Abstract
In response to wounding and pathogens, jasmonate (JA) serves as a signal molecule for both induction and repression of gene expression. To examine defense-regulated gene repression in Arabidopsis (Arabidopsis thaliana), we have identified a nonclassical arabinogalactan protein (AGP) gene, AGP31, and show that its mRNA decreased to about 30% of its original level within 8 h in response to methyl JA (MeJA) treatment of whole 7-d-old seedlings. Wounding and abscisic acid treatment had similar effects. MeJA suppression primarily depends on the action of the JA-signaling protein, COI1, as shown by much lower MeJA suppression in coi1-1 mutant plants. The main mechanism of mRNA suppression by MeJA is repression of transcription, as shown by nuclear run-on experiments. The AGP31 protein shares features with several known and putative nonclassical AGPs from other species: a putative signal peptide, a histidine-rich region near the N terminus followed by a repetitive proline-rich domain, and a cysteine-rich C-terminal PAC (for proline-rich protein and AGP, containing cysteine) domain. Positive Yariv reagent interaction demonstrated that the protein is an AGP. Monosaccharide analysis of purified AGP31 indicated it is a galactose-rich AGP. Expression of an AGP31-enhanced green fluorescent protein fusion protein in transgenic cells revealed that the AGP31 protein was localized to the cell wall. AGP31 promoter-β-glucuronidase reporter gene analysis showed expression in the vascular bundle throughout the plant, except in the flower. In the flower, β-glucuronidase staining occurred throughout the pistil, except in the stigma. The strong preferential expression in vascular tissues suggests that AGP31 may be involved in vascular tissue function during both the defense response and development.
Plants respond to insect attack, mechanical damage, and pathogen infection by regulating specific and overlapping sets of genes to provide wound-healing and protective functions. These responses widely involve both gene activation and repression. Both of these processes appear to be important in coordinating the antagonistic and synergistic interplay of different signaling pathways and downstream effectors to address biotic stress (Lorenzo et al., 2003). For example, defense-induced salicylic acid promotes expression of various defense genes, such as pathogenesis-related proteins, while repressing the expression of genes under the control of the jasmonate (JA)-signaling pathway (Li et al., 2004). Recently, a bacterial elicitor has been shown to mediate repression of auxin signaling and enhance disease resistance through induced microRNA accumulation in Arabidopsis (Arabidopsis thaliana; Navarro et al., 2006).
Positive and negative regulation of gene expression, along with diverse nongenomic processes, contribute to the dynamic remodeling of the plant cell wall during responses to herbivory, wounding, and infection. Changes in the wall often play a central role in the defense outcome (Schulze-Lefert, 2004). Callose formation at infection sites increased susceptibility to several fungal pathogens in Arabidopsis (Nishimura et al., 2003). Tomato (Lycopersicon esculentum) plants treated by wounding, methyl JA (MeJA), or systemin application released peptides from a cell wall Hyp-rich glycoprotein that stimulated two well-known defense responses, extracellular alkalinization, and accumulation of proteinase inhibitors (Nishimura et al., 2003; Narvaez-Vasquez et al., 2005). Among roles identified for cell wall proteins, rapid oxidative cross-linking of selected cell wall proteins to strengthen the cell wall against pathogen attack has been widely documented in many plants. Hydroxy/Pro-rich proteins (HRGPs), such as extensins and Pro-rich proteins (PRPs), have been identified as undergoing cross-linking, particularly those rich in Tyr (Bradley et al., 1992; Brown et al., 1998). Tyr residues in these proteins have long been postulated to form intermolecular linkages responsible for insolubilization of the proteins in the wall during the defense response. A recent study using a model extensin peptide system expressed in tobacco (Nicotiana tabacum) cells has identified the formation of diisodityrosine intermolecular cross-links in vitro (Held et al., 2004). Besides oxidative cross-linking of extensins and PRPs, oxidative cross-linking was also demonstrated by formation of high-Mr complexes of plasma membrane-associated arabinogalactan proteins (AGPs) after wounding in sugar beet (Beta vulgaris) leaves (Kjellbom et al., 1997). It has been hypothesized that other HRGPs and PRPs with lower Tyr content do not cross-link and may not contribute to the defense response (Sheng et al., 1991). The cucumber (Cucumis sativus) PRP1 remained soluble after wounding, elicitor, and hydrogen peroxide treatments and was instead suggested to participate in the deposition of protective silica at infection sites (Kauss et al., 2003).
In a number of cases, changes in HRGP and PRP gene expression have been found to correlate with the presumed function or lack of function in protection during the plant defense response. Accordingly, specific HRGP and PRP mRNAs whose encoded proteins perform postulated or demonstrated adaptive roles are induced by defense-related stimuli (Bradley et al., 1992; Kauss et al., 2003; Pearce and Ryan, 2003). In contrast, several mRNAs encoding low-Tyr HRGPs and PRPs are down-regulated (Sauer et al., 1990; Sheng et al., 1991). For the latter group, it has been postulated that the genes are down-regulated to reduce synthesis of unneeded proteins to conserve energy and perhaps enhance the cell wall structure (Sheng et al., 1991; Mehdy, 1994). Whereas down-regulation of numerous genes during plant defense responses has been observed, particularly with the advent of microarray technology, the molecular mechanisms generating down-regulation are poorly understood. The mechanism of down-regulation of low-Tyr PRP mRNA has been examined for the French bean (Phaseolus vulgaris) gene, PvPRP1. PvPRP1 mRNA was destabilized in elicitor-treated cells such that mRNA half-life decreased from about 60 h in unelicited cells to approximately 45 min in elicited cells. In contrast, there was no change in the rate of PvPRP1 transcription (Zhang et al., 1993). Whereas progress has been made in identifying an RNA-binding protein that binds to the PvPRP1 mRNA in vitro (Zhang and Mehdy, 1994), the lack of facile genetic approaches, a sequenced genome, and many resources, such as mutant insertion lines in the French bean system, hamper elucidation of the regulatory pathway. Identification of a PRP gene in Arabidopsis homologous to the bean PvPRP1 gene may provide a good candidate to examine defense-regulated gene repression in the model Arabidopsis system.
In this study, a low-Tyr PRP was identified in Arabidopsis, whose mRNA was down-regulated by various defense-related stimuli: wounding, MeJA, and abscisic acid (ABA) treatments. MeJA repression of the mRNA level was COI1 dependent and occurred at the level of transcription. This PRP is better classified as a nonclassical AGP on the basis of its hallmark reactivity with β-glucosyl Yariv reagent and having an extensive carbohydrate moiety rich in Gal and Ara. Hence, the gene was named AGP31. Analysis of AGP31 promoter-GUS reporter gene expression in transgenic plants revealed a striking association of expression with the vascular tissue throughout the plant, except for more widespread expression in flowers. The strong preferential expression in vascular tissues suggests that AGP31 may be involved in vascular tissue function during both the defense response and development.
RESULTS
AGP31 Encodes a Nonclassical AGP
A BLASTP search was conducted to identify Arabidopsis proteins with features and protein organization similar to the French bean PvPRP1 protein, including low Tyr content (generally <5%). The gene with the greatest similarity throughout the encoded protein length was At1g28290. Using reverse transcription-PCR, we obtained a cDNA clone spanning the complete open reading frame of At1g28290. DNA sequencing confirmed the annotation of At1g28290 (http://www.arabidopsis.org). At1g28290 has one intron and a 208-nucleotide 3′-untranslated region (UTR) deduced from the cDNA sequence. At1g28290 encodes a 359-amino acid Pro-rich protein with four distinct domains: a putative signal peptide followed by a His-rich domain, a middle repetitive Pro-rich domain, and a C-terminal non-Pro-rich domain (Fig. 1A). In the Pro-rich domain, there are only five types of amino acids, with 45.9% Pro, 18.2% Lys, 15.7% Val, 6.3% Ala, and 3.8% Tyr. These amino acids form different exact repeat units. There are two long repeats with 31 amino acids overlapping with four shorter 20-amino acid repeats (Fig. 1A). These repeats are essentially composed of different variants of the basic repeat unit PP(A/V/T)(K/Y). This degenerate quadripeptide is different from the typical pentapeptide PPVX(K/T) of other PRPs, such as several AtPRPs in Arabidopsis (Fowler et al., 1999) and ENOD11 in Medicago (Journet et al., 2001), and is characteristic of a class of nonclassical AGPs and AGP-like proteins. Also in common with these proteins, the At1g28290 protein has a C-terminal region with six well-conserved Cys residues that has been named PAC (for PRP and AGP, containing Cys; Baldwin et al., 2001). This domain also exists in the OLE E 1 pollen allergen in olive (Olea europa).
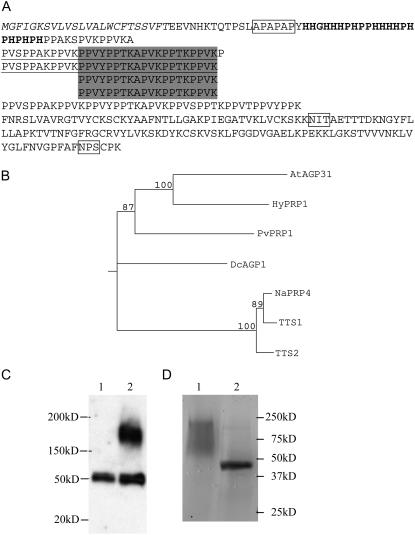
Figure 1.
Characterization of AGP31. A, Structure of AGP31. Shaded regions are four exact repeat units. Underlined regions are two longer repeat units. The smallest repeat unit is PPXX. The italicized region is the predicted signal peptide; the bold region is the His-rich region. Boxed regions are the predicted APAPAP module for arabinogalactan attachment and N-glycosylation sites. B, Neighbor-joining tree generated from the alignment of full-length amino acid sequences of the homologs of AGP31. Numbers indicate the percentage of 1,000 bootstrap replicates. C, Western blot showing that AGP31 exists in AGP precipitation of cell wall protein. Lane 1, AGP fraction from wild-type plants; lane 2, AGP fraction from transgenic plants expressing 35S∷AGP31∷myc. D, Purified AGP31. AGP31 was isolated from high-salt eluted cell wall protein by a Ni-NTA column and deglycosylated by TFMS. Approximately 2 μg of native and deglycosylated AGP31 were separated by SDS-PAGE on a 15% gel and stained by Coomassie Blue. Lane 1, Native AGP31; lane 2, deglycosylated AGP31.
The protein sequence of At1g28290 suggested that it may encode a nonclassical AGP. Preceding the His-rich domain, there is a module APAPAP and there are six single AP duplets scattered in the PRP domain (Fig. 1A). Increasing evidence suggests that these AP modules are sites of carbohydrate attachment in AGPs (Tan et al., 2004). In addition to these putative O-glycosylation sites, there are two putative N-glycosylation sites in the PAC region (Fig. 1A). Based on these analyses, we postulated that this protein might be a nonclassical AGP.
Further supporting evidence was provided using the PAC domain sequence to BLAST the GenBank database. This identified several putative homologs among different species. Among these putative homologs, we searched for proteins that have similar structure: predicted signal sequence, N-terminal His-rich stretch, a Pro-rich region with the repeat PPXX, and a PAC domain. After applying these criteria, we identified several proteins, PvPRP1 (French bean), HyPRP1 (cotton [Gossypium hirsutum]), DcAGP1 (carrot [Daucus carota]), NaPRP4, NtTTS1, and NtTTS2 (tobacco), which we consider to be homologs of At1g28290 (Fig. 1B). Among these homologs, DcAGP1, NaPRP4, NtTTS1, and NtTTS2 have been shown to be bona fide nonclassical AGPs (Cheung et al., 1995; Sommer-Knudsen et al., 1996; Baldwin et al., 2001). In the Arabidopsis genome, there are two genes, AtAGP30 and At2g34700, showing similarity with At1g28290. AtAGP30 (van Hengel and Roberts, 2003) shows the highest similarity with At1g28290 but it has no His-rich domain. At2g34700 has only the PAC domain without His- and Pro-rich domains. Based on these analyses, we concluded that At1g28290 is the only gene in this nonclassical AGP subclass in the Arabidopsis genome.
To determine whether At1g28290 is a bona fide AGP, we used the 35S promoter to overexpress the At1g28290 protein fused with a 6×-myc tag at the C-terminal end in transgenic plants. A cell wall fraction was isolated from 7-d-old seedlings. Proteins released by high salt extraction of this fraction were incubated with β-glucosyl Yariv reagent, a compound that selectively precipitates AGPs. Western-blot analysis of the AGP fraction using an anti-myc antibody detected the fusion protein and an endogenous, nonspecific 50-kD protein (Fig. 1C). This result demonstrates that At1g28290 is a bona fide AGP. The myc fusion protein detected by the antibody ran by SDS-PAGE as a broad high-molecular-mass band of about 170 to 200 kD, whereas the predicted molecular mass of the mature At1g28290 protein alone is 38 kD. In addition to this experiment, we isolated the myc fusion protein from the high-salt cell wall protein fraction by nickel nitrilotriacetic acid agarose (Ni-NTA) affinity purification, capitalizing on the fact that there is a His-rich stretch near the N terminus of At1g28290. The resulting enriched protein fraction spotted on nitrocellulose stained positively with Yariv reagent (data not shown). Based on these analyses, we concluded that At1g28290 encodes a nonclassical AGP and named it AGP31.
AGP31 Is a Gal-Rich AGP
To examine its carbohydrate composition and likely hydroxylation of Pro residues, we purified native AGP31 protein from transgenic plants overexpressing native AGP31. Transgenic lines were generated by introducing a genomic fragment that contains about 3 kb upstream of translation start codon and about 700 bp downstream of the translation stop codon of the AGP31 gene. Due to the nature of Agrobacterium transformation, multiple copies of fragments are frequently introduced into plants and can exhibit higher mRNA levels. Plants of a transgenic line that showed a higher mRNA level than wild type grown in liquid Murashige and Skoog medium were used to isolate the high-salt eluted cell wall protein. AGP31 was purified by a Ni-NTA metal affinity column from high-salt eluted cell wall protein and an aliquot subjected to trifluoromethanesulfonic (TFMS) acid hydrolysis, a treatment that removes the majority of carbohydrate in glycoproteins. The Coomassie Blue-stained gel showed that acid hydrolysis resulted in a dramatic shift from the broad high-molecular-mass band to a single prominent band of 45 kD in the deglycosylated sample (Fig. 1D). It is noteworthy that the innermost Asn-linked N-acetylglucosamine of an N-linked oligosaccharide is resistant to TFMS treatment (Edge, 2003). The observed higher than predicted molecular mass migration may be due to nonuniform association of SDS with HRGP as suggested by others (Desai et al., 1983; Bosch et al., 2001) and possibly some residual N-linked sugars. The identity of the band was identified by matrix-assisted laser-desorption time-of-flight (MALDI-TOF) mass spectrometry (MS), which covered 61% of the AGP31 amino acid sequence. No other protein was identified by MALDI-TOF MS from the single band. Together, these findings demonstrate that AGP31 is a glycoprotein and that a highly purified preparation of AGP31 has been obtained.
Amino acid composition analysis of purified, deglycosylated AGP31 showed approximately equal amounts of Pro and Hyp (Table I). Another nonclassical AGP, NaPRP4, which is the homolog of AGP31 in Nicotiana alata, also has about equal amounts of Pro and Hyp (Sommer-Knudsen et al., 1996). Monosaccharide composition analysis showed that purified AGP31 contains mostly Gal (80.9%), with lesser amounts of Ara (13.5%), Xyl (2.1%), Fuc (1.3%), and Man (1.3%; Table II). In addition, very low amounts of Glc (0.5%) and GlcUA (0.5%) were detected. Again, AGP31 is very similar to NaPRP4, which has a glycosyl composition of 83% Gal, 6.8% Ara, 3.5% Xyl, and 1.5% Man (Sommer-Knudsen et al., 1996). Besides AGP31's interaction with the Yariv reagent, glycosyl composition analysis provided independent confirmation that AGP31 is an AGP.
Table I.
Amino acid composition of AGP31
| Amino Acid | Experimentally Determined Composition | Predicted Composition |
|---|---|---|
| mol % | ||
| Hyp | 14.5 | |
| Pro | 13.1 | 26.9 |
| Lys | 14.0 | 14.9 |
| Val | 11.4 | 12.2 |
| Ala | 6.6 | 6.6 |
| Thr | 6.0 | 6.9 |
| His | 5.6 | 4.5 |
| Gly | 5.4 | 3.9 |
| Glu | 4.7 | 1.8 |
| Ser | 3.8 | 4.2 |
| Tyr | 3.7 | 3.9 |
| Leu | 3.6 | 4.2 |
| Asp | 3.5 | 0.9 |
| Phe | 2.0 | 2.7 |
| Arg | 1.2 | 1.2 |
| Ile | 0.9 | 0.6 |
| Met | 0.1 | 0 |
| Cys | Not determined | 4.5 |
Table II.
Monosaccharide composition of AGP31
| Glycosyl Residue | mol % |
|---|---|
| Gal | 80.9 |
| Ara | 13.5 |
| Xyl | 2.1 |
| Fuc | 1.3 |
| Man | 1.3 |
| GlcUA | 0.5 |
| Glc | 0.5 |
AGP31 mRNA Is Repressed by Wounding, MeJA, and ABA
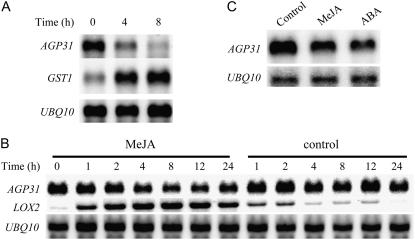
To investigate AGP31 gene expression during the defense response, the leaves of 4-week-old plants were mechanically wounded and directly wounded leaves were harvested at various times. RNA-blot analysis shows a decrease in AGP31 mRNA level over the 8-h time course (Fig. 2A). The 1.35-kb size of the AGP31 transcript matches the expected size based on the cDNA sequence. The GLUTATHIONE S-TRANSFERASE1 (GST1) transcript was also hybridized as a known wounding-induced mRNA, whereas the UBIQUITIN10 (UBQ10) transcript was monitored to show equal loading. Because JA and its derivatives are well-known secondary messengers induced by wounding, we also examined the effect of MeJA. MeJA treatment resulted in a decrease in AGP31 mRNA to about 30% of the original level by 8 h (Fig. 2B). After 12 h, the AGP31 mRNA level began to recover but remained below its original level through the 24-h time course. In contrast, control plants treated with the solvent exhibited only a slight decrease in AGP31 mRNA. The marked induction of LIPOXYGENASE2 (LOX2) mRNA, a known MeJA-regulated mRNA, demonstrated effective MeJA exposure. Control plants treated with solvent showed low-level induction of LOX2 mRNA, especially at early time points. However, compared with MeJA-treated plants, the response was much lower and diminished after 4 h.
Figure 2.
Effects of wounding, MeJA, and ABA treatments on AGP31 and reference mRNA levels. A, Northern-blot analysis of RNA from wounded leaves of 4-week-old plants. About one-third of the leaves were pressed with pliers and wounded leaves were harvested for RNA at given time points. The same blot hybridized with the AGP31 probe was washed and rehybridized with GST1 and then again with the UBQ10 probe to show equal loading. B, Northern-blot analysis of RNA from MeJA-treated 7-d-old plants. Seedlings were grown in petri dishes, sprayed with 500 μm MeJA in 0.1% ethanol, 0.01% Tween 20, or solvent control (0.1% ethanol, 0.01% Tween 20), and whole seedlings were harvested at given time points. The same blot hybridized with the AGP31 probe was washed and rehybridized with LOX2 and then again with the UBQ10 probe. C, Northern-blot analysis of RNA from ABA-treated 7-d-old plants. Seedlings were grown in petri dishes, transferred to assay plates containing 0.1% ethanol solvent control, 50 μm MeJA, and 10 μm ABA, and whole seedlings were harvested at 8 h. The same blot hybridized with the AGP31 probe was washed and rehybridized with the UBQ10 probe.
ABA has been shown to be involved in some plant defense responses and is widely active in the response to water stress and other abiotic stresses. To determine the expression pattern of AGP31 mRNA in response to ABA, we treated seedlings with ABA and harvested after 8 h. Northern-blot analysis showed that AGP31 mRNA was also repressed by ABA treatment (Fig. 2C).
Repression of AGP31 mRNA Is Primarily COI1 Dependent
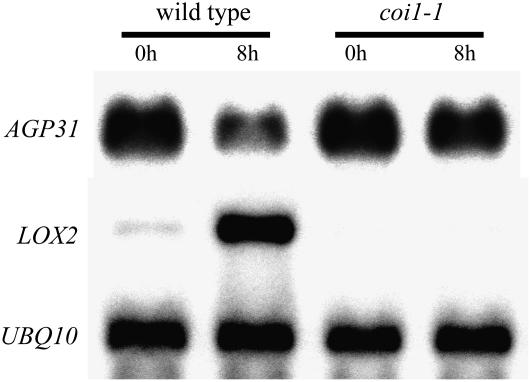
COI1 has been shown to play a pivotal role in JA signaling. Based on microarray data, it has been estimated that about one-half of JA signaling involves the action of this protein, which encodes an F-box protein as part of the E3 ubiquitin ligase complex (Devoto et al., 2005). Although slight repression was still observed, AGP31 mRNA repression in response to MeJA was dramatically reduced in the coi1-1 mutant compared to the response in wild-type plants, indicating the AGP31 mRNA repression is mainly dependent on the COI1 pathway (Fig. 3). Interestingly, the basal level of AGP31 mRNA is similar in untreated plants of both genetic backgrounds.
Figure 3.
Effects of MeJA treatment on AGP31, LOX2, and UBQ10 mRNA levels in wild-type and coi1-1 plants. Northern-blot analysis of RNA from 7-d-old plants, either wild type or coi1-1, transferred to plates containing 50 μm MeJA and whole seedlings were harvested at 8 h. The same blot hybridized with the AGP31 probe was washed and rehybridized with LOX2 and then again with the UBQ10 probe.
Repression of AGP31 mRNA Occurs Primarily at the Transcriptional Level
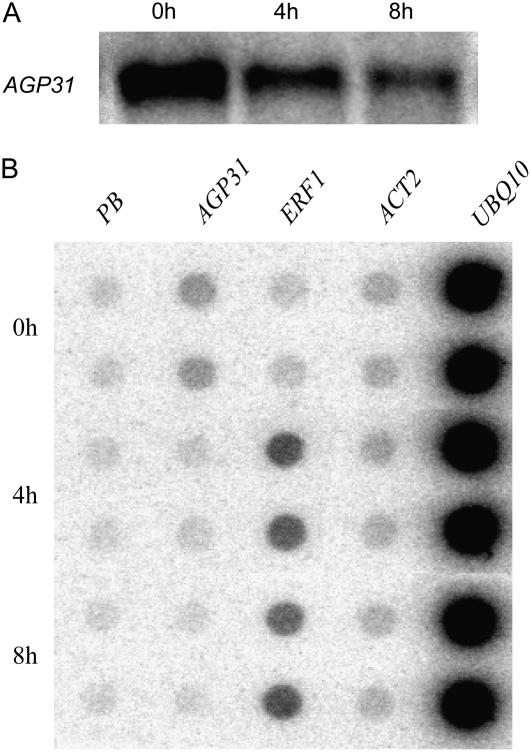
In French bean, repression of PvPRP1 mRNA is due to mRNA stability regulation with no change in the rate of transcription (Zhang et al., 1993). To evaluate these two parameters for AGP31 regulation, we performed a nuclear run-on assay to measure the transcription rate of the AGP31 gene after MeJA treatment. Because Arabidopsis cultured cells can provide a large quantity of homogeneous cells, the run-on assay was carried out with cultured cell nuclei. Figure 4A shows that MeJA treatment of cultured cells resulted in AGP31 mRNA repression similar to the response in whole plants. Nuclei from the same batches of cells were used for run-on transcription assays. The results show that AGP31 mRNA repression reflected decreased transcription rate (Fig. 4B). Moreover, the magnitude of transcription reduction (to about 30% of the original level) correlates well with the observed reduction in the steady-state mRNA level to approximately 30% of the original level. A known MeJA-induced gene, ETHYLENE RESPONSE FACTOR1 (ERF1), showed increased transcription, whereas the rates of ACTIN2 and UBQ10 transcription were essentially unchanged.
Figure 4.
AGP31 mRNA steady-state levels and transcription rates of different genes in Arabidopsis cultured cells treated with MeJA. A, Northern-blot analysis of AGP31 mRNA in MeJA-treated (50 μm) cultured cells harvested at the given time points. B, Nuclei were isolated from the same batch of 1-week cultured Arabidopsis suspension cells as shown in A at the given time points. After incubation with 32P-UTP to produce radiolabeled run-off transcripts, RNA was purified from nuclei and hybridized to duplicate spots for each linearized plasmid DNA: PB, pBluescript vector; ACT2, ACTIN2.
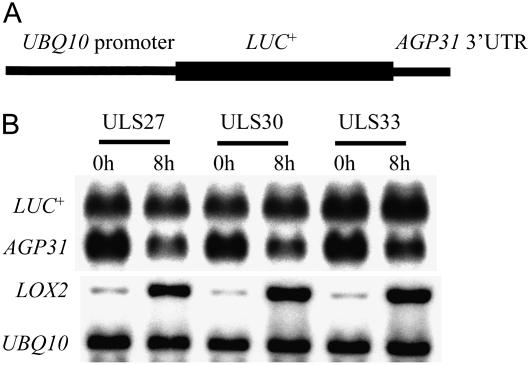
To investigate whether mRNA stability regulation may also play a role, the AGP31 3′-UTR was inserted downstream of a LUC+ reporter coding sequence. Agrobacterium-mediated transformation generally results in the concatenated insertion of several T-DNA copies at a single locus. To avoid possible generation of double-stranded RNAs and RNA interference processes that could variously impact the steady-state mRNA levels, transgenic plants were screened to identify single-copy insertions by Southern blot as described by Forsbach et al. (2003). Analysis of three lines of transgenic plants containing single-copy insertions showed that the LUC+ reporter mRNA level remained approximately constant in MeJA-treated plants, whereas the endogenous AGP31 and LOX2 mRNAs showed expected regulation (Fig. 5). The results suggest that MeJA-induced down-regulation of AGP31 mRNA does not involve mRNA destabilization through the 3′-UTR sequence. Whereas mRNA stability regulation commonly operates through the 3′-UTR (Bevilacqua et al., 2003), the evidence does not rule out a role for another part of the transcript or the 3′-UTR plus another sequence in conferring MeJA-induced mRNA stability regulation. Together with the run-on data, our results support predominant regulation by repression of transcription.
Figure 5.
Effect of MeJA on the levels of LUC+ reporter mRNA containing the 3′-UTR of AGP31 in transgenic seedlings. A, The construct structure of UBQ10 promoter∷LUC+∷AGP31 3′-UTR gene. B, Northern-blot analysis of RNA from transgenic plants carrying a UBQ10 promoter∷LUC+∷AGP31 3′-UTR construct. Seven-day-old seedlings of three single-insertion lines were sprayed with 500 μm MeJA and harvested at the given times. The same blot was sequentially hybridized to the following probes: 3′-UTR of AGP31 (to detect LUC+ and endogenous AGP31 mRNA), LOX2, and UBQ10.
AGP31 Is Localized in the Cell Wall
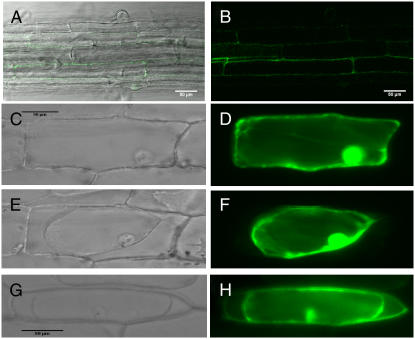
AGP31 protein is predicted to be a cell wall or secreted protein based on its putative signal sequence and past findings of these locations for numerous AGPs. To establish the subcellular localization of AGP31, enhanced GFP (eGFP) was fused to the C-terminal end of AGP31 to form an AGP31∷eGFP fusion construct under the control of the 35S promoter. The constructs were stably expressed in Arabidopsis transgenic plants and also bombarded into onion (Allium cepa) epidermal cells to express the fusion protein transiently. Figure 6, A and B, shows confocal microscope views of the same root section from stable transgenic Arabidopsis plants. The eGFP signal indicated that the fusion protein was localized to the cell periphery (Fig. 6, A and B). In bombarded onion epidermal cells, the eGFP-alone construct showed intracellular localization, including a commonly observed presence in the nuclei (Fig. 6, C and D). To distinguish between cell wall and cell membrane locations, the transiently transformed cells were treated with 0.8 m mannitol to induce plasmolysis. As shown in Figure 6E, mannitol treatment effectively contracted the plasma membrane from the cell periphery in a cell that expressed eGFP alone. The corresponding fluorescence image (Fig. 6F) showed that eGFP fluorescence was also contracted with the cytoplasm. In contrast, in a plasmolyzed transgenic cell expressing the AGP31-eGFP fusion protein, a significant portion of the fusion protein was found at the cell periphery (Fig. 6H), indicating cell wall localization. Some fusion protein was also seen more faintly inside the cell and at the plasma membrane, which would be consistent with intermediate locations of the protein during its biogenesis. The AGP31-myc fusion protein was also highly enriched in the high-salt elution fraction from partially purified cell walls (data not shown). In work from other researchers, AGP31 was also identified in a cell wall proteomics study (Feiz et al., 2006) and this is consistent with our data indicating that AGP31 is a cell wall protein.
Figure 6.
Subcellular localization of AGP31-GFP fusion protein. eGFP was fused with the C terminus of AGP31 and stably transformed into Arabidopsis (A and B) or introduced biolistically into onion epidermal cells (G and H). Control is eGFP alone introduced into onion epidermal cells biolistically (C–F). C and D, Unplasmolyzed cell. E to H, Plasmolyzed cells. The indicated bar scales are used for A and B, C to F, and G and H. A, Overlay of fluorescence image of B on bright-field image. C, E, and G, Bright-field image. B, Confocal image excited by 488-nm light. D, F, and H, Fluorescence images excited by blue light.
AGP31 Is Expressed Primarily in the Vascular Bundle and in the Flower
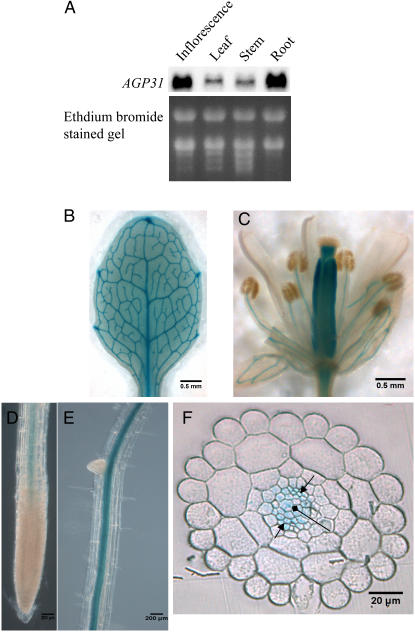
Toward understanding the biological function of this protein during defense and normal development, we characterized the AGP31 mRNA expression level in roots, leaves, stems, and inflorescences of Arabidopsis plants. RNA-blot analysis showed that AGP31 mRNA was expressed in all parts of the plant, with the roots and inflorescences having the highest expression levels (Fig. 7A). To determine the expression pattern of the AGP31 gene more precisely, we made transgenic plants harboring the bacterial uidA gene (GUS) under the control of a 1.4-kb fragment extending upstream from the AGP31 start codon. This construct is termed the AGP31 promoter-GUS reporter construct (PAGP31∷GUS). The GUS expression pattern based on histochemical staining for GUS activity revealed that the AGP31 promoter-GUS reporter gene was predominantly expressed in the vascular bundles throughout the plants (Fig. 7, B–F). In flowers, apart from staining vascular bundles in sepals and stamen filaments, GUS was also expressed throughout pistils, except for stigmas (Fig. 7C). In roots, GUS stain was predominant in vascular bundles without appearance in the root tip regions (Fig. 7, D and E). Cross section of the root showed GUS activity was broadly distributed through the vascular bundle, including the phloem and primary xylem (Fig. 7F).
Figure 7.
Localization AGP31 gene in different tissues. An approximately 1.4-kb genomic fragment upstream of the AGP31 ATG start codon was fused with GUS reporter gene and transformed into wild-type Arabidopsis. A, Northern blot of AGP31 mRNA in different plant parts of 4-week-old plants. B, Leaf of 10-d-old plant. C, Flower in stage 15 to 16. D and E, Roots of 7-d-old seedling. F, Cross section of root. Arrow, Phloem; diamond arrow, xylem.
DISCUSSION
Wounding and MeJA have been shown to induce dozens of genes while repressing an approximately equal number of other genes (Devoto et al., 2005). However, the mechanisms of gene repression are poorly understood. Defense-related remodeling of the plant cell wall involves activation of some cell wall protein genes, whereas others are repressed, providing an opportunity to understand molecular regulatory mechanisms that have evolved in the context of altering cell wall properties for improved disease resistance. In this study, we have identified a nonclassical AGP gene, AGP31, whose mRNA is down-regulated by MeJA, wounding, and ABA. We further showed that MeJA suppression is primarily due to transcriptional repression and the signaling pathway involves a central JA regulator, COI1. In addition, AGP31 expression during normal development showed a striking association with the vascular tissue and the pistil exclusive of the stigma.
As part of our interest in understanding gene repression during the defense response, we identified the At1g28290 gene in Arabidopsis as a homolog to the known French bean elicitor and wounding down-regulated PRP gene, PvPRP1 (Sheng et al., 1991; Zhang et al., 1993; Zhang and Mehdy, 1994). More broadly, AGP31 belongs to a subclass of nonclassical AGPs discussed by Roberts and coworkers (Baldwin et al., 2001). This subclass includes a cotton PRP (HyPRP1), a carrot AGP (DcAGP1), a French bean PRP (PvPRP1), two tobacco AGPs localized in the transmitting tissue of the style (NtTTS1, NtTTS2), as well as the Nicotiana alata AGP, NaPRP4/GaRSGP. Although PvPRP1 (Sheng et al., 1991) and HyPRP1 were previously identified as PRPs in their database annotations, their features suggest that they may also be AGPs in this class. There are four common features of this family: a putative signal peptide, a His-rich domain near the N terminus of the mature protein after presumed cleavage of the signal peptide, a Pro-rich domain with the unique repeat PPXX, and a C-terminal domain, called PAC, which has six well-conserved Cys residues (Baldwin et al., 2001). This PAC region has homology to the pollen allergen OLE E 1.
Although the functions of these domains remain to be clarified, there is some evidence for roles in diverse aspects of plant development and defense. The His-rich domain usually functions in metal binding in other proteins. For example, the His-rich domain of citrus dehydrin binds Cu2+ (Hara et al., 2005). In soybean (Glycine max), the His-rich domain is necessary for Ni2+ binding to Eu3, a Ni2+-binding GTPase that is required for urease activity (Freyermuth et al., 2000). Therefore, we postulate that the His-rich domain in AGP31, which we have shown can bind to Ni2+ in vitro, may possess metal ion-binding activity in the cell wall in vivo. Alternatively, the His-rich domain may interact with pectin, as has been previously postulated (Baldwin et al., 2001). In rice (Oryza sativa), genes containing the OLE E 1 domain also have other domains involved in various aspects of plant physiology, such as auxin response, defense response, and ion binding (Jiang et al., 2005). The tomato LeLAT52 protein has only the OLE E 1 domain and has been shown to interact with a Leu repeat receptor kinase and is essential for pollen hydration and pollen tube growth (Tang et al., 2002).
It is worth noting that, although members of the nonclassical AGP subclass discussed in this article have similar overall structures, differences in their spatial expression and regulation suggest there may be different and common functions. For example, carrot DcAGP1 mRNA is unaffected by wounding (Baldwin et al., 2001), whereas AGP31 and bean PvPRP1 mRNAs are both down-regulated (Sheng et al., 1991; Baldwin et al., 2001). Tobacco TTS1 and TTS2 mRNAs are highly localized to the transmitting tract of the style and have been implicated in pollen tube growth guidance (Cheung et al., 1995). On the other hand, AGP31 is primarily expressed in vascular tissues throughout the plant and broadly in the pistil. Because AGP31 appears to be the only member of this subclass of nonclassical AGPs in Arabidopsis based on our database searches, analysis of its function during defense and development may be less complicated by gene redundancy.
The physiological functions of nonclassical AGPs have remained more elusive than individual classical AGPs whose functions have begun to be uncovered by genetic analyses (Yang et al., 2007). This is partially due to difficulties in identifying nonclassical AGPs due to their highly variable C-terminal regions (Schultz et al., 2002). Surprisingly, in Arabidopsis, only one other nonclassical AGP has been previously identified, AtAGP30. In comparison to AGP31, AtAGP30 is missing the His-rich domain but possesses a similar repetitive Pro-rich domain and a PAC domain. This gene is expressed primarily in the atrichoblasts of root epidermal cells (van Hengel et al., 2004). A transposon insertional mutant of AtAGP30 exhibited resistance to ABA inhibition of seed germination and abnormal somatic embryogenesis (van Hengel and Roberts, 2003). ABA is a phytohormone involved in plant stress responses. Here, we showed AGP31 was repressed by ABA treatment, suggesting it might function in stress responses. Interestingly, two chimeric AGPs, xylogen 1 and xylogen 2, stimulate the organization of xylem elements in zinnia (Zinnia elegans) and Arabidopsis. Xylogen 1 and xylogen 2 are apparently chimeras of nonspecific lipid transferase protein and AGP (Motose et al., 2004).
A common feature among all AGPs is that they are highly glycosylated cell wall or secreted proteins in which the carbohydrate moiety usually accounts for about 90% of AGP mass. The carbohydrate exists predominantly as type II arabinogalactans and shows distinctive binding to a class of synthetic phenylazo dyes. Binding one member of this class of dyes, β-glucosyl Yariv, is a diagnostic parameter used to classify proteins as AGPs. Our findings that AGP31 can be precipitated by Yariv reagent and that it is a glycoprotein with abundant Gal (80.9%) and substantial Ara (13.5%) establish it as a bona fide AGP. Western-blot analysis of the AGP31-myc tag protein from transgenic whole seedlings revealed a much higher molecular mass protein (170–200 kD) than expected based on the deduced amino acid sequence (38 kD), consistent with substantial glycosylation, at least at the stage of development used in these experiments. Compared with classical AGPs, AGP31 and NaPRP4 have less Hyp and Gal is the predominant monosaccharide residue. For example, in LeAGP1 (Zhao et al., 2002) and gum arabic glycoprotein (Goodrum et al., 2000), most Pros are hydroxylated and there are about equal amounts of Ara and Gal.
Using the promoter-GUS fusion technique, we showed that expression of AGP31 was localized to vascular bundles throughout the plant, including phloem and primary xylem cells. AGP31 mRNA repression by MeJA and ABA correlates spatially with the vascular tissue being a major site for biosynthesis of both phytohormones (Stenzel et al., 2003; Koiwai et al., 2004). During the defense response, we postulate that AGP31 may not participate in the oxidative cross-linking of cell wall proteins and hence its synthesis is curtailed. Oxidative cross-linking in the vascular system as part of the defense response has been documented in vascular parenchyma cells in tomato in response to wounding, systemin, and MeJA (Orozco-Cardenas et al., 2001).
We conclude that down-regulation of AGP31 mRNA by MeJA is achieved by the combination of repression of transcription and a relatively short mRNA half-life (less than 4 h is suggested by the kinetics of mRNA loss). The mRNA half-life may be constitutively short or regulated from a longer half-life found in untreated cells. However, the finding that the AGP31 3′-UTR did not impart MeJA-induced instability to a LUC+ reporter transcript makes it more likely that mRNA stability regulation is not a major factor because a common region controlling mRNA stability is the 3′-UTR (Bevilacqua et al., 2003). The predominant transcriptional repression of AGP31 contrasts with the mRNA stability regulation that down-regulates the homologous PvPRP1 mRNA in French bean. The different regulation mechanisms may reflect the different functions of these homologs in different plants.
Transcription factors mediating AGP31 transcriptional repression by MeJA are unknown. A number of Arabidopsis transcription factors have been identified that repress several JA-inducible genes, including ERF4 (McGrath et al., 2005), WRKY70 (Li et al., 2004), and JIN1/MYC2 (Lorenzo et al., 2004). The WRKY70 transcript was repressed by JA with kinetics similar to the loss of the AGP31 mRNA (Li et al., 2004). The observed repression of AGP31 mRNA by ABA also may be due to transcriptional and/or posttranscriptional processes. An ABA-regulated transcriptional repressor, AtERF7, has been characterized (Song et al., 2005). There is also precedent for posttranscriptional regulation by ABA (Borsani et al., 2005; Nishimura et al., 2005). ABA repressed another group of AGP mRNAs: fasciclin-like AGP mRNAs encoding FLA1, 2, and 8 in Arabidopsis (Johnson et al., 2003). Identifying the function of AGP31 in diverse tissues and during defense, and identifying the signaling pathway leading to gene repression will be aided by the analysis of AGP31 mutants and use of other Arabidopsis genetic resources.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown on agar plates with 0.5× Murashige and Skoog salts, 1% Suc, and 0.8% phytoagar (Bio World) under continuous light at 20°C to 23°C. The wild-type plant is ecotype Columbia. The coi1-1 mutant was a gift from Dr. J.G. Turner (University of East Anglia). Arabidopsis cell culture was obtained from Dr. W.F. Thompson (North Carolina State University).
Cell Wall Protein Isolation
The cell wall fractions were isolated from 8-g 11-d seedlings according to Feiz et al. (2006), with some modifications. Briefly, plants were ground into powder in liquid nitrogen and extracted with 2 volumes of extraction buffer (5 mm sodium acetate, pH 4.0, 0.4 m Suc with Roche protease inhibitor cocktail, 0.1 g polyvinylpolypyrrolidone/g tissue) at 4°C for 30 min. Extracts were centrifuged at 1,000g for 15 min at 4°C. After removing the supernatant, pellets were sequentially washed with 5 mm sodium acetate, pH 4.0, with 0.6 m Suc and 1 m Suc, respectively. Finally, pellets were washed with 5 mm sodium acetate (at least five times) and considered to be the cell wall fraction.
AGP Isolation
AGPs were isolated from high-salt eluted cell wall proteins according to Schultz et al. (2000) with some modifications. The high-salt eluted cell wall proteins were extracted by incubating the cell wall fraction with high-salt extraction buffer (5 mm sodium acetate, pH 4.0, 1 m sodium chloride, protease inhibitor) at 4°C for 30 min. Then the samples were centrifuged at 5,000g for 15 min at 4°C, the supernatants were transferred to new tubes and dialyzed against water overnight at 4°C. The dialyzed cell wall proteins were freeze dried and dissolved in 1 mL 1% sodium chloride. To isolate the AGP fraction from cell wall protein, Yariv reagent was added to the cell wall protein at 1 mg/mL and incubated at 4°C overnight. To collect the Yariv-AGP complex, mixtures were centrifuged at 10,000g for 1 h at 4°C. The resulting pellets were washed three times with 1% sodium chloride and once with methanol. After air drying at room temperature, pellets were dissolved in minimal dimethyl sulfoxide. To release AGP from Yariv reagent, water and sodium hydrosulfite were added until the solution became pale yellow. Solutions were then dialyzed against water overnight at 4°C and freeze dried. Finally, AGP fractions were dissolved in water and concentrations were determined by gel diffusion assay using gum arabic as standard (van Holst and Clarke, 1985).
AGP31 Isolation
To isolate native AGP31, a transgenic plant overexpressing native AGP31 was generated by transforming plants with Agrobacterium carrying an EcoRI and HindIII genomic fragment from bacterial artificial chromosome F3H6 (obtained from The Arabidopsis Information Resource). This fragment contains 3,130 bp upstream of AGP31 translation start codon and 701 bp downstream of AGP31 translation stop codon, respectively. A transgenic plant overexpressing AGP31 was used to isolate native AGP31. Plants were grown in Murashige and Skoog liquid medium supplemented with 1% Suc and full-strength Murashige and Skoog vitamins (Czako et al., 1993). Thirty-gram plants were used to isolate cell wall protein as described above. The high-salt eluted cell wall protein was dialyzed against water overnight. One-tenth volume of lysis buffer (100 mm NaH2PO4, pH 8.0, 500 mm imidozone, 3 m NaCl) was added to the cell wall protein fraction. AGP31 was isolated from cell wall protein by a Ni-NTA column (Ni-NTA Fast Start kit; Qiagen) following the manufacturer's instructions. Finally, isolated AGP31 was dialyzed against water and concentrated by Amicon Ultra-15 (Millipore).
AGP31 Deglycosylation
Isolated AGP31 was deglycosylated by the TFMS acid method using the Glyco-Profile IV kit (Sigma). Deglycosylated AGP31 and native AGP31 were separated by SDS-PAGE on 15% Tris-Gly gel.
Protein Identification
Protein bands were excised from Coomassie Blue-stained gel and digested by trypsin. MALDI-TOF MS was carried out by the core facility service of the University of Texas at Austin to identify the protein bands.
Amino Acid Composition Analysis
Amino acid analysis was carried out by AAA Service Laboratory as described (Roach and Gehrke, 1970; Simpson et al., 1976).
Monosaccharide Composition Analysis
Monosaccharide composition analysis was carried out by the Complex Carbohydrate Research Center as described (Merkle and Poppe, 1994).
Western Blot
AGP (7.5 μg) from wild-type and transgenic plants was loaded on 4% to 15% gradient gel and blotted to nitrocellulose membrane (Bio-Rad) by a semidry transfer unit (Bio-Rad). Membranes were blocked in 5% bovine serum albumin in phosphate-buffered saline + 0.5% Triton 100. The primary antibody, anti-myc polyclonal antibody (Sigma), was diluted 1:1,000, and the secondary antibody was diluted 1:25,000. The signal was captured using the enhanced chemiluminescent method following the manufacturer's instructions (Pierce).
Wounding, MeJA, and ABA Treatments
Four-week-old plants grown in soil were used for wounding treatment. About one-third of the leaves were pressed with pliers and only wounded leaves were used for RNA extraction. For MeJA treatment, 11-d-old plants grown vertically in standard agar plates (0.5× Murashige and Skoog salts, 1% Suc, and 0.8% phyto agar) were sprayed with 500 μm MeJA or transferred to 50 μm MeJA plates (0.5× Murashige and Skoog salts, 1% Suc, and 0.8% phyto agar, 50 μm MeJA). For ABA treatment, 11-d-old plants were transferred to 10 μm ABA plates (0.5× Murashige and Skoog salts, 1% Suc, and 0.8% phyto agar, 10 μm ABA). After treatment for the indicated times, whole plants were harvested and frozen in liquid nitrogen.
RNA Isolation and Northern Blot
Total RNA was isolated from plant and cell culture samples according the protocol of Ahn (2000). Ten micrograms of total RNA were separated on 1.2% formaldehyde agarose gels and blotted to ζ-probe (Bio-Rad) membranes. Blots were hybridized following the manufacturer's instructions. Probes were amplified by PCR from genomic DNA or reverse-transcribed cDNA and labeled by the DECAprime II kit (Ambion). Signals were quantitated by the National Institutes of Health ImageJ program.
Nuclei Isolation and Run-On Assay
Nuclei were isolated from 10-g Arabidopsis cell culture according to the previously described protocol (Yu et al., 1998). Nuclei in vitro transcription was carried out using 50 μL nuclei in a 100-μL reaction with 100 mm NH4SO4, 5 mm MgCl2, 500 μm ATP, GTP, and CTP, 30 μm UTP, 20 μL α-32P-UTP (3,000 Ci/mmol, 10 μCi/μL; Perkin-Elmer), and 80 units RNAsin (Promega). The mixture was incubated at 30°C for 30 min. The reaction was stopped by extracting the reaction using the RNA extraction buffer used in RNA isolation. Subsequently, labeled RNA was extracted as total RNA isolation. Five micrograms of linearized plasmid DNAs were blotted onto a ζ-probe membrane using a BIO DOT apparatus (Bio-Rad). Membranes were prehybridized in 50% formamide, 7% SDS, 0.25 m NaCl, 0.125 m phosphate buffer, pH 7.2, for 1 h, then membranes were hybridized at 42°C in the same solution with 1 × 106 cpm/mL in vitro-transcribed RNA overnight. Blots were then sequentially washed with 2× SSC/0.1% SDS, 0.5× SSC/0.1% SDS, and 0.1× SSC/0.1% SDS. Finally, blots were wrapped in parafilm and exposed to the phosphor screen for 48 h. Signals were quantitated by the National Institutes of Health ImageJ program.
Bombardment of Onion Epidermal Cells
35S∷GFP was generated by cloning the eGFP coding region into the KpnI and BamHI sites of pCHF3. 35S∷AGP31∷eGFP was generated by cloning the eGFP coding region into the BamHI site of 35S∷AGP31∷myc to replace the 6×-myc fragment. Constructs were bombarded into onion (Allium cepa) epidermal cell using a PDS-1,000/He particle delivery system (Bio-Rad) as described (Arnim, 2002). After bombardment, cells were incubated for 18 h in the dark at room temperature before being examined under the microscope.
Transgenic Plants and GUS Staining
About 1.4 kb upstream of the translation start site of the AGP31 promoter were amplified by PCR from genomic DNA using primers 5′-CCAGAATTCGCAATTACGCTCAAAGTCTCC-3′ and 5′-CGCGAGCTCTTTGTTTTGTTTTTGGGTTA-3′ and cloned into EcoRI and SacI sites of binary vector pCHF3 (Borevitz et al., 2000) to replace the 35S promoter in pCHF3. The resulting construct is called PAGP31-pCHF3. Then, a BamHI-XbaI fragment containing the GUS coding region from PKYLX71 (Schardl et al., 1987; Lloyd et al., 1992) was cloned into the PAGP31-pCHF3 to generate the construct harboring AGP31 promoter∷GUS (PAGP31∷GUS). For the 35S∷AGP31∷myc fusion protein construct, the cDNA of AGP31 was amplified by PCR using primers 5′-CCCGGTACCATGGGTTTCATTGGTAAGA-3′ and 5′-TTTGGATCCTTTGGGGCAAGACGG-3′ from cDNA to remove the stop codon and cloned into KpnI and BamHI sites of pCHF3, then a fragment encoding 6×-myc was cloned in frame to the C terminus of the AGP31 at the BamHI site. The UBQ10 promoter was amplified from genomic DNA using primers 5′-TTTAGATCTGAATTCTGTCCCGACGGTGGTGT-3′ and 5′-CGTCCATGGCTTGATCACGGTAGAGAGAATT-3′. The UBQ10 promoter∷Luc+∷AGP31-3′-UTR construct was made by cloning the UBQ10 promoter and the 208-bp 3′-UTR of AGP31 in front of and behind the luciferase coding region (Promega), respectively. All construct sequences were confirmed by DNA sequencing. Constructs were transformed into plants using the floral-dip method (Clough and Bent, 1998). The GUS staining procedure was carried out as described in Bomblies (2000).
Acknowledgments
We would like to thank Dr. Maria Person and Ms. Michelle Gadush at the Core Facilities of the University of Texas at Austin for their excellent technical assistance in MALDI-MS. This project is supported in part by the Department of Energy-funded (DE–FG09–93ER–20097) Center for Plant and Microbial Complex Carbohydrates.
This work was supported by the University of Texas at Austin Office of Vice President for Research (grant to M.C.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mona C. Mehdy (mmehdy@mail.utexas.edu).
Open Access articles can be viewed online without a subscription.
References
- Ahn JH (2000) RNA extraction for northern blot and RT-PCR. In D Weigel, J Glazebrook, eds, Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 172–173
- Arnim Av (2002) Subcellular localization of GUS- and GFP-tagged protein in onion epidermal cells. In D Weigel, J Glazebrook, eds, Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 252–257
- Baldwin TC, Domingo C, Schindler T, Seetharaman G, Stacey N, Roberts K (2001) DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol Biol 45 421–435 [DOI] [PubMed] [Google Scholar]
- Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A (2003) Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol 195 356–372 [DOI] [PubMed] [Google Scholar]
- Bomblies K (2000) Whole mount GUS staining. In D Weigel, J Glazebrook, eds, Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 243–245
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Knudsen JS, Derksen J, Mariani C (2001) Class III pistil-specific extensin-like proteins from tobacco have characteristics of arabinogalactan proteins. Plant Physiol 125 2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70 21–30 [DOI] [PubMed] [Google Scholar]
- Brown KC, Yu Z, Burlingame AL, Craik CS (1998) Determining protein-protein interactions by oxidative cross-linking of a glycine-glycine-histidine fusion protein. Biochemistry 37 4397–4406 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82 383–393 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Czako M, Wilson J, Yu X, Marton L (1993) Sustained root culture for generation and vegetative propagation of transgenic Arabidopsis thaliana. Plant Cell Rep 12 603–606 [DOI] [PubMed] [Google Scholar]
- Desai NN, Allen AK, Neuberger A (1983) The properties of potato (Solanum tuberosum) lectin after deglycosylation by trifluoromethanesulphonic acid. Biochem J 211 273–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58 497–513 [DOI] [PubMed] [Google Scholar]
- Edge ASB (2003) Deglycosylation of glycoproteins with trifluoromethanesulphonic acid: elucidation of molecular structure and function. Biochem J 376 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiz L, Irshad M, Pont-Lezica RF, Canut H, Jamet E (2006) Evaluation of cell wall preparations for proteomics: a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsbach A, Schubert D, Lechtenberg B, Gils M, Schmidt R (2003) A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol Biol 52 161–176 [DOI] [PubMed] [Google Scholar]
- Fowler TJ, Bernhardt C, Tierney ML (1999) Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol 121 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyermuth SK, Bacanamwo M, Polacco JC (2000) The soybean Eu3 gene encodes an Ni-binding protein necessary for urease activity. Plant J 21 53–60 [DOI] [PubMed] [Google Scholar]
- Goodrum LJ, Patel A, Leykam JF, Kieliszewski MJ (2000) Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry 54 99–106 [DOI] [PubMed] [Google Scholar]
- Hara M, Fujinaga M, Kuboi T (2005) Metal binding by citrus dehydrin with histidine-rich domains. J Exp Bot 56 2695–2703 [DOI] [PubMed] [Google Scholar]
- Held MA, Tan L, Kamyab A, Hare M, Shpak E, Kieliszewski MJ (2004) Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J Biol Chem 279 55474–55482 [DOI] [PubMed] [Google Scholar]
- Jiang SY, Jasmin PX, Ting YY, Ramachandran S (2005) Genome-wide identification and molecular characterization of Ole_e_I, Allerg_1 and Allerg_2 domain-containing pollen-allergen-like genes in Oryza sativa. DNA Res 12 167–179 [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) The fasciclin-like arabinogalactan proteins of Arabidopsis: a multigene family of putative cell adhesion molecules. Plant Physiol 133 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14 737–748 [DOI] [PubMed] [Google Scholar]
- Kauss H, Seehaus K, Franke R, Gilbert S, Dietrich RA, Kroger N (2003) Silica deposition by a strongly cationic proline-rich protein from systemically resistant cucumber plants. Plant J 33 87–95 [DOI] [PubMed] [Google Scholar]
- Kjellbom P, Snogerup L, Stohr C, Reuzeau C, McCabe PF, Pennell RI (1997) Oxidative cross-linking of plasma membrane arabinogalactan proteins. Plant J 12 1189–1196 [DOI] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T (2004) Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol 134 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Walbot V, Davis RW (1992) Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258 1773–1775 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdy MC (1994) Active oxygen species in plant defense against pathogens. Plant Physiol 105 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle RK, Poppe I (1994) Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol 230 1–15 [DOI] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429 873–878 [DOI] [PubMed] [Google Scholar]
- Narvaez-Vasquez J, Pearce G, Ryan CA (2005) The plant cell wall matrix harbors a precursor of defense signaling peptides. Proc Natl Acad Sci USA 102 12974–12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T (2005) Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 44 972–984 [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Narvaez-Vasquez J, Ryan CA (2001) Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13 179–191 [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Ryan CA (2003) Systemic signaling in tomato plants for defense against herbivores: isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J Biol Chem 278 30044–30050 [DOI] [PubMed] [Google Scholar]
- Roach D, Gehrke CW (1970) The hydrolysis of proteins. J Chromatogr 52 393–404 [DOI] [PubMed] [Google Scholar]
- Sauer N, Corbin DR, Keller B, Lamb CJ (1990) Cloning and characterization of a wound-specific hydroxyproline-rich glycoprotein in Phaseolus vulgaris. Plant Cell Environ 13 257–266 [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61 1–11 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A (2002) Using genomic resources to guide research directions: the arabinogalactan protein gene family as a test case. Plant Physiol 129 1448–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P (2004) Knocking on the heaven's wall: pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr Opin Plant Biol 7 377–383 [DOI] [PubMed] [Google Scholar]
- Sheng J, D'Ovidio R, Mehdy MC (1991) Negative and positive regulation of a novel proline-rich protein mRNA by fungal elicitor and wounding. Plant J 1 345–354 [DOI] [PubMed] [Google Scholar]
- Simpson RJ, Neuberger MR, Liu TY (1976) Complete amino acid analysis of proteins from a single hydrolysate. J Biol Chem 251 1936–1940 [PubMed] [Google Scholar]
- Sommer-Knudsen J, Clarke AE, Bacic A (1996) A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J 9 71–83 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan CA, Wasternack C (2003) Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato—amplification in wound signalling. Plant J 33 577–589 [DOI] [PubMed] [Google Scholar]
- Tan L, Qiu F, Lamport DT, Kieliszewski MJ (2004) Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J Biol Chem 279 13156–13165 [DOI] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S (2002) A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Barber C, Roberts K (2004) The expression patterns of arabinogalactan-protein AtAGP30 and GLABRA2 reveal a role for abscisic acid in the early stages of root epidermal patterning. Plant J 39 70–83 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K (2003) AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J 36 256–270 [DOI] [PubMed] [Google Scholar]
- van Holst GJ, Clarke AE (1985) Quantification of arabinogalactan-protein in plant extracts by single radial gel diffusion. Anal Biochem 148 446–450 [DOI] [PubMed] [Google Scholar]
- Yang J, Sardar HS, McGovern KR, Zhang Y, Showalter AM (2007) A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. Plant J 49 629–640 [DOI] [PubMed] [Google Scholar]
- Yu X, Sukumaran S, Marton L (1998) Differential expression of the Arabidopsis nia1 and nia2 genes: cytokinin-induced nitrate reductase activity is correlated with increased nia1 transcription and mRNA levels. Plant Physiol 116 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Mehdy MC (1994) Binding of a 50-kD protein to a U-rich sequence in an mRNA encoding a proline-rich protein that is destabilized by fungal elicitor. Plant Cell 6 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sheng J, Liu Y, Mehdy MC (1993) Fungal elicitor-induced bean proline-rich protein mRNA down-regulation is due to destabilization that is transcription and translation dependent. Plant Cell 5 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DT, Kieliszewski MJ (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31 431–444 [DOI] [PubMed] [Google Scholar]