Abstract
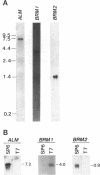
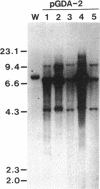
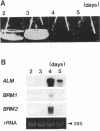
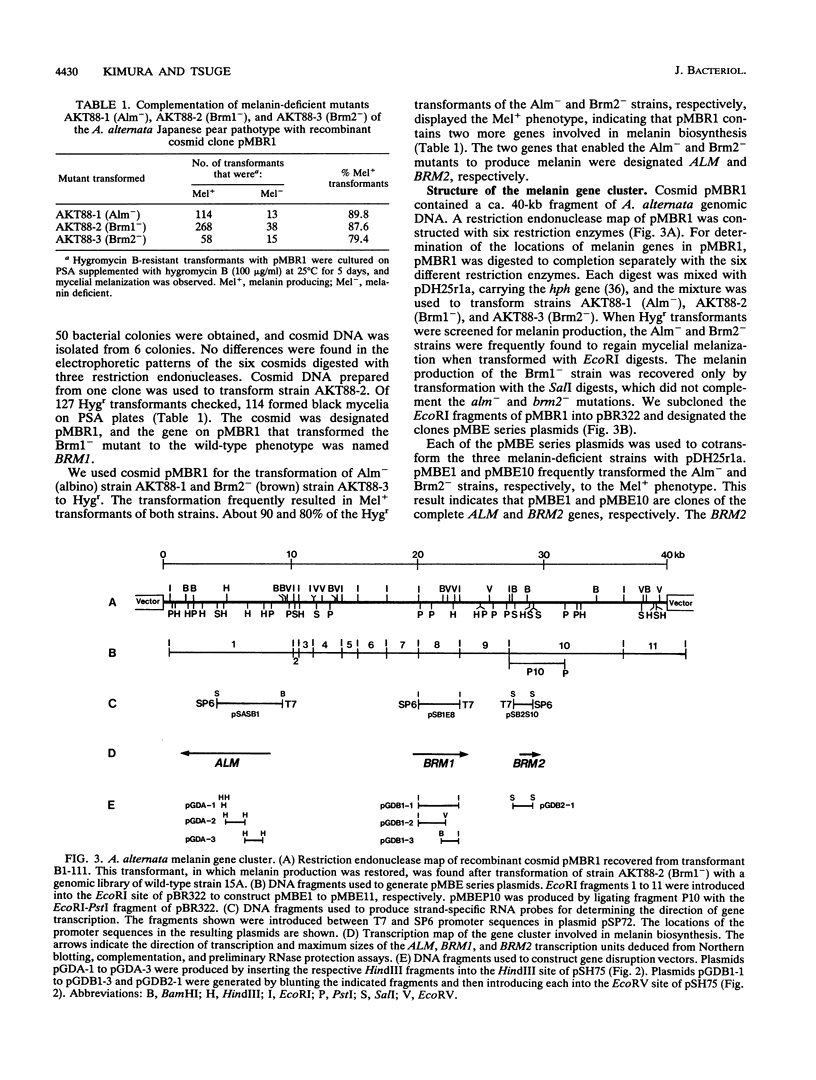
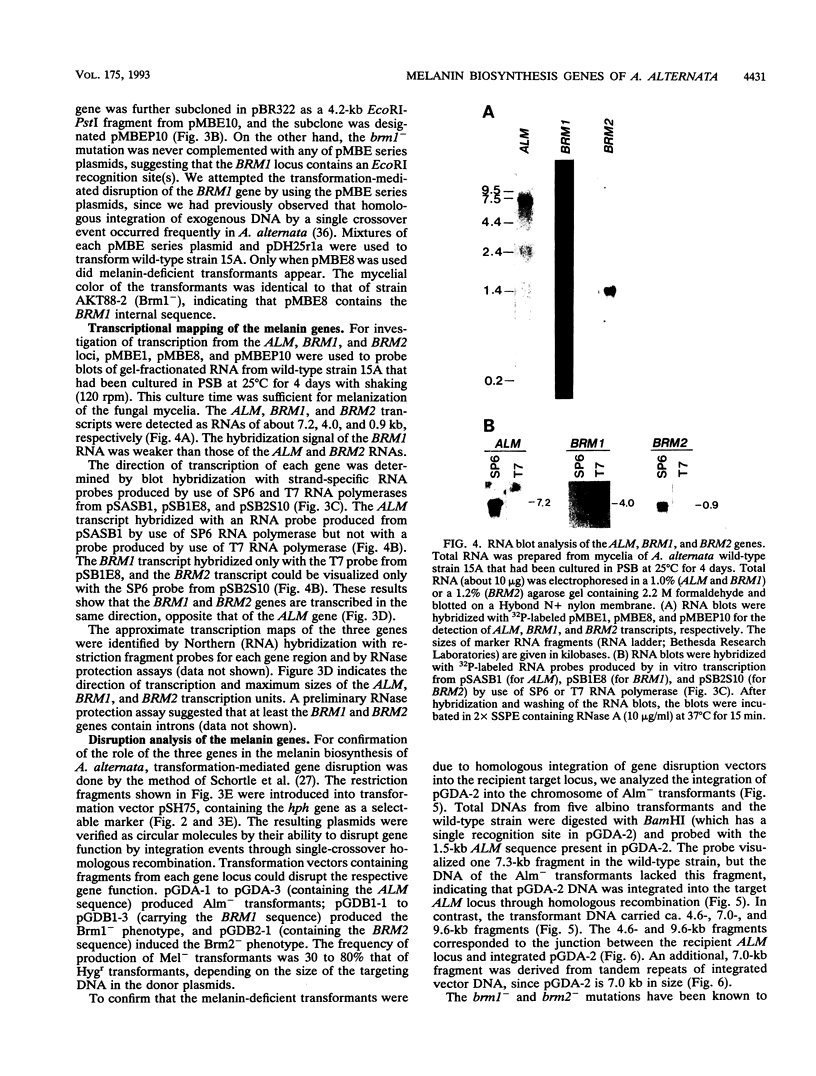
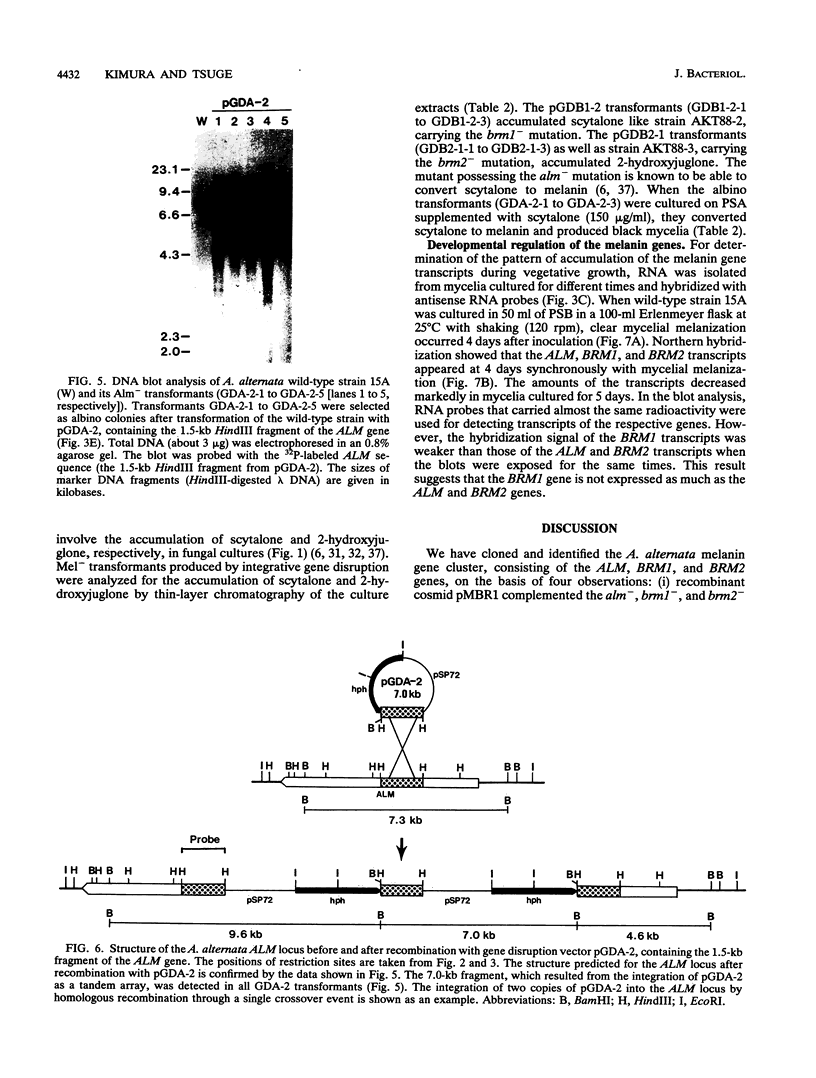
The filamentous fungus Alternaria alternata produces melanin, a black pigment, from acetate via 1,8-dihydroxynaphthalene. To isolate a fungal gene required for melanin biosynthesis, we transformed an A. alternata Brm1- (light brown) mutant with the DNA of a wild-type strain genomic library constructed by use of a cosmid carrying the hygromycin B phosphotransferase gene. When hygromycin B-resistant transformants were screened for melanin production, 1 of 1,363 transformants appeared to regain melanin production, as judged by black pigmentation of the cultured mycelia. The cosmid, named pMBR1, was recovered by packaging nuclear DNA of the melanin-producing transformant into lambda phage. The gene on pMBR1 that enables the Brm1- mutant to produce melanin was designated BRM1. In addition to the BRM1 gene, pMBR1 was found to carry two more genes involved in melanin biosynthesis. These two genes, designated ALM and BRM2, transformed A. alternata Alm- (albino) and Brm2- (brown) mutants, respectively, to the wild-type phenotype. The three genes are located within a ca. 30-kb genomic region in the order ALM-BRM1-BRM2. Analysis of the gene transcripts indicated approximate sizes of 7.2, 4.0, and 0.9 kb for ALM, BRM1, and BRM2, respectively. The BRM1 and BRM2 transcripts are generated from the same strand, but the ALM transcript is generated from the opposite strand. The three mRNA species accumulate in cultured mycelia of the wild-type strain synchronously with mycelial melanization. The essential roles of the three genes in melanin biosynthesis were confirmed by transformation-mediated gene disruption experiments.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arst H. N., Jr Regulation of gene expression in Aspergillus nidulans. Microbiol Sci. 1984 Sep;1(6):137–141. [PubMed] [Google Scholar]
- Ballance D. J. Sequences important for gene expression in filamentous fungi. Yeast. 1986 Dec;2(4):229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Beck J., Ripka S., Siegner A., Schiltz E., Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur J Biochem. 1990 Sep 11;192(2):487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- Bell A. A., Puhalla J. E., Tolmsoff W. J., Stipanovic R. D. Use of mutants to establish (+)-scytalone as an intermediate in melanin biosynthesis by Verticillium dahliae. Can J Microbiol. 1976 Jun;22(6):787–799. doi: 10.1139/m76-115. [DOI] [PubMed] [Google Scholar]
- Cullen D., Leong S. A., Wilson L. J., Henner D. J. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene. 1987;57(1):21–26. doi: 10.1016/0378-1119(87)90172-7. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fincham J. R. Transformation in fungi. Microbiol Rev. 1989 Mar;53(1):148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Baum J., Geever R., Huiet L., Patel V., Tyler B. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol Rev. 1985 Sep;49(3):338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritz L., Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983 Nov;25(2-3):179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Sherman D. H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney E. J., Hamer J. E., Roberti K. A., Yelton M. M., Timberlake W. E. Primary structure of the trpC gene from Aspergillus nidulans. Mol Gen Genet. 1985;199(1):37–45. doi: 10.1007/BF00327506. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambosek J., Leach J. Recombinant DNA in filamentous fungi: progress and prospects. Crit Rev Biotechnol. 1987;6(4):357–393. doi: 10.3109/07388558709089387. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Shortle D., Haber J. E., Botstein D. Lethal disruption of the yeast actin gene by integrative DNA transformation. Science. 1982 Jul 23;217(4557):371–373. doi: 10.1126/science.7046050. [DOI] [PubMed] [Google Scholar]
- Stipanovic R. D., Bell A. A. Pentaketide metabolites of Verticillium dahliae. II. Accumulation of naphthol derivatives by the aberrant-melanin mutant BRM-2. Mycologia. 1977 Jan-Feb;69(1):164–172. [PubMed] [Google Scholar]
- Tsuge T., Kobayashi H., Nishimura S. Organization of ribosomal RNA genes in Alternaria alternata Japanese pear pathotype, a host-selective AK-toxin-producing fungus. Curr Genet. 1989 Oct;16(4):267–272. doi: 10.1007/BF00422113. [DOI] [PubMed] [Google Scholar]
- Tsuge T., Nishimura S., Kobayashi H. Efficient integrative transformation of the phytopathogenic fungus Alternaria alternata mediated by the repetitive rDNA sequences. Gene. 1990 Jun 15;90(2):207–214. doi: 10.1016/0378-1119(90)90181-p. [DOI] [PubMed] [Google Scholar]
- Wheeler M. H., Bell A. A. Melanins and their importance in pathogenic fungi. Curr Top Med Mycol. 1988;2:338–387. doi: 10.1007/978-1-4612-3730-3_10. [DOI] [PubMed] [Google Scholar]
- Wiesner P., Beck J., Beck K. F., Ripka S., Müller G., Lücke S., Schweizer E. Isolation and sequence analysis of the fatty acid synthetase FAS2 gene from Penicillium patulum. Eur J Biochem. 1988 Oct 15;177(1):69–79. doi: 10.1111/j.1432-1033.1988.tb14346.x. [DOI] [PubMed] [Google Scholar]
- Yelton M. M., Timberlake W. E., Hondel C. A. A cosmid for selecting genes by complementation in Aspergillus nidulans: Selection of the developmentally regulated yA locus. Proc Natl Acad Sci U S A. 1985 Feb;82(3):834–838. doi: 10.1073/pnas.82.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]