Abstract
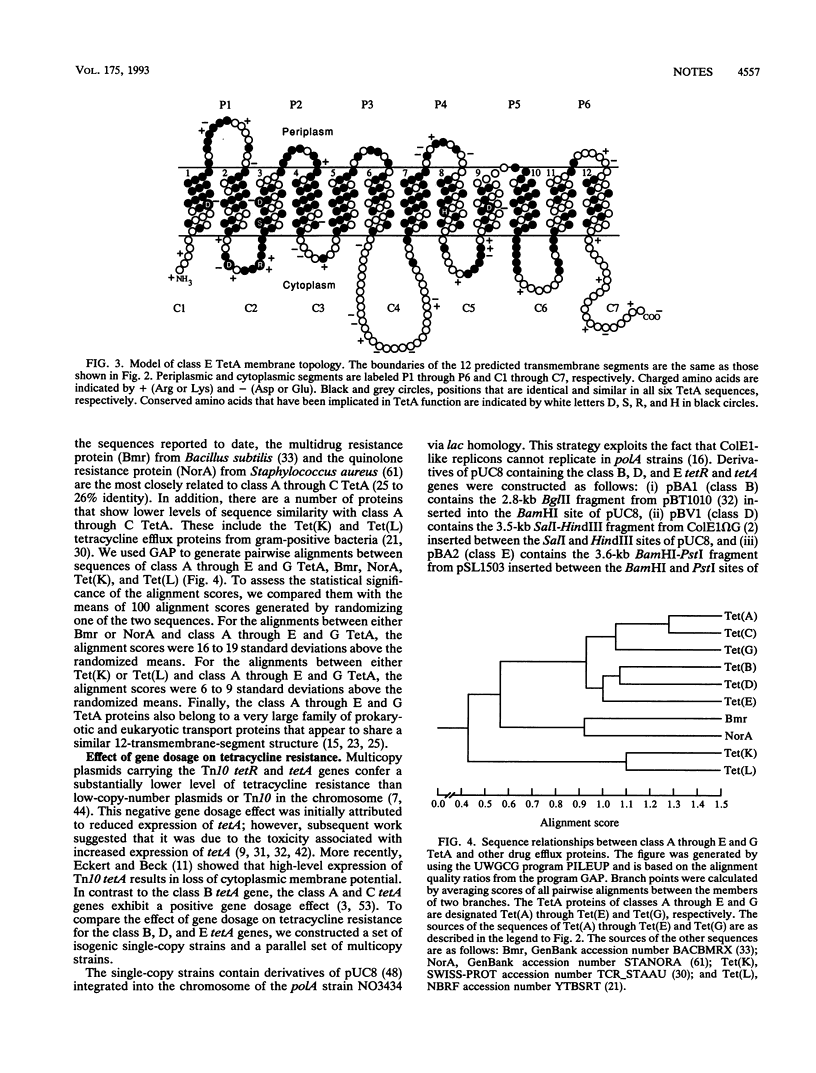
We determined the nucleotide sequence of the class E tetA gene on plasmid pSL1456 from Escherichia coli SLH1456A. The deduced amino acid sequence of the class E TetA protein shows 50 to 56% identity with the sequences of five related TetA proteins (classes A through D and G). Hydrophobicity profiles identify 12 putative transmembrane segments with similar boundaries in all six TetA sequences. The N-terminal alpha domain of the six sequences is more highly conserved than the C-terminal beta domain; the central hydrophilic loop connecting the alpha and beta domains is the least conserved region. Amino acid residues that have been shown to be important for class B (Tn10) TetA function are conserved in all six TetA sequences. Unlike the class B tetA gene, the class D and E tetA genes do not exhibit a negative gene dosage effect when present on multicopy plasmids derived from pACYC177.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allard J. D., Bertrand K. P. Membrane topology of the pBR322 tetracycline resistance protein. TetA-PhoA gene fusions and implications for the mechanism of TetA membrane insertion. J Biol Chem. 1992 Sep 5;267(25):17809–17819. [PubMed] [Google Scholar]
- Allard J. D., Gibson M. L., Vu L. H., Nguyen T. T., Bertrand K. P. Nucleotide sequence of class D tetracycline resistance genes from Salmonella ordonez. Mol Gen Genet. 1993 Feb;237(1-2):301–305. doi: 10.1007/BF00282811. [DOI] [PubMed] [Google Scholar]
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Hawkey P. M., Hinton M. Tetracyclines, molecular and clinical aspects. J Antimicrob Chemother. 1992 Mar;29(3):245–277. doi: 10.1093/jac/29.3.245. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli: specific complementation of argininosuccinate lyase (argH) mutations. J Mol Biol. 1978 Apr 25;120(4):517–532. doi: 10.1016/0022-2836(78)90351-0. [DOI] [PubMed] [Google Scholar]
- Coleman D. C., Foster T. J. Analysis of the reduction in expression of tetracycline resistance determined by transposon Tn10 in the multicopy state. Mol Gen Genet. 1981;182(1):171–177. doi: 10.1007/BF00422786. [DOI] [PubMed] [Google Scholar]
- Curiale M. S., McMurry L. M., Levy S. B. Intracistronic complementation of the tetracycline resistance membrane protein of Tn10. J Bacteriol. 1984 Jan;157(1):211–217. doi: 10.1128/jb.157.1.211-217.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. W., Bertrand K. P. Promoter mutations affecting divergent transcription in the Tn10 tetracycline resistance determinant. J Mol Biol. 1985 Aug 20;184(4):599–610. doi: 10.1016/0022-2836(85)90306-7. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B., Beck C. F. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J Bacteriol. 1989 Jun;171(6):3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B., Beck C. F. Topology of the transposon Tn10-encoded tetracycline resistance protein within the inner membrane of Escherichia coli. J Biol Chem. 1989 Jul 15;264(20):11663–11670. [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Griffith J. K., Baker M. E., Rouch D. A., Page M. G., Skurray R. A., Paulsen I. T., Chater K. F., Baldwin S. A., Henderson P. J. Membrane transport proteins: implications of sequence comparisons. Curr Opin Cell Biol. 1992 Aug;4(4):684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- Gutterson N. I., Koshland D. E., Jr Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986 Nov;5(11):3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P. J., Maiden M. C. Homologous sugar transport proteins in Escherichia coli and their relatives in both prokaryotes and eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):391–410. doi: 10.1098/rstb.1990.0020. [DOI] [PubMed] [Google Scholar]
- Hickman R. K., Levy S. B. Evidence that TET protein functions as a multimer in the inner membrane of Escherichia coli. J Bacteriol. 1988 Apr;170(4):1715–1720. doi: 10.1128/jb.170.4.1715-1720.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen W., Schollmeier K., Gatz C. Control of expression of the Tn10-encoded tetracycline resistance operon. II. Interaction of RNA polymerase and TET repressor with the tet operon regulatory region. J Mol Biol. 1984 Jan 15;172(2):185–201. doi: 10.1016/s0022-2836(84)80037-6. [DOI] [PubMed] [Google Scholar]
- Hillen W., Schollmeier K. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 1983 Jan 25;11(2):525–539. doi: 10.1093/nar/11.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Ikeda T., Tomizuka N., Furukawa K. Nucleotide sequence of the tetracycline resistance gene of pTHT15, a thermophilic Bacillus plasmid: comparison with staphylococcal TcR controls. Gene. 1985;37(1-3):131–138. doi: 10.1016/0378-1119(85)90265-3. [DOI] [PubMed] [Google Scholar]
- Levy S. B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992 Apr;36(4):695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B., McMurry L. M., Burdett V., Courvalin P., Hillen W., Roberts M. C., Taylor D. E. Nomenclature for tetracycline resistance determinants. Antimicrob Agents Chemother. 1989 Aug;33(8):1373–1374. doi: 10.1128/aac.33.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger M. D., Saier M. H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993 Jan;18(1):13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Marshall B., Morrissey S., Flynn P., Levy S. B. A new tetracycline-resistance determinant, class E, isolated from Enterobacteriaceae. Gene. 1986;50(1-3):111–117. doi: 10.1016/0378-1119(86)90315-x. [DOI] [PubMed] [Google Scholar]
- McMurry L. M., Stephan M., Levy S. B. Decreased function of the class B tetracycline efflux protein Tet with mutations at aspartate 15, a putative intramembrane residue. J Bacteriol. 1992 Oct;174(19):6294–6297. doi: 10.1128/jb.174.19.6294-6297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez B., Tachibana C., Levy S. B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980 Mar;3(2):99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- Mojumdar M., Khan S. A. Characterization of the tetracycline resistance gene of plasmid pT181 of Staphylococcus aureus. J Bacteriol. 1988 Dec;170(12):5522–5528. doi: 10.1128/jb.170.12.5522-5528.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed H. S., Bertrand K. P. Mutations in multicopy Tn10 tet plasmids that confer resistance to inhibitory effects of inducers of tet gene expression. J Bacteriol. 1983 Aug;155(2):557–564. doi: 10.1128/jb.155.2.557-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed H. S., Nguyen T. T., Bertrand K. P. Multicopy Tn10 tet plasmids confer sensitivity to induction of tet gene expression. J Bacteriol. 1983 Aug;155(2):549–556. doi: 10.1128/jb.155.2.549-556.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyfakh A. A., Bidnenko V. E., Chen L. B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. T., Postle K., Bertrand K. P. Sequence homology between the tetracycline-resistance determinants of Tn10 and pBR322. Gene. 1983 Nov;25(1):83–92. doi: 10.1016/0378-1119(83)90170-1. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H., Watanabe K., Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992 Mar;56(1):229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983 May-Jun;22(2-3):277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- Postle K., Nguyen T. T., Bertrand K. P. Nucleotide sequence of the repressor gene of the TN10 tetracycline resistance determinant. Nucleic Acids Res. 1984 Jun 25;12(12):4849–4863. doi: 10.1093/nar/12.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R. A., Levy S. B., Heinrikson R. L., Kézdy F. J. Gene duplication in the evolution of the two complementing domains of gram-negative bacterial tetracycline efflux proteins. Gene. 1990 Mar 1;87(1):7–13. doi: 10.1016/0378-1119(90)90489-e. [DOI] [PubMed] [Google Scholar]
- Rubin R. A., Levy S. B. Tet protein domains interact productively to mediate tetracycline resistance when present on separate polypeptides. J Bacteriol. 1991 Jul;173(14):4503–4509. doi: 10.1128/jb.173.14.4503-4509.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers A. A., Speer B. S., Shoemaker N. B. New perspectives in tetracycline resistance. Mol Microbiol. 1990 Jan;4(1):151–156. doi: 10.1111/j.1365-2958.1990.tb02025.x. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Chang C. H., Stevens F. J. The functions of tryptophan residues in membrane proteins. Protein Eng. 1992 Apr;5(3):213–214. doi: 10.1093/protein/5.3.213. [DOI] [PubMed] [Google Scholar]
- Smith L. D., Bertrand K. P. Mutations in the Tn10 tet repressor that interfere with induction. Location of the tetracycline-binding domain. J Mol Biol. 1988 Oct 20;203(4):949–959. doi: 10.1016/0022-2836(88)90120-9. [DOI] [PubMed] [Google Scholar]
- Sørum H., Roberts M. C., Crosa J. H. Identification and cloning of a tetracycline resistance gene from the fish pathogen Vibrio salmonicida. Antimicrob Agents Chemother. 1992 Mar;36(3):611–615. doi: 10.1128/aac.36.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. P., Greenberg J., Rownd R. H. Generation of miniplasmids from copy number mutants of the R plasmid NR1. J Bacteriol. 1977 Dec;132(3):986–995. doi: 10.1128/jb.132.3.986-995.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar K., Ernst A., Hillen W. Identification and nucleotide sequence of the class E tet regulatory elements and operator and inducer binding of the encoded purified Tet repressor. Mol Gen Genet. 1988 Dec;215(1):76–80. doi: 10.1007/BF00331306. [DOI] [PubMed] [Google Scholar]
- Unger B., Becker J., Hillen W. Nucleotide sequence of the gene, protein purification and characterization of the pSC101-encoded tetracycline resistance-gene-repressor. Gene. 1984 Nov;31(1-3):103–108. doi: 10.1016/0378-1119(84)90199-9. [DOI] [PubMed] [Google Scholar]
- Unger B., Klock G., Hillen W. Nucleotide sequence of the repressor gene of the RA1 tetracycline resistance determinant: structural and functional comparison with three related Tet repressor genes. Nucleic Acids Res. 1984 Oct 25;12(20):7693–7703. doi: 10.1093/nar/12.20.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wada K., Wada Y., Doi H., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1981–1986. doi: 10.1093/nar/19.suppl.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters S. H., Rogowsky P., Grinsted J., Altenbuchner J., Schmitt R. The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res. 1983 Sep 10;11(17):6089–6105. doi: 10.1093/nar/11.17.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K., Schraml S., Shales S. W., Schmitt R. Tetracycline resistance transposon Tn1721: recA-dependent gene amplification and expression of tetracycline resistance. J Bacteriol. 1981 Sep;147(3):851–859. doi: 10.1128/jb.147.3.851-859.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Adachi K., Sawai T. Orientation of the carboxyl terminus of the transposon Tn10-encoded tetracycline resistance protein in Escherichia coli. FEBS Lett. 1990 Jun 4;265(1-2):17–19. doi: 10.1016/0014-5793(90)80872-g. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Akasaka T., Ono N., Someya Y., Nakatani M., Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. Roles of the aspartyl residues located in the putative transmembrane helices. J Biol Chem. 1992 Apr 15;267(11):7490–7498. [PubMed] [Google Scholar]
- Yamaguchi A., Ono N., Akasaka T., Noumi T., Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon, Tn10. The role of the conserved dipeptide, Ser65-Asp66, in tetracycline transport. J Biol Chem. 1990 Sep 15;265(26):15525–15530. [PubMed] [Google Scholar]
- Yamaguchi A., Ono N., Akasaka T., Sawai T. Serine residues responsible for tetracycline transport are on a vertical stripe including Asp-84 on one side of transmembrane helix 3 in transposon Tn10-encoded tetracycline/H+ antiporter of Escherichia coli. FEBS Lett. 1992 Jul 28;307(2):229–232. doi: 10.1016/0014-5793(92)80773-a. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Someya Y., Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. The role of a conserved sequence motif, GXXXXRXGRR, in a putative cytoplasmic loop between helices 2 and 3. J Biol Chem. 1992 Sep 25;267(27):19155–19162. [PubMed] [Google Scholar]
- Yamaguchi A., Udagawa T., Sawai T. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J Biol Chem. 1990 Mar 25;265(9):4809–4813. [PubMed] [Google Scholar]
- Yoshida H., Bogaki M., Nakamura S., Ubukata K., Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990 Dec;172(12):6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Aoki T. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol Immunol. 1992;36(10):1051–1060. doi: 10.1111/j.1348-0421.1992.tb02109.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992 May 20;225(2):487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]




