Abstract
Thioredoxin reductase, lipoamide dehydrogenase, and glutathione reductase are members of the pyridine nucleotide–disulfide oxidoreductase family of dimeric flavoenzymes. The mechanisms and structures of lipoamide dehydrogenase and glutathione reductase are alike irrespective of the source (subunit Mr ≈55,000). Although the mechanism and structure of thioredoxin reductase from Escherichia coli are distinct (Mr ≈35,000), this enzyme must be placed in the same family because there are significant amino acid sequence similarities with the other two enzymes, the presence of a redox-active disulfide, and the substrate specificities. Thioredoxin reductase from higher eukaryotes on the other hand has a Mr of ≈55,000 [Luthman, M. & Holmgren, A. (1982) Biochemistry 21, 6628–6633; Gasdaska, P. Y., Gasdaska, J. R., Cochran, S. & Powis, G. (1995) FEBS Lett 373, 5–9; Gladyshev, V. N., Jeang, K. T. & Stadtman, T.C. (1996) Proc. Natl. Acad. Sci. USA 93, 6146–6151]. Thus, the evolution of this family is highly unusual. The mechanism of thioredoxin reductase from higher eukaryotes is not known. As reported here, thioredoxin reductase from human placenta reacts with only a single molecule of NADPH, which leads to a stable intermediate similar to that observed in titrations of lipoamide dehydrogenase or glutathione reductase. Titration of thioredoxin reductase from human placenta with dithionite takes place in two spectral phases: formation of a thiolate–flavin charge transfer complex followed by reduction of the flavin, just as with lipoamide dehydrogenase or glutathione reductase. The first phase requires more than one equivalent of dithionite. This suggests that the penultimate seleno-cysteine [Tamura, T. & Stadtman, T.C. (1996) Proc. Natl. Acad. Sci. USA 93, 1006–1011] is in redox communication with the active site disulfide/dithiol. Nitrosoureas of the carmustine type inhibit only the NADPH reduced form of human thioredoxin reductase. These compounds are widely used as cytostatic agents, so this enzyme should be studied as a target in cancer chemotherapy. In conclusion, three lines of evidence indicate that the mechanism of human thioredoxin reductase is like the mechanisms of lipoamide dehydrogenase and glutathione reductase and differs fundamentally from the mechanism of E. coli thioredoxin reductase.
Thioredoxin reductases (TrxR) are homodimeric flavoenzymes occurring, or implied by gene sequence to occur, in a wide variety of organisms—prokaryotes, eukaryotes, and the archaeon Methanococcus jannaschii (1–2). The three-dimensional structure of TrxR from Escherichia coli (eTrxR) is known, and its mechanism has been studied extensively (3–4). On the basis of significant amino acid sequence similarities and the presence of a redox-active disulfide and the substrate specificities (pyridine nucleotides and disulfide compounds), eTrxR has been placed in the pyridine nucleotide–disulfide oxidoreductase family that includes lipoamide dehydrogenase (LipDH) and glutathione reductase (GR) (5–6). The three-dimensional structures of the latter enzymes are known (7–9). Their mechanisms have been studied extensively and have been found to be very similar to each other over a wide range of organisms. Their structures and mechanisms are rather different from those of eTrxR. In particular, the subunit Mr are 35,000 for eTrxR and 55,000 for LipDH and GR (6).
TrxR isolated from various mammalian tissues were found to have a subunit Mr of ≈55,000 (10). Moreover, complete DNA sequences have been published for TrxR from humans (1, 11) and Plasmodium falciparum (ref. 12 and references therein). The deduced amino acid sequences of these high Mr TrxR are much more closely related to the sequences of GR from humans (35% identity) or E. coli (40% identity) than to the sequence of eTrxR (24% identity) (1, 13). Low Mr TrxR have been found not only in E. coli but also in the anaerobe Eubacterium acidoaminophilum, the archaeon Methanococcus jannaschii, and the facultative phototroph Rhodobacter sphaeroides Y, as well as a number of fungi and plants (1, 2, 14–17, and references in ref. 1). In addition, a TrxR-like enzyme has been isolated from Salmonella typhimurium that, together with AhpC, acts as an alkyl hydroperoxide reductase. AhpC is a 22-kDa protein that, like Trx, has a redox-active disulfide/dithiol (18, 19). A reductase having similar activity has been isolated from Amphibacillus xylanus (20). These reductases are homologous with low Mr TrxR (19). On the basis of the presently available database, high and low Mr TrxR are mutually exclusive in the same organism. It is, however, premature to assign the two different forms of TrxR to individual branching points of the phylogenetic tree.
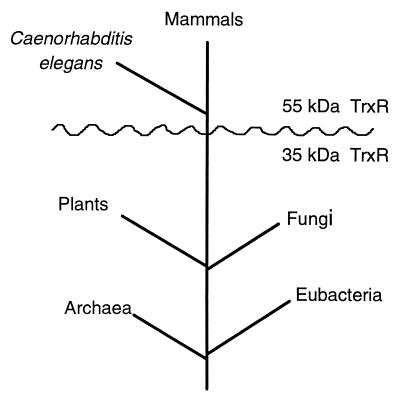
In any case, the evolution of TrxR has followed at least two courses (Fig. 1). One pathway can be explained by the continuous Apollonian divergent evolution of the ancestor of LipDH, GR, and high Mr TrxR. In contrast, for the development of low Mr TrxR, the same modules, i.e., the NADPH-binding and the FAD-binding domains, as in the high Mr counterpart are used, but the active site and subunit interface exhibit a completely different design. In addition, the catalytic activity is accompanied by domain movements in low Mr TrxR (3). Thus, the evolution of the TrxR is characterized by radical Dionysian divergence from an ancestral structure followed by convergent evolution to a common function (Fig. 1). It will be interesting to study which selecting forces might have governed this unique development of protein function. Two courses also have been followed, for example, in the evolution of methionine synthetase, but in this case, E. coli contains both cobalamin-dependent and -independent forms of methionine synthetase (ref. 21 and C. W. Goulding and R. G. Matthews, personal communication).
Figure 1.
Phylogenetic tree. The distances between branch points are not intended to reflect time quantitatively. A gene sequence from the worm (Caenorhabditis elegans) is homologous with the sequence of hTrxR (1).
All members of the pyridine nucleotide–disulfide oxidoreductase family have a redox-active disulfide/dithiol adjacent to the FAD, on the re side of the flavin ring in the low Mr TrxR, and on the si side in the rest of the family. A fascinating aspect of at least some high Mr TrxR is that their sequence includes a cysteine–selenocysteine (Cys–SeCys) dipeptide near the carboxyl terminus (11, 22).
The mechanism of high Mr TrxR from eukaryotes has been investigated only in preliminary studies (10, 23). The long wavelength band observed on reduction of LipDH and GR by substrate also was seen when rat liver TrxR was reduced (10), but this telling detail was not discussed. The long wavelength band is indicative of a thiolate–flavin charge transfer complex as a stable intermediate in the two electron-reduced enzyme (EH2) (24). It is stable because the redox potential of the Eox–EH2 couple is high relative to that of the EH2–EH4 couple (25). In contrast in eTrxR, the potentials are approximately the same, and the enzyme is reduced by 2 mol of substrate without accumulation of a unique intermediate (26–27). In the present study, we examined the redox properties of hTrxR and found that its mechanism is like that of LipDH and GR and is quite easily distinguished from the mechanism of eTrxR.
MATERIALS AND METHODS
TrxR from Human Placenta (hTrxR).
hTrxR was purified according to Oblong et al. (23) with modifications (S.G., K.B., and H.S., unpublished work). As a final step, the enzyme was eluted from a 2′5′-ADP–Sepharose column using 0.7 mM NADP+ in 50 mM Tris·HCl/1 mM EDTA buffer of pH 7.6; NADP+ was subsequently removed by washing with this buffer without NADP+ in a Centriprep 30 (Amicon).
Isolated TrxR (≈1.0 mg from one placenta of 550 g) had a specific activity of 35 units/mg when tested in the 5,5′-dithiobis(2-nitrobenzoate) reduction assay (23, 28). Enzyme concentration was estimated by measuring the A463 nm and applying an assumed extinction coefficient of 11.3 mM−1·cm−1 per subunit of Eox. As revealed by SDS/PAGE analysis, the preparation was >95% pure and had—as an apoprotein—an apparent subunit Mr of ≈57,000, which corresponds to a value of 58,000 for the FAD-containing subunit. Disulfide reductases tend to exhibit a 5–10% higher Mr in SDS/PAGE analyses than in amino acid sequence analyses, so we recommend using a Mr value of 55,000 (1, 11).
The selenium content as determined by atomic absorption spectroscopy was 0.93 ± 0.02 mol/mol subunit. 0.98 ± 0.06 mol FAD/mol TrxR subunit was found using the apoGR complementation test (29). An enzyme of 35 units/mg was used in the dithionite titration experiment (see below). A slightly less pure preparation (32 units/mg) was used to measure the presteady state kinetics (see below).
Equilibrium Titration with Dithionite.
Approximately 17 μM of enzyme in 1 ml of 50 mM potassium phosphate/2 mM EDTA, pH 7.4, containing 0.1 equivalents relative to enzyme-bound FAD (eq) methyl viologen as a carrier of reducing equivalents was degassed by eight cycles of vacuum and pure nitrogen gas and titrated with a solution of anaerobic dithionite, which had been standardized by titrating lumiflavin-3-acetic acid (30). Spectra were recorded at 25°C using a Perkin–Elmer spectrophotometer. At 3 points in the first phase, the titration was suspended to allow thorough equilibration of reducing equivalents between all redox-active components of the enzyme.
Presteady State Reaction Kinetics with NADPH.
The rapid reaction spectrophotometer as well as the methods of data analysis have been described by Rietveld et al. (31). At the beginning of this experiment, we were not aware of possible interference from NADP+ left over from the purification of the enzyme. We estimated the amount spectrally to be <2 eq NADP+/subunit. If the Kd was very low, as we suspected, then the effect may be significant. We chose not to remove it because it appeared to stabilize the enzyme. Thus, 16 μM of degassed enzyme (in 50 mM potassium phosphate/2 mM EDTA, pH 7.4) containing ≈2 eq of NADP+ in one syringe was rapidly mixed at 4°C with degassed NADPH (in the same buffer) in the second syringe. Because of the small amount of enzyme available, each concentration of NADPH (1, 2, or 5 eq, relative to enzyme-bound FAD) was mixed with enzyme only four times. The kinetics were followed at A463 nm for two of the mixings and at A540 nm for the other two mixings using the photomultiplier detector. Spectra were obtained in all four mixings using the diode array detector. The enzyme also was mixed twice with buffer to establish the starting point of the enzyme absorbance.
Modification by Carmustine [1,3-bis(2-chloroethyl)-1-nitrosourea] (BCNU).
When working with the cytostatic agent carmustine, the precautions described by Becker and Schirmer (32) were taken. BCNU, a kind gift of Professor Eisenbrand, German Cancer Research Center, Heidelberg, was dissolved in ethanol; a 40-mM solution was found to be stable at −20°C for at least 2 weeks. hTrxR (1.6 units/ml; corresponding to 1 nmol subunit/ml) was incubated at 37°C in the assay buffer (100 mM potassium phosphate/2 mM EDTA, pH 7.4) with various concentrations of BCNU in the presence or absence of 100 μM of NADPH. Controls contained an equal volume of ethanol instead of BCNU. At given time points, aliquots were taken, diluted 100-fold, and assayed for residual activity.
RESULTS AND DISCUSSION
The feature in the mechanisms of GR and LipDH that distinguishes them from the mechanism of eTrxR is the formation of a distinct EH2 species that predominates at equilibrium in titrations with dithionite or NAD(P)H in the first two enzymes but not in eTrxR (25, 33–34). This EH2-species has long wavelength absorbance centered around 540 nm, beyond the region in which Eox and EH4 absorb, that is due to a thiolate–flavin charge transfer complex (24). It is our hypothesis that hTrxR has a mechanism similar to that of GR and distinct from that of eTrxR. The basis of this hypothesis, as discussed in the Introduction, is the similarity in molecular weight and the sequence homology between hTrxR and GR. Therefore, hTrxR should behave in reductive titrations as does GR, forming the thiolate–flavin charge transfer complex as a distinct EH2 intermediate. This mechanism implies reaction with only a single molecule of NADPH in each catalytic cycle. Moreover, in reaction with a strong reductant like dithionite, there will be two spectral phases, manifested by formation and disappearance of a long wavelength band at 540 nm. No such band forms in reductions of eTrxR where Eox is converted to EH4 by two molecules of NADPH via E(FADH−)(S2) and E(FAD)(SH)2, neither of which has 540-nm absorbance. It is not clear why E(FAD)(SH)2 has no thiolate–flavin charge transfer absorbance, but the juxtaposition of the thiolate donor to the flavin ring acceptor appears to be unfavorable (26).
Equilibrium Titration with Dithionite.
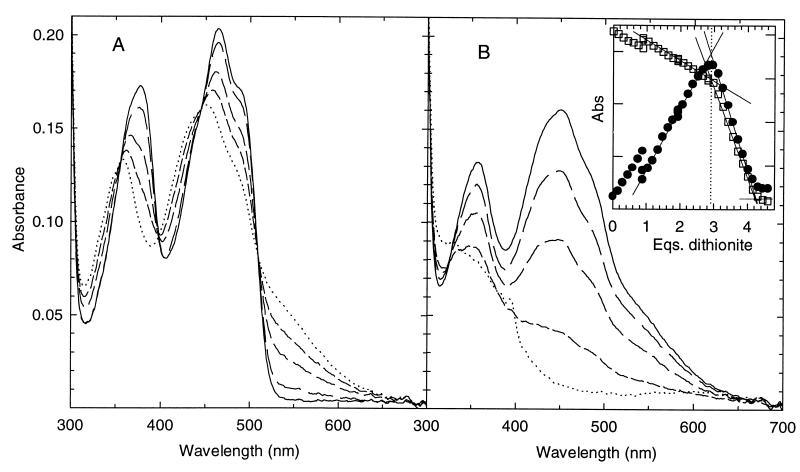
The spectrum of hTrxR shown in Fig. 2 is similar to that of rat liver TrxR (10). Fig. 2 shows the two distinct phases in the titration of hTrxR with dithionite. The clear set of four isosbestics (A) indicates conversion of one absorbing species to another, Eox and EH2. Formation of the band at 540 nm due to the thiolate–flavin charge transfer complex is complete in this phase, and the final spectrum is typical of these complexes in other enzymes such as GR and LipDH. Absorbance at 540 nm disappears as the titration continues and the flavin is reduced (B). In the final few spectra, peaks appearing at 600, 395, and 385 nm are due to accumulation of the methyl viologen radical; methyl viologen was added as an electron carrier because the direct reaction of dithionite with the enzyme is very slow. Absence of increasing absorbance at 340 nm shows that NADP+ was absent in this enzyme preparation.
Figure 2.
Titration of oxidized hTrxR with sodium dithionite. (A) Representative spectra showing the emergence of the EH2 species during the first phase of titration. Spectra are in order from solid line to dotted: 18 μM of oxidized enzyme before addition of dithionite, then 0.9, 1.5, 2.1, and 2.7 eq. (B) Representative spectra showing the emergence of the EH4 species during the second phase of titration. Spectra start with the solid line; equivalents of dithionite added are: 2.8, 3.2, 3.5, 4.0, and 4.4. Note the 0.1 equivalent of methyl viologen radical in the last spectrum with the two major absorbance bands, 394 and 600 nm. (Inset) Observed absorbance changes upon titration of hTrxR with sodium dithionite. Conditions are as in Materials and Methods. Note the equivalent points of 0.9, 1.9, and 2.8 where several data points are shown representing longer equilibration times of 50, 50, and 40 minutes, respectively. For all other data points, spectra were recorded after 3–7 min of equilibration time. A540 nm (closed circles) = 0.02 and A463 nm (open squares) = 0.05 for each marking on the left and right y axes, respectively.
Plots of absorbance at 463 and 540 nm as a function of the added dithionite show that the first phase requires between two and three times the amount required for the second phase (Fig. 2B, Inset). The absorbance at 540 nm rises and then falls whereas that at 463 nm falls and then decreases more steeply. The transition points are clear, and the end points are fairly distinct despite absorbance by the methyl viologen radical. Three to four two-electron equivalents of dithionite are required for the complete titration, and two to three of these are required in the phase in which the 540-nm absorbance is rising. This indicates that at least two redox-active species are reacting in this phase.
It was known from a preliminary experiment with enzyme of lower specific activity (32 units/mg) that the increase at 540 nm after each addition of dithionite was initially larger but relaxed to the equilibrium values shown here. This pattern of hTrxR reduction is very reminiscent of titration studies on mercuric ion reductase. This closely related enzyme has three redox-active groups capable of interaction with each other, namely FAD and two redox-active Cys pairs (35). Thus, in the titration of both mercuric ion reductase and hTrxR, the initial absorbance at 540 nm is high after each addition of dithionite and relaxes to an equilibrium value as electrons distribute themselves among the redox-active groups. Furthermore, 2 eq of dithionite are consumed as the 540-nm absorbance forms, and 1 eq reacts as the flavin is reduced (35). In titrations of this kind, the group having the least negative redox potential remains reduced when, after each addition of dithionite, equilibrium is approached. The flow of electrons in mercuric ion reductase is presumed to be from the flavin to the adjacent disulfide and from the nascent dithiol to the distal disulfide because this has the highest redox potential. We suggest that the excess electrons taken up by hTrxR react with the SeCys oxidized either by reaction with the nearby Cys or by conversion to CysSeOH. The enzyme reoxidizes upon admission of air in two phases: rapidly to EH2 and more slowly to Eox. Similar kinetics of reoxidation are observed with mercuric reductase and GR.
Presteady State Reaction Kinetics with NADPH.
The reduction of GR takes place in three phases: (i) binding of NADPH with formation of an NADPH–FAD charge transfer complex, (ii) reduction of FAD with formation of an FADH−–NADP+ charge transfer complex, and (iii) reduction of the disulfide by the reduced flavin with formation of the thiolate–flavin charge transfer complex while NADP+ is still bound to the enzyme (31, 36). In the presence of excess NADPH, NADP+ is displaced. The NADPH–FAD charge transfer complex absorbs maximally at 580 nm, and its formation is largely complete in the 4-ms dead time of the rapid reaction mixer (stopped-flow). Flavin reduction is fast and best observed at 680 nm as the FADH−–NADP+ charge transfer complex forms (ref. 31; Fig. 3, Inset, curve 3). Flavin reoxidation and formation of the thiolate–FAD charge transfer complex is also a fast process as it is observed at 540 nm. We argued in a study on GR (31) that flavin reoxidation is rapid enough to largely obscure flavin reduction at 460 nm because one process causes a decrease and the other an increase in absorbance.
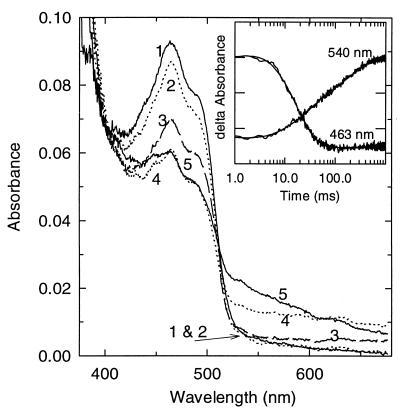
Figure 3.
Rapid reaction of hTrxR with NADPH. Anaerobic reduction of 8 μM of enzyme with 5 eq of NADPH (after mixing). Times in milliseconds for each spectrum are as follows: 1, oxidized; 2, 9; 3, 31; 4, 63; and 5, 415. (Inset) Rapid reaction kinetic traces observed at 463 and 540 nm for the reduction of hTrxR by 5 eq of NADPH. Observed traces with fits to a sum of three exponentials. A540 nm = 0.06–0.10 (0.01 for each marking) on the left axis and A463 nm = 0–0.04 (0.01 for each marking) on the right y axis.
Fig. 3 shows the spectra recorded during the time course of the experiment with 5 eq of NADPH. Spectrum 1 is that of Eox. The pattern appears much like that seen in the reduction of GR with only one molecule of NADPH reacting with the enzyme (cf. figure 3 of ref. 31). The pattern is completely different from that seen with eTrxR, in which two molecules of NADPH react (ref. 37 and B. W. Lennon and C. H. Williams, unpublished work). Spectrum 5 is predominantly that of the thiolate–flavin charge transfer complex (24). Please note, from the early work of Massey et al. (38) on LipDH, that the main peak of the thiolate–flavin charge transfer complex was distinctly blue-shifted relative to Eox. This also applies to GR (33–34). In spectrum 4, the A680 nm is higher than in spectrum 5, which indicates the presence of the FADH−–NADP+ charge transfer complex (39). This species is not favored under the conditions shown because the NADP+ is displaced by excess NADPH. Between 55 and 400 ms, the absorbance in the 460-nm region did not decrease further than shown in the spectrum. Thiolate–flavin charge transfer complex formation is observed at 540 nm between 50 and 400 ms as a slower increase in the “triangular” shape characteristic of that species. Thiolate–FAD charge transfer complex formation can also be inferred from 50 to 400 ms as a small increase in absorbance at 430–445 nm although the changes in this region are thought to be primarily associated with the displacement of NADP+ by NADPH.
There are two minor differences between this experiment and the one with GR (cf. figure 3 of ref. 31). First, a small amount of NADP+ (≈2 eq from spectral measurements; see Materials and Methods) was present in the starting sample. The NADP+ must dissociate before the NADPH can bind, so the early spectra (2 and 3) differ from those in Rietveld et al. (31). Second, the spectra shown here are for 5 eq of NADPH as opposed to the 1 eq used in the experiment with GR (figure 3 of ref. 31). Excess NADPH is required because hTrxR is more difficult to reduce than GR; this is consistent with results of Tamura and Stadtman (11). The excess NADPH displaces the bound NADP+ so that less 680-nm absorbance is observed (curve 5) than in the experiment with GR. A further potential complication is the Cys–SeCys ensemble, which appears to be reduced by the active center dithiol.
Turning now to the kinetics at single wavelengths, recall (Materials and Methods) that only duplicate experiments could be carried out with each concentration of NADPH observing at 463 nm and similarly at 540 nm. Because of the low enzyme concentration, the signal-to-noise ratio is low, and the data for the duplicate experiments were averaged. The kinetic traces are shown in the Inset to Fig. 3. The control mixing of enzyme with buffer alone showed that no absorbance change was missed at the beginning (data not shown). The data were fitted to three exponentials (Materials and Methods). Before proceeding with the interpretation, a difference between the present study and the investigation of the GR reductive half reaction (31) should be mentioned. In the study on GR (31), the wavelength monitored was 440 nm as opposed to 463 nm in the present work. The decrease in absorbance observed at 463 nm in the main phase reflects primarily the combined rates of flavin reduction and flavin reoxidation and formation of the thiolate–flavin charge transfer complex. Measurements at 440 nm, on the other hand, are thought to include a contribution from the displacement of NADP+ by NADPH. The first phase, representing formation of the Michaelis complex, is largely complete in the dead time, and rates derived from this phase are not reliable, particularly in the presence of enzyme-bound NADP+. Interpretations of the data from observations at 540 nm are complex (see below). The rates determined for the second phase from data at 463 and 540 nm are hyperbolically dependent on NADPH concentration, as expected. The apparent maximal kobs at saturating NADPH is 113 s−1 and 65 s−1 from 463 nm and 540 nm data, respectively. The rates are different because the two wavelengths are differentially sensitive to the several steps that contribute to this apparent maximal kobs. The absorbance decrease due to FAD reduction and the increase due to flavin reoxidation are the primary processes detected at 463 nm. The absorbance decrease due to loss of the FADH−–NADP+ charge transfer complex and the increase due to the formation of the thiolate–flavin charge transfer complex are detected at 540 nm. Moreover, the thiolate–flavin charge transfer complex absorbance is enhanced at 540 nm as NADPH displaces NADP+ (40). The apparent Kd values of the Michaelis constant for NADPH determined from data at 463 and 540 nm were 20–30 μM. The rates were somewhat slower than those measured for GR. The rate of the third phase, observed mainly at 540 nm, was ≈5 s−1, and this rate did not appear to be dependent on the NADPH concentration. Based on very preliminary steady state data (NADPH–TrxR in assays not coupled to any terminal acceptor), it is likely that a rate of 5 s−1 at 4°C is significant relative to kcat.
Clearly, the reaction of hTrxR with only the single molecule of NADPH needed to form the thiolate–flavin charge transfer complex as seen here (Fig. 3, spectrum 4) is different from the reduction of eTrxR by two molecules of NADPH that reduce both the flavin and the redox-active disulfide (ref. 37 and B. W. Lennon and C. H. Williams, unpublished work).
Carmustine (BCNU) as a Probe for the Two Electron-Reduced Form of hTrxR.
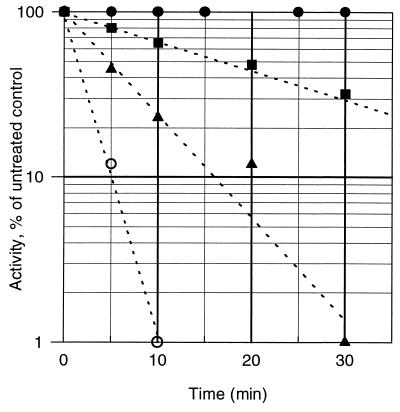
The widely used cytostatic agent BCNU is an established in vivo probe for the EH2 state of GR. In vitro, this nitrosourea drug, or one of its degradation products, also inactivates two electron-reduced LipDH (32). Consequently, we tested the effect of BCNU on hTrxR. Guided by the presteady state experiments (Fig. 3), we chose conditions that favor the Eox and EH2 forms, respectively. Fig. 4 shows that, for EH2, the half-life of inactivation by 500 μM BCNU is ≈5 min under quasiphysiologic conditions. Eox (control in the absence of NADPH), in contrast to EH2, resists modification for hours even at millimolar concentrations of BCNU.
Figure 4.
Inactivation of two electron-reduced hTrxR by BCNU. HTrxR (1.6 units/ml; corresponding to 1 nmol subunit/ml) was incubated at 37°C in hTrxR assay buffer (100 mM potassium phosphate/2 mM EDTA, pH 7.4) with various concentrations of BCNU in the presence of 100 μM of NADPH (control without NADPH, closed circles). Aliquots were diluted 100-fold and assayed for residual activity. The apparent half-times of inactivation and BCNU concentrations are: open circles, 1.7 min, 1 mM; closed triangles, 5 min, 500 μM; and closed squares, 17 min, 100 μM. Because the decomposition of BCNU to chloroethyl isocyanate and other reactive species (t1/2 = 58 min under these conditions) is often rate-limiting (32), the kinetics of EH2 inactivation can be more complex than suggested by this figure.
The fact that hTrxR and GR have very similar mechanisms suggests that inhibition of hTrxR must be taken into account when BCNU is used as an apparently specific inhibitor of GR in in vivo experiments (32). Moreover, TrxR can be considered a potential target of nitrosourea drugs in chemotherapy. This aspect has been studied by Schallreuter et al. (41) using TrxR from melanoma cells. The specific activity of their preparations was not sufficient to allow detailed mechanistic studies. These and other authors emphasize that the reactions catalyzed by TrxR and GR provide the bulk of the reducing equivalents for the biosynthesis of deoxyribonucleotides from ribonucleotides (41–43). Inhibition of this process, which is crucial for DNA synthesis, is assumed to contribute to the cytostatic properties of nitrosourea drugs (32). With respect to the importance of TrxR from tumor cells, it is noteworthy that the levels of TrxR activity in three tumor transformed cell lines (Novikoff ascites, lung adenocarcinoma, and Hela cells) were found to be ≈10 times higher than the levels of enzyme reported in bovine liver, human placenta, and T cells (ref. 22 and references therein). Possible roles of TrxR in malignant cells include synthesis of deoxyribonucleotides and the protection from natural killer lysin (44). On the basis of our data, the mechanism of TrxR inhibition by BCNU derivatives can now be studied in molecular detail.
CONCLUSIONS
The redox potentials of the flavin and the active center disulfide–dithiol couples are separated by more than 60 mV in GR and LipDH (25, 45). In contrast, these potentials in eTrxR are separated by only 12 mV (26–27). This leads to differences in the mechanisms of GR and LipDH when compared with the mechanism of eTrxR (6). The most easily observed difference is that LipDH and GR form an intermediate that is a thiolate–flavin charge transfer complex that can be detected by its absorbance at 540 nm, where Eox and EH4 do not absorb. This intermediate results from reaction with only 1 eq of NADPH. Excess NADPH binds to this EH2 intermediate, enhancing its absorbance at 540 nm, but fails to reduce the enzyme further because the redox potential of the NADP+–NADPH couple is high (more positive) relative to that of the EH2–EH4 couple. On the other hand, eTrxR is fully reduced by a slight excess of NADPH.
We have exploited these differences in the present study to show that the mechanism of hTrxR is like the mechanisms of LipDH and GR and is thus quite easily distinguished from the mechanism of eTrxR. The presence of a third potentially redox-active component, SeCys, in hTrxR complicates the data analysis somewhat, but the reduction of the thiolate–flavin charge transfer complex by 1 eq of dithionite is clear. The fact that the formation of the thiolate–flavin charge transfer complex appears to require two to three eq of dithionite suggests that the Cys–SeCys sequence, penultimate to the C terminus, can receive reducing equivalents from the active center dithiol as is the case for the Cys–Cys sequence near the C terminus of mercuric ion reductase (35). The variability in the dithionite required most likely stems from variations in the redox state of the Cys–SeCys, which could include species with the equivalent of a disulfide bond between the sulfur and the selenium or a sulfenic acid and its SeCys equivalent. Thus, an uptake of two to three equivalents would be required to reduce the active center disulfide and the Cys–SeCys depending on the latter’s level of oxidation. With respect to an essential role of SeCys, it is worth mentioning that high Mr TrxR of the malaria parasite Plasmodium falciparum has a penultimate CysXXXXCys motif. According to the deduced amino acid sequence, the enzyme does not contain SeCys, however (12, 46).
Formation of the thiolate–flavin charge transfer complex upon reduction of hTrxR with excess NADPH implies that NADPH does not reduce EH2 to EH4 (under these conditions). one molecule of NADPH has reacted. Again, the pattern of reduction is reminiscent of that observed with GR in which the rates of FAD reduction and reoxidation are similar so that there is practically no net absorbance change at 463 nm as the thiolate–flavin charge transfer complex is formed (31). The reaction of eTrxR with 2 eq of NADPH to form EH4 is in stark contrast to the observation with hTrxR. These results, taken together with the dithionite titration, leave little doubt that hTrxR and GR have similar mechanisms. This will now form the basis for the further study of hTrxR. This continuation is of special interest because of the potential of TrxR as a cancer chemotherapeutic target and because of the highly unusual evolution—structural divergence and functional convergence—of the two types of TrxR.
Acknowledgments
The authors are grateful to Ms. Donna Veine and Mrs. Irene König for preparing the proteins and for help with the manuscript and to Dr. Scott B. Mulrooney for help with sequence alignments used in deducing Fig. 1. This research was supported by the Department of Veterans Affairs (C.H.W.), by National Institute of General Medical Sciences Grant GM21444 (C.H.W.), by the Deutsche Forschungsgemeinschaft (R.H.S.; Schi 102/7–5), and by the Bundesministerium für Forschung und Technologie (K.B.; Research Focus Tropical Medicine, 01 KA 9301).
ABBREVIATIONS
- TrxR
thioredoxin reductases
- BCNU
1,3-bis(2-chloroethyl)-1-nitrosourea
- EH2
2-electron reduced enzyme, primarily thiolate-flavin charge transfer complex, EH4, 4-electron reduced enzyme with reduced FAD and an active site dithiol
- Eox
oxidized enzyme containing an active site disulfide
- eq
equivalents relative to enzyme bound FAD
- eTrxR
TrxR from E. coli
- GR
glutathione reductase
- hTrxR
TrxR from human placenta
- LipDH
lipoamide dehydrogenase
- SeCys
selenocysteine. It should be noted that oxidized enzyme (Eox), 2-electron reduced enzyme (EH2), and 4-electron reduced enzyme (EH4) refer only to the Cys pair/FAD-ensemble at the catalytic site. Other titratable redox centers of the enzyme are not included in these definitions
References
- 1.Gasdaska P Y, Gasdaska J R, Cochran S, Powis G. FEBS Lett. 1995;373:5–9. doi: 10.1016/0014-5793(95)01003-w. [DOI] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Waksman G, Krishna T S R, Sweet R M, Williams C H, Jr, Kuriyan J. J Mol Biol. 1994;236:800–816. [PubMed] [Google Scholar]
- 4.Williams C H., Jr FASEB J. 1995;9:1267–1276. doi: 10.1096/fasebj.9.13.7557016. [DOI] [PubMed] [Google Scholar]
- 5.Schirmer R H, Schulz G E. In: Coenzymes and Cofactors. Dolphin D, Poulson R, Avramovic O, editors. IIB. New York: Wiley; 1987. pp. 333–379. [Google Scholar]
- 6.Williams C H., Jr . In: Chemistry and Biochemistry of Flavoenzymes. Müller F, editor. III. Boca Raton, FL: CRC; 1992. pp. 121–211. [Google Scholar]
- 7.Schulz G E, Schirmer R H, Sachsenheimer W, Pai E F. Nature (London) 1978;273:120–124. doi: 10.1038/273120a0. [DOI] [PubMed] [Google Scholar]
- 8.Karplus P A, Schulz G E. J Mol Biol. 1987;95:701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- 9.Mattevi A, Schierbeek A J, Hol W G J. J Mol Biol. 1991;220:975–994. doi: 10.1016/0022-2836(91)90367-f. [DOI] [PubMed] [Google Scholar]
- 10.Luthman M, Holmgren A. Biochemistry. 1982;21:6628–6633. doi: 10.1021/bi00269a003. [DOI] [PubMed] [Google Scholar]
- 11.Tamura T, Stadtman T C. Proc Natl Acad Sci USA. 1996;93:1006–1011. doi: 10.1073/pnas.93.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller S, Gilberger T W, Färber P M, Becker K, Schirmer R H, Walter R D. Mol Biochem Parasitol. 1996;80:215–219. doi: 10.1016/0166-6851(96)02694-1. [DOI] [PubMed] [Google Scholar]
- 13.Becker K, Färber P M, von der Lieth C W, Müller S. In: Flavins and Flavoproteins. Stevenson K J, Massey V, Williams C H Jr, editors. XII. Calgary, Canada: University Press; 1997. , in press. [Google Scholar]
- 14.Freudenberg W, Andreesen J R. J Bacteriol. 1989;171:2209–2215. doi: 10.1128/jb.171.4.2209-2215.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clement-Metral J D, Höög J-O, Holmgren A. Eur J Biochem. 1986;161:119–129. doi: 10.1111/j.1432-1033.1986.tb10131.x. [DOI] [PubMed] [Google Scholar]
- 16.Speranza M L, Ronchi S, Minchiotti L. Biochem Biophys Acta. 1973;327:274–281. doi: 10.1016/0005-2744(73)90410-5. [DOI] [PubMed] [Google Scholar]
- 17.Florencio F J, Yee B C, Johnson T C, Buchanan B B. Arch Biochem Biophys. 1988;266:496–507. doi: 10.1016/0003-9861(88)90282-2. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson F S, Morgan R W, Christman M F, Ames B N. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]
- 19.Tartaglia L A, Storz G, Brodsky M H, Lai A, Ames B N. J Biol Chem. 1990;265:10535–10540. [PubMed] [Google Scholar]
- 20.Niimura Y, Poole L, Massey V. J Biol Chem. 1995;270:25645–25650. doi: 10.1074/jbc.270.43.25645. [DOI] [PubMed] [Google Scholar]
- 21.González J C, Peariso K, Penner-Hahn J E, Matthews R G. Biochemistry. 1996;35:12228–12234. doi: 10.1021/bi9615452. [DOI] [PubMed] [Google Scholar]
- 22.Gladyshev V N, Jeang K T, Stadtman T C. Proc Natl Acad Sci USA. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oblong J E, Gasdaska P Y, Sherrill K, Powis G. Biochemistry. 1993;32:7271–7277. doi: 10.1021/bi00079a025. [DOI] [PubMed] [Google Scholar]
- 24.Massey V, Ghisla S. Ann NY Acad Sci. 1974;227:446–465. [Google Scholar]
- 25.Matthews R G, Williams C H., Jr J Biol Chem. 1976;251:3956–3964. [PubMed] [Google Scholar]
- 26.O’Donnell M E, Williams C H., Jr J Biol Chem. 1983;258:13795–13805. [PubMed] [Google Scholar]
- 27.O’Donnell M E, Williams C H., Jr J Biol Chem. 1984;259:2243–2251. [PubMed] [Google Scholar]
- 28.Holmgren A, Björnstedt M. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 29.Krauth-Siegel R L, Enders B, Henderson G B, Fairlamb A H, Schirmer R H. Eur J Biochem. 1987;164:123–128. doi: 10.1111/j.1432-1033.1987.tb11002.x. [DOI] [PubMed] [Google Scholar]
- 30.Williams C H, Jr, Arscott L D, Matthews R G, Thorpe C, Wilkinson K D. In: Methods in Enzymology: Vitamins and Coenzymes. McCormick D B, Wright L D, editors. New York: Academic; 1979. pp. 185–198. [DOI] [PubMed] [Google Scholar]
- 31.Rietveld P, Arscott L D, Perham R N, Williams C H., Jr Biochemistry. 1994;33:13888–13895. doi: 10.1021/bi00250a043. [DOI] [PubMed] [Google Scholar]
- 32.Becker K, Schirmer R H. Methods Enzymol. 1995;251:173–190. doi: 10.1016/0076-6879(95)51120-2. [DOI] [PubMed] [Google Scholar]
- 33.Coleman R F, Black S. J Biol Chem. 1965;240:1796–1803. [PubMed] [Google Scholar]
- 34.Massey V, Williams C H., Jr J Biol Chem. 1965;240:4470–4480. [PubMed] [Google Scholar]
- 35.Miller S M, Moore M J, Massey V, Williams C H, Jr, Distefano M D, Ballou D P, Walsh C T. Biochemistry. 1989;28:1194–1205. doi: 10.1021/bi00429a037. [DOI] [PubMed] [Google Scholar]
- 36.Huber P W, Brandt K G. Biochemistry. 1980;19:4568–4575. doi: 10.1021/bi00561a005. [DOI] [PubMed] [Google Scholar]
- 37.Williams C H, Jr, Prongay A J, Lennon B W, Kuriyan J. In: Flavins and Flavoproteins 1990. Curti B, Ronchi S, Zanetti G, editors. Berlin: de Gruyter; 1991. pp. 497–504. [Google Scholar]
- 38.Massey V, Gibson Q H, Veeger C. Biochem J. 1960;77:341–351. doi: 10.1042/bj0770341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massey V, Palmer G. J Biol Chem. 1962;237:2347–2358. [PubMed] [Google Scholar]
- 40.Williams C H., Jr . In: The Enzymes. Boyer P D, editor. New York: Academic; 1976. pp. 89–173. [Google Scholar]
- 41.Schallreuter K U, Gleason F K, Wood J M. Biochem Biophys Acta. 1990;1054:14–20. doi: 10.1016/0167-4889(90)90199-n. [DOI] [PubMed] [Google Scholar]
- 42.Holmgren A. J Biol Chem. 1979;254:3672–3678. [PubMed] [Google Scholar]
- 43.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 44.Andersson M, Holmgren A, Spyrou G. J Biol Chem. 1996;271:10116–10120. doi: 10.1074/jbc.271.17.10116. [DOI] [PubMed] [Google Scholar]
- 45.Veine D M, Arscott L D, Williams C H., Jr . In: Flavins and Flavoproteins 1993. Yagi K, editor. Berlin: de Gruyter; 1994. pp. 496–500. [Google Scholar]
- 46.Becker K, Müller S, Keese M A, Walter R D, Schirmer R H. Biochem Soc Trans. 1996;24:67–72. doi: 10.1042/bst0240067. [DOI] [PubMed] [Google Scholar]