Abstract
Eleven anti-HIV Env monoclonal antibodies (MAbs) were isolated from mice immunized with soluble Env proteins derived from the clade B Env, SF162, or ΔV2 (a derivative of SF162 lacking the V2 loop). All six anti-gp120 MAbs studied, neutralized SF162 and their activities were dependent by the glycosylation patterns of the V1, V2 or V3 loops. Only one anti-gp120 MAb (an anti-V3 MAb) displayed cross-neutralizing activity, which was influenced by the type of V1 loop present on the target heterologous viruses. None of the five anti-gp41 MAbs studied displayed anti-SF162 neutralizing activity. Our studies indicate that the current limitation of soluble HIV Env gp140 immunogens to elicit robust cross-reactive neutralizing antibody responses is not only due to the elicitation of high titers of homologous antibodies, but also due to the elicitation of antibodies whose epitopes are naturally occluded, or not present, on the virion-associated Env.
Keywords: HIV, Monoclonal antibodies, Neutralization, escape, V1 loop, V3 loop, GP41, HIV Env trimers, gp140 proteins
INTRODUCTION
Soluble trimeric gp140 HIV Env proteins are currently being tested as potential immunogens to elicit broadly-reactive neutralizing antibodies (NAbs) against primary HIV isolates and several groups have shown that trimeric gp140 proteins are more effective in eliciting NAbs against heterologous HIV isolates than soluble monomeric gp120 immunogens (Barnett et al., 2001; Beddows et al., 2005; Earl et al., 2001; Grundner et al., 2005; Kim et al., 2005; Li et al., 2006; Yang, Wyatt, and Sodroski, 2001). Overall however, the potential of the currently available trimeric gp140 proteins to elicit broadly reactive neutralizing antibody (NAb) responses is limited in that the antibodies elicited by such constructs can neutralize only a handful of heterologous primary isolates (Barnett et al., 2001; Beddows et al., 2005; Earl et al., 2001; Grundner et al., 2005; Kim et al., 2005; Li et al., 2006; Yang, Wyatt, and Sodroski, 2001). We believe that by examining the immunogenic properties of gp140 constructs we will be able to optimize their design and improve their abilities to elicit cross-reactive NAbs.
We have been designing soluble trimeric gp140 proteins derived from the CCR5-tropic primary clade B HIV-1 isolate SF162, which is susceptible to neutralization by broadly reactive NAbs, such as b12, 447-52D, 2G12, 2F5 and 4E10 (Binley et al., 2004; Saunders et al., 2005). We hypothesized that since the epitopes recognized by these NAbs are exposed on the SF162 Env, SF162 Env-derived gp140 constructs may elicit such antibodies upon immunization. However, the SF162gp140 protein elicits cross-reactive NAbs with a narrow breadth of cross-reactivity (Derby et al., 2006). Deletion of the second hypervariable region improves the ability of this protein (termed ΔV2gp140) to elicit NAbs but only against certain isolates (Barnett et al., 2001; Burke et al., 2006), but not against others (Derby et al., 2006; Xu et al., 2006). In contrast, deletion of the third hypervariable region from SF162gp140 (termed ΔV3gp140) abrogates its ability to elicit cross-reactive NAbs (Derby et al., 2006). Our initial analysis of the immunogenic properties of SF162gp140-derived proteins indicated that the majority of the NAbs elicited by such constructs target the first hypervariable region (V1 loop) (Derby et al., 2006). This may explain the narrow breadth of NAb responses elicited by these immunogens since the V1 loop is highly variable. The above-mentioned immunogenicity results were obtained using polyclonal macaque sera in peptide competition neutralization and ELISA assays (Derby et al., 2006). Such experiments provide a general information on the epitopes recognized by the various types of antibodies present in the polyclonal responses to Env immunogens, but they provide little information on how antibodies with specific epitope specificities bind to their targets and how the kinetics of such interactions influence the neutralization potential of the antibodies. Such information can be however obtained with the use of monoclonal antibodies (MAbs) isolated from the immunized animals.
MAbs have been isolated previously by other groups from animals immunized with oligomeric HIV Env immunogens and their epitopes have been defined (Broder et al., 1994; Earl et al., 1997). Here, we focused our efforts to better understand the molecular details of the interaction of such antibodies with their targets on the virion surface. Specifically, we investigated how the binding properties of individual gp140-elicited antibodies, such as their association and dissociation rates and affinity constants, affect their neutralization potentials. We also examined how the binding of these MAbs to their epitopes on the virion surface is influenced by the positioning of the variable Env regions and by the glycosylation pattern of Env.
Our results indicate that the gp41 portion of our gp140 constructs is immunogenic, but the elicited antibodies have no neutralizing activity. The V1 loop is also highly immunogenic on these gp140 constructs, but it elicits only homologous neutralizing antibodies. In contrast, the V3 loop on these gp140 constructs is less immunogenic than the V1 loop but is capable of eliciting NAbs with a narrow breadth of cross-reactivity, limited in part due to epitope masking by the V1 region on heterologous primary isolates. The results of this study have implications for optimizing the presentation of conserved neutralization epitopes on the next generation of HIV Env gp140 vaccine constructs.
RESULTS
Gp120 and gp41 specificity of MAbs
In this study, two Env gp140 constructs (SF162gp140 and ΔV2gp140) and two methods of immunization (recombinant HIV Env protein and DNA-prime followed by recombinant protein boost) were used to elicit anti-HIV Env antibodies in mice. Eleven MAbs were cloned from hybridomas, all of which are of either IgG1 or IgG2α with κ light chains (Table 2).
TABLE 2.
Mouse monoclonal antibodies characterized in this study generated by immunization with gp140s.
| ELISA Reactivity with Env Antigensa |
Affinity (nM) Determined by Biacoreb |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAb | Ig Subclass |
Subunit | Epitope | Immunogen | Immunization | SF162 t-gp140c |
ΔV2 t-gp140 |
ΔV2 m-gp140 |
SF162 m-gp120 |
HxB2 gp41 |
SF162 t-gp140 |
ΔV2 t-gp140
|
||||
| ka (× 104) | kd (× 10−4) | KD (nM) | ka (×104) | kd (×10−4) | KD (nM) | |||||||||||
| P1H6 | IgG1 κ | gp120 | V1 | ΔV2gp140 | DNA-Protein | +++d | +++ | +++ | +++ | − | nd | nd | nd | nd | nd | nd |
| P3C8 | IgG1 κ | gp120 | V1 | ΔV2gp140 | Protein | +++ | +++ | +++ | +++ | − | nd | nd | nd | nd | nd | nd |
| P3B2 | IgG1 κ | gp120 | V1 | ΔV2gp140 | Protein | +++ | +++ | +++ | +++ | − | nd | nd | nd | nd | nd | nd |
| P4D7 | IgG1 κ | gp120 | V1 | ΔV2gp140 | Protein | ++ | ++ | ++ | ++ | − | nd | nd | nd | nd | nd | nd |
| P3E1 | IgG2α κ | gp120 | V3 | SF162gp140 | Protein | +++ | +++ | +++ | +++ | − | 25.60 | 0.73 | 0.29 | 12.90 | 0.76 | 0.59 |
| P3C5 | IgG2α κ | gp120 | V3 | SF162gp140 | DNA-Protein | ++ | ++ | ++ | ++ | − | nd | nd | nd | nd | nd | nd |
| b12 | IgG1 κ | gp120 | CD4BS | HIV infection | nae | ndf | nd | nd | nd | nd | 11.30 | 0.40 | 0.35 | 15.70 | 0.97 | 0.62 |
| P1G9 | IgG1 κ | gp41 | cluster I | SF162gp140 | DNA-Protein | + | + | − | − | ++ | 1.13 | 1.62 | 14.30 | 8.26 | 1.79 | 21.67 |
| P2D2 | IgG2α κ | gp41 | C-HR | ΔV2gp140 | DNA-Protein | ++ | ++ | − | − | ++ | 2.01 | 3.00 | 14.90 | 2.12 | 3.38 | 15.94 |
| P3G9 | IgG2α κ | gp41 | unknown | SF162gp140 | Protein | + | + | − | − | ++ | 2.15 | 3.14 | 14.60 | 1.64 | 3.59 | 21.89 |
| P4A3 | IgG2α κ | gp41 | unknown | SF162gp140 | DNA-Protein | +++ | +++ | − | − | +++ | 7.12 | 0.94 | 1.30 | 4.70 | 0.95 | 2.02 |
| P4C2 | IgG1 κ | gp41 | unknown | SF162gp140 | DNA-Protein | ++ | ++ | − | − | ++ | 4.50 | 1.35 | 3.00 | 2.74 | 2.23 | 8.14 |
| 2F5 | IgG3 κ | gp41 | MPER | HIV infection | na | nd | nd | nd | nd | nd | 2.58 | 2.17 | 8.41 | 2.00 | 1.33 | 6.65 |
Data reported are the average of two independent experiments.
Values are the average of three independent experiments.
‘t’ indicates trimeric and ‘m’ indicates monomeric.
ELISA half-maximal binding values (μg/ml) for Env proteins are denoted by + symbols. +++ = 0.001–0.01; ++ = 0.01–0.1; + = 0.1–1.0; − = no binding.
na indicates not applicable.
nd indicates not determined in this study.
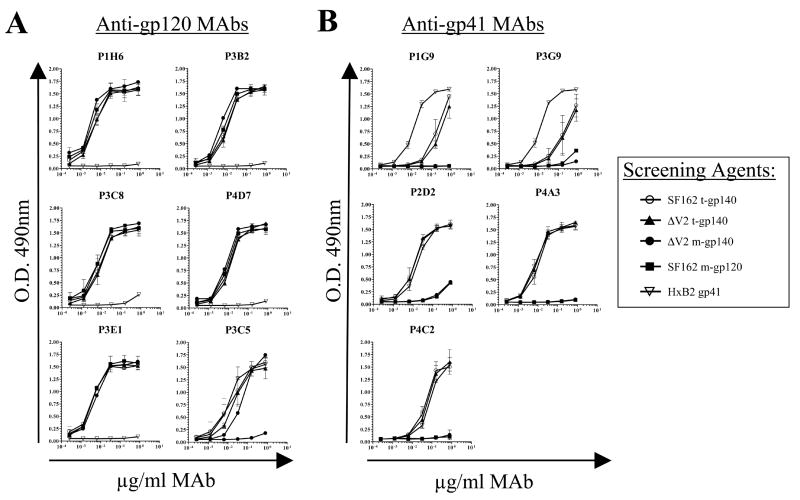
Initial screening for epitope-specificity was performed by ELISA using the SF162gp120 and HxB2gp41 proteins as screening agents (Fig. 1). Six MAbs were specific for gp120 while 5 were specific for gp41 (Fig. 1 and Table 2). Four of the six anti-gp120 MAbs were derived by immunization with ΔV2gp140 while four of the five anti-gp41 MAbs were derived by immunization with SF162gp140 (Table 2). Four anti-gp41 MAbs were elicited by DNA prime plus recombinant protein boost and only one was elicited by recombinant protein immunization alone. Two anti-gp120 MAbs were elicited by DNA prime plus recombinant protein boost and the remaining four were elicited by recombinant protein immunization alone. The epitopes of these 11 MAbs were further characterized by peptide and chimeric protein ELISA (Table 1).
Figure 1. Recognition of Env protein antigens by MAbs.
The binding of the six anti-gp120 (A) and five anti-gp41 (B) MAbs to various Env protein antigens was determined by ELISA as described in the Materials and Methods section. The following five Env proteins were used: trimeric SF162 gp140 (open circles), trimeric ΔV2 gp140 (filled triangles), monomeric ΔV2 gp140 (filled circles), monomeric SF162 gp120 (filled squares), and HxB2 gp41 (open inverted triangles). Data are the average of two independent experiments. The error bars represent standard deviations of the mean.
TABLE 1.
Epitope mapping using peptides and chimeric proteins
| Peptide Namea |
Isolate | Region | Peptide Sequenceb | % SF162 Identity |
P1H6 | P3C8 | P3B2 | P4D7 |
|---|---|---|---|---|---|---|---|---|
| V1 ‘whole’ | HIV-1SF162 | V1 | TNLKNATNTK SSNWKEMDRGEIK | 100 | +++c | +++ | +++ | +++ |
| V1V2-CP | HIV-1SF162 | V1V2 | CTNLKNATNTK SSNWKEMDRGEIKNC…d | 100 | ++++ | ++++ | +++ | +++ |
| 7438 | SHIVSF162P3 | V1 | KLTPLCVTLHCTNLE | 93 | − | − | − | − |
| 7439 | SHIVSF162P3 | V1 | LCVTLHCTNLENATN | 93 | − | − | − | − |
| 7441 | SHIVSF162P3 | V1 | NLENATNTT SSNWKE | 87 | ++ | − | − | − |
| 7443 | SHIVSF162P3 | V1 | T SSNWKEMNRGEIKN | 87 | +++ | − | − | − |
| 7444 | SHIVSF162P3 | V1 | WKEMNRGEIKNCSFN | 87 | − | ++ | ++ | +/− |
| 7445 | SHIVSF162P3 | V1 | NRGEIKNCSFNVTTS | 87 | − | − | − | − |
| 8795 | HIV-1 B cons. | V1 | NCTDLMNATNTTNSS | 73 | − | − | − | − |
| 8786 | HIV-1 B cons. | V1 | LMNATNTTNSSSGEK | 60 | − | − | − | − |
| 8797 | HIV-1 B cons. | V1 | TNTTNSSSGEKMEKG | 47 | − | − | − | − |
| 8798 | HIV-1 B cons. | V1 | NSSSGEKMEKGEIKN | 53 | − | − | − | − |
| 8799 | HIV-1 B cons. | V1 | GEKMEKGEIKNCSFN | 66 | − | − | − | − |
|
| ||||||||
| P3E1 | P3C5 | 447D | ||||||
|
| ||||||||
| V3 crown | HIV-1SF162 | V3 | CKSITIGPGRAFYATGDC | 100 | +++ | + | +++ | |
| V3-CP | HIV-1JR-CSF | V3 | CTR PSNNTRKSIHIGPGRAFYTTGEIIGDIRQAHC | 89 | ++++ | + | ++++ | |
| 8837 | HIV-1 B cons. | V3 | TRPNNNTRKSIHIGP | 93 | − | − | − | |
| 8838 | HIV-1 B cons. | V3 | NNTRKSIHIGPGRAF | 93 | +++ | − | ++ | |
| 8839 | HIV-1 B cons. | V3 | KSIHIGPGRAFYTTG | 87 | ++ | − | ++ | |
| 8840 | HIV-1 B cons. | V3 | IGPGRAFYTTGEIIG | 87 | − | − | − | |
| 8841 | HIV-1 B cons. | V3 | RAFYTTGEIIGDIRQ | 87 | − | − | − | |
| 6285 | HIV-1MN | V3 | NYNKRKRIHIGPGRA | 73 | − | − | +++ | |
| 6286 | HIV-1MN | V3 | RKRIHIGPGRAFYTT | 80 | +++ | − | ++ | |
| 7483 | SHIVSF162P3 | V3 | NNTRKSIPIGPGKAF | 87 | − | − | − | |
| 7484 | SHIVSF162P3 | V3 | KSIPIGPGKAFYATG | 87 | − | − | − | |
|
| ||||||||
| P1G9 | P2D2 | |||||||
|
| ||||||||
| 7555 | SHIVSF162P3 | cluster I | LLGIWGCSGKLICTT | 100 | − | − | ||
| 7556 | SHIVSF162P3 | cluster I | WGCSGKLICTTAVPW | 100 | ++ | − | ||
| 7557 | SHIVSF162P3 | cluster I | GKLICTTAVPWNASW | 100 | + | − | ||
| 7558 | SHIVSF162P3 | cluster I | CTTAVPWNASWSNKS | 100 | − | − | ||
| 7561 | SHIVSF162P3 | C-HR | NKSLDQIWNNMTWME | 100 | − | − | ||
| 7562 | SHIVSF162P3 | C-HR | DQIWNNMTWMEWERE | 100 | − | − | ||
| 7563 | SHIVSF162P3 | C-HR | NNMTWMEWEREIGNY | 93 | − | ++ | ||
| 7564 | SHIVSF162P3 | C-HR | WMEWEREIGNYTNLI | 93 | − | − | ||
Numbers refer to the catalog number in the NIH ARRRP for Env 15-mer peptides; ‘whole’ refers to the entier V1 loop; CP refers to a chimeric MLV antigen expressing the V1V2 region of SF162 (V1V2-CP) or the V3 region of JR-CSF (V3-CP).
Amino acids differing from the amino acid present in SF162 are underlined.
ELISA half-maximal binding affinities (μg/ml) for peptides and CP are denoted by + symbols. ++++ = <0.01; +++ = 0.01–0.1; ++ = 0.1–1.0; + = 1.0–10.0; +/− = >10.0; − = no binding.
Only the V1 portion of the V1V2 insert is shown.
Epitope-specificity of the anti-gp120 MAbs
All six anti-gp120 MAbs recognized peptides derived from the variable regions of gp120. Four MAbs (P1H6, P3B2, P3C8 and P4D7) targeted the V1 loop, as indicated by their recognition of the V1 ‘whole’ peptide which represents the entire V1 loop of SF162 (Table 1). Finer mapping of these V1 epitopes using overlapping peptides derived from the SHIVSF162P3 Env and the HIV-1 clade B consensus Env indicated that the epitopes were all located in the central region of the V1 loop but differed slightly from each other. P1H6 bound to peptides 7441 (NLENATNTTSSNWKE) and 7443 (TSSNWKEMNRGEIKN), but not to any of the other V1 peptides tested, suggesting that its minimal epitope is the SSNWKE motif of the SF162 V1 loop. Specificity of P1H6 for SSNWKE is consistent with its lack of reactivity with V1 peptides derived from the HIV-1 clade B consensus, which do not contain this sequence. P3C8, P3B2, and P4D7 all bound to just one peptide, 7444 (WKEMNRGEIKNCSFN). The inability of these MAbs to bind to peptide 7445 (NRGEIKNCSFNVTTS) is consistent with their inability to recognize peptides derived from the HIV-1 clade B consensus V1, which contains GEIKNC. Thus, the minimal epitopes of P3C8, P3B2, and P4D7 are most likely all located within the N-terminal portion of peptide 7444, WKEM(D/N)R.
All four anti-V1 MAbs bound to the chimeric protein containing the V1V2 region of SF162 on the MLV background (V1V2-CP) with greater affinity than the V1 peptides (Table 1), suggesting that the binding of these MAbs is partially dependent on the conformation of V1.
The presence of the V3 loop is not required for the binding of these four anti-V1 MAbs to their epitopes even though V1 and V3 loops interact on the Env trimer (Chen et al., 2005) since the MAbs were able to interact with an SF162gp140 protein that lacks the central region of the V3 loop (ΔV3gp140) (Derby et al., 2006; Saunders et al., 2005) (data not shown).
The remaining two anti-gp120 MAbs, P3E1 and P3C5, both bound to the crown of the V3 loop (Table 1). P3E1 bound to the SF162 V3 peptide and to the chimeric protein which expresses the heterologous V3 loop of the JR-CSF Env (V3-CP) (differs from that of SF162 V3 at four amino acid positions) with a higher (by over 2 Log10) affinity than P3C5 (Table 1).
Because P3E1 displayed cross neutralizing activity (see below), we mapped its epitope more precisely using overlapping V3 peptides derived from the HIV-1 clade B consensus, HIV-1 MN, and SHIVSF162P3 (Table 1). In parallel, we examined the binding of the well-characterized broadly neutralizing anti-V3 human MAb, 447-52D, to the same peptides. The minimal epitope of 447-52D is GPGR (Zolla-Pazner et al., 2004), and 447-52D bound only to V3 peptides from the three isolates (clade B consensus, MN, and SHIVSF162P3) which contain the GPGR motif (Table 1). One exception was noted: 447-52D did not recognize the B consensus 8840 peptide (IGPGRAFYTTGRIIG). Like 447-52D, P3E1 recognized the two B consensus peptides that express the GPGR motif (8838 [NNTRKSIHIGPGRAF] and 8839 [KSIHIGPGRAFYTTG]) (Table 1), and like 447-52D, P3E1 could not bind to peptide 8840 (IGPGRAFYTTGEIIG). P3E1 also recognized the MN peptide 6286 (RKRIHIGPGRAFYTT) but, unlike 447-52D, did not recognize peptide 6285 (NYNKRKRIHIGPGRA). These observations suggest that the presence of a phenylalanine (F) at the C-terminus of the GPGR motif may be important for P3E1 binding. P3E1 was incapable of recognizing the V3 peptides spanning the same region of the SHIVSP162P3 Env which contains the GPGK instead of the GPRG motif, suggesting that the P3E1 epitope requires the presence of arginine (R) in the GPGX motif, like 447-52D. These results indicate that that the P3E1 epitope is within the V3 crown sequence IGPGRAF.
In contrast to P3E1, the second anti-V3 MAb, P3C5, recognized with low affinity only the SF162 V3 peptide and the V3-CP, and did not bind to the heterologous or consensus V3 peptides tested here (Table 1).
We examined the binding of these anti-gp120 MAbs to trimeric SF162gp140, trimeric ΔV2gp140, monomeric ΔV2gp140, and monomeric SF162gp120 to determine whether binding of these MAbs to their epitopes in V1 and V3 depended on the presence of V2 or on the oligomeric structure of the Env protein (Fig. 1A). These anti-gp120 MAbs had similar affinities for all of the proteins used, suggesting that their binding does not depend on the presence of the V2 loop or on the oligomerization state of Env.
Epitope-specificity of the anti-gp41 MAbs
Using overlapping peptides, we mapped the epitopes of two of the five anti-gp41 MAbs to the surface-exposed C-terminal side of the gp41 ectodomain (Table 1). This is anticipated since the bulk of the N-terminal side of gp41 is buried within the Env trimer (Lu, Blacklow, and Kim, 1995) and is involved in gp41-gp41 contacts (Center, Kemp, and Poumbourios, 1997; Center et al., 2004; Poumbourios et al., 1997). P1G9 recognizes an epitope in the immunodominant cluster I region overlapping the disulfide loop. P1G9 binding was detected to peptides 7556 (WGCSGKLICTTAVPW) and 7557 (GKLICTTAVPWNASW), suggesting that the minimal epitope of P1G9 is likely contained within the GKLICTTAVPW sequence, which is immediately C-terminal to the disulfide loop and contains an immunodominant epitope (Palacios-Rodriguez et al., 2007; Zolla-Pazner, 2004). The second anti-gp41 MAb (P2D2) bound to peptide 7563 (NNMTWMEWEREIGNY) but not to peptides 7561 (NKSLDQIWNNMTWME), 7562 (DQIWNNMTWMEWERE), or 7564 (WMEWEREIGNYTNLI), indicating that the epitope is contained within the sequence NNMTWMEWEREREIGNY in the C-heptad repeat (C-HR) region of gp41. So far we have been unable to map the epitopes of the remaining three anti-gp41 MAbs, P3G9, P4A3, and P4C2 using peptides.
Interestingly, and contrary to what we observed with the anti-gp120 MAbs, the four anti-gp41 MAbs recognized the trimeric ΔV2gp140 protein while they did not recognize the monomeric ΔV2gp140 protein (Fig. 1B). It appears therefore that their epitopes are affected by the state of Env oligomerization.
Neutralizing potentials of the isolated MAbs
i. Neutralization of SF162
Five (P3E1, P1H6, P3C8, P4D7 and P3B2) of the six anti-gp120 MAbs neutralized SF162 potently (IC50 between 0.2 and 1 μg/ml) (Table 3 and data not shown). The sixth MAb, P3C5, neutralized SF162 with significantly reduced potency; at the highest MAb concentration tested (25 μg/ml), 80% inhibition of infection was not achieved. In contrast, none of the anti-gp41 MAbs neutralized SF162, suggesting that although the epitopes of these MAbs are better exposed on the soluble trimeric gp140 Env than the soluble monomeric gp140 Env (Fig. 1B), they are not accessible on virion-associated Env spikes.
TABLE 3.
Neutralizing Potential of MAbs
| IC50 Neutralization titer (μg/ml)a |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | P1H6 (V1) |
P3C8 (V1) |
P3B2 (V1) |
P4D7 (V1) |
P3E1 (V3) |
P3C5 (V3) |
447-52D (V3) |
P1G9 (cluster I) |
P2D2 (C-HR) |
P3G9 (gp41) |
P4A3 (gp41) |
P4C2 (gp41) |
2F5 (MPER) |
| SF162 | 0.3 | 0.6 | 1.0 | 0.8 | 0.2 | 16.0 | 0.2c | - | - | - | - | - | 1.0c |
| ΔV1 | -b | - | - | - | 4.0 | - | 1c | - | - | - | - | - | 2.0c |
| ΔV2 | 0.003 | 0.003 | 0.01 | 0.08 | <0.001 | 1.0 | 0.01c | - | - | 25.0 | - | - | <0.01c |
| ADA | - | - | - | - | - | - | - | - | - | - | - | - | nd |
| ADA/SF162 V1 | ndd | nd | nd | nd | - | nd | - | nd | nd | nd | nd | nd | nd |
| JRFL | - | - | - | - | - | - | 15.0 | - | - | - | - | - | nd |
| JRFL/SF162 V1 | nd | nd | nd | nd | 10 | nd | 1.5 | nd | nd | nd | nd | nd | nd |
| YU2 | - | - | - | - | - | - | - | - | - | - | - | - | nd |
| YU2/SF162 V1 | nd | nd | nd | nd | - | nd | 2.0 | nd | nd | nd | nd | nd | nd |
| HxB2 | - | - | - | - | - | - | 0.2 | - | - | - | - | - | nd |
| HxB2/SF162 V1 | nd | nd | nd | nd | - | nd | 0.01 | nd | nd | nd | nd | nd | nd |
| 89.6 | - | - | - | - | 6.0 | - | 0.001 | - | - | - | - | - | nd |
| 89.6/SF162 V1 | nd | nd | nd | nd | 0.1 | nd | <0.001 | nd | nd | nd | nd | nd | nd |
| SS1196.01 | - | - | - | - | 3.5 | - | nd | nd | nd | nd | nd | nd | nd |
| 6535.3 | - | - | - | - | 16.5 | - | nd | nd | nd | nd | nd | nd | nd |
| 7165.18 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| 6101.1 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| QH0692.42 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| QH0515.1 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| AC10.0.29 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| PVO.4 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| TRO.11 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| 5768.4 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| 3988.25 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
| BG1168.1 | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd |
Values reported are the average of 3 independent experiments.
- indicates 50% neutralization not achieved at the highest MAb concentration tested, 25μg/ml.
Values are from Saunders, et al. 2005.
nd: not done.
ii. Neutralization of ΔV2 and ΔV1 viruses
Although our binding studies with recombinant SF162gp140 and ΔV2gp140 proteins indicated that the MAbs bound equally well to these two proteins (Fig. 1), our previous neutralization data implicated the V2 loop in preventing the neutralizing action of certain anti-gp41 MAbs (Saunders et al., 2005). We thus compared the abilities of the anti-gp41 MAbs to neutralize the SF162 and ΔV2 viruses (Stamatatos and Cheng-Mayer, 1998). Although the anti-gp41 MAbs were all unable to neutralize SF162, one MAb, P3G9, weakly neutralized ΔV2 (Table 3) (IC50 = 25 μg/ml). What is interesting is that this MAb was elicited following immunization with the SF162gp140 immunogen and not the ΔV2gp140 immunogen (Table 2).
In contrast to the resistance of the ΔV2 virus to neutralization by 4 out of 5 of the anti-gp41 MAbs tested here, the ΔV2 virus was significantly more susceptible (between 10 and 300-fold) to neutralization by all of the anti-gp120 MAbs examined, irrespective of their epitopes on the V1 or V3 loops (Table 3). These results are in agreement with previous studies from our group, which show that the ΔV2 virus is more sensitive than SF162 to neutralization by diverse anti-gp120 MAbs (Saunders et al., 2005).
To further examine the mechanisms of neutralization of these MAbs, we examined their abilities to neutralize an SF162-derived virus whose Env lacks the V1 loop (Saunders et al., 2005). As anticipated, the anti-V1 MAbs could not neutralize the ΔV1 virus (Table 3). In contrast however, to what we observed with the ΔV2 virus, the anti-V3 MAbs P3E1 and P3C5 neutralized ΔV1 with approximately 20-fold (P3E1) and 3.5-fold (P3C5) reduced potency compared to SF162 (Table 3). This observation suggests that the exposure and/or conformation of these V3 epitopes is affected by the positioning of the V1 loop. We were unable to detect neutralization of ΔV1 by the anti-gp41 MAbs (Table 3). Thus, the neutralizing potentials of the anti-V1 and anti-V3 MAbs discussed here are affected by the presence of the V2 loop, and the neutralizing potentials of the anti-V3 MAbs are in addition affected by the V1 loop. In contrast, the V2 loop affected (but only marginally) the neutralizing potential of only one anti-gp41 MAb characterized here. Overall, the neutralizing potentials of the MAbs discussed here are affected by the presence of the V1 and V2 loops in a similar manner to that observed previously with human MAbs isolated from HIV infected patients (Saunders et al., 2005).
iii. Neutralization of ‘deglycosylated’ SF162
Certain N-linked glycans present within or at the base of the V1, V2 and V3 loops affect the neutralization phenotype of diverse HIV-1 clade B isolates and modulate the neutralizing potential of NAbs (Blay et al., 2006; Losman et al., 2001; Ly and Stamatatos, 2000; Malenbaum et al., 2000; McCaffrey et al., 2004; Wei et al., 2003). Here we examined the role that glycans in these regions might play in protecting the epitopes of the newly identified anti-V1, anti-V3, and anti-gp41 MAbs (Table 4). We compared the neutralization susceptibility of SF162 with the susceptibilities of SF162-derived viruses which express Envs mutated to remove specific sites for N-linked glycosylation (termed GM viruses) (Ly and Stamatatos, 2000; McCaffrey et al., 2004). GM viruses are named according to the position of the N → Q mutation in the NXT/S motif using the SF162 numbering system. A ten fold difference in IC50 between SF162 and each GM virus was considered significant.
TABLE 4.
Neutralization of SF162 expressing deglycosylated Envs.
| Fold change in neutralization sensitivity compared with SF162a |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1V2b | V3 | V4 | C4V5 | |||||||||||||
| 154 | 186 | 195 | 293 | 299 | 329 | 382 | 388 | 392 | 398 | 401 | 438 | 454 | ||||
| NCSFKVTTSIRNKMQKEYALFYKLDVVPIDNDNTSYKLINCNTS | NCTRPNNNTRKSITIGPGRAFYATGDIIGDIRQAHCNIS | CNSTQLFNSTWNNTIGPNNTNGT | NITGLLLTRDGGKEISNTT | |||||||||||||
| MAb | Epitope | GM154c | GM186 | GM195 | GM293 | GM299 | GM329 | GM382 | GM388 | GM392 | GM398 | GM401 | GM438 | GM454 | ||
| P1H6 | V1 | 30.0 | 15.0 | 150.0 | 3.5 | 106.3 | 229.7 | −2.9 | 2.4 | −14.6 | 4.3 | 1.4 | 1.3 | 2.0 | ||
| P3C8 | V1 | 250.0 | 12.5 | 250.0 | 1.4 | 225.0 | 15.0 | −15.0 | 1.5 | −9.4 | 1.8 | 1.1 | −1.2 | −1.4 | ||
| P3B2 | V1 | 31.0 | 5.2 | 44.3 | 2.0 | 39.0 | 18.0 | −19.0 | −1.0 | −6.0 | 2.6 | −1.2 | 1.1 | 1.5 | ||
| P4D7 | V1 | 20.0 | 2.0 | 10.0 | 1.4 | 31.0 | 13.9 | −36.0 | −1.0 | −16.0 | 1.6 | −1.3 | 1.2 | 1.0 | ||
| P3E1 | V3 | 50.0 | −2.7 | 10.0 | 2.5 | 24000.0 | 120.0 | −2.5 | 3.2 | −5.0 | 4.4 | −1.3 | −1.5 | 1.0 | ||
| P3C5 | V3 | 4.0 | −9.7 | 1.0 | 1.4 | 203.0 | 10.9 | −1.6 | 1.1 | −1.4 | 1.5 | 1.4 | 1.2 | 1.4 | ||
Increase in neutralization sensitivity determined at the IC50. Changes greater than 10 fold are shown in bold. Negative numbers indicate a fold decrease in IC50 values.
GM154, GM186, and GM195 were replication competent viruses generated in PBMC, and neutralization of these viruses was compared with neutralization of PMBC-produced replication competent SF162 virus. In all other cases, pseudoviruses were used.
All GM viruses are named for the position of the N→Q mutation according to SF162 numbering.
Elimination of the conserved V3 loop glycan at position 299 within the N-terminal side of the V3 loop dramatically enhanced the neutralization susceptibility of SF162 to the anti-V3 MAbs (203 fold for P3C5 and over 20,000 fold for P3E1) while deletion of the glycan at position 329 (immediately C-terminal to the V3 loop) led to significant but somewhat less pronounced enhancement in neutralization susceptibility (10.9 fold for P3C5 and 120 fold for P3E1). In contrast, deletion of the glycan at position 293 (immediately N-terminal to the V3 loop) had a smaller effect on the susceptibility of SF162 to neutralization by P3E1 and P3C5.
The glycan present at position 186 within the V2 loop also had little, if any, role in protection of SF162 from the anti-V3 MAbs. The glycans in V1 (position 154) and C-terminal to the V2 loop (position 195) played a more significant role, particularly for MAb P3E1 (50 fold increase for position 154 and 10 fold increase for position 195). As previously reported for the human anti-V3 MAbs 447-52D and 391–95 (Saunders et al., 2005), the glycan at position 154 was more important for the protection from anti-V3 MAbs than the glycan at position 195.
The glycans at positions 154 and 195 in V1V2 were also involved in regulating the neutralizing potential of the anti-V1 MAbs discussed here (observed neutralization increases between 10 and 250 fold) (Table 4). Furthermore, as we described for anti-V3 MAbs, neutralization by all of the anti-V1 MAbs was enhanced strongly by deletion of the V3 glycan at position 299 (ranging from 31 to 225 fold), somewhat less so by deletion of the glycan at position 329 (ranging from 14 to 230 fold), and only slightly or not at all by deletion of the glycan at position 293 (ranging from 1.4 to 3.5 fold). P1H6, which is directed to an epitope slightly N-terminal to the epitopes of the other 3 anti-V1 MAbs, was the only MAb which neutralized the virus lacking the glycan at position 329 more potently than the virus lacking the glycan at position 299, suggesting that the former glycan may preferentially shield the SSNWKE epitope over the WKEMDR epitope of V1.
We also examined whether glycans present in the V4–V5 region of Env, such as those at positions 382, 388, 392, 398, 401, 438, and 454 (McCaffrey et al., 2004), were involved in modulating the neutralizing activities of the newly isolated MAbs (Table 4). Our results indicate that these glycans have only modest effects on the neutralizing potentials of the anti-V1 and anti-V3 MAbs and that these effects are strongest for the glycans at positions 382 and 392. In these cases, removal of a glycan rendered the virus more resistant to neutralization. Overall therefore, the neutralizing potentials of the anti-V1 and anti-V3 MAbs were influenced by the presence of N-linked glycosylation sites in both the V1V2 and V3 loops.
The anti-gp41 MAbs were unable to neutralize any of the GM viruses studied here (data not shown).
iv. Neutralization of heterologous isolates
To further examine the neutralization properties of the anti-gp120 MAbs, we evaluated their abilities to neutralize heterologous clade B HIV-1 isolates, including a lab-adapted isolate (HxB2) and several well-characterized primary isolates from both chronic and primary infection (Li et al., 2005) (Table 3).
Despite their potent neutralizing activities against SF162, the V1-specific MAbs were not capable of neutralizing any of the heterologous isolates tested. In fact, the only MAb with cross-neutralizing activity was the anti-V3 MAb, P3E1, which neutralized 89.6, SS1196.1, and 6535.3. The resistance of all the other isolates to neutralization by P3E1 was not due to the absence of the minimal P3E1 epitope, which was mapped to IGPGRAF (see above) and which is present in many of the isolates which were resistant to P3E1 neutralization ((Li et al., 2005) and data not shown). Instead, the P3E1 epitope may be shielded on these isolates by other regions of the viral Env, such as the V1V2 loop (Krachmarov et al., 2006; Pinter et al., 2004; Saunders et al., 2005). To test specifically whether or not the V1 loops of heterologous isolates masked the P3E1 epitope, we evaluated the ability of P3E1 to neutralize viruses expressing chimeric Env constructed by replacing the V1 loop of each isolate with that of SF162. We examined the neutralization of such chimeras on the background of 89.6, JRFL, ADA, YU2, and HxB2 in parallel with their wild type parent virus. As control we used the well-characterized anti-V3 MAb, 447-52D (Table 3). P3E1 was able to neutralize the chimeric 89.6/SF162V1 and JRFL/SF162V1 viruses with significantly enhanced potency compared to the corresponding wild type viruses, indicating that on these two primary isolates, their V1 loop masks the exposure of the P3E1 epitope in V3. Neutralization of the 89.6/SF162V1 and JRFL/SF162V1 chimeric viruses by 447-52D was also enhanced compared to the wild type. However, 447-52D, but not P3E1, also neutralized the chimeric YU2/SF162V1 and HxB2/SF162V1 viruses more efficiently than their wild type counterparts.
Affinity for soluble trimeric gp140 proteins by surface plasmon resonance using Biacore
The neutralizing potential of anti-HIV MAbs has been linked with their binding affinities for the HIV Env (Fouts et al., 1997). However, the observed differences in the neutralizing potentials of the anti-gp120 and anti-gp41 MAbs described here could not be explained by their reactivities with trimeric gp140 by ELISA (Fig. 1). We hypothesized that differences in the association or dissociation rates of MAb-Env interactions could explain the inability of the anti-gp41 MAbs to neutralize SF162. Thus, we investigated the binding kinetics of the anti-gp41 MAbs with soluble trimeric SF162gp140 and ΔV2gp140 proteins by Biacore surface plasmon resonance (SPR). As controls for these studies, we used the broadly neutralizing human MAbs 2F5 (anti-gp41) and b12 (anti-gp120), and the P3E1 mouse MAb described here since it neutralized SF162 very potently and displayed a modest breadth of neutralization.
Lack of neutralizing activity by the anti-gp41 MAbs could not be attributed to either a slow association with or a rapid dissociation from trimeric SF162gp140 as MAbs P4A3 and P4C2 both had faster on rates and more stable off rates than the human MAb 2F5 (Table 2), and yet only 2F5 neutralizes SF162 ((Saunders et al., 2005) and Table 3). Furthermore, there was no correlation between the ability of P3G9 to neutralize ΔV2, but not SF162, and enhanced binding of this MAb to the trimeric ΔV2gp140 protein over the trimeric SF162gp140 protein.
A difference in binding kinetics was observed, however, when all anti-gp41 MAbs were compared with the two anti-gp120 MAbs, P3E1 and b12, which neutralize SF162 very efficiently (Table 3 and (Binley et al., 2004; Saunders et al., 2005)). P3E1 and b12 had higher affinities for SF162gp140 and ΔV2gp140 than any of the anti-gp41 MAbs (Table 2). P3E1 and b12 also bound with faster on rates and slower off rates than the anti-gp41 MAbs. Differences in the kinetics of MAb binding to soluble trimeric gp140 Env proteins, however, cannot explain why b12 and 2F5 have a broader neutralizing potential than P3E1. Breadth of neutralization may therefore be more directly related to the accessibility of the epitope than with the kinetics of binding to soluble gp140 proteins.
DISCUSSION
In the present study, we describe the generation and characterization of eleven MAbs elicited in response to immunization with soluble trimeric SF162gp140 and ΔV2gp140 Env immunogens. Our results suggest that there is a limited repertoire of highly immunogenic regions on the gp120 portion of our SF162-derived soluble trimeric gp140 proteins, namely: the V1 and V3 loops. In contrast, a more diverse repertoire of epitopes is recognized by the anti-gp41 MAbs elicited by these immunogens. However, regardless of the epitopes they recognize, all the anti-gp41 MAbs isolated here lacked neutralizing activities. Because our gp140 immunogens are soluble and not membrane-bound, regions of gp41 which are not normally accessible on the virion-associated Env gp160 may have elicited these anti-gp41 MAbs. The high immunogenicity of regions of the gp41 ectodomain that are not accessible on native virions may divert the ability of our gp140 constructs to elicit NAbs. However, even if the anti-gp41 MAbs were able to bind to the virion surface, the poor binding kinetics (slow on rates and fast off rates) of many of these MAbs may hinder their ability to neutralize. Interestingly, the binding of the anti-gp41 MAbs to their epitopes was greater in the context of the trimeric gp140 than the monomeric gp140 while such a differential binding was not observed for the anti-gp120 MAbs. So, our studies indicate that the binding properties of the anti-gp120 and anti-gp41 antibodies elicited by our soluble trimeric gp140 immunogens differ significantly.
Although all the anti-gp120 MAbs displayed neutralizing activity against SF162, only one MAb, the anti-V3 MAb P3E1, displayed cross-neutralizing activity. P3E1 bound more efficiently to its epitope (within the IGPGRAF V3 loop motif) when the V3 loop was presented in the context of a protein rather than as a peptide. Thus, P3E1-binding to IGPGRAF appears to be affected by the conformational state of the V3 loop. Also, the ability of P3E1 to neutralize certain heterologous isolates (such as JRFL and 89.6) improved when the V1 loops from the Env of these isolates was replaced by that of SF162. A similar observation was made for the broadly neutralizing anti-V3 MAb 447-52D although this MAb was more potent in neutralizing the chimeric viruses than P3E1. Therefore the accessibility of the V3 loop to NAbs is affected by the nature of the V1 loop. Since the V1 and V3 loops are not closely spaced within the monomeric HIV Env (Chen et al., 2005; Kwong et al., 1998), our results indicate that the V1 loop of one Env protomer affects the exposure of V3 loop epitopes on an adjoining Env protomer within the same trimeric spike. Although P3E1 and 447-52D recognize overlapping epitopes, the observation that 447-52D has a broader neutralizing activity than P3E1 is most likely related to differences in the binding constraints created by the positioning of the V1 loop on these two MAbs. Our results indicate that in part these constraints are imposed by the glycosylation pattern of the V1 loop. Additional constraints are created, however, by the glycosylation pattern of the V3 loop itself. Our results are in agreement with those of others (Krachmarov et al., 2006; Pinter et al., 2004), indicating that the V1V2 region influences the accessibility of anti-V3 NAbs, but our study highlights the important involvement of the V1 loop itself in this regulation. Our observations strongly suggest that improving the neutralizing potential of vaccine-elicited anti-V3 MAbs will depend on our ability to overcome several obstacles related to the positioning and glycosylation pattern of the V1 loop.
An obvious question is why does the neutralizing ability of anti-V3 MAbs (such as P3E1 or 447-52D (Saunders et al., 2005)) not improve when the V1 loop is deleted from the SF162 Env (ΔV1 virus)? After all, deletion of the V2 loop renders HIV more susceptible to neutralization by anti-V3 NAbs (Cao et al., 1997; Saunders et al., 2005). Most likely, deletion of the V1 loop results in the reorganization of many Env regions within each protomer of the trimeric Env spike and these reorganizations do not necessarily result in an enhanced exposure of the V3 loop. In fact, they may result in its further occlusion. This is clearly contrary to the way that the V2 loop affects the neutralizing activity of anti-V3 NAbs. Our understanding of the interaction between the V1, V2 and V3 loops will greatly improve when a crystal structure of the entire trimeric Env containing the V1, V2, and V3 variable loops becomes available.
Four out of the six anti-gp120 MAbs isolated so far recognize the V1 loop. This observation suggests that the V1 loop is highly immunogenic on our gp140 immunogens and supports results from immunogenicity studies conducted with sera from SF162gp140-immunized macaques (Derby et al., 2006). All four of the anti-V1 MAbs recognized the central region of the V1 loop just C-terminal to the conserved SS dipeptide, but N-terminal to the conserved GEIKNC motif. The region of V1 just C-terminal to the SS dipeptide has previously been identified as immunodominant on soluble trimeric YU2-derived Env constructs (Li et al., 2006). We were unable to examine whether the positioning of the V3 loop alters the neutralizing properties of these anti-V1 MAbs, since deletion of the V3 loop from the SF162 Env abrogates its fusogenic potential (Saunders et al., 2005), but their binding was not abrogated by the absence of the V3 loop from the SF162 Env (data not shown). Clearly their neutralizing potential was affected by the V2 loop and the glycosylation patterns of the V1V2 and V3 regions. We have not yet determined whether deletion of the V2 loop and elimination of N-linked glycosylation sites from heterologous Env will improve the cross-neutralizing potentials of these anti-V1 MAbs. However, in contrast to the observations made with the anti-V3 MAb P3E1, we do not believe that the absence of cross-neutralizing activity of the anti-V1 MAbs is related to the occlusion of their epitopes on heterologous Envs, but rather to the fact that their epitopes are highly variable among HIV isolates.
Our studies contribute to the understanding of the immunogenic properties of currently evaluated gp140 Env immunogens, and they provide additional information on the interaction of gp140-elicted antibodies with homologous and heterologous isolates. They highlight that the limitation of soluble HIV Env gp140 immunogens to elicit robust cross-reactive neutralizing antibody responses is not only due to the elicitation of high titers of homologous antibodies, but also to the elicitation of significant titers of antibodies whose epitopes although exposed on these immunogens, are naturally occluded, or not present, on the virion-associated Env. Such information could be used to improve the ability of gp140 immunogens to elicit cross-reactive neutralizing antibody responses.
MATERIALS AND METHODS
Monoclonal antibodies
Eleven mouse MAbs were generated and characterized in this study. The human MAbs, IgG1b12 (Burton et al., 1994), directed to the CD4 binding site, and 2F5 (Zwick et al., 2001), directed to the MPER of gp41, were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP). MAb 447-52D (Conley et al., 1994; Gorny et al., 1992; Gorny et al., 1993), directed to the GPGR motif at the crown of the V3 loop of gp120, and MAb 1418 (Conley et al., 1994; Gorny et al., 1992; Gorny et al., 1993), directed to the parvovirus glycoprotein, were provided by Susan Zolla-Pazner and Mirek Gorny (New York University, New York, NY). MAb 17b (Sullivan et al., 1993), directed to a CD4-induced (CD4i) epitope, was provided by James Robinson (Tulane University, New Orleans, LA) and IgG X5 (Moulard et al., 2002), directed to a different CD4i epitope, was provided by Dimiter Dimitrov (National Cancer Institute, Frederick, MD).
Proteins and peptides
CHO-produced recombinant Env proteins: monomeric SF162gp120, trimeric SF162gp140, trimeric ΔV2gp140, monomeric ΔV2gp140, monomeric US4gp140 (clade B), monomeric SF2gp120 (clade B), trimeric TV1gp140 (clade C), trimeric MJ4gp140 (clade C), and four domain soluble CD4 (sCD4) were provided by Novartis Vaccine & Diagnostics (Emeryville, CA) (Srivastava et al., 2003a). P. pastoris produced recombinant gp41 ectodomain lacking the fusion peptide, membrane-spanning, and cytoplasmic domains (aa 541–682) from the HxB2 isolate (HxB2gp41) was purchased from Viral Therapeutics (Ithaca, NY).
Chimeric proteins expressing the V1V2 region of SF162 Env (V1V2-CP), or the V3 region of JR-CSF Env (V3-CP) on the MLV envelope glycoprotein gp70 background (Kayman et al., 1994) were provided by Abraham Pinter (Public Health Research Institute, Newark, NJ).
Peptides derived from the V1 and V2 variable regions of SF162 Env were purchased from Sigma-Genosys (The Woodlands, TX). An SF162 V3 peptide was provided by Novartis. Peptides spanning the V1 and V3 regions of the HIV-1 clade B consensus Env sequence (Catalog #9480) and the SHIVSF162P3 Env sequence (Catalog #7619) were obtained from the ARRRP. V3 peptides from HIV-1 MN (Catalog #6451) were also obtained from the ARRRP. Two gp41 peptides spanning the epitopes of 2F5 and 4E10 were provided by John Mascola and Richard Wyatt (VRC/NIH, Bethesda, MD), and a set of 43 overlapping peptides encompassing the entire gp41 ectodomain derived from the SHIVSF162P3 gp41 Env was obtained from the ARRRP (Catalog # 7619). The sequences of all above-mentioned peptides are reported in Table 1.
Mice and immunizations
8–12 week old RBF/DnJ mice were immunized as follows. Mouse 1L was immunized with the recombinant trimeric SF162gp140 protein; mouse 1R was immunized with the recombinant trimeric ΔV2gp140 protein; mouse 2L was immunized with a DNA vector expressing SF162gp140 and subsequently with the recombinant trimeric SF162gp140 protein as a boost; and mouse 2R was immunized with a DNA vector expressing ΔV2gp140 and subsequently with the recombinant trimeric ΔV2gp140 protein as a boost. The construction and characterization of SF162gp140 and ΔV2gp140 plasmid DNA and recombinant proteins has been described extensively elsewhere (Barnett et al., 2001; Srivastava et al., 2003a; Stamatatos, Lim, and Cheng-Mayer, 2000). Mouse 1L received 11 immunizations with recombinant SF162gp140 (5–50 μg). During the 1st, 2nd and 9th immunizations, the protein was mixed with Ribi adjuvant. No adjuvant was used during the remaining immunizations. Mouse 1R was immunized 13 times with recombinant ΔV2gp140 (5–50 μg). Ribi adjuvant was used during the 1st, 2nd, 9th and 12th immunizations. Mouse 2L was immunized twice with a DNA vector expressing SF162gp140 and subsequently 8 times with the recombinant trimeric SF162gp140 protein. Mouse 2R was immunized twice with a DNA vector expressing ΔV2gp140 and subsequently 9 times with the recombinant trimeric ΔV2gp140 protein. For mice 2L and 2R, Ribi adjuvant was used during both DNA immunizations and during the first immunization with recombinant protein. No adjuvant was used during the remaining protein immunizations.
Hybridomas and monoclonal antibodies
Sera were collected throughout the immunization schedule and monitored for the presence of anti-Env antibodies. Three days following the final immunization, the mice were euthanized and their spleens were harvested. Splenocytes were fused with a myeloma cell line. Splenocytes were fused with FOX NY myeloma cells to generate hybridomas secreting anti-HIV Env antibody. Between 20–30 hybridoma supernatants were screened for reactivity with HIV Env proteins by ELISA. The soluble protein antigens used for ELISA screening were SF162gp140, SF162gp120, US4gp140, SF2gp120, HxB2gp41, TV1gp140, and MJ4gp140. Two to four hybridomas displaying the broadest cross-reactivity by ELISA were selected for further characterization.
Selected hybridomas were cloned by limiting dilution, and the secreted antibodies were purified on protein A/G agarose columns (Pierce, Rockford, IL) according to the manufacturer’s instructions and as previously described (Derby et al., 2006). Briefly, hybridoma supernatants were concentrated by centrifugation using Centricon-80 30kDa cut-off filters, and the concentrate was diluted in binding buffer pH 8.0 (Pierce) and applied to pre-equilibrated protein A/G agarose columns. Columns were washed with binding buffer, and bound IgG was eluted with imidazole, pH 2.0 (Pierce). Those fractions containing the highest concentrations of protein (by UV absorbance at 280nm) that reacted with SF162gp140 by ELISA were pooled, buffer-exchanged in PBS to remove elution buffer salts, concentrated, and sterile-filtered. The concentration of the final product was determined by UV absorbance at 280nm and adjusted to 2 mg/ml in PBS.
MAb-epitope specificity and affinity determination by ELISA
MAbs were serially diluted into microtiter ELISA plates (Immulon 2HB, Thermo electron, Waltham, MA), which had been coated overnight with one of the following proteins (50 ng/well): gp140, gp120, gp41, V1V2-CP, or V3-CP; or with Env-derived peptides (100 ng/well), and MAb binding was determined as previously described (Barnett et al., 2001; Srivastava et al., 2003b; Xu et al., 2006). Half-maximal MAb binding titers were determined as the μg/ml concentration of MAb at which the OD490 was half that at saturation.
BIACORE surface plasmon resonance
Surface plasmon resonance studies were performed on either a Biacore X or Biacore 3000 (Biacore, Uppsala, Sweden) using Biacore 3000 Control Software (Biacore). Approximately 10,000 response units (RU) of a capture antibody, either goat-anti-mouse IgG (Biorad, Hercules, CA) or goat-anti-human IgG (Biorad), were immobilized on CM5 chips (Biacore) by standard amine coupling according to the manufacturer’s instructions. For kinetic analysis, approximately 400 RU of each MAb were then captured onto the surface. Soluble trimeric SF162gp140 or ΔV2gp140 proteins in HBS-EP buffer (Biacore) were then injected over the MAbs in a concentration series ranging from 500nM to 1.56nM in two-fold serial dilutions. Kinetic injections were performed for 3 minutes at a flow rate of 20 μl/min followed by 60 minutes of dissociation. Between protein injections, chips were regenerated down to the goat-anti-mouse or goat-anti-human IgG surface by 10 second injections of glycine pH 1.5 buffer at a flow rate of 100 μl/min. All assays were performed in triplicate, and the data were analyzed using the Biaevaluation 3.1 Software (Biacore) and fitted according to a Langmuir 1:1 with or without drifting baseline model.
Viruses. (a) Single round competent viruses
Luciferase reporter viruses capable of only a single round of replication were generated in HEK 293T cells as previously described (McCaffrey et al., 2004; Saunders et al., 2005). In most cases, viruses were concentrated by centrifugation at 3,000 rpm through an Amicon Ultra-15 100kD cut-off filter (Millipore) to enhance their infectious titers. Single round competent viruses expressing Env proteins from the following isolates were generated: SF162, variable loop-deleted variants of SF162 (ΔV1 lacking the V1 loop and ΔV2 lacking the V2 loop) (Saunders et al., 2005), and SF162 variants lacking specific N-linked glycosylation sites around the V3 loop (GM293, GM299, GM329), the V4 loop (GM382, GM388, GM392, GM398, GM401), and the C4V5 region (GM438, GM454) (McCaffrey et al., 2004). Single round competent viruses were also generated that expressed heterologous Env proteins: HxB2 (a T cell line adapted clade B virus); 89.6, ADA, JRFL, and YU2 (clade B primary isolates from chronic infection); and 3988.25, 5768.4, 6101.1, 6535.3, 7165.18, AC10.0.29, BG1168.1, QH0515.1, QH0692.42, PVO.4, SS1196.1, TRO.11, REJO4541.67, and RHPA4259.67 (clade B primary isolates collected within 3 months following HIV infection) (Li et al., 2005). (b) Replication competent viruses: In some experiments, replication competent SF162, GM154, GM186 and GM195 viruses were used. The generation of such viruses was previously reported (Ly and Stamatatos, 2000).
HIV Neutralization assays
Neutralization assays were performed in TZM-bl cells as previously described (Derby et al., 2006). Percent neutralization at each MAb concentration was calculated based on the reduction in virus entry determined from the reduction in cell-associated luciferase (relative light units, RLU) in the presence of MAb relative to the entry in the absence of MAb as described mathematically here: ((RLUcells+virus − RLUcells+virus+MAb)/RLUcells+virus)×100.
Acknowledgments
These studies were supported by RO1 AI47708 (L.S.). N.R.D. was also supported by NIH training grant AI007509. We would also like to acknowledge the J.B. Pendleton Charitable Trust; the M. J. Murdock Charitable Trust; and the UW CFAR Humoral Immunity Core (NIAID P30 AI27757). We thank all those who contributed materials used in these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnett SW, Lu S, Srivastava I, Cherpelis S, Gettie A, Blanchard J, Wang S, Mboudjeka I, Leung L, Lian Y, Fong A, Buckner C, Ly A, Hilt S, Ulmer J, Wild CT, Mascola JR, Stamatatos L. The Ability of an Oligomeric Human Immunodeficiency Virus Type 1 (HIV- 1) Envelope Antigen To Elicit Neutralizing Antibodies against Primary HIV-1 Isolates Is Improved following Partial Deletion of the Second Hypervariable Region. J Virol. 2001;75(12):5526–40. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddows S, Schulke N, Kirschner M, Barnes K, Franti M, Michael E, Ketas T, Sanders RW, Maddon PJ, Olson WC, Moore JP. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79(14):8812–27. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–52. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay WM, Gnanakaran S, Foley B, Doria-Rose NA, Korber BT, Haigwood NL. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J Virol. 2006;80(2):999–1014. doi: 10.1128/JVI.80.2.999-1014.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder CC, Earl PL, Long D, Abedon ST, Moss B, Doms RW. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc Natl Acad Sci USA. 1994;91:11699–11703. doi: 10.1073/pnas.91.24.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Derby NR, Kraft Z, Saunders CJ, Dai C, Llewellyn N, Zharkikh I, Vojtech L, Zhu T, Srivastava IK, Barnett SW, Stamatatos L. Viral evolution in macaques coinfected with CCR5- and CXCR4-tropic SHIVs in the presence or absence of vaccine-elicited anti-CCR5 SHIV neutralizing antibodies. Virology. 2006;355(2):138–51. doi: 10.1016/j.virol.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thorton GB, Parren PHI, Sawyer LSW, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ER, Bryson YJ, Cao Y, Moore JP, Ho DD, Barbas CFI. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center RJ, Kemp BE, Poumbourios P. Human Immunodeficiency virus type 1 and 2 envelope glycoproteins oligomerize through conserved sequences. J Virol. 1997;71:5706–5711. doi: 10.1128/jvi.71.7.5706-5711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center RJ, Lebowitz J, Leapman RD, Moss B. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J Virol. 2004;78(5):2265–76. doi: 10.1128/JVI.78.5.2265-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–41. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Gorny MK, Kessler JA, 2nd, Boots LJ, Ossorio-Castro M, Koenig S, Lineberger DW, Emini EA, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J Virol. 1994;68(11):6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby NR, Kraft Z, Kan E, Crooks ET, Barnett SW, Srivastava IK, Binley JM, Stamatatos L. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J Virol. 2006;80(17):8745–62. doi: 10.1128/JVI.00956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Broder CC, Doms RW, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Sugiura W, Montefiori DC, Broder CC, Lee SA, Wild C, Lifson J, Moss B. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J Virol. 2001;75(2):645–53. doi: 10.1128/JVI.75.2.645-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts TR, Binley JM, Trkola A, Robinson JE, Moore JP. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu J-Y, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu J-Y, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- Grundner C, Li Y, Louder M, Mascola J, Yang X, Sodroski J, Wyatt R. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology. 2005;331(1):33–46. doi: 10.1016/j.virol.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J Virol. 1994;68(1):400–10. doi: 10.1128/jvi.68.1.400-410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Qiao ZS, Montefiori DC, Haynes BF, Reinherz EL, Liao HX. Comparison of HIV Type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res Hum Retroviruses. 2005;21(1):58–67. doi: 10.1089/aid.2005.21.58. [DOI] [PubMed] [Google Scholar]
- Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–35. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Decker JM, Johnson RW, Bibollet-Ruche F, Wei X, Mulenga J, Allen S, Hunter E, Hahn BH, Shaw GM, Blackwell JL, Derdeyn CA. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol. 2006;80(11):5211–8. doi: 10.1128/JVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J Virol. 2005;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman B, Bolmstedt A, Schonning K, Bjorndal A, Westin C, Fenyo EM, Olofsson S. Protection of neutralization epitopes in the V3 loop of oligomeric human immunodeficiency virus type 1 glycoprotein 120 by N-linked oligosaccharides in the V1 region. AIDS Res Hum Retroviruses. 2001;17(11):1067–76. doi: 10.1089/088922201300343753. [DOI] [PubMed] [Google Scholar]
- Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2(12):1075–82. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Ly A, Stamatatos L. V2 loop Glycosylation of the human Immunodeficiency Virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J Virol. 2000;74:6769–6776. doi: 10.1128/jvi.74.15.6769-6776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenbaum SE, Yang D, Cavacini L, Posner M, Robinson J, Cheng-Mayer C. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J Virol. 2000;74(23):11008–16. doi: 10.1128/jvi.74.23.11008-11016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey RA, Saunders C, Hensel M, Stamatatos L. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J Virol. 2004;78(7):3279–95. doi: 10.1128/JVI.78.7.3279-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PW, Burton DR, Dimitrov DS. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci U S A. 2002;99(10):6913–8. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Rodriguez Y, Gazarian T, Rowley M, Majluf-Cruz A, Gazarian K. Collection of phage-peptide probes for HIV-1 immunodominant loop-epitope. J Microbiol Methods. 2007;68(2):225–35. doi: 10.1016/j.mimet.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–15. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poumbourios P, Wilson KA, Center RJ, El Ahmar W, Kemp BE. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic alpha-helical/leucine zipper-like sequence. J Virol. 1997;71(3):2041–9. doi: 10.1128/jvi.71.3.2041-2049.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CJ, McCaffrey RA, Zharkikh I, Kraft Z, Malenbaum SE, Burke B, Cheng-Mayer C, Stamatatos L. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J Virol. 2005;79(14):9069–80. doi: 10.1128/JVI.79.14.9069-9080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Stamatatos L, Kan E, Vajdy M, Lian Y, Hilt S, Martin L, Vita C, Zhu P, Roux KH, Vojtech L, D CM, Donnelly J, Ulmer JB, Barnett SW. Purification, Characterization, and Immunogenicity of a Soluble Trimeric Envelope Protein Containing a Partial Deletion of the V2 Loop Derived from SF162, an R5-Tropic Human Immunodeficiency Virus Type 1 Isolate. J Virol. 2003a;77(20):11244–11259. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, VanDorsten K, Vojtech L, Barnett SW, Stamatatos L. Changes in the Immunogenic Properties of Soluble gp140 Human Immunodeficiency Virus Envelope Constructs upon Partial Deletion of the Second Hypervariable Region. J Virol. 2003b;77(4):2310–20. doi: 10.1128/JVI.77.4.2310-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization resistant, clade B HIV-1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Lim M, Cheng-Mayer C. Generation and structural analysis of soluble oligomeric envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV-1 isolates. AIDS Res and Human Retroviruses. 2000;16:981–994. doi: 10.1089/08892220050058407. [DOI] [PubMed] [Google Scholar]
- Sullivan N, Thali M, Furman C, Ho DD, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67:3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Xu R, Srivastava IK, Kuller L, Zarkikh I, Kraft Z, Fagrouch Z, Letvin NL, Heeney JL, Barnett SW, Stamatatos L. Immunization with HIV-1 SF162-derived Envelope gp140 proteins does not protect macaques from heterologous simian-human immunodeficiency virus SHIV89.6P infection. Virology. 2006;349(2):276–89. doi: 10.1016/j.virol.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Yang X, Wyatt R, Sodroski J. Improved Elicitation of Neutralizing Antibodies against Primary Human Immunodeficiency Viruses by Soluble Stabilized Envelope Glycoprotein Trimers. J Virol. 2001;75(3):1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4(3):199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–8. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]