al., 2007).
The Medicago gene that we selected as encoding a potential MtLAR had been identified as such informatically by previous workers (Bogs et al., 2005; Paolocci et al., 2007). The recombinant enzyme exhibits the same in vitro catalytic activity as the LAR from D. uncinatum (Tanner et al., 2003), namely, the conversion of 3H-leucocyanidin to a compound with the same retention time and UV absorption characteristics as (+)-catechin, although, even with scaled-up reactions, we were unable to recover sufficient product for rigorous identification with determination of stereochemistry. This has yet to be achieved for the product of any of the recombinant LARs described to date (Bogs et al., 2005; Pfeiffer et al., 2006; Takos et al., 2006; Paolocci et al., 2007). The in vitro activity of the purified recombinant MtLAR was very low (only 40 pmol min−1 mg−1 protein) compared to the activity purified from Desmodium leaves (Tanner et al., 2003). However, a recent report failed to detect any activity for recombinant Lotus LAR when assayed as a single protein with leucocyanidin as substrate; weak activity was detected by a thin-layer chromatography assay only if the leucocyanidin was generated in situ through the activity of recombinant DFR in a coupled assay (Paolocci et al., 2007).
The precise function of MtLAR in PA biosynthesis in Medicago requires further clarification because of (1) the apparent lack of catechin units in Medicago seed coat PAs, although the seed coats express LAR; (2) the similar expression of MtLAR in flowers and in seed coats, although flowers accumulate much lower levels of PAs and do not appear to accumulate free catechin; (3) the poor correlation of MtLAR transcript levels with PA accumulation in the seed coat, in contrast to the good correlation in the cases of ANS and ANR, and particularly DFR1 and F3′H that produce upstream precursors for PA biosynthesis through LAR; and (4) the lack of production of catechin, and the reduction of soluble PA levels, in flowers of transgenic tobacco expressing MtLAR, in contrast to the predictable effects of genetically modifying expression of ANS in Medicago (this work) and ANR in tobacco (Xie et al., 2006). In the latter case, ectopic expression of MtANR in tobacco flowers resulted in production of considerable levels of both PAs and free epicatechin. Furthermore, a previous study in which Desmodium LAR was ectopically expressed in tobacco failed to report accumulation of catechin (Tanner et al., 2003) and we were unable to show catechin production in tobacco expressing Desmodium LAR.
In the absence of evidence that proteins encoded by LAR-like genes actually catalyze a different reaction from the conversion of leucocyanidin to (+)-catechin, we remain cautious as to the true in vivo function of MtLAR. We have yet to generate transgenic Medicago plants in which LAR is down-regulated, although it is not clear what the PA phenotype of these plants would be because Medicago PAs appear to contain insignificant levels of catechin. We cannot at present rule out the possibility that a subset of insoluble PAs containing catechin units are made in Medicago or that some type of metabolic channeling exists in vivo such that LAR does not properly couple with endogenous pathway enzymes (such as DFR; see below) in transgenic plants expressing a foreign LAR, thereby accounting for the lack of catechin production in transgenic plants overexpressing the enzyme, although this is not a problem with transgenic expression of ANR for epicatechin production (Xie et al., 2006). The reduction in PA levels in tobacco flowers expressing MtLAR could likewise be explained by incorrect insertion of a heterologous functional LAR into a metabolic channel for PA formation or by LAR having alternate or additional functions that divert flux from PA biosynthesis. The D. uncinatum LAR protein was shown to be inhibited by naringenin, dihydroflavonols, and flavonols (Tanner et al., 2003), so it is possible that enzyme activity may be blocked in vivo in transgenic plants in which pools of flavonoid intermediates are higher than in the natural situation.
Metabolic Channeling at the Branch Point for PA Biosynthesis?
Because of the instability and expense of leucoanthocyanidin, the first detection of LAR activity in crude plant cell extracts utilized coupled assays in which dihydroflavonol was converted by endogenous DFR to leucoanthocyanidin and then on to catechin (Stafford and Lester, 1985; Kristiansen, 1986). A similar coupled assay with recombinant enzymes was shown to be essential for detection of LAR activity from L. corniculatus (Paolocci et al., 2007). A protein complex from Onobrychis viciifolia could produce catechin from dihydroflavonol (Singh et al., 1997), although the exact nature of the protein components was not reported. These observations are consistent with a recently proposed model for metabolic channeling in flavonoid biosynthesis, whereby PA-specific enzymes, such as LAR, ANS, and ANR, possibly along with DFR, might form a complex through which intermediates are channeled directly into the formation of PAs. A separate channel would be envisaged to produce anthocyanins (Winkel, 2004).
In such a model, the flavonoid biosynthetic pathway is seen as localized as one or two ER-associated multienzyme complexes (Stafford, 1974). Such localization has been confirmed for some phenylpropanoid/flavonoid pathway enzymes (Rasmussen and Dixon, 1999; Liu and Dixon, 2001; Achnine et al., 2004). However, our results suggest that LAR, ANS, and ANR are all localized to the cytosol. This is consistent with the fact that none of these enzymes has clear signal peptides or membrane-targeting domains. Nevertheless, these experiments were performed by particle bombardment of single-enzyme constructs into tobacco leaf epidermal cells, not into the natural milieu of the endothelial layer of the Medicago seed coat. Our data suggest that none of the enzymes alone has membrane localization, but cannot rule out possible complex formation when all are present together in the same cell. Furthermore, we cannot rule out the possibility of associations between DFR and LAR, which specifically channel common precursors away from anthocyanin synthesis and into PA formation. In this respect, it is interesting to note the presence of two distinct DFR genes in M. truncatula. Although the specificity of the corresponding enzymes has been determined and exhibits differences as regards to the A- and B-ring substitution patterns of the dihydroflavonol substrates (Xie et al., 2004a), the exact stereochemistry of the products has yet to be determined. This may be important for understanding LAR function.
In summary, our results provide the information on PA content and composition required to allow for the development of M. truncatula as a model for functional genomic approaches to understanding PA biosynthesis, with translational opportunities for quality improvement in alfalfa. Future studies will address the function of LAR in Medicago through direct forward and reverse genetic approaches.
MATERIALS AND METHODS
Plant Material
Medicago truncatula ‘Jemalong A17’ plants were grown in Metro-mix 350 (Scotts), with an 18-h light/25°C and 6-h dark/22°C photoperiod in the greenhouse. Individual tissues were harvested from greenhouse material, separated, and frozen in liquid nitrogen until further processing. Transgenic tobacco (Nicotiana tabacum) and M. truncatula (‘R108’) were grown under the same conditions and leaf tissue for anthocyanin analysis was collected 1 month after the Medicago seedlings were transferred from medium to soil. To collect immature seed, the pollination date of each flower was recorded, the flower labeled, and individual pods were then collected at either 10, 12, 16, 20, 24, or 36 dap. Seeds from the same day after pollination were then separated, pooled, and frozen in liquid nitrogen.
Extraction and Quantification of PAs, Anthocyanins, and Flavonols
For extraction of PAs, tissues were ground in liquid nitrogen and 0.25- to 1.0-g batches were extracted with 5 mL of extraction solution (70% acetone:0.5% acetic acid) by vortexing followed by sonication at 30°C for 30 min. Following centrifugation at 2,500g for 10 min, residues were reextracted twice as above. Pooled supernatants were then extracted with 30 mL of chloroform, and the aqueous supernatant reextracted twice with chloroform and three times with hexane. Samples were freeze dried and resuspended in extraction solution to a final concentration of 3 g of original sample/mL. Samples were spun briefly, transferred to another tube, and soluble PA content determined using DMACA reagent with catechin standards. In brief, aliquots of samples or standards (2.5 μL) were mixed with 197.5 μL of DMACA reagent (0.2% [w/v] DMACA in methanol-3 n HCl [1:1]) in microplate wells; for blanks, the same samples were replaced with 2.5 μL of methanol-3 n HCl. Samples, blanks, and standards were read within 15 min on a Wallac Victor 2 plate reader equipped with a 640-nm emission filter. Blanks were subtracted from samples and PA content calculated as catechin equivalents.
For measurement of insoluble PAs, the residues from the above tissue extractions were dried in air for 2 d, and 1 mL butanol-HCl reagent was then added and the mixture sonicated at room temperature for 60 min, followed by centrifugation at 2,500g for 10 min. Supernatants were transferred to cuvettes for determination of absorption at 550 nm and samples were then boiled for 1 h. After cooling to room temperature, the A550 was recorded again and the first value subtracted from the second. Absorbance values were converted into PA equivalents using a standard curve (2.5, 5, 10, 20, and 40 μg) of procyanidin B1 (Indofine).
Normal-phase HPLC analysis of soluble PAs was performed using an HP 1100 system equipped with a diode array detector. Samples were separated on a 250 × 4.6-mm Luna 5-μm silica column, and postcolumn derivatization was accomplished using a separate HPLC pump (model 426; Alltech), which was used to deliver the DMACA reagent (1% DMACA in 1.5 m H2SO4 in methanol) to a mixing tee where effluent from the column and the reagent combined and passed through an 8-m coil of 0.2-mm-i.d. PEEK tubing prior to detection at 640 nm (Peel and Dixon, 2007).
Soluble PAs from seeds at different developmental stages were purified on a Sephadex LH-20 column (Pharmacia) using 50% (v/v) methanol, followed by 70% acetone to elute the PAs. Acetone was removed by rotary evaporation and the aqueous phase was freeze dried. One hundred micrograms of purified PA samples were then analyzed by the phloroglucinolysis procedure, with detection of the PA degradation products by HPLC, as described previously (Kennedy and Jones, 2001).
For analysis of anthocyanin levels, 5 mL of methanol:0.1% HCl was added to 0.5 g of ground tissue and sonicated for 1 h, followed by shaking overnight at 120 rpm. After centrifugation at 2,500g for 10 min, 1 mL of water was added to 1 mL of extract, followed by addition of 1 mL of chloroform to remove chlorophyll. Absorption of the clear supernatant was then measured at 530 nm. Total anthocyanin concentration was calculated using the molar absorbance of cyanidin-3-O-glucoside.
For analysis of flavonols, 100 mg of plant tissue was extracted with 3 mL 80% methanol, sonicated for 1 h, and allowed to stand overnight at 4°C. The extract was centrifuged to remove debris and the supernatant dried under nitrogen. Dried samples were incubated with 3 mL of 1 n HCl at 90°C for 2 h and extracted twice with 3 mL of ethyl acetate. Ethyl acetate extracts were pooled, dried under nitrogen, and resuspended in 200 μL of methanol, 20 μL of which were used for reverse-phase HPLC analysis as described below.
Isolation of LAR and ANS cDNA from M. truncatula
Total RNA from developing seeds of M. truncatula ‘Jemalong A17’ was isolated using modified cetyl-trimethyl-ammonium bromide (CTAB) extraction buffer (2% CTAB, 2% polyvinylpyrrolidone, 100 mm Tris-HCl, pH 8.0, 20 mm EDTA, 1.4 m NaCl, 0.5% [w/v] spermidine, and 3% [v/v] β-mercaptoethanol) as described previously (Jaakola et al., 2001) with minor modifications. RNA was purified and concentrated using RNeasy MinElute cleanup kits (Qiagen). First-strand cDNA was synthesized from 4 μg total RNA in a total volume of 20 μL using SuperScript III reverse transcriptase (Invitrogen) after DNaseI treatment (Invitrogen) according to the manufacturer's instructions. The ORF of MtLAR was obtained by RT-PCR (GenBank accession no. BN000703) with 2 μL cDNA from seeds, using 5′-ATGGCACCATCATCATCACCAAC-3′ as forward primer (the start codon is underlined) and 5′-TCAACAGGAAGCTGTGATTGG-3′ as reverse primer (the stop codon is underlined) with Pfu DNA polymerase (Stratagene) in a total volume of 50 μL. The PCR reaction was carried out at 94°C for 5 min; 35 cycles of 94°C for 30 s; 58°C for 30 s; and 72°C for 2 min; followed by a final extension of 72°C for 7 min. The A tail was added to the PCR product, which was then cloned into pGEM-T easy vector according to the manufacturer's instructions (Promega), and the correct reading frame of the resulting construct (pGEMT-MtLAR) confirmed by sequencing.
The putative EST sequence (BM812824) of M. truncatula ANS was chosen by sequence comparison and alignment from M. truncatula EST sequences. Full-length MtANS cDNA was cloned by RACE with cDNA from seeds, using the SMART RACE cDNA amplification kit (CLONTECH). Primers used for 3′-RACE were 5′-GAGTTAGCAAACATAGGTAACATCTTTG-3′ and 5′-GTCTATATGGCCCAAGACACCTGCTGA-3′ (nested primer); primers used for 5′-RACE were 5′-CTTCCATAACCTTGAATCTTCCCAGAAC-3′ and 5′-CTTCTGCAGCTTTCTTAAGCTTCTCTC-3′ (nested primer). The ORF of MtANS was generated using 5′-ATGGGAACGGTGGCTCAAAGAG-3′ (start codon underlined) and 5′-TCATTTTTTGGGATCATCTTTCTTC-3′ (stop codon underlined) with pfu DNA polymerase in a total volume of 50 μL, according to the manufacturer's instructions. PCR conditions and cloning of the product into pGEM-T easy vector were as for MtLAR.
The pIs and molecular weights of the MtLAR and MtANS proteins were calculated using the pI/molecular weight calculation tools at www.expasy.org.
DNA Gel-Blot Analysis of MtLAR
Thirty micrograms of genomic DNA, extracted with SDS buffer from young M. truncatula ‘Jemalong A17’ plants, were digested with BamHI (no site within the LAR ORF), EcoRI (two sites), and EcoRV or HindIII (one site). The digested products were then size fractionated on a 0.8% agarose gel, blotted onto nylon membrane, and hybridized and washed as described by Sambrook et al. (1989) under stringent conditions. The probe used was the ORF region of MtLAR, labeled with 32P-dCTP using the DECAprime II random priming DNA labeling kit (Ambion) according to the manufacturer's instructions.
In Vitro Expression of Recombinant MtLAR and MtANS Proteins
The ORF of the MtLAR gene was PCR amplified from pGEMT-MtLAR using pfu DNA polymerase (Stratagene) and the following primers: MtLAR-BF (5′-GGATCCATGGCACCATCATCATCACCAAC-3′; the BamHI site is in italics and the start codon single underlined) and MtLAR-SR (5′-GAGCTCTCAACAGGAAGCTGTGATTGG-3′; the SacI site is in italics and the stop codon single underlined). It was then subcloned into the BamHI and SacI sites of the pQE30 expression vector (Qiagen). After confirmation by sequencing, the resulting vector pQE-MtLAR was transformed into Escherichia coli strain M15 for protein induction by 1 mm isopropyl β-d-thiogalactopyranoside (IPTG). Recombinant MtLAR protein with a 6×-His tag at the N terminus was purified using a Magne-His kit (Promega) and the protein concentration measured by the Bradford method (Bradford, 1976).
The ORF of the MtANS gene was amplified from pGEMT-MtANS, subcloned into the pQE30 expression vector, and expressed in E. coli using the same strategy as for MtLAR. Primers were MtANS-BF, 5′-GGATCCATGGGAACGGTGGCTCAAAGAG-3′ and MtANS-SR, 5′-GAGCTCTCATTTTTTGGGATCATCTTTCTTC-3′.
Assay of LAR and ANS Activities
3H-3,4-cis-Leucocyanidin was synthesized as described elsewhere (Tanner and Kristiansen, 1993). Assay of recombinant MtLAR protein with 3H-3,4-cis-leucocyanidin was carried out in a final volume of 100 μL containing 10% (w/v) glycerol, 100 mm KPi (pH 7.0), 4 mm dithiothreitol (DTT), 0.5 mm NADPH, 0.4 μm 3H-leucocyanidin (288 mCi/mmol), and 20 μg of purified recombinant MtLAR protein. The reaction was initiated by the addition of enzyme and incubated at 30°C for 1 h. The assay was terminated by the addition of 10 μL of methanol followed by centrifugation. Products were analyzed by HPLC, monitoring at 280 nm. Products eluting at retention times between 19.5 to 22.5 min were collected (0.5 min/tube) and the fractions subjected to liquid scintillation counting. Crude protein extract from induced M15 containing empty vector was used as a control.
ANS activity for production of cyanidin from leucocyanidin was determined in a final volume of 100 μL containing 20 mm KPi (pH 7.0), 200 mm NaCl, 10 mm maltose, 5 mm DTT, 4 mm sodium ascorbate, 1 mm 2-oxoglutaric acid, 0.4 mm FeSO4, 0.4 μm 3H-3,4-cis-leucocyanidin, and 20 μg of purified recombinant MtANS protein. After incubation at 30°C for 1 h, the reaction was terminated by the addition of 1 μL of 36% HCl followed by 10 μL of methanol. Samples were centrifuged and then subjected to HPLC analysis with monitoring at 530 nm. Products eluting at retention times from 33.5 to 40 min were collected (0.5 min/tube) and subjected to scintillation counting. Crude protein extract from induced M15 containing empty vector was used as a control.
For assay of recombinant MtANS with dihydroquercetin as substrate, reaction mixtures (100 μL) contained 20 mm KPi (pH 7.0), 5 mm DTT, 4 mm sodium ascorbate, 1 mm 2-oxoglutaric acid, 0.4 mm FeSO4, 0.2 μm dihydroquercetin, and 20 μg of purified recombinant MtANS. After incubation at 30°C for 1 h, reactions were stopped with 10 μL methanol, centrifuged, and subjected to HPLC analysis. Crude protein extract from induced M15 containing empty vector was used as a control.
Reverse-phase HPLC analysis of enzymatic products (and flavonols; see above) used an Agilent HP1100 HPLC with the following gradient: solvents A (1% phosphoric acid) and B (acetonitrile) at 1 mL/min flow rate: 0 to 5 min, 5% B; 5 to 10 min, 5% to 10% B; 10 to 25 min, 10% to 17% B; 25 to 30 min, 17% to 23% B; 30 to 65 min, 23% to 50% B; 65 to 79 min, 50% to 100% B; 79 to 80 min, 100% to 5% B. Data were collected at 254, 280, and 530 nm. Identifications were based on comparison of chromatographic behavior and UV spectra with those of authentic standards.
Transformation of Tobacco with MtLAR
The ORF of MtLAR was amplified with the primers MtLAR-BF and MtLAR-SR and then cloned into the BamHI and SacI sites of the binary vector pBI121. The resulting vector pBI121-MtLAR, with MtLAR driven by the cauliflower mosaic virus 35S promoter was transferred into Agrobacterium tumefaciens strain LBA4404 by electroporation. Transgenic tobacco ‘Xanthi’ plants were generated by Agrobacterium-mediated transformation of leaf discs on Murashige and Skoog medium supplied with 100 mg/L kanamycin using standard protocols (Horsch et al., 1988). Only one bud was picked from each explant to ensure independent transformants.
Transformation of M. truncatula with Antisense MtANS
The MtANS ORF in the antisense orientation was amplified with the primers MtANS-SF (5′-GAGCTCATGGGAACGGTGGCTCAAAGAG-3′; the SacI site is in italics and the start codon is single underlined) and MtANS-BR (5′-GGATCCTCATTTTTTGGGATCATCTTTCTTC-3′; the BamHI site is in italics and the stop codon is single underlined). The amplified PCR product was cloned into the BamHI and SacI sites of the binary vector pBI121, and the resulting vector pBI121-MtANSanti, with MtANS in the antisense orientation driven by the 35S promoter, was transferred into A. tumefaciens strain LBA4404 by electroporation. Transgenic M. truncatula ‘R108’ plants were generated by Agrobacterium-mediated transformation of cotyledons as described previously (Wright et al., 2006). Each transgenic plant was from a separate cotyledonary explant.
Analysis of LAR, ANS, and ANR Transcript Levels by RT-PCR in Transgenic Plants
Total RNA from leaves, flowers, and seeds of transgenic M. truncatula lines was isolated using the CTAB method (Jaakola et al., 2001) and purified and concentrated using the RNeasy MinElute cleanup kit (Qiagen). Total RNA from leaves and flowers of transgenic tobacco plants was isolated using the RNeasy Plant mini kit (Qiagen). cDNA was synthesized from 4 μg of total RNA in a total volume of 20 μL using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions, diluted 10-fold, and 5 μL were used in subsequent RT-PCR reactions. The primers used for MtANS were MtANS-F1 and MtANS-R1, MtLAR-F1 and MtLAR-R1 were used for MtLAR, and MtANR-F (5′-TGCAAACAAAACATCTCACCTCATAG-3′, forward) and MtANR-R (5′-AATTTCCACGCAGCCTTTTCAG-3′, reverse) were used for MtANR. Optimized PCR conditions were 94°C for 5 min; 23 cycles of 94°C for 30 s; 55°C for 30 s; and 72°C for 2 min; followed by a final extension of 72°C for 7 min. Actin was used as internal standard using the primers actin-F (5′-GATATGGAAAAGATCTGGCATCAC-3′, forward) and actin-R (5′-TCATACTCGGCCTTGGAGATCCAC-3′, reverse). For RT-PCR analysis of MtANS with cDNA from leaves, the number of PCR cycles was increased to 25. PCR products were analyzed by electrophoresis of 10-μL aliquots of reactions on 1% agarose gels in Tris-acetic acid-EDTA buffer, and visualized with SYBR DNA stain dye (Molecular Probes).
Analysis of Flavonoid/PA Pathway Gene Transcript Levels by Quantitative Real-Time RT-PCR
Total RNA from pods, seeds, seed coats, and seeds without coat was isolated using the CTAB method, and all RNA was purified and concentrated and first-strand cDNA synthesized, as described above. Primers for quantitative real-time RT-PCR were designed using Primer3 software (http://www.frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The forward and reverse primers for MtLAR amplification were MtLAR-RTF (5′-GGAATAGCCGCAGAAAATTG-3′) and MtLAR-RTR (5′-CATGGCAACAAAGCTCTCAA-3′); primers for MtANR amplification were MtANR-RTF (5′-GCAGTTTCTATCGGGTTCAA-3′) and MtANR-RTR (5′-CTGAGGGTATCGTTTGCTGA-3′); and primers for MtANS amplification were MtANS-RTF (5′-GGTTGGAAGGTGGAAGGTTA-3′) and MtANS-RTR (5′-CCCATTTGCCCTCATAGAAA-3′). Each primer pair gave a single PCR product. Quantitative real-time RT-PCR and analysis were as described by Modolo et al. (2007), except that the PCR reactions were performed in a 10-μL reaction volume containing 5 μL of SYBR Green master mix reagent (Applied Biosystems), 1 μL of 1:10 dilution of cDNA (reverse transcribed from 2 μg total RNA treated with DNaseI), and 200 nm of gene-specific primer pair.
Microarray Analysis
Affymetrix microarray analysis of various Medicago tissues and six seed development stages will be described elsewhere (V.A. Benedito, I. Torres-Jerez, J. Murray, A. Andriankaja, S. Allen, K. Kakar, M. Wandrey, R. Thomson, T. Ott, S. Moreau, A. Niebel, J. He, X. Zhao, Y. Tang, and M.K. Udvardi, unpublished data). Data showing the normalized relative transcript levels of M. truncatula flavonoid and PA biosynthetic genes at various stages of seed development were extracted for this study. For normalization of the transcript level at different seed development stages, the highest average value for each gene was taken as 100 and the relative transcript level of each gene at other times calculated accordingly. Mean transcript levels were determined from three biological replicates. Statistical analysis of data will be described (V.A. Benedito, I. Torres-Jerez, J. Murray, A. Andriankaja, S. Allen, K. Kakar, M. Wandrey, R. Thomson, T. Ott, S. Moreau, A. Niebel, J. He, X. Zhao, Y. Tang, and M.K. Udvardi, unpublished data). Briefly, for each Affymetrix array hybridized, the resulting .cel file was exported from GeneChip Operating Software, version 1.4 (Affymetrix), and imported into Robust Multiarray Average for global normalization. Presence/absence call for each probe set was obtained using dCHIP. Gene selections based on Student's t test and associative t test were made using Matlab (MathWorks). A Bonferroni correction P-value threshold of 1 × 10−5 was used. Affymetrix microarray analysis of total RNA from Medicago seed coats was conducted as above with three biological replicates.
Subcellular Localization of PA Pathway Enzyme-EGFP/YFP Fusions by Confocal Microscopy
For construction of fusions of the MtANS, MtLAR, and MtANR ORFs to EGFP/YFP (CLONTECH), a two-step recombinant PCR strategy was applied. In the first step, the ORF of each gene was amplified using pGEMT-MtLAR/MtANS or pSE380-BAN (Xie et al., 2003) as templates, with a forward primer to introduce an XhoI restriction site at the 5′ end of the target gene, and a reverse primer containing reverse complementary sequences from the 3′ end of the target gene and the 5′ end of the EGFP ORFs. Similarly, EGFP/YFP fusions were amplified using the corresponding forward primers with reverse complementarities to the target genes, and one reverse primer that introduced a BamHI restriction site at the 3′ end. Primers designed for MtLAR were MtLAR-XF (5′-CCCTCGAGATGGCACCATCATCATCACCAACC-3′, forward; the XhoI site is in italics and the start codon is single underlined) and MtLAR-R (5′-CTTGCTCACCATACAGGAAGCTGTGATTGG-3′, reverse) for MtLAR gene amplification with an EGFP linker, and EGFP-MtLAR-F (5′-ACAGCTTCCTGTATGGTGAGCAAGGGCGAG-3′, forward) and EGFP-BR (5′-CGGGATCCTTACTTGTACAGCTCGTC-3′, reverse; the BamHI site is in italics and the stop codon is single underlined) for EGFP gene amplification with an MtLAR linker. Similarly, the four primers used for MtANS-YFP were MtANS-XF (5′-CTCGAGATGGGAACGGTGGCTCAAAGAG-3′, forward; the XhoI site is in italics and the start codon is single underlined), MtANS-R (5′-CTTGCTCACCATTTTTTTGGGATCATCTTTCTTC-3′, reverse), YFP-MtANS-F (5′-GATGATCCCAAAAAAATGGTGAGCAAGGGCGAG-3′, forward), and EGFP-BR. Primers for the MtANR constructs were MtANR-XF (5′-CTCGAGATGGCTAGTATCAAACAAATAGAA-3′, forward; the XhoI site is in italics and the start codon is single underlined), MtANR-R (5′-CTTGCTCACCATCTTGATCCCCTGAGTCTT-3′, reverse), EGFP-MtANR (5′-CAGGGGATCAAGATGGTGAGCAAGGGCGAG-3′), and EGFP-BR.
The resulting target genes and EGFP/YFP ORF fusion fragments were further purified and served as templates in second-round PCR using the 5′ end forward primer of the target gene (with the XhoI site) and EGFP 3′ end reverse primer (EGFP-BR with the BamHI site). PCR was performed using high-fidelity Pfu DNA polymerase (Stratagene).The resulting EGFP/YFP fusions were then subcloned into pGEM-T easy vector followed by sequencing to ensure that no mutations were introduced. Constructs were digested with XhoI and BamHI, and the resulting enzyme-EGFP/YFP fusions inserted into the corresponding sites of the shuttle vector pRTL2 (Restrepo et al., 1990) under the control of the double 35S promoter. The pRTL2-EGFP (Tian and Dixon, 2006) and pRTL2-C4H MA-EGFP (Achnine et al., 2004) constructs were used as controls for cytosolic and ER membrane localization.
Plasmid DNAs (5 μg) harboring EGFP, C4H MA-EGFP, or target genes fused with EGFP/YFP, under the control of the double 35S promoter were transiently expressed in young tobacco leaf ‘Xanthi Cornell’ epidermal cells as described previously (Liu and Dixon, 2001). Tobacco leaves were bombarded with plasmid DNA coated to gold particles at 900 psi and were examined 24 h later using a Leica SP2 AOBS confocal imaging system (Leica Microsystems). EGFP/YFP-expressing cells were imaged using the 488/514-nm line of an argon laser and emission was detected at 509/527 nm.
Confocal images were collected and assembled using Adobe Photoshop 5.0 L.E. (Adobe Systems).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BN000703 and EF544389.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. DNA gel-blot analysis of LAR gene copy number in M. truncatula.
Supplemental Figure S2. Alignment of the deduced amino acid sequences of ANS genes.
Supplemental Figure S3. Phylogenetic tree of several classes of 2-ODDs involved in flavonoid biosynthesis.
Supplemental Figure S4. HPLC analysis of the products from incubation of recombinant MtANS protein with dihydroquercetin.
Supplemental Figure S5. Subcellular localization of MtANS, MtLAR, and MtANR in tobacco leaf epidermal cells by particle bombardment.
Supplemental Figure S6. RT-PCR analysis of MtLAR transcript levels in transgenic tobacco plants constitutively expressing MtLAR.
Supplemental Figure S7. RT-PCR analysis of transcripts encoding PA biosynthetic genes in transgenic M. truncatula plants expressing an antisense MtANS transgene.
Supplemental Figure S8. HPLC analysis of flavonoids from control M. truncatula and transgenic MtANS antisense line A10.
Supplementary Material
Acknowledgments
We thank Dr. Aline Valster and Dr. Elison Blancaflor for assistance with confocal microscopy; Jack Blount for assistance with HPLC analysis; Kristy Richerson for growth and propagation of plants; Dr. Yuhong Tang for assistance with microarray analysis; Stacy Allen and Dr. Vagner Benedito for assistance with quantitative real-time PCR experiments; Dr. Xianzhi He, Dr. Guodong Wang, Dr. Rui Zhao, and Dr. Luzia Modolo for assistance with Medicago seed harvest; and Dr. Stephen Temple and Dr. Elison Blancaflor for critical reading of the manuscript.
This work was supported by the Samuel Roberts Noble Foundation and Forage Genetics International. The confocal microscope used in this study was funded by the National Science Foundation (equipment grant no. DBI–0400580).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard A. Dixon (radixon@noble.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin P, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35 624–636 [DOI] [PubMed] [Google Scholar]
- Achnine L, Blancaflor EB, Rasmussen S, Dixon RA (2004) Co-localization of l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16 3098–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts RJ, Barry TN, McNabb WC (1999) Polyphenols and agriculture: beneficial effects of proanthocyanidins in forages. Agric Ecosyst Environ 75 1–12 [Google Scholar]
- Antony U, Chandra TS (1998) Antinutrient reduction and enhancement in protein, starch, and mineral availability in fermented flour of finger millet (Eleusine coracana). J Agric Food Chem 46 2578–2582 [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG (2000) Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology 148 187–197 [DOI] [PubMed] [Google Scholar]
- Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol 139 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Britsch L, Dedio J, Saedler H, Forkmann G (1993) Molecular characterization of flavanone 3-β-hydroxylases: consensus sequence, comparison with related enzymes and the role of conserved histidine residues. Eur J Biochem 217 745–754 [DOI] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Scheix T, et al (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA 103 14959–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Jung HY, Kim HS, Cho HG (2000) PhyloDraw: a phylogenetic tree drawing system. Bioinformatics 16 1056–1058 [DOI] [PubMed] [Google Scholar]
- Cos P, De Bruyne T, Hermans N, Apers S, Berghe DV, Vlietinck AJ (2004) Proanthocyanidins in health care: current and new trends. Curr Med Chem 11 1345–1359 [DOI] [PubMed] [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19 387–398 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Sharma SB, Xie D (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165 9–28 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Sumner LW (2003) Legume natural products: understanding and manipulating complex pathways for human and animal health. Plant Physiol 131 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas GB, Stienezen M, Waghorn GC, Foote AG, Purchas RW (1999) Effect of condensed tannins in birdsfoot trefoil (Lotus corniculatus) and sulla (Hedysarum coronarium) on body weight, carcass fat depth, and wool growth of lambs in New Zealand. NZJAR (N Z J Agric Res) 42 55–64 [Google Scholar]
- Harborne JB, Williams CA (2001) Anthocyanins and other flavonoids. Nat Prod Rep 18 310–333 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT (1988) Leaf disc transformation. In SB Gelvin, R Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–9
- Jaakola L, Pirttila AM, Halonen M, Hohtola A (2001) Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol 19 201–203 [DOI] [PubMed] [Google Scholar]
- Jende-Strid B (1993) Genetic control of flavonoid biosynthesis in barley. Hereditas 119 187–204 [Google Scholar]
- Kennedy JA, Jones GP (2001) Analysis of proanthocyanidin cleavage products following acid-catalysis in the presence of excess phloroglucinol. J Agric Food Chem 49 1740–1746 [DOI] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, U S, Carne AF, Lee RE, Lobanenkov VV, Goodwin GH (1997) Molecular weight abnormalities of the CTCF transcription factor: CTCF migrates aberrantly in SDS-PAGE and the size of the expressed protein is affected by the UTRs and sequences within the coding region of the CTCF gene. Nucleic Acids Res 25 466–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koupai-Abyazani MR, McCallum J, Muir AD, Lees GL, Bohm BA, Towers GHN, Gruber MY (1993) Purification and characterization of a proanthocyanidin polymer from seed of alfalfa (Medicago sativa cv. beaver). J Agric Food Chem 41 565–569 [Google Scholar]
- Kristiansen KN (1986) Conversion of (+)-dihydroquercetin to (+)-2,3-trans-3,4-cis-leucocyanidin and (+)-catechin with an enzyme extract from maturing grains of barley. Carlsberg Res Commun 51 51–60 [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57 405–430 [DOI] [PubMed] [Google Scholar]
- Liu CJ, Dixon RA (2001) Elicitor-induced association of isoflavone O-methyltransferase with endomembranes prevents formation and 7-O-methylation of daidzein during isoflavonoid phytoalexin biosynthesis. Plant Cell 13 2643–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacin R, Britsch L (1997) Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3-β-hydroxylase. Eur J Biochem 249 748–757 [DOI] [PubMed] [Google Scholar]
- Marles MAS, Ray H, Gruber MY (2003) New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry 64 357–383 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang X, Dixon RA (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64 499–518 [DOI] [PubMed] [Google Scholar]
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, De Vos CHR, van Tunen AJ, Verhoeyen ME (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19 470–474 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Geurts R (2001) Medicago truncatula, going where no plant has gone before. Trends Plant Sci 6 552–554 [DOI] [PubMed] [Google Scholar]
- Paolocci F, Robbins MP, Madeo L, Arcioni S, Martens S, Damiani F (2007) Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis: structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus. Plant Physiol 143 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel GJ, Dixon RA (2007) Detection and quantification of engineered proanthocyanidins in transgenic plants. Natural Product Commun (in press)
- Pfeiffer J, Kuhnel C, Brandt J, Duy D, Punyasiri P, Forkmann G, Fischer T (2006) Biosynthesis of flavan 3-ols by leucoanthocyanidin 4-reductases and anthocyanidin reductases in leaves of grape (Vitis vinifera L.), apple (Malus × domestica Borkh.) and other crops. Plant Physiol Biochem 44 323–334 [DOI] [PubMed] [Google Scholar]
- Prescott AG, John P (1996) Dioxygenases: molecular structure and role in plant metabolism. Annu Rev Plant Physiol Plant Mol Biol 47 245–271 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Dixon RA (1999) Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11 1537–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AM, Reddy VS, Scheffler BE, Wienand U, Reddy AR (2007) Novel transgenic rice overexpressing anthocyanidin synthase accumulates a mixture of flavonoids leading to an increased antioxidant potential. Metab Eng 9 95–111 [DOI] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224 96–107 [DOI] [PubMed] [Google Scholar]
- Saito K, Kobayashi M, Gong Z, Tanaka Y, Yamazaki M (1999) Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase: molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J 17 181–189 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Singh S, McCallum J, Gruber MY, Towers GHN, Muir AD, Bohm BA, Koupai-Abyazani MR, Glass ADM (1997) Biosynthesis of flavan-3-ols by leaf extracts of Onobrychis viciifolia. Phytochemistry 44 425–432 [Google Scholar]
- Stafford HA (1974) Possible multienzyme complexes regulating the formation of C6-C3 phenolic compounds and lignins in higher plants. In VC Runeckles, ed, Metabolism of Secondary Plant Products Including Regulation. Academic Press, New York, pp 53–79
- Stafford HA, Lester HH (1985) Flavan-3-ol biosynthesis: the conversion of (+)-dihydromyrecetin to its flavan-3,4-diol (leucodelphinidin) and to (+)-gallocatechin by reductases extracted from tissue cultures of Ginkgo biloba and Pseudotsuga menziesii. Plant Physiol 78 791–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Ratet P, Mysore KS (2005) Insertional mutagenesis: a Swiss army knife for functional genomics of Medicago truncatula. Trends Plant Sci 10 229–235 [DOI] [PubMed] [Google Scholar]
- Takos AM, Ubi BE, Robinson SP, Walker AR (2006) Condensed tannin biosynthesis genes are regulated separately from other flavonoid biosynthesis genes in apple fruit skin. Plant Sci 170 487–499 [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278 31647–31656 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Kristiansen KN (1993) Synthesis of 3,4-cis-[3H]-leucocyanidin and enzymatic reduction to catechin. Anal Biochem 209 274–277 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Dixon RA (2006) Engineering isoflavone metabolism with an artificial bifunctional enzyme. Planta 224 496–507 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Sobey WJ, Aplin RT, Hassan A, Hassan A, Firmin JL, Schofield CJ, Prescott AG (2000) Are anthocyanidins the immediate products of anthocyanidin synthase? Chem Comm 24 2473–2474 [Google Scholar]
- Wang H, Li G, Chen R (2006) Fast neutron bombardment (FNB) induced deletion mutagenesis for forward and reverse genetic studies in plants. In JT da Silva, ed, Floriculture, Ornamental and Plant Biotechnology; Advances and Topical Issues, Vol 1. Global Science Books, Isleworth, UK, pp 629–639
- Wilmouth RC, Turnbull JJ, Welford RW, Clifton IJ, Prescott AG, Schofield CJ (2002) Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10 93–103 [DOI] [PubMed] [Google Scholar]
- Winkel BSJ (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55 85–107 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E, Dixon RA, Wang ZY (2006) Medicago truncatula transformation using cotyledon explants. In K Wang, ed, Agrobacterium Protocols, Vol 343. Human Press, Totowa, NJ, pp 129–136 [DOI] [PubMed]
- Xie D, Jackson LA, Cooper JD, Ferreira D, Paiva NL (2004. a) Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol 134 979–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Sharma SB, Dixon RA (2004. b) Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch Biochem Biophys 422 91–102 [DOI] [PubMed] [Google Scholar]
- Xie D, Sharma SR, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299 396–399 [DOI] [PubMed] [Google Scholar]
- Xie D, Sharma SB, Wright E, Wang Z, Dixon RA (2006) Engineering plants for introduction of health-beneficial proanthocyanidins. Plant J 45 895–907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.